Abstract
Here we describe a new strategy, HILAQ (Heavy Isotope Labeled Azidohomoalanine Quantification), to rapidly quantify the molecular vulnerability profile to oxytosis, which is an oxidative stress-induced programed cell death pathway that has been reported to be involved in aging and neurodegenerative diseases. HILAQ was able to quantify 1,962 Newly Synthesized Proteins (NSPs) after 1h pulse labeling in HEK293T cell line, while 353 proteins were quantified using the previously published QuaNCAT protocol. HILAQ was successfully applied to the HT22 oxytosis model. 226 proteins were found to have a 2-fold change in abundance, and 108 proteins were enriched in the cell death pathway, demonstrating the utility of HT22 cells as a tool to study the molecular details of cell death involved in neurodegenerative diseases. The HILAQ strategy simplifies the analysis of newly synthesized proteomes through the use of isobaric labels and achieves higher sensitivity than previously published methods.
Introduction
Measuring how proteomes respond to perturbations or disease is crucial to understand the underlying mechanisms involved. Typically, strategies to measure cellular response have used techniques to measure relative gene or protein expression between perturbed and non-perturbed states and these methods have limitations in terms of scale, dynamic range and pathway efficacy. The details of biochemical mechanisms are difficult to infer from expression analysis in part because of limitations associated with presumptions about co-regulation having mechanistic importance. A new method has been developed that uses a bioorthogonal non-canonical amino acid that can be inserted into newly synthesized proteins (NSPs) and exploited for enrichment and measurement. Azidohomoalanine (AHA, Dietrich et al) is an analog of methionine that can be readily incorporated into proteins using cell’s normal translational machinery simply by adding AHA to media for cultured cells or to food pellets for animals(1). Proteins with incorporated AHA can be enriched using click chemistry (2). To date, most analyses using AHA have been qualitative, but recently Eichelbaum et al used AHA labeling combined with metabolic stable isotope labeling to quantitate low-abundance, newly secreted proteins by mass spectrometry (MS)(3), as well as to quantitate new proteins synthesized after T-cell stimulation (QuaNCAT)(4). In the QuaNCAT method, AHA is exploited to enrich new proteins using avidin beads and heavy isotope labeled amino acids (AA) confirm proteins as newly synthesized and allows quantification in the MS. We propose a new strategy for quantification of NSPs using a heavy isotope labeled AHA molecule (Fig. 1a), which is synthesized with heavy stable isotopes (heavy-AHA or hAHA). This strategy, HILAQ (Heavy Isotope Labeled Azidohomoalanine Quantification), enables NSPs enrichment, confirmation and quantification using a single stable isotope labeled molecule.
Figure 1.
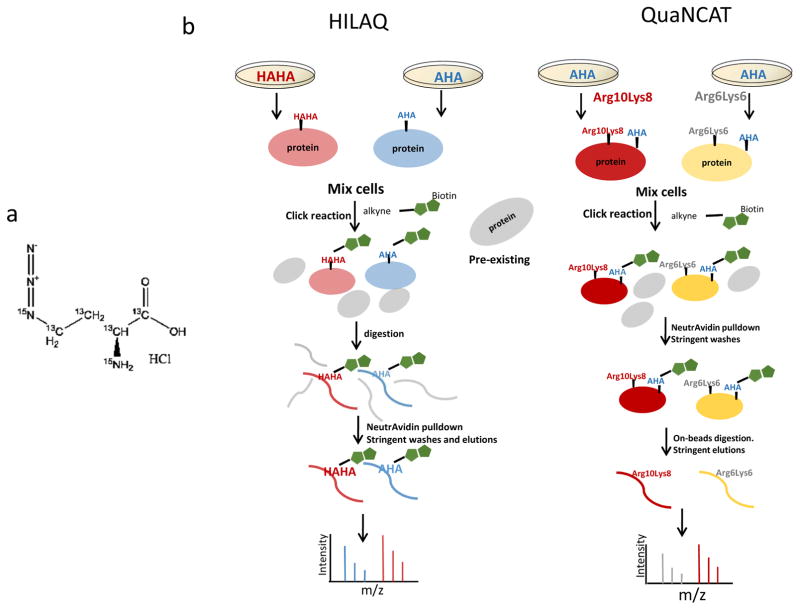
