Abstract
Objective
We have previously demonstrated a brain-machine interface (BMI) neuroprosthetic system that provided continuous control of functional electrical stimulation (FES) and restoration of grasp in a primate model of spinal cord injury (SCI). Predicting intended EMG directly from cortical recordings provides a flexible high-dimensional control signal for FES. However, no peripheral signal such as force or EMG is available for training EMG decoders in paralyzed individuals.
Approach
Here we present a method for training an EMG decoder in the absence of muscle activity recordings; the decoder relies on mapping behaviorally-relevant cortical activity to the inferred EMG activity underlying an intended action. Monkeys were trained at a 2D isometric wrist force task to control a computer cursor by applying force in the flexion, extension, ulnar, and radial directions and execute a center-out task. We used a generic muscle-to-force model based on muscle pulling directions to relate each target force to an optimal EMG pattern that attained the target force while minimizing overall muscle activity. We trained EMG decoders during the target hold periods using a gradient descent algorithm that compared EMG predictions to optimal EMG patterns.
Main results
We tested this method both offline and online. We quantified both the accuracy of offline force predictions and the ability of a monkey to use these real-time force predictions for closed-loop brain-control. We compared both offline and online results to those obtained with several other force decoders, including an optimal decoder computed from concurrently measured neural and force signals.
Significance
This novel approach to training an adaptive EMG decoder could make a brain-control FES neuroprosthesis an effective tool to restore the hand function of paralyzed individuals. Clinical implementation would make use of individualized muscle-to-force models. Broad generalization could be achieved by including data from multiple grasping tasks in the training of the neuron-to-muscle decoder. Our approach would make it possible for persons with SCI to grasp objects with their own hands, using near-normal motor intent.
Keywords: Spinal Cord Injury, Brain Machine Interface, Adaptive Decoder
INTRODUCTION
With relatively few exceptions, Brain-Machine Interface (BMI) research involves the development of decoders that use signals recorded from the brain to predict the kinematics of movements attempted by intact subjects [2–12]. In the earliest experiments, decoders were trained by computing a map from brain activity to actual movement, an approach that cannot be implemented for a paralyzed individual. For BMIs to be useful clinically, it is important to develop techniques to compute decoders without relying on the user’s ability to make actual movements. To solve this problem, several groups have developed “observation-based” decoders with intact monkey subjects [13, 14] as well as paralyzed people [15–19]. In this approach, maps relate neural activity not to actual limb movement but rather to the observed trajectory of a cursor or robot arm that the subject attempts to mimic. By this means, any of a wide variety of supervised learning algorithms can be used to compute a decoder without requiring actual limb movement.
While brain control of a robotic limb would clearly provide significant benefit to individuals paralyzed by SCI, restored control of their own muscles would undoubtedly have even greater impact on their overall physiological and psychological well-being [20, 21]. Among those people with tetraplegia, the great majority identifies the return of hand function as their most critical need [22, 23]. Muscles can be made to contract through electric stimulation, a clinically used procedure referred to as functional electrical stimulation (FES) [20, 24]. However, people with high level SCI have very few available options to provide voluntary control of this electrical stimulation. One successful approach is to use the remaining control of proximal limb muscles as a trigger for patterned FES [25]. A limited number persons have used EEG to control FES using implanted [26] or surface electrodes [27]. Quite recently, another group used intracortical signals recorded from an implanted microelectrode array to control FES in a human with a C5-C6 spinal cord lesion [28]. The system enabled six different attempted movements, each detected from the cortical recordings with a different decoder, and effected by a set of surface electrodes that were selected and calibrated at the beginning of each session. Although the surface stimulation significantly limited its performance, it was a dramatic demonstration of the potential of this technology.
We recently demonstrated a novel BMI that controls multi-channel FES by continuously predicting the intended activity (EMG) of each of several muscles temporarily paralyzed by peripheral nerve block [29, 30]. In those proof-of-concept experiments, the decoders were trained using both neural and EMG signals collected concurrently prior to the paralysis. Training an EMG decoder without access to actual EMG signals is more challenging than training an observation-based kinematic decoder. The mapping of the high-dimensional EMG activity to movement is redundant, and any particular pattern of EMG activity cannot be explicitly presented to subjects in any meaningful way for them to observe and imitate.
Fortunately, the relationship between endpoint forces and patterns of EMG activity is reliably predictable, particularly for simple, well-constrained tasks [31, 32]. Here, we have investigated whether it is possible to train a decoder by inferring the level of muscle activity that a monkey is likely to be using as it attempts simple isometric contractions. We have adopted a 2D isometric wrist task similar to that used in a variety of motor control studies [33–35]. During this highly stereotypical task, the patterns of muscle activity corresponding to any given target force are approximately similar, even across monkeys. We tested if, by mapping neural data recordings to these inferred patterns of muscle activity, we could train a decoder capable of extracting useful muscle activation signals from neural data.
Unfortunately, the large amount of covariation among muscles in such a simple isometric task implies that generalization to other tasks is likely to be quite poor. Our ultimate strategy is to use a robust training scenario that includes a wide range of simple tasks, each involving a different muscles-to-force map, with the goal of developing a single M1 to EMG decoder that remains accurate across a wide range of tasks. In this study, we have shown that it is possible to train an EMG decoder through a combination of behavioral cues and inferred patterns of EMG activity. We have successfully tested this method offline by measuring the accuracy of force predictions obtained from a generic EMG-to-force model based on muscle pulling directions. We have also verified a monkey’s ability to use this EMG decoder online by controlling a computer cursor using the same EMG-to-force transformation, and found that his performance was as good as that achieved with a linear decoder trained using actual muscle signals.
METHODS
Overview
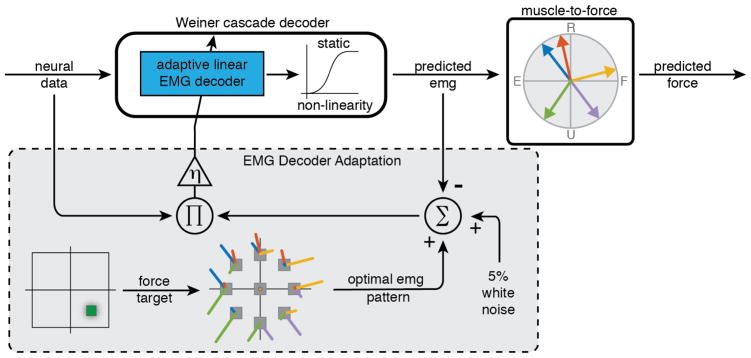
Our overall goal was to develop a method that would be applicable to paralyzed individuals for training neural decoders to predict intended muscle activation. In this scenario the calculation of the decoder parameters could not involve measurement of any output related information such as EMG, force, or movement. We relied instead on an isometric force task that allowed us to infer the motor intent, in terms of spatiotemporal EMG patterns, from the behavioral cues provided to the monkeys. Specifically, we associated an optimal spatial pattern of wrist muscle activity with each presented force target. We used a gradient descent learning algorithm to identify the decoder parameters that led to a map from neural recordings to these inferred optimal EMG patterns. We tested the performance of the resulting decoders both offline and online, by transforming the predicted patterns of muscle activity into force through an additional static muscle-to-force map based on muscle pulling directions. In a clinical application, the same EMG predictions could instead be used to drive FES.
Behavioral task
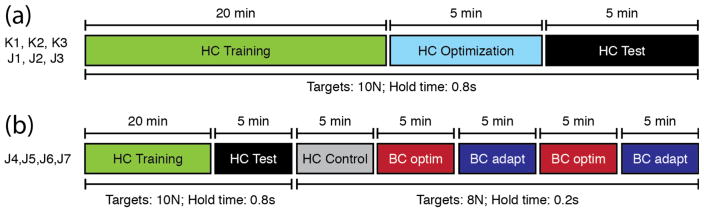
Two monkeys were trained at a 2D isometric wrist force task (figure 1(a)) for which the relationship between muscle activity and force is relatively simple and well characterized. Each monkey’s left arm was positioned in a splint so as to immobilize the forearm in an orientation midway between supination and pronation (with the thumb upwards). A small box was placed around the monkey’s open left hand, incorporating a 6 DOF load cell (20E12, 100N, JR3 Inc., CA) aligned with the wrist joint. The box was padded to comfortably constrain the monkey’s hand and minimize its movement within the box. The monkeys controlled the position of a cursor displayed on a monitor by the force they exerted on the box. Flexion /extension force moved the cursor right and left, respectively, while forces along the radial /ulnar deviation axis moved the cursor up and down. Prior to placing the monkey’s hand in the box, the force was nulled in order to place the cursor in the center target. Being supported at the wrist, the weight of the monkey’s hand alone did not significantly move the cursor when the monkey was at rest. The gain of the system was calibrated so that a 1N force applied to the wrist device would move the cursor by 1cm. Targets were displayed either at the center of the screen (zero force), or equally-spaced along a ring around the center target. All targets were squares with 4cm sides (figure 1(b)).
Figure 1.
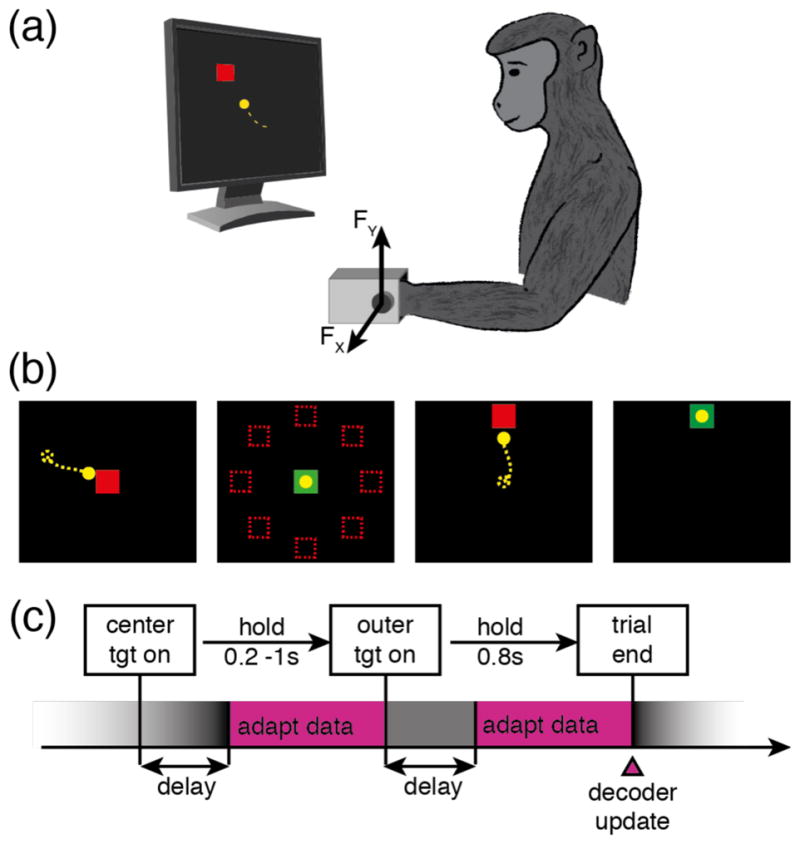
Experimental setup for computing adaptive decoders of EMG activity during production of isometric wrist forces. (a) The monkey controlled a cursor on a computer monitor. The position of the cursor was a function of the 2D force measured about the monkey’s left wrist, as he applied flexion/extension and radial/ulnar deviation isometric forces within a padded metal box mounted on a load cell. (b) The monkey performed a center-out task by applying the appropriate force to move the cursor to one of eight outer targets uniformly distributed around the center. (c) During two time windows in each trial the adaptation algorithm assumed that the monkey’s neural activity corresponded to the motor intent of holding the cursor in the center and outer targets, respectively (adapt data, pink rectangles).
The monkeys performed a center-out task that began with the appearance of a center target (figure 1(c)). They were allowed two seconds to move to the center target, which they were required to hold for a time randomly chosen from a uniform distribution between 0.2s and 1.0s. A successful center hold triggered the appearance of one of eight possible outer targets, chosen in a block-randomized fashion. The monkeys were allowed another two seconds after target onset to move the cursor to the outer target. The required hold time for the outer target was 0.8s. Successful trials ended with the delivery of a liquid reward. Failure to reach a target within the allowed two seconds or to remain within a target as required resulted in an aborted (center target) or failed (outer target) trial. Successive trials were separated by a two-second inter-trial interval.
During collection of the data used for decoder adaptation and offline prediction, the outer targets were located 10cm from the center, thus requiring a 10N force to be attained. For all data used to compute online behavioral performance (hand control, brain-control with the optimal decoder, brain-control with the adapted decoder), the distance from center to outer targets was reduced to 8cm and the target hold time to 0.2s. These parameters were chosen to allow the monkeys to receive sufficient rewards to remain motivated, while avoiding performance saturation effects.
Cortical implant
In each monkey, we implanted arrays composed of 96, 1.5 mm microelectrodes in a 10 by 10 grid (Blackrock Microsystems) in the wrist /hand area of the primary motor cortex (M1) contralateral to the hand used for the task. We determined the implant site with reference to sulcal patterns and by using intraoperative surface cortical stimulation to evoke hand and wrist muscle contractions.
All surgeries were performed under isoflurane gas anesthesia (1–2%) except during cortical stimulation, for which the monkeys were transitioned to reduced isoflurane (0.25%) in combination with remifentanil (0.25 μg/kg bolus + 0.25 to 0.4 μg/kg/min continuous infusion). The monkeys were administered antibiotics, anti-inflammatories, and analgesics for several days after surgery. All surgical and experimental procedures were consistent with the guide for the care and use of laboratory animals and approved by the institutional animal care and use committee of Northwestern University.
Data collection
We recorded force and neural signals using the Cerebus system (Blackrock Microsystems, Inc.). We did not attempt to identify spikes belonging to individual neurons, but instead used a simple multi-unit signal, registering a spike whenever an electrode signal exceeded -5 times the RMS amplitude for that electrode, as calculated at the beginning of every session. We computed firing rates as the number of spikes detected in each electrode during each time bin, divided by the 50ms bin duration. We excluded neural channels for which the firing rate, averaged over the entire training dataset, fell below 0.5 Hz. The mean number of remaining channels was 78±8 (mean±1SD).
We fitted an exponential sinusoidal tuning curve to the firing rate data, and identified a preferred direction (PD) for each of the multi-unit signals included in our analyses by assuming a lag of 150ms between neural activity and force. We used bootstrapping with 100 repetitions and a generalized linear model with a Poisson noise model [36] to compute the statistics leading to the identification of the PD.
Neuron-to-muscle decoder
The decoder we used to map neural activity into muscle activity was a Wiener cascade, consisting of a dynamic linear stage followed by a fixed static nonlinearity, as we have done in previous studies [37]. The linear portion of the decoder was parameterized by a set of weights w that mapped N neural inputs {nk}, 1 ≤ k ≤ N into EMG predictions {Em}, 1 ≤ m ≤ M for each of the M=5 muscles included in our muscle-to-force model. The decoder used 500ms of neural firing rate history divided into L=10 time bins. Unlike previous our implementations, in this case we computed the weights of the linear decoder adaptively, using gradient descent; these methods are described in greater detail below. The resulting Wiener cascade model is:
| Eq. 1 |
where wm0 is a bias term for muscle m, l is the time lag, nk,t is the firing rate of neuron k at time bin t , and wmk,l is the weight that quantifies the effect of the firing activity of neuron k at time (t − l) on the EMG signal of muscle m at time (t) . The sigmoidal nonlinearity is:
| Eq. 2 |
Sigmoid curves are known to provide a good fit to the relation between cortical activation and muscle recruitment levels [38]. In addition, a sigmoidal nonlinearity constrains the predicted output to the range from 0 to 1. The outputs of the neuron-to-muscle Wiener cascade thus corresponded to the normalized amplitude of the predicted muscle activity.
Muscle-to-force model
In clinical applications, the pulling directions and maximal forces of available muscles could be obtained empirically for each person. This individual characterization is particularly important, given the expected variability due to specific nature of each person’s injury. For simplicity, in our monkey experiments we relied on published values of the pulling directions for the five major wrist muscles, obtained from electrical stimulation in monkeys [39], to develop a generic muscle-to-force model (figure 2(a)). These pulling directions are similar those reported for humans with an isometric force device [32]. Considering the 0˚ direction to the right (flexion for left hand), with angles increasing counterclockwise, these directions were 15˚, 103˚, 128˚, 235˚, and 307˚ for flexor carpi radialis (FCR), extensor carpi radialis longus and brevis (ECRl, ECRb), extensor carpi ulnaris (ECU), and flexor carpi ulnaris (FCU), respectively (figure 2(a)). We assigned each muscle a maximal force magnitude of 15N; a normalized maximal EMG prediction of 1 thus corresponds to a 15N force output. These virtual muscle properties ensured that there existed a combination of muscle activation to produce each target force (figure 2(b)). In a clinical setting the maximal force would be measured empirically by stimulating each muscle. See the discussion for more details.
Figure 2.
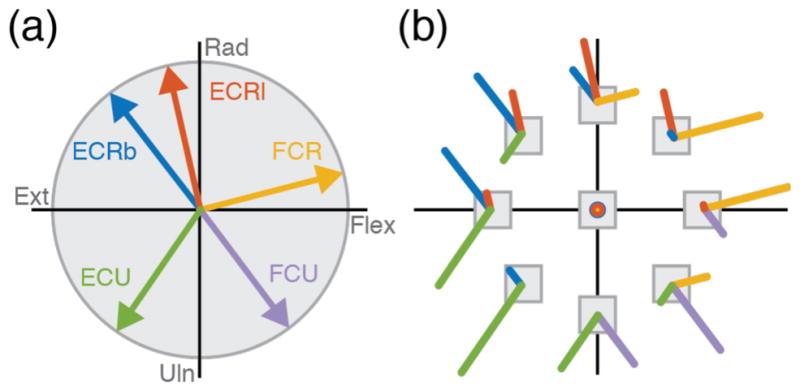
Wrist muscle pulling directions and optimal muscle activation patterns. (a) Vectors indicate the direction of force exerted by each of the five main wrist muscles modeled in this study: Extensor Carpi Radialis longus (ECRl), Extensor Carpi Radialis brevis (ECRb), Extensor Carpi Ulnaris (ECU), Flexor Carpi Ulnaris (FCU), Flexor Carpi Radialis (FCR). Derived from [1]. (b) Optimal patterns of muscle activity needed to produce a force corresponding to each of eight targets using a minimal activation of the muscles shown in (a).