Chemical structure of heavy AHA and schematic workflows. (a) Two 14N and four 12C are substituted with 15N and 13C in heavy AHA. (b) Workflows of HILAQ and QuaNCAT. Cells are metabolically labeled with “heavy” AHA (HAHA) or “light” AHA (AHA) in the HILAQ strategy. In the QuaNCAT strategy, cells are metabolically labeled with a combination of “light” AHA, 13C615N2-L-lysine (Lys8) and 13C615N4-L-arginine (Arg10) or “light” AHA, 13C6-L-lysine (Lys6) and 13C6-L-arginine (Arg6). After 1h pulse labeling, plate one and two are mixed at 1:1(wt/wt) for each protocol. For both strategies, a click reaction is performed on the mixtures followed by protein precipitation. For QuaNCAT samples, AHA proteins are enriched by neutravidin beads, followed by on-bead tryptic digestion to obtain NSP peptides. The HILAQ samples are processed similarly except that the neutravidin enrichment step is performed on peptides after tryptic digestion, followed by AHA peptide elution from the beads.
In this paper we compare HILAQ with QuaNCAT, and we report the results of our study employing HILAQ to quantify NSPs in a cell line model of non-receptor oxidative glutamate toxicity or oxytosis. Glutamate is a major neurotransmitter in the brain, but excessive extracellular glutamate can result in oxidative stress leading to cell death. Glutamate toxicity has been implicated in many neurological diseases, including cerebral ischemia, Huntington’s disease, Alzheimer’s disease (5), and amyotrophic lateral sclerosis (6). Glutamate induces toxicity through two mechanisms: excitotoxicity or oxytosis. In excitotoxicity, excess extracellular glutamate overstimulates ionotropic glutamate receptors leading to cell death initiated by an influx of calcium ions (7, 8). In oxytosis or non-receptor oxidative glutamate toxicity, excess glutamate inhibits a cystine/glutamate antiporter, preventing cystine import which is the rate limiting step of glutathionine (GSH) synthesis. GSH is one of the cell’s most important antioxidants to neutralize reactive oxygen species (ROS). ROS produces oxidative stress through modification of proteins, lipids and DNA, and it triggers downstream signaling pathways, generating increased intracellular calcium which leads to cell death (9). This is a novel form of programmed cell death with characteristics of both necrosis and apoptosis, and has been described as necroptosis (10). Increased oxidative stress and depletion of GSH is associated with aging and neurodegenerative diseases, such as Alzheimer’s Disease (11, 12). The immortalized hippocampal cell line, HT22, is an in vitro model of oxytosis due to its lack of ionotropic glutamate receptors and high sensitivity to extracellular glutamate(13). It has been used as an important research tool to study the molecular details of oxytosis and investigate neuroprotective pathways. For example, HT22 cells selected for resistance to oxytosis were discovered to be less vulnerable to amyloid-beta toxicity, which is hypothesized to contribute to the pathogenesis of Alzheimer’s disease (14). Thus, studying oxytosis may hold potential for identifying treatments for complex neurodegenerative disorders (15).
Materials and Methods
Cell preparation
HEK293T or HT22 cells were maintained in DMEM (Gibco) supplemented with 10% FBS (Invitrogen) and 1% penicillin/streptomycin (Gibco) in a humidified incubator at 37 °C and 5% CO2. Before labeling, cells were washed with warm PBS (Gibco) twice and incubated in labeling media (Hank’s Balanced Salt Solution with Glutamax, 10% dialyzed FBS, 4mM MgCl2, 4mM CaCl2, 1mM Sodium Pyruvate, 1% penicillin/streptomycin) for 30 min. For the HILAQ strategy, cells were incubated in labeling media with either 1mM “Heavy” AHA (13C4, 15N2, Cambridge Isotope Laboratories) (plate one ) or 1mM “Light” AHA (Cambridge Isotope Laboratories) (plate two) for 1h.