This model is similar in concept to the virtual biomechanical model presented by de Rugy et al. to reconstruct force from EMG [1]. The predicted force, F̂, can be determined by adding the force vectors of figure 2(a), each scaled by the corresponding predicted EMG, namely:
| Eq. 3 |
where H is the 5x2 matrix of the x and y components of each of the pulling vectors (figure 2a) that map EMG to force, HT is its 2x5 transpose, and E is a pattern of activation of the five muscles. Given this muscle-to-force model, we determined optimal patterns of muscle activity for each force target included in the decoder training procedure. We used the MATLAB optimization routine fmincon to solve the underdetermined force-to-muscle relationship by minimizing a regularized cost function in the M-dimensional space of muscle activities E = {Em}, 1 ≤ m ≤ M . Each component of the M -dimensional EMG vector E was constrained to the interval between 0 and 1. The cost function C(E) minimizes the force error (figure 2(b)) while also minimizing the sum squared EMG activity:
| Eq. 4 |
Here, C(E) is the cost, E is the M = 5 dimensional vector of normalized EMG amplitudes, FT is the target force, and penalizes large values of EMG. The performance of the adaptive decoders discussed below and summarized in figure 3 was insensitive over several orders of magnitude to the specific value of the multiplier λ used to infer target EMG patterns. We therefore chose to use λ =1 in all experiments with the adaptive decoder reported here. The optimal EMG patterns that resulted from solving equation 4 were constrained to values between 0 and 1 and then processed through the inverse of the sigmoid of equation 2, to obtain target EMG patterns for training the linear stage of the neuron-to-muscle decoder.
Figure 3.
Adaptive EMG decoder structure and the stochastic gradient training process. At the end of each trial, the linear decoder weights were modified according to the contribution of each neuronal unit to the error in the predicted EMG in comparison to pre-determined optimal EMG for the corresponding force target. During the testing phase, the neuron-to-EMG decoder weights were fixed, and the EMG predictions were transformed into force predictions by a vector sum of the force contribution of each muscle, as determined by the muscle-to-force model.
The muscle-to-force model described in equation 3, in combination with the minimization of the cost function in equation 4, allowed us to identify optimal EMG patterns for each force target that served as a supervisory learning signal in the training of the neuron-to-muscle decoder. The muscle-to-force model was also used as a surrogate to FES, to compute the force that would be generated by the predicted EMG (figure 3, top of diagram). This transformation allowed us to evaluate the accuracy of the adaptive EMG decoder in terms of force output.
Adaptive decoder training and evaluation
The training of the weights w that parameterize the decoder was accomplished using stochastic gradient descent; weights were updated only once per trial. The optimal EMG patterns associated with each target (including a zero EMG activity pattern for the center target) were used as the supervisory signal (figure 3, dashed rectangle). In order to improve the search and avoid local minima traps, on each trial we added ±5% noise from a uniform distribution to the amplitude of each component of the optimal EMG vector. Weights were initialized with normally distributed random values with a mean of 0 and a standard deviation of 10−4. This range was chosen to approximately match the weight values obtained through regression of neural firing rates to actual EMG activity.
It is important to note that the adaptation process used data only from a limited time window beginning shortly after the target appearance, when we could reasonably infer that the motor intent of the monkey corresponded to the instructed target force (pink rectangles in figure 1(c)). This “adaptation onset delay” was one of the training parameters that we optimized across all datasets. Although the full training datasets included 20 minutes of data each, the data actually used for adaptation, limited to the target presentation period shifted by the delay, averaged only 4.5±0.5min for the 10 datasets.
The decoder adaptation process was developed offline but designed to be applicable to online decoder training. All calculations and decoder updates were causal, and could thus be performed in real-time while a subject performs or attempts to perform the task. To this end, our approach was to “play back” previously recorded neural and force data. We updated the neuron-to-EMG decoder weights once per trial, according to equation 5. A learning rate η = 5x10−7 was selected through a systematic parameter search. The equations for the update of the weights at each trial follow from using backpropagation through time [40]:
| Eq. 5 |
After training the decoder, we evaluated its performance by testing its predictions on novel test data recorded immediately after the data used for training. We computed the mean square Euclidean error (MSE) between the actual and predicted two dimensional force signals as a measure of accuracy.
Comparison with neuron-to-force decoders
Our adaptive EMG decoder was trained using only knowledge of the force targets. In order to better interpret the accuracy of this decoder, we compared its performance to that of several other decoders (summarized below in Table 1) that were computed either with or without adaptation, and with varying amounts and features of force-related signals. We used comparisons to these other decoders to understand the characteristics and limitations of the adaptive EMG decoder. One of the decoders used for comparison was an “optimal-force” decoder computed between neural activity and actual force measured during the entire 20 minutes of training data. This decoder is optimal, in the sense that it is the best least-squares linear mapping between the high-dimensional neural space and the measured force signal. Whether it would also allow a monkey to achieve the best online performance is an important empirical question. At the opposite extreme, we computed an “optimal-target” decoder using only knowledge of the target force during the same limited segments of data used to train the EMG decoder. An intermediate case is that of the “adaptive-force” decoder, computed using gradient descent on actual force signals, but only during the restricted target-related periods and with the same adaptation rate used for the adaptive EMG decoder.
Table 1.
Summary of EMG and force decoders. In order to assess the effectiveness of the adaptive EMG decoder, we compared it to an “optimal force” decoder based on regression between neurons and actual continuous force data. In order to identify the impact of limiting the decoder training to short data segments and using a gradient descent instead of a regression, we also tested “optimal target” and “adaptive force” decoders.
| Decoder name | Training method | Supervisory signal | Data segments used | Rationale for testing |
|---|---|---|---|---|
| Optimal force | Regression | Actual force | Entire dataset (continuous) | Provides an upper bound performance level for force prediction or control. |
| Adaptive EMG | Gradient descent | Inferred EMG patterns | Target hold periods | Clinically applicable decoder. Provides muscle activation signals based only on motor intent without measuring EMG. |
| Optimal target | Regression | Target force | Target hold periods | Comparison to Adaptive EMG decoder revealed little difference between regression and adaptation, or inferred EMG rather than measured force. |
| Adaptive force | Gradient descent | Actual force | Target hold periods | Given the minimal effect of adaptation seen above, comparison to Optimal Force decoder revealed the large effect of using limited force training data. |
Data analysis
We used three datasets from each of two monkeys for the offline analysis (K1 to J3; figure 4(a)). Each of these six datasets included 30 minutes of data divided into 20 minutes for training, five minutes for parameter optimization, and five minutes for testing. After training the decoders, we evaluated the MSE between actual and predicted testing data. The testing data were also used to evaluate the force prediction accuracy of the four different types of decoders. Six months later, we conducted four online brain-control sessions with monkey J (J4 to J7; figure 4(b)). These sessions involved a 20-minute data segment for decoder training and five minutes for offline testing of force predictions. They also included five blocks of five minutes each for measuring the monkey’s behavioral performance: one block of hand control, two blocks of brain control with the optimal force decoder, and two blocks of brain control using the adaptive EMG decoder. These online test sessions used the closer targets (8N) and shorter hold times (0.2s) indicated in figure 4(b), and the adaptation parameters that had been previously optimized offline.
Figure 4.
Summary of the 10 datasets used for offline prediction and online control. (a) Three datasets from each of two monkeys, each consisting of a total of 30 minutes, divided into training, optimization, and test subsets. (b) Four datasets from a single monkey to study brain control comprised 25 minutes of hand control for training and offline analyses, and an additional 25 minutes of center-out behavior, including both hand-control (HC) and alternating five minute blocks of brain-control (BC).
We quantified online behavioral performance using five different metrics: trial time, path length, success rate, failure rate, and abort rate. Trial time included the duration of the movement starting from outer target onset and including the outer target hold time. The path length was the sum of all the Euclidean distances between successive time points along the path until the end of the trial; this was computed only for successful trials, abort and failed trials were excluded. The success, abort, and failure rates were the number of successful, aborted (failure to hold center target), and failed (failure to hold outer target) trials, respectively, divided by the total number of trials.
RESULTS
Adaptation parameter search
We systematically varied the adaptation parameters (adaptation onset delay and learning rate) while testing the resulting prediction performance in order to determine their optimal values across datasets. There was typically a small but consistent peak in performance for delays between 500ms and 650ms (figure 5(a)). Longer delays reduced decoder performance significantly, presumably because of the reduced amount of available training data. A delay of 600ms provided the lowest MSE across all datasets. The decoding performance for datasets for which this value was suboptimal was reduced by 15% in the worst case (figure 5(b)).
Figure 5.