For the QuaNCAT strategy, cells were incubated in labeling media with either “medium” 0.5mM L-lysine(13C6, Cambridge Isotope Laboratories), 0.5mM “medium” L-Arginine (13C6, Cambridge Isotope Laboratories) (plate one) or 1mM “Light” AHA or “Heavy” L-lysine (13C6, 15N2, Cambridge Isotope Laboratories), “Heavy” L-Arginine (13C6, 15N4, Cambridge Isotope Laboratories) (plate two), and 1mM “Light” AHA for 1h.
For each HILAQ or QuaNCAT biological replicate, 2 plates (1 and 2) of 7×106Cells were washed with warmed Phosphate-buffered saline (PBS), resuspended in PBS with protease and phosphatase inhibitor cocktail (Roche), then sonicated using a tip sonicator. Protein concentration was determined with a Pierce BCA protein assay (Life Technologies) prior mixing the cells 1:1(wt/wt).
Click chemistry
For each biological replicate, 2mg of a 1:1 mixture was centrifuged. The supernatants were divided into 5 aliquots. The pellets were dissolved with 0.5% sodium dodecyl sulfate (SDS) in 200ul PBS, sonicated, heated on 100 °C for 10min and divided into 5 aliquots after cooling to room temperature. A click reaction was performed on each aliquot as previously published(16). Briefly, for each click reaction, the following reagents were added in this order: 1) 30 μl of 1.7mM Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA), 2) 8 μl of 50 mM Copper Sulfate, 3) 8 μl of 5mM Biotin-Alkyne (C21H35N3O6S, Click Chemistry Tools), and 4) 8 μl of 50 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP). PBS was then added to a final volume of 400 μl and reactions were incubated for 1 hour at room temperature. All reactions were combined and Trichloroacetic acid (TCA) was added to 25%. TCA precipitation was performed overnight on shaker at 4°C.
QuaNCAT (Biotin protein enrichment and on-beads digestion)
Precipitated proteins were dried at room temperature and resuspended in 200ul 8M Urea with 20ul 20% SDS. 150ul neutravidin agarose resin (ThermoScientific) was added and samples were incubated for 2 h at room temperature while rotating. Supernatants were removed after centrifugation at 500 × g for 30 sec. The resin was then washed with 1ml PBS, then PBS with 5% acetonitrile, and a final wash of PBS. The resin was centrifuged at 500 × g for 30 sec, and supernatant was removed for each wash. The resin was resuspended in 50 μl 8M Urea and 50 μl 0.2% MS compatible surfactant ProteaseMAX (Promega) in 50mM ammonium bicarbonate, then reduced, alkylated, and digested with trypsin as previously described(17). The resin + digest solution was centrifuged at 500 × g for 2 min and supernatant was transferred to a new tube. 100 ul 50 mM ammonium bicarbonate was added to the resin and the sample was centrifuged at 500 × g for 2 min. The supernatant was removed and combined with the first supernatant. 5% formic acid was added to the combined supernatant before centrifugation at 13,000 × g for 30 min. Supernatant was transferred to a new tube, and loaded on MudPIT column.
HILAQ (Digestion and biotin peptide enrichment)
Precipitated pellets were resuspended in 50 μl 8M Urea and 50ul 0.2% MS compatible surfactant ProteaseMAX (Promega) in 50 mM ammonium bicarbonate, then reduced, alkylated, and digested with trypsin as previously described(17). The digest was then centrifuged at 13,000 × g for 10min. The supernatant was transferred to a new tube and the pellet was resuspended with PBS and centrifuged at 13,000 × g for 10min. Supernatants were combined and 150 μl of neutravidin agarose resin (Thermo Scientific) was added. The resin was incubated with the peptides for 2 h at room temperature while rotating, and then washed as in the QuaNCAT protocol. The peptides were eluted with two treatments of 150 μl 80% acetonitrile, 0.2% formic acid, and 0.1% TFA on shaker for 5 min at room temperature, and another two times on shaker at 70 °C. All elutions were transferred to a single new tube. Prior to MS analysis, the elutions were dried with a speed-vac and dried peptides were re-solubilized in Buffer A (5% ACN, 95% water, 0.1% formic acid).
Mass spectrometry
Soluble peptides were pressure-loaded onto a 250-μm i.d capillary with a kasil frit containing 2 cm of 10 μm Jupiter C18-A material (Phenomenex) followed by 2 cm 5 μm Partisphere strong cation exchanger (Whatman). This column was washed with Buffer A after loading. A 100 μm i.d capillary with a 5 μm pulled tip packed with 15 cm 4 μm Jupiter C18 material (Phenomenex) was attached to the loading column with a union and the entire split-column (loading column–union–analytical column) was placed in line with an Agilent 1100 quaternary HPLC (Palo Alto). The sample was analyzed using MudPIT, using a modified 11-step separation described previously (18). The buffer solutions used were Buffer A, 80% acetonitrile/0.1% formic acid (Buffer B), and 500 mM ammonium acetate/5% acetonitrile/0.1% formic acid (Buffer C). Step 1 consisted of a 10 min gradient from 0–10% Buffer B, a 50 min gradient from 10–50% Buffer B, a 10 min gradient from 50–100% Buffer B, and 20 min 100% Buffer A. Step 2 consisted of 1 min of 100% Buffer A, 4 min of 20% Buffer C, a 5 min gradient from 0–10% Buffer B, a 80 min gradient from 10–45% Buffer B, a 10 min gradient from 45–100% Buffer B, and 10 min 100% Buffer A. Steps 3–9 had the following profile: 1 min of 100% Buffer A, 4 min of X% Buffer C, a 5 min gradient from 0–15% Buffer B, a 90 min gradient from 15–45% Buffer B, and 10 min 100% Buffer A. The Buffer C percentages (X) were 30, 40, 50, 60, 70, 80, 100% for the steps 3–9, respectively. In the final two steps, the gradient contained: 1 min of 100% buffer A, 4 min of 90% buffer C plus 10% B, a 5 min gradient from 0–10% buffer B, a 80 min gradient from 10–45% buffer B, a 10 min gradient from 45–100% buffer B, and 10 min 100% buffer A. As peptides eluted from the microcapillary column, they were electrosprayed directly into an Orbitrap Elite mass spectrometer (ThermoFisher) with the application of a distal 2.4 kV spray voltage. A cycle of one full-scan FT mass spectrum (300–1600 m/z) at 240,000 resolution followed by 20 data-dependent IT MS/MS spectra at a 35% normalized collision energy was repeated continuously throughout each step of the multidimensional separation. Application of mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur data system.
Data analysis
1) Identification and quantification
Both MS1 and MS2 (tandem mass spectra) were extracted from the XCalibur data system format (.RAW) into MS1 and MS2 formats using in house software (RAW_Xtractor)(19). Tandem mass spectra remaining after filtering were searched with Prolucid (20) against the UniProt_Human_03-25-2014 concatenated to a decoy database in which the sequence for each entry in the original database was reversed(21). All searches were parallelized and performed on a Beowulf computer cluster consisting of 100 1.2 GHz Athlon CPUs (22). No enzyme specificity was considered for any search. The following modifications were searched for HAHA analysis: a static modification of 57.02146 on cysteine for all analyses, a differential modification of 452.2376 on methionine for AHA-biotin-alkyne or 458.2452 for HAHA-biotin-alkyne. For QuaNCAT: a static modification of 57.02146 on cysteine, a differential modification of 6.0201 on arginine and lysine for medium label, or 8.0142 on lysine and 10.0083 on arginine for the heavy label was used for all analyses. The QuaNCAT-pep search was the same as QuaNCAT except the addition of a static modification of 452.2376 on methionine for AHA-biotin-alkyne. Prolucid