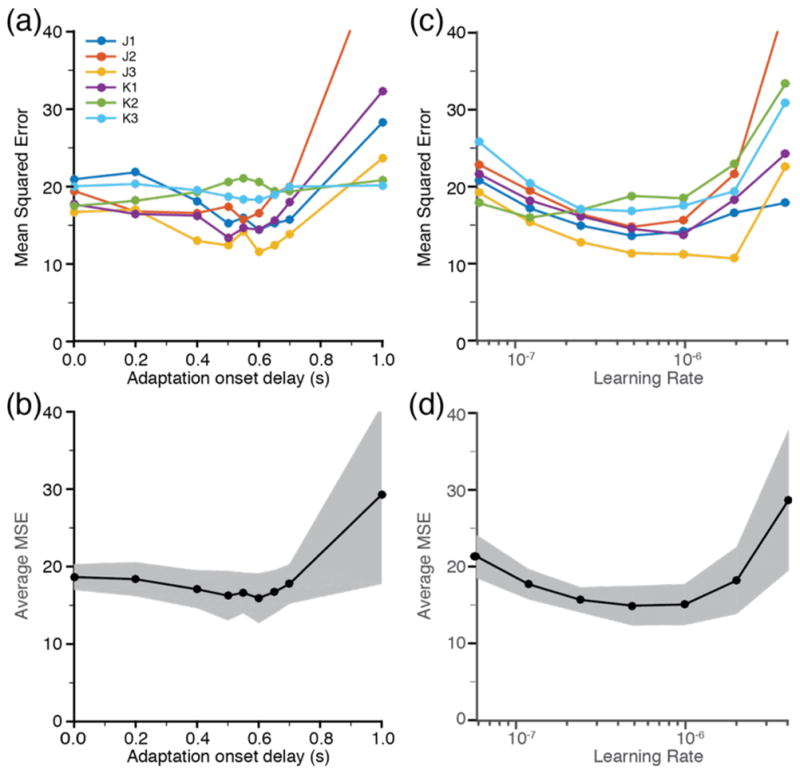
Optimization of decoder adaptation parameters. (a) Mean squared Euclidean error of the offline force predictions for each of the six data sets in figure 4(a), as a function of the delay between target appearance and the onset of training data sampling. (b) Mean and one standard deviation of the results in (a) across all datasets. (c) Individual and (d) mean error as a function of the learning rate parameter. The values leading to the minimal overall MSE were a delay of 600ms and a learning rate of 5x10e−7.
We tested learning rates η of 0.625, 1.25, 2.5, 5, 10, 20, and 40 (in multiples of 10−7). The learning rates leading to the lowest MSE ranged from 2.5x10−7 to 2x10−6 (figure 5(c)) over individual datasets. The single best learning rate across datasets was 5x10−7 (figure 5(d)); as for the delay parameter, this value was suboptimal by only 15% in the worst case.
Performance dependence on training duration
Figure 6 shows how the performance of the decoders varied with amounts of training data. After two and a half minutes of continuous data (figures 6(a) and 6(b), cyan lines), the decoder weights had begun to capture the neuron-to-muscle mapping, and most force bursts were predicted in the correct direction. However, these predictions still had very low magnitude; the weights had not yet substantially changed from their initial random small values, resulting in near zero EMG (and thus force) predictions. After 20 minutes (figures 6(a) and 6(b), blue lines), the predictions had improved dramatically, matching the actual force (figures 6(a) and 6(b), grey lines) both in magnitude and in direction. In this example, the MSE of the radial /ulnar deviation force predictions (6.2) was better that of the flexion / extension force predictions (11.1).
Figure 6.
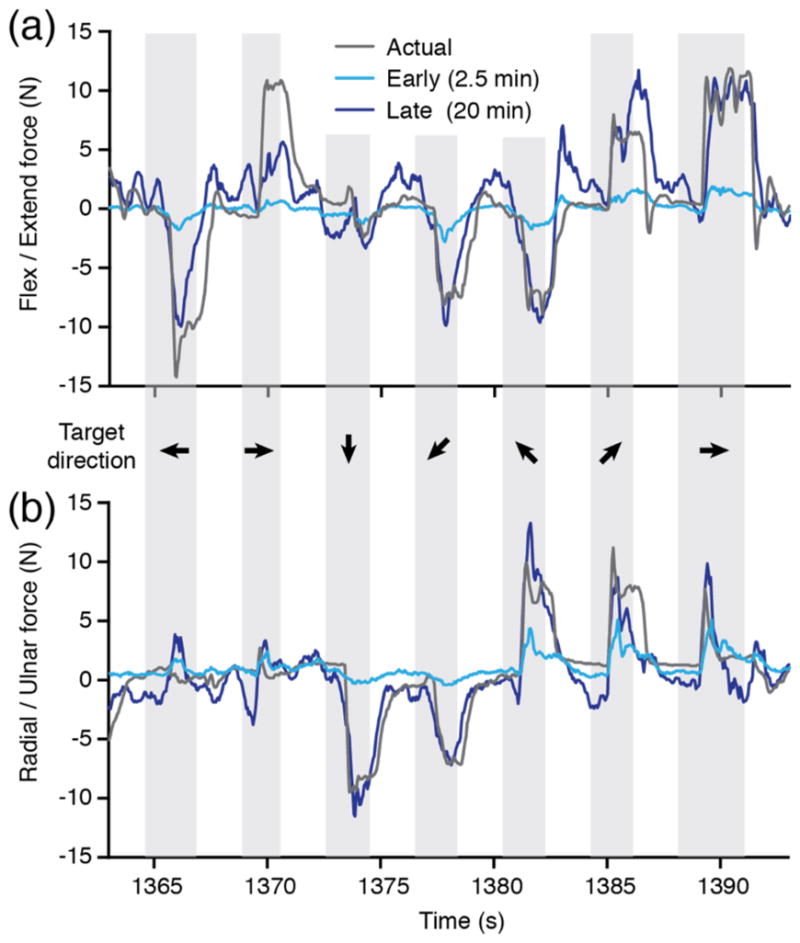
Improvement in offline force prediction from early to late adaptation. (a) Cyan lines indicate predicted flexion / extension force following decoder training with only 2.5 minutes of data (34 trials). Blue lines indicate the improved prediction with 20 minutes of data (283 trials); they matched the actual force (grey) more closely. (b) Similar results for radial / ulnar deviation force predictions. All data are from dataset K3.
For most datasets, as in this example, both the adaptive and optimal decoders predicted radial/ulnar deviation forces better than flexion/extension forces. Overall, MSE for the adaptive decoder averaged 6.9±1.5 and 8.9±1.6 for R/U and F/E, respectively. Two possible explanations are that 1) F/E forces may simply be more difficult to predict, or 2) the neural data contained more information about R/U. Failure of the decoders to predict forces generated by the long finger flexor and extensor muscles (note the failure to predict the peak at 1370 seconds in figure 6(a)) may have contributed to greater errors along this axis. However, there was also compelling evidence that the F/E axis was underrepresented in the neural recordings.
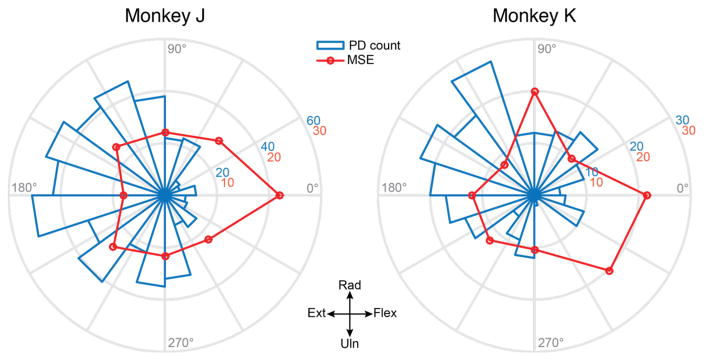
Figure 7 shows polar histograms of PDs for each monkey superimposed on the average MSE of force predictions for each target. The MSE was calculated from 600ms after target onset until the end of target hold. The histograms show the number of neural channels having PDs within each 18° bin. There were fewer PDs in the flexion direction, in close correspondence with the overall spatial pattern of the MSE: the average MSE was higher for targets in directions that were poorly represented in the PD distribution. The scarcity of neural channels with PDs in the flexion direction provides a plausible explanation for the higher accuracy for the prediction of forces in the radial and ulnar directions.
Figure 7.
Non-uniform distribution of the preferred directions of neural recordings and offline force prediction accuracy. Polar histograms (blue) show the total number of neural signals with a PD within each of 20 bins dividing the 2D force space, across all datasets for each monkey. There was a marked paucity of signals with PDs near the 0° (flexion) axis for both monkeys, as well as in the 270–360° quadrant for monkey K. The MSE of the optimal force predictions was higher during the presentation of targets falling in these directions (red curve).
EMG predictions
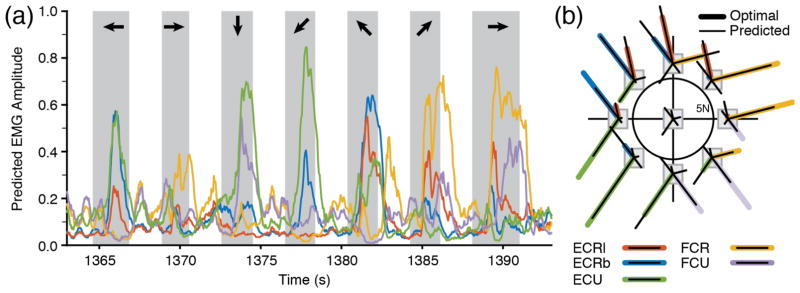
Figure 8(a) shows predicted EMG amplitude, a quantity equivalent to a rectified and filtered EMG signal. The signal predicted for each muscle was strongly modulated in a target-dependent manner. These signals, when applied to the static muscle model shown in figure 2, produced the forces shown in figure 6. In clinical use, these signals might serve to control stimulation intensity in an FES prosthesis used by a spinal cord injured person. In the clinical setting, the muscle model corresponding to that of figure 2 would not be a generic one, but one specifically determined for each individual to reflect the force that could actually be generated by the stimulation of each muscle.
Figure 8.
Accuracy of offline EMG predictions following adaptation. (a) Predicted EMG signals were well modulated following each target appearance (gray shaded rectangles) and were predictive of the target location (black arrows). This data segment corresponds to the force signals in figure 6. (b) During target presentation, the mean EMG magnitudes (thin black lines) matched with great accuracy the optimal EMG associated with each target (thick colored lines).
Figure 8(b) shows the average EMG magnitude predicted for each muscle during presentation of all possible targets. The average predictions (thin black lines) roughly match the magnitude of the optimal EMG used to train the decoders (thick colored lines). For the 10 test datasets used in this study, the overall correlation across all targets and all muscles between the magnitudes of predicted and optimal EMG was ρ = 0.88 (Spearman rank pairwise correlation).
Adaptive decoder performance in offline force predictions
As a benchmark for evaluating the performance of the adaptive EMG decoder, we compared it to the performance of several other decoders. These included both fixed and adaptive decoders that were trained with differing amounts and types of force-related signals (see Methods and Table 1). Figure 9(a) shows the evolution of performance with increasing offline adaptation for dataset J7. Prediction error generally decreased as a function of training time, with most of the improvement occurring within the first 10 minutes. With between 15 and 20 minutes of training data, the performance of all decoders approached a plateau, but did not consistently saturate. As anticipated, the force prediction accuracy of the “optimal force” decoder, which was computed to directly map neural data into continuously recorded force, was significantly better than that of the adaptive EMG decoder (figure 9(a), red and blue lines respectively). To understand whether this difference was due to the different amount of training data or to the use of adaptive methods, we tested two other neuron-to-force decoders. We compared the adaptive EMG decoder to an “optimal target” decoder that was computed without adaptation, using only the instructed target force training data that was available to the adaptive EMG decoder. With 20 minutes of behavior data, the performance of these two decoders was very similar (figure 9(a), purple and blue lines), suggesting that the difference in performance between the optimal force and adaptive EMG decoders was not due to the adaptive training. We also tested the effect of the inferred EMG error measure by comparing an “adaptive force” decoder trained with actual force signals measured during the same target hold periods that were used for the adaptive EMG decoder (figure 9(a), green line). The performance of this decoder was no different than that of the adaptive EMG decoder. This result suggests that the use of inferred (as opposed to measured) EMG for the adaptive EMG decoder was not a significant source of error.
Figure 9.
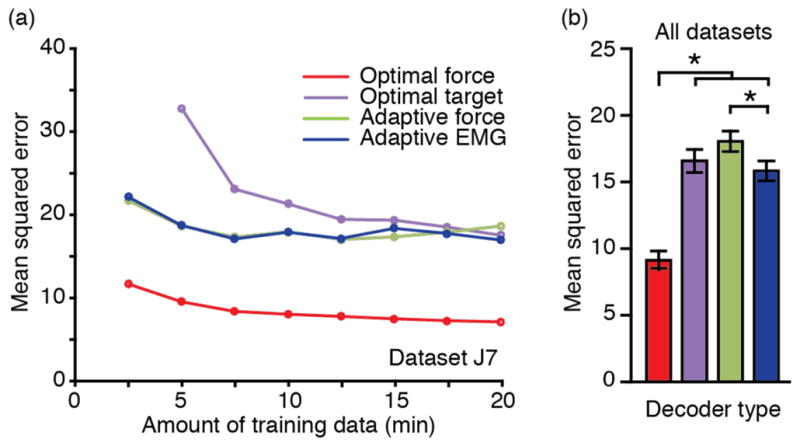
Comparison of different decoders performance. (a) Effect of training time on prediction accuracy for different types of force decoding approaches (Dataset J7). There was little additional improvement beyond 15 minutes. (b) With 20 minutes of training data, the optimal force decoder (red) provided the most accurate predictions across all datasets. The adaptive EMG approach (blue) led to force prediction accuracy comparable to the “optimal target” (purple) and slightly better than the “adaptive force” (green) decoders. Error bars represent 2*SEM.
The performance across all datasets was similar to that shown in figure 9(a) for dataset J7, with the optimal force decoder significantly better than all others. We conclude that this superior performance was due to the much larger amount of force-related data available to that decoder. Surprisingly, we found that given 20 minutes of training data, the adaptive EMG decoder was slightly, but significantly better than the adaptive force decoder (p<0.0005, figure 9(b)). The optimal target decoder was intermediate in performance, not substantially different from either the adaptive EMG or the adaptive force decoders.
Adaptive decoder performance during online brain control
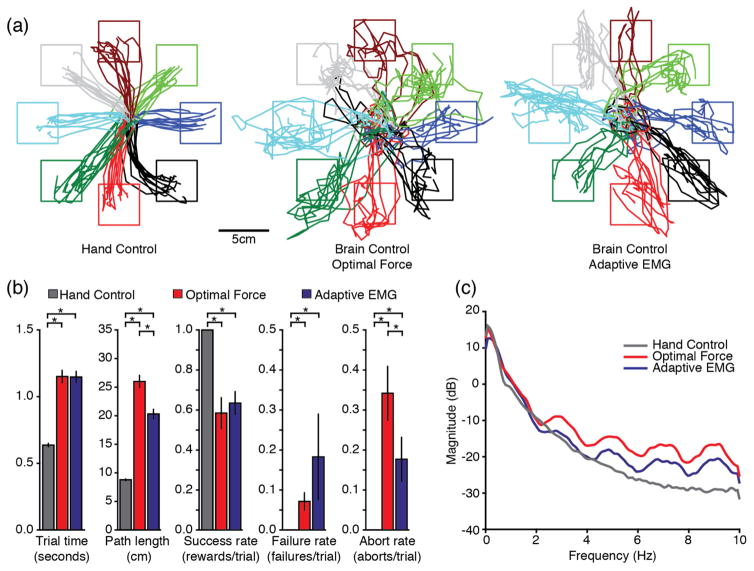
In a later set of experiments that took place six months after the last offline dataset was recorded, we evaluated monkey J’s ability to perform a center-out task in an online, brain control (BC) setting. The data presented in figure 10 illustrate the quality of the control achieved during hand control (HC) along with that achieved with both the adaptive EMG decoder and the optimal force decoder. Figure 10(a) shows cursor trajectories for all successful trials performed during a five-minute block from dataset J6, for each of the three conditions. The center-out trajectories were highly stereotypical during HC. The path to some targets was consistently curved (e.g., the lower right target), but most paths were straight and smooth. The trajectories during the two BC conditions were more variable (figure 10(a), middle and right panels). Note that for these three conditions, we reduced the distance to the outer targets from 10N to 8N, and the hold time for outer targets from 0.8s to 0.2s, in contrast to the behavioral parameters used for offline decoder training and testing (see Methods).
Figure 10.
Summary of online cursor control performance with optimal force and adaptive EMG decoders. (a) The cursor trajectories during BC conditions (middle and rightmost panels) were more jagged than during HC (leftmost panel). (b) Most performance metrics were comparable for the adaptive EMG (blue bars) and the optimal force (red bars) decoders, but neither was as good as during HC (grey bars). Error bars represent 2*SEM. (c) The main reason for these differences appeared to be related to the greater cursor jitter present in the optimal force decoder predictions, observable as increased power in the higher frequencies. Note that the 2Hz periodic fluctuations in the force power spectra during both BC conditions are an artifact of using a 500ms decoder window length.
We quantified performance in these three conditions using five behavior metrics (figure 10(b)). As expected, performance during HC was better by all measures when compared to BC (p<0.001 for all comparisons, paired t-test). Surprisingly, the monkey’s performance with the optimal force decoder in terms of trial time and success rate was no better than that achieved with the adaptive EMG decoder, despite its poorer offline prediction accuracy (figure 9(b)). However, the similar overall performance masked differences in the nature of control: cursor movement appeared to be relatively fast and jittery with the optimal decoder, while smoother but slower with the adaptive EMG decoder. These qualitative observations are reflected in several performance metrics. The higher jitter with the optimal force decoder led to a significantly longer path length. The similar trial time despite the shorter path when using the adaptive EMG decoder reflects a slower speed. The greater jitter also led to more aborted trials for the optimal force decoder, as it was more difficult for the monkey to hold the cursor in the center target for the longer (up to 1 sec) hold time. On the other hand, although the effect did not reach statistical significance, the monkey tended to fail more often at reaching and holding the outer targets when using the adaptive EMG decoder, perhaps because of its lower prediction accuracy.