results were assembled and filtered using the DTASelect (version 2.0) program(23, 24). DTASelect 2.0 uses a linear discriminant analysis to dynamically set XCorr and DeltaCN thresholds for the entire dataset to achieve a user-specified false discovery rate (FDR). In DTASelect, the modified peptides were required to be partially tryptic, less than 5ppm deviation from peptide match, and a FDR at the spectra level of 0.01. The FDRs are estimated by the program from the number and quality of spectral matches to the decoy database. For all datasets, the protein FDR was < 1% and the peptide FDR was < 0.5%. The MS data was quantified (i.e. generate heavy/light ratios) using the software, pQuant(25), which uses the DTASelect and MS1 files as the input. pQuant assigns a confidence score to each heavy/light ratio from zero to one. Zero, the highest confidence, means there is no interference signal and a confidence score of one means the peptide signals are corrupted by interference signals (i.e. very noisy). For this analysis, the median of peptide ratios with sigma less than or equal 0.5 were taken to calculate protein ratios. Each protein isoform was given a ratio and defined as “leading”, “sameset” and “subset”. “Subset” means the peptides belonging to this protein is the subset of those of the “Leading” protein. “Sameset” means the peptides belonging to this protein is the same set as those of the “Leading” protein. The ratio of protein isoform quantified by most peptides were selected as “leading protein” to represent the ratio of the protein groups in the final dataset. Only ratios with sigma less than or equal 0.5 were counted for statistics.
2) Pathway analysis
Proteins quantified using the HILAQ or QuaNCAT methods were input to the online tool PANTHER (http://pantherdb.org/), and pathway oncology analysis revealed the pathways where proteins were enriched for each strategy. Protein hits in every pathway were compared and visualized in Excel.
NSP that were observed to have more than 2 fold change between HT22 and Hek293T cell lines were the input for functional pathway analysis performed by the software Ingenuity pathway analysis (IPA) (26). 108 Proteins enriched in cell death related pathways were analyzed by STRING and visualized by Cytoscape. Colors were defined to show fold of change. Shapes were defined to indicate how this protein can affect cell death according to IPA database.
Results and Discussion
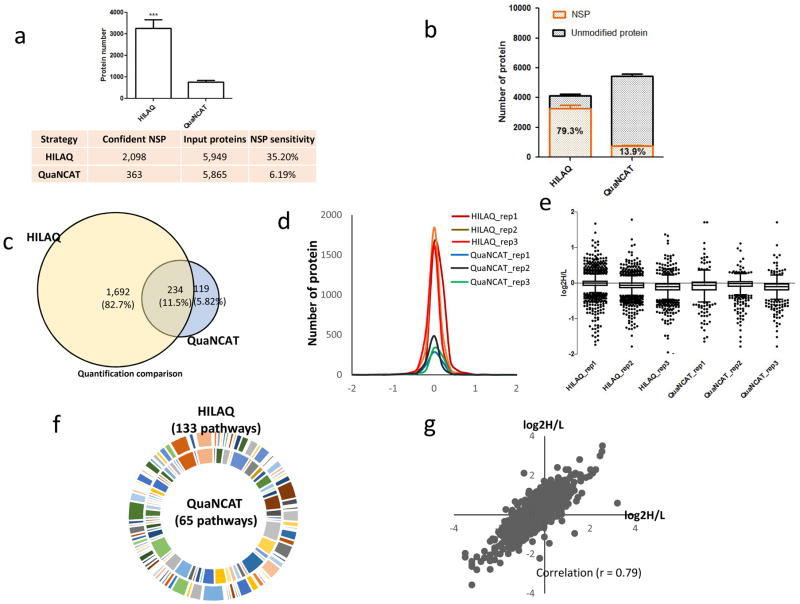
To determine how the HILAQ protocol is different from the QUANCAT protocol, HEK293T cells were starved for 30 min in medium depleted of amino acids. Then, for each protocol, two plates of cells were pulsed with different modified AAs for 1h (Fig. 1b). For the QuaNCAT protocol, Plate 1 was pulsed with two “medium” SILAC AA (i.e. 13C6-R and 13C6-K) and AHA, and Plate 2 was pulsed with two “heavy” SILAC AA (i.e. 13C615N4-R and 13C615N2-K) and AHA. For the HILAQ protocol, the plates were pulsed with AHA (Plate 1) or HAHA (Plate 2). After mixing Plate 1 and Plate 2 1:1(wt/wt) for each protocol, 2 mg of the mixture was employed for the subsequent steps. For both strategies, a click reaction (2, 16) was performed on the mixtures, followed by protein precipitation. For QuaNCAT samples, AHA proteins were enriched by neutravidin beads, followed by on-bead tryptic digestion (17) to obtain NSP peptides. The HILAQ samples were processed similarly except that the DidBIT (Direct identification of Biotin Tags) protocol was