During BC there was a greater high-frequency jitter, with an amplitude of roughly 1cm, riding on a slower, more deliberate cursor trajectory. This jitter appeared to be of smaller magnitude when using the adaptive EMG decoder. The difference in jitter magnitude is reflected in the force power spectra (figure 10(c)). The force predicted using either decoder contained more power in frequencies between 2Hz and 8Hz than did the actual force; this effect was larger for the optimal force decoder.
DISCUSSION
Summary
We have demonstrated the ability to make accurate predictions of intended EMG that can be used for online brain control, without the need to first measure actual EMGs. The approach is similar to that used previously for “observation based” decoders of kinematic information [15, 41], but the EMG scenario is more difficult because of the high-dimensional nature of the EMG signal from multiple correlated muscles. We made use of the fact that the mapping from EMG (or muscle force) to endpoint force is largely preserved across subjects, at least for simple, highly constrained contractions like the isometric wrist task used in this study. This observation allowed us to use gradient descent to train EMG decoders that minimized both EMG error and total muscle activity. An important application of this approach is as a controller of functional electrical stimulation (FES) used to restore movement in paralyzed individuals. It may also be possible to use predictions like these to provide muscle activation inputs to a prosthetic limb controller that emulates the natural musculoskeletal architecture and dynamics of an actual limb.
Quality of the EMG predictions and clinical implications
There are many reasons to expect the EMG predictions produced by this adaptive approach to be, at best, of modest accuracy. The decoders were trained with limited data. The EMG error signal was not measured, but inferred from the instructed target force and a generic muscle-to-force model. To resolve the underdetermined nature of the inverse map from force to muscle activity, we searched for solutions that minimized total muscle activity. In spite of these limitations, both force and EMG predictions were quite good. For force predictions, the details of the muscle-to-force model were not critical, since we used the same model not only to infer the EMG patterns needed to train the decoder, but also to produce force output from the predicted EMG. In a clinical application, the EMG predictions could be used as FES commands, as we have done before [29, 30]. In that case, using an accurate model of the muscle to force mapping, determined individually for each person (as described below) would be important. Here, we chose to develop and validate the adaptation algorithm under the simpler scenario of force simulation instead of force production via FES.
Despite the additional complications related to making EMG (as opposed to force) predictions, those too, were quite accurate. The bursts of predicted EMG in figure 8(a) were appropriately timed and with at least qualitatively the right target-dependent magnitude. It is worth noting that these patterns were not identical for repeated targets (e.g., the 2nd and last movements in this example), presumably reflecting the monkey’s varied effort. In spite of this variability, the intended target was readily discernable from the overall pattern of predicted muscle activity. In summary, the EMG predictions were similar to those observed during “natural” movements.
In a clinical setting, the muscles actually available and their response to stimulation will vary for any given individual. The muscle-to-force model would thus need to be obtained empirically through the process of electrode characterization, which maps each person’s muscle-to-force output [42–44]. This will determine the details of the patterns of muscle activity for any given endpoint force. A pattern that conforms to the constraints of a particular person while minimizing total activation (thereby also minimizing fatigue) is clearly optimal. In this respect, our EMG predictions were very successful. Across all data sets, their target-dependent magnitude (an example is shown in figure 8(b)) was very close to the optimal pattern used in the learning algorithm (figures 2(b) and 3).
The online brain control experiments provided a more direct demonstration of the usability of the adaptive EMG decoder than did the accuracy of offline force and EMG prediction. The monkey performed well when using the adaptive EMG decoder, in spite of its lower offline accuracy than the optimal force decoder. This similar online performance suggests that our novel adaptive training approach is clinically viable. However, a similar overall performance in terms of simple behavior metrics ignores differences in the quality of control: there was more jitter for the optimal force decoder, but larger prediction errors for the adaptive EMG decoder. The optimal force decoder is so named because it is the optimal linear map between neural activity and endpoint force. However, this decoder may not yield optimal online control, as has been noted before [45–48]. Remaining inaccuracies in the forward map from M1 to output, alterations in the somatosensory feedback, and the ability of the user to correct errors may all cause an “optimal” offline decoder to produce less than optimal performance upon transition from hand to brain control. One possibility that is relevant to our experiment is the observation that simply adding a low pass filter to improve cursor stability despite causing slightly less accurate offline predictions, might have improved target acquisition.
What were the limitations of the adaptive decoder?
Even better online performance would presumably have been achieved if the adaptive EMG decoder had provided more accurate offline force predictions. In addition to its use of actual force signals, the optimal force decoder had access to substantially more data than did the adaptive EMG decoder, for which training data was restricted to the limited fraction of time that the monkey maintained force within each target. We computed two additional decoders in an effort to understand this performance difference, a target-only version of the optimal decoder, and an adaptive version of the force decoder. Both alternate decoders performed significantly more poorly than the optimal force decoder (figure 9). The similarity between the optimal target and adaptive EMG decoders suggests that the limited information available from the target force was a critical factor in limiting their performance. We were surprised to find that the accuracy of the adaptive force decoder, which was trained with actual force data, did not surpass that of the adaptive EMG decoder. This observation supports the use of inferred EMG patterns for decoder training, and highlights the importance of the amount of training data.
These observations suggest that further improvements in decoder performance could be achieved by providing training data not restricted to the limited target-hold periods. While this is technically challenging, it would also address one of the other main limitations: the decoder currently has no means to learn the dynamics of the neuron / EMG, relation since it is trained during only steady-state force application. Here we assumed that the intended levels of EMG activity would be reached at a short delay after target appearance and would remain fixed until the end of the target hold. By restricting training data to the static hold periods, we were able to use a muscle-to-force model that did not include dynamics. A potential way to remove the limitations of this static decoding would be to use a target tracking task together with inferred EMGs that vary temporally as well as spatially. While this continuous approach could appear more effective at providing larger amounts of training data, it also has the added complexity of having to infer optimal EMG patterns under dynamic conditions, and may be more prone to failure in matching the motor effort consistently.
Performance of all the decoders may have been affected by a non-uniform distribution of PDs, which significantly underrepresented flexion direction forces (figure 7). This systematic bias in PDs was unexpected. Such a bias could have arisen artifactually, had there been a force offset in our device. Since we nulled the force prior to each session, such an offset would have to have arisen from a passive mechanical effect as the monkeys inserted their hand into the device which biased muscle activation (and M1 activity) toward excess extension. Although we cannot completely rule out this possibility, is seems very unlikely that it could have been the sole effect, as it would have had to be large, systematic across sessions and monkeys, yet undetected by the experimenters.
Similar, though smaller asymmetries have been described earlier [49–51]. There is reason to believe that the flexor muscles receive disproportionate input from other sources, including the reticular formation [52]. This asymmetry is thought to underlie the characteristic flexor tone following stroke [53, 54]. The implications of this apparent disparity for cortical BMIs have not been directly investigated.
What is the role of user adaptation?
This study has focused on the development of a decoder adaptation algorithm that can be applied to unobserved EMG errors, in an effort to develop a decoder with properties as close as possible to those that can be achieved with an optimally trained decoder with access to measured EMG from an intact monkey. In principle, this “biomimetic” approach should allow the monkey or human subject to generate FES command signals simply by making a normal motor effort. In practice, prediction errors will arise and FES commands will not exactly match the intended action.
Fortunately, the user can also adapt and compensate for some of these discrepancies, through a motor learning process that is presumably like that of achieving normal reaching movements when visual feedback is rotated [55, 56] or when the arm is perturbed by predictable forces [57, 58]. Several studies have shown that as brain control performance increases, measured by any number of different metrics, the firing rate and spatial tuning characteristics of neurons also change. This is true during the initial adaptation to brain control, but also in response to spatial distortions of the decoder [2, 6, 11, 59, 60]. One experiment in particular, extended the classic visual rotation experiments by rotating the effects of 25 or 50% of the neurons in the decoder about a common axis, causing the monkeys to make misdirected movements [60]. Accuracy gradually improved over the course of single experimental sessions, as the monkeys learned to compensate for the distortion. The authors found that in addition to the monkeys’ overall re-aiming strategy, there was evidence of “local” learning within the subset of rotated neurons that was greater than that of the unrotated neurons.
A more recent experiment investigated whether all such decoder distortions can be learned equally well [4]. The answer was very clearly “no”. In this case, the decoder map was perturbed within the 90-dimensional space defined by neural activity. Only when the perturbation did not disrupt the natural coactivation among neurons (i.e., when it remained within the 10D subspace spanned by the activity exhibited by the neural population prior to the perturbation), were monkeys able to learn to use the new decoder within a single session. A similar effect was observed in an experiment using muscle rather than neural inputs. Human subjects were better able to learn distorted maps that nonetheless respected natural muscle synergies than those that did not [61].
These results all point to the conclusion that a decoder that reflects the natural patterns of coactivation among neurons can be learned much more readily than one that does not. The goal of our proposed decoder adaptation is to assure that the adaptation requirements imposed on an individual remain feasible, similar to those that allow monkeys to adapt to perturbations within a single session. Presumably the more accurate the decoder, the simpler, and more intuitive will be the user adaptation. Further performance improvements could be achieved through user adaptation, combined with continued decoder adaptation and retraining [2, 9, 12, 15].