followed (17), in which the neutravidin enrichment step is performed on peptides after tryptic digestion. After AHA peptides were enriched by neutravidin beads, they were eluted with 80% acetonitrile, 0.2% formic acid, and 0.1% TFA (1). Peptides from all samples were identified using a ThermoFisher Elite Orbitrap mass spectrometer using the online peptide separation MudPIT(18). For each protocol, MS data from independent biological replicates was extracted and searched(27),(28) against a human protein database. For HILAQ analysis, mass shifts on methionine, representing either AHA-biotin-alkyne or HAHA-biotin-alkyne, were used in the database search to identify NSP. For QuaNCAT analysis, mass shifts on arginine and lysine were used in the database search to identify NSP. 2,098 NSP were confidently identified from all three biological replicates of HILAQ samples, while 363 NSP were obtained from all three biological QuaNCAT samples (Supplementary Fig. 1). There were 256 NSP identified in common from both methods (Supplementary Fig. 2). Next, the protein samples were digested without any click reaction or avidin enrichment, then analyzed by MudPIT to determine a baseline proteome measurement. These MS datasets were searched identically without any mass shifts. The number of total peptides (i.e. HILAQ-43163 and QuaNCAT-43357) and proteins (i.e. HILAQ-5949 and QuaNCAT-5865) identified were similar for both methods. There were 5330 proteins identified in baseline of both strategies (Supplementary Fig. 3). We defined NSP detection sensitivity as the number of NSP over this baseline. The NSP detection sensitivity was significantly increased from 6.19% using QuaNCAT to 35.2% using HILAQ (Fig. 2a). In addition, after affinity purification, NSP typically comprised 13.9% of the identified proteins in QuaNCAT and increased to 79.3% in HILAQ (Fig. 2b and supplementary Table 1), indicating that HILAQ enrichment is more robust. Quantification of the identified NSP showed similar quantification efficiencies for the two strategies (Supplementary Table 2). Consistent with the large increase in NSP identifications, there were 82.7% more proteins quantified by HILAQ. (Fig. 2c). NSP were quantified by an average of 8.78 peptides using the HILAQ protocol compared with 3.40 peptides using QuaNCAT (supplementary Table 1). The Gaussian distribution pattern of protein ratios and (Fig. 2d) the dispersion of protein ratios for the HILAQ and QuaNCAT strategies were similar (Fig. 2e), suggesting that HILAQ and QuaNCAT have equal accuracy. Pathway analysis was performed on the proteins quantified with the HILAQ or QuaNCAT methods. Fig. 2f shows that proteins quantified by HILAQ were enriched in 133 pathways. This is twice as many as with QuaNCAT, and suggests that HILAQ is a more informative strategy (supplementary Fig. 4). In addition, we compared our HILAQ dataset with a previously published protein turnover dataset in HEK293T cell line (PMID: 18988847). This study defined a Protein Stability Index (PSI) as measure of protein stability, with a higher PSI indicating higher protein stability and lower protein turnover rate. In the HILAC dataset, we found that the top 10 proteins (identified by the highest spectral counts) have slightly higher PSI than the 10 proteins identified with least spectral counts, suggesting proteins of lower abundance among the NSP might have relatively higher protein turnover activity. Besides using different heavy molecules, the QuaNCAT protocol employs