What needs to be done to make the adaptive decoder clinically viable?
We evaluated the accuracy of force predictions after training decoders with a range of training parameters. The use of globally optimal values of these parameters resulted in only a very small decrease in prediction accuracy for individual datasets. Finding that a single set of training parameters could be used across subjects is important, since a detailed parameter optimization process based on the accuracy of force prediction would not be possible with a paralyzed person. However, in this study we used only a subset of the muscles that produce torque about the wrist, and we used the same subset to develop decoders for both monkeys. Another simplifying feature of our study was to arbitrarily assign a maximal force of 15N to each muscle. In paralyzed people, a universal muscle-to- force model is likely to be neither appropriate nor necessary. The force evocable from each muscle will differ among individuals because of the variable nature of spinal cord injury. Denervation due to motoneuron damage at the site of the injury renders muscles essentially nonexcitable electrically. Greater levels of hypertonia and muscle spasms in some individuals may also make the control of particular muscles and movements more difficult. Although computing an accurate muscle model for each person will be necessary, this process is feasible and would be very similar to the “grasp patterning” currently used to customize fixed stimulus patterns for each person who received the Freehand prosthesis [43].
More critical to the success of our approach is the question of its ability to generalize to more complex tasks, such as object grasping and manipulation [33]. Clearly, adaptive training using a single muscle-to-force model based on 2D wrist movement cannot be expected to generalize to independent finger use. However, we have shown previously that isometric wrist EMG predicted with an optimal linear filter decoder generalized better across forearm posture than did a decoder similarly trained to predict endpoint force [33]. We have made similar observations for reaching movements across different arm postures and external loads [62]. If the actual mapping between M1 activity and EMG were linear, an EMG decoder trained with a robust data set should generalize well. However, the question of this linearity remains to be conclusively demonstrated. In addition, the need for a simple, constrained muscle-to-force map from which to infer intended EMG poses a challenge. A potential solution is to collect training data while an individual imagines or attempts to perform a series of tasks, each with well-defined mechanics: palmar grasp, key grip, and precision grip, as well as the wrist task used here. Each device or task must be associated with its corresponding muscle-to-force model, determined by stimulating each implanted electrode and measuring the corresponding endpoint force for each device or task. A single neuron-to-muscle decoder would then be trained using data collected while the person imagined activating all the devices, using their jointly derived optimal muscle activation pattern.
To the extent that the map from M1 to muscles is well preserved across these various motor behaviors, it should be possible to train a single decoder that would function well for a range of functionally important motor behaviors. Should the mapping prove to be significantly nonlinear, or significantly context dependent, a more complex decoder would be required. In principle, a nonlinear or state-dependent decoder could be trained with the same approach, although it would undoubtedly require significantly more data. This would be true, however, for both a clinically relevant, adaptive decoder, or one computed directly from recorded neural activity and EMG under normal movement conditions.
Conclusions
We have shown that an adaptive EMG decoder trained without access to output signals that would be unavailable from a paralyzed person performs well for both offline predictions and online control. This proof of principle, although for a very simple motor behavior, suggests that cortically controlled FES that incorporates an accurate decoder of intended muscle activity is a clinically viable application of BMI technology. Rather than use actual muscle stimulation, we used a simple model of the relation between muscle activity and endpoint force for the online experiments in this study. Exactly the same approach could be used to control stimulation intensity, as we have done in the past [30]. This will be a critical next step in the further development of this method, along with the use of a more robust training set to develop decoders that generalize across a wider range of useful motor tasks.
References
- 1.de Rugy A, Loeb GE, Carroll TJ. Muscle Coordination Is Habitual Rather than Optimal. The Journal of Neuroscience. 2012;32(21):7384–7391. doi: 10.1523/JNEUROSCI.5792-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor DM, Tillery SI, Schwartz AB. Direct cortical control of 3D neuroprosthetic devices. Science. 2002;296(5574):1829–32. doi: 10.1126/science.1070291. [DOI] [PubMed] [Google Scholar]
- 3.Velliste M, et al. Cortical control of a prosthetic arm for self-feeding. Nature. 2008;453(7198):1098–1001. doi: 10.1038/nature06996. [DOI] [PubMed] [Google Scholar]
- 4.Sadtler PT, et al. Neural constraints on learning. Nature. 2014;512(7515):423–426. doi: 10.1038/nature13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serruya MD, et al. Instant neural control of a movement signal. Nature. 2002;416(6877):141–2. doi: 10.1038/416141a. [DOI] [PubMed] [Google Scholar]
- 6.Kim SP, et al. Neural control of computer cursor velocity by decoding motor cortical spiking activity in humans with tetraplegia. J Neural Eng. 2008;5(4):455–76. doi: 10.1088/1741-2560/5/4/010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hauschild M, et al. Cognitive signals for brain–machine interfaces in posterior parietal cortex include continuous 3D trajectory commands. Proceedings of the National Academy of Sciences. 2012;109(42):17075–17080. doi: 10.1073/pnas.1215092109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilja V, et al. A high-performance neural prosthesis enabled by control algorithm design. Nat Neurosci. 2012;15(12):1752–1757. doi: 10.1038/nn.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan JM, et al. Intention estimation in brain–machine interfaces. Journal of Neural Engineering. 2014;11(1):016004. doi: 10.1088/1741-2560/11/1/016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carmena JM, et al. Learning to Control a Brain-Machine Interface for Reaching and Grasping by Primates. PLoS Biol. 2003;1(2):193–208. doi: 10.1371/journal.pbio.0000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganguly K, Carmena JM. Emergence of a stable cortical map for neuroprosthetic control. PLoS Biol. 2009;7(7):e1000153. doi: 10.1371/journal.pbio.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orsborn AL, et al. Closed-Loop Decoder Adaptation on Intermediate Time-Scales Facilitates Rapid BMI Performance Improvements Independent of Decoder Initialization Conditions. Neural Systems and Rehabilitation Engineering, IEEE Transactions on. 2012;20(4):468–477. doi: 10.1109/TNSRE.2012.2185066. [DOI] [PubMed] [Google Scholar]
- 13.Tkach D, Reimer J, Hatsopoulos NG. Observation-based learning for brain-machine interfaces. Curr Opin Neurobiol. 2008;18(6):589–594. doi: 10.1016/j.conb.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wahnoun R, He J, Helms Tillery SI. Selection and parameterization of cortical neurons for neuroprosthetic control. J Neural Eng. 2006;3(2):162–71. doi: 10.1088/1741-2560/3/2/010. [DOI] [PubMed] [Google Scholar]
- 15.Hochberg LR, et al. Neuronal ensemble control of prosthetic devices by a human with tetraplegia. Nature. 2006;442(7099):164–1kim71. doi: 10.1038/nature04970. [DOI] [PubMed] [Google Scholar]
- 16.Hochberg LR, et al. Reach and grasp by people with tetraplegia using a neurally controlled robotic arm. Nature. 2012;485(7398):372–5. doi: 10.1038/nature11076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collinger JL, et al. High-performance neuroprosthetic control by an individual with tetraplegia. The Lancet. 2013;381(9866):557–564. doi: 10.1016/S0140-6736(12)61816-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wodlinger B, et al. Ten-dimensional anthropomorphic arm control in a human brain−machine interface: difficulties, solutions, and limitations. Journal of Neural Engineering. 2015;12(1):016011. doi: 10.1088/1741-2560/12/1/016011. [DOI] [PubMed] [Google Scholar]
- 19.Aflalo T, et al. Decoding motor imagery from the posterior parietal cortex of a tetraplegic human. Science. 2015;348(6237):906–910. doi: 10.1126/science.aaa5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ragnarsson KT. Functional electrical stimulation after spinal cord injury: current use, therapeutic effects and future directions. Spinal Cord. 2008;46(4):255–74. doi: 10.1038/sj.sc.3102091. [DOI] [PubMed] [Google Scholar]
- 21.Nightingale EJ, et al. Benefits of FES gait in a spinal cord injured population. Spinal Cord. 2007;45(10):646–657. doi: 10.1038/sj.sc.3102101. [DOI] [PubMed] [Google Scholar]
- 22.Anderson KD. Targeting recovery: priorities of the spinal cord-injured population. J Neurotrauma. 2004;21(10):1371–83. doi: 10.1089/neu.2004.21.1371. [DOI] [PubMed] [Google Scholar]
- 23.French JS, Anderson-Erisman KD, Sutter M. What do Spinal Cord Injury Consumers Want? A Review of Spinal Cord Injury Consumer Priorities and Neuroprosthesis from the 2008 Neural Interfaces Conference. Neuromodulation. 2010;13(3):229–231. doi: 10.1111/j.1525-1403.2009.00252.x. [DOI] [PubMed] [Google Scholar]
- 24.Peckham PH, Knutson JS. Functional electrical stimulation for neuromuscular applications. Annu Rev Biomed Eng. 2005;7:327–60. doi: 10.1146/annurev.bioeng.6.040803.140103. [DOI] [PubMed] [Google Scholar]
- 25.Kilgore KL, et al. An implanted upper-extremity neuroprosthesis. Follow-up of five patients. J Bone Joint Surg Am. 1997;79(4):533–41. doi: 10.2106/00004623-199704000-00008. [DOI] [PubMed] [Google Scholar]
- 26.Lauer RT, Peckham PH, Kilgore KL. EEG-based control of a hand grasp neuroprosthesis. Neuroreport. 1999;10(8):1767–71. doi: 10.1097/00001756-199906030-00026. [DOI] [PubMed] [Google Scholar]
- 27.Pfurtscheller G, et al. 'Thought'--control of functional electrical stimulation to restore hand grasp in a patient with tetraplegia. Neurosci Lett. 2003;351(1):33–6. doi: 10.1016/s0304-3940(03)00947-9. [DOI] [PubMed] [Google Scholar]
- 28.Bouton CE, et al. Restoring cortical control of functional movement in a human with quadriplegia. Nature. 2016 doi: 10.1038/nature17435. [DOI] [PubMed] [Google Scholar]
- 29.Pohlmeyer EA, et al. Toward the Restoration of Hand Use to a Paralyzed Monkey: Brain-Controlled Functional Electrical Stimulation of Forearm Muscles. PLoS ONE. 2009;4(6):e5924. doi: 10.1371/journal.pone.0005924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ethier C, et al. Restoration of grasp following paralysis through brain-controlled stimulation of muscles. Nature. 2012;485(7398):368–371. doi: 10.1038/nature10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fagg AH, Shah A, Barto AG. A computational model of muscle recruitment for wrist movements. J Neurophysiol. 2002;88(6):3348–58. doi: 10.1152/jn.00621.2002. [DOI] [PubMed] [Google Scholar]
- 32.de Rugy A, Davoodi R, Carroll TJ. Changes in wrist muscle activity with forearm posture: implications for the study of sensorimotor transformations. J Neurophysiol. 2012;108(11):2884–2895. doi: 10.1152/jn.00130.2012. [DOI] [PubMed] [Google Scholar]
- 33.Oby ER, Ethier C, Miller LE. Movement representation in the primary motor cortex and its contribution to generalizable EMG predictions. J Neurophysiol. 2013;109(3):666–78. doi: 10.1152/jn.00331.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmer SS, Fetz EE. Discharge properties of primate forearm motor units during isometric muscle activity. J Neurophysiol. 1985;54:1178–1193. doi: 10.1152/jn.1985.54.5.1178. [DOI] [PubMed] [Google Scholar]
- 35.Shalit U, et al. Descending systems translate transient cortical commands into a sustained muscle activation signal. Cerebral Cortex. 2012;22(8):1904–1914. doi: 10.1093/cercor/bhr267. [DOI] [PubMed] [Google Scholar]
- 36.Truccolo W, et al. A point process framework for relating neural spiking activity to spiking history, neural ensemble, and extrinsic covariate effects. J Neurophysiol. 2005;93(2):1074–89. doi: 10.1152/jn.00697.2004. [DOI] [PubMed] [Google Scholar]
- 37.Westwick DT, et al. Identification of Multiple-Input Systems with Highly Coupled Inputs: Application to EMG Prediction from Multiple Intracortical Electrodes. Neural Comput. 2006;18(2):329–55. doi: 10.1162/089976606775093855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devanne H, Lavoie BA, Capaday C. Input-output properties and gain changes in the human corticospinal pathway. Experimental Brain Research. 1997;114(2):329–38. doi: 10.1007/pl00005641. [DOI] [PubMed] [Google Scholar]
- 39.Hoffman DS, Strick PL. Step-tracking movements of the wrist. IV. Muscle activity associated with movements in different directions. J Neurophysiol. 1999;81(1):319–333. doi: 10.1152/jn.1999.81.1.319. [DOI] [PubMed] [Google Scholar]
- 40.Werbos PJ. Backpropagation through time: what it does and how to do it. Proceedings of the IEEE. 1990;78(10):1550–1560. [Google Scholar]
- 41.Tkach D, Reimer J, Hatsopoulos NG. Congruent activity during action and action observation in motor cortex. J Neurosci. 2007;27(48):13241–50. doi: 10.1523/JNEUROSCI.2895-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hincapie JG, et al. Musculoskeletal model-guided, customizable selection of shoulder and elbow muscles for a C5 SCI neuroprosthesis. IEEE Trans Neural Syst Rehabil Eng. 2008;16(3):255–63. doi: 10.1109/TNSRE.2008.922681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kilgore KL, et al. Synthesis of hand grasp using functional neuromuscular stimulation. Biomedical Engineering, IEEE Transactions on. 1989;36(7):761–770. doi: 10.1109/10.32109. [DOI] [PubMed] [Google Scholar]
- 44.Schearer EM, et al. System identification for 3D force control of a human arm neuroprosthesis using functional electrical stimulation. Robotics and Automation (ICRA), 2012 IEEE International Conference on; 2012. [Google Scholar]
- 45.Koyama S, et al. Comparison of brain–computer interface decoding algorithms in open-loop and closed-loop control. Journal of Computational Neuroscience. 2010;29(1–2):73–87. doi: 10.1007/s10827-009-0196-9. [DOI] [PubMed] [Google Scholar]
- 46.Chase SM, Schwartz AB, Kass RE. Bias, optimal linear estimation, and the differences between open-loop simulation and closed-loop performance of spiking-based brain-computer interface algorithms. Neural Networks. 2009;22(9):1203–1213. doi: 10.1016/j.neunet.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cunningham JP, et al. A closed-loop human simulator for investigating the role of feedback control in brain-machine interfaces. J Neurophysiol. 2011;105(4):1932– 1949. doi: 10.1152/jn.00503.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dangi S, et al. Design and Analysis of Closed-Loop Decoder Adaptation Algorithms for Brain-Machine Interfaces. Neural Computation. 2013;25(7):1693–1731. doi: 10.1162/NECO_a_00460. [DOI] [PubMed] [Google Scholar]
- 49.Sinkjaer T, et al. Synaptic linkages between red nucleus cells and limb muscles during a multi-joint motor task. Experimental Brain Research. 1995;102:546–550. doi: 10.1007/BF00230659. [DOI] [PubMed] [Google Scholar]
- 50.Clough JFM, Kernell D, Phillips CG. The distribution of monosynaptic excitation from the pyramidal tract and from primary spindle afferents to motoneurones of the baboon’s hand and forearm. J Physiology (London) 1968;198:145–166. doi: 10.1113/jphysiol.1968.sp008598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fetz EE, et al. Sensory and motor responses of precentral cortex cells during comparable passive and active joint movements. J Neurophysiol. 1980;43:1070– 1089. doi: 10.1152/jn.1980.43.4.1070. [DOI] [PubMed] [Google Scholar]
- 52.Baker SN. The primate reticulospinal tract, hand function and functional recovery. J Physiol. 2011;589(23):5603–5612. doi: 10.1113/jphysiol.2011.215160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaaimi B, et al. Changes in descending motor pathway connectivity after corticospinal tract lesion in macaque monkey. Brain. 2012;135(7):2277–2289. doi: 10.1093/brain/aws115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ellis MD, et al. Neck rotation modulates flexion synergy torques, indicating an ipsilateral reticulospinal source for impairment in stroke. Journal of Neurophysiology. 2012;108(11):3096–3104. doi: 10.1152/jn.01030.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harris CS. Adaptation to displaced vision: visual, motor, or proprioceptive change? Science. 1963;140(3568):812–813. doi: 10.1126/science.140.3568.812. [DOI] [PubMed] [Google Scholar]
- 56.Krakauer JW, et al. Learning of Visuomotor Transformations for Vectorial Planning of Reaching Trajectories. The Journal of Neuroscience. 2000;20(23):8916–8924. doi: 10.1523/JNEUROSCI.20-23-08916.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shadmehr R, Mussa-Ivaldi FA. Adaptive representation of dynamics during learning of a motor task. J Neurosci. 1994;14(5 Pt 2):3208–24. doi: 10.1523/JNEUROSCI.14-05-03208.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lackner JR, Dizio P. Rapid adaptation to Coriolis force perturbations of arm trajectory. J Neurophysiol. 1994;72(1):299–313. doi: 10.1152/jn.1994.72.1.299. [DOI] [PubMed] [Google Scholar]
- 59.Musallam S, et al. Cognitive control signals for neural prosthetics. Science. 2004;305(5681):258–62. doi: 10.1126/science.1097938. [DOI] [PubMed] [Google Scholar]
- 60.Jarosiewicz B, et al. Functional network reorganization during learning in a brain-computer interface paradigm. Proc Natl Acad Sci U S A. 2008;105(49):19486–91. doi: 10.1073/pnas.0808113105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berger DJ, et al. Differences in adaptation rates after virtual surgeries provide direct evidence for modularity. The Journal of Neuroscience. 2013;33(30):12384–12394. doi: 10.1523/JNEUROSCI.0122-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cherian A, Krucoff MO, Miller LE. Motor cortical prediction of EMG: evidence that a kinetic brain-machine interface may be robust across altered movement dynamics. J Neurophysiol. 2011;106(2):564–575. doi: 10.1152/jn.00553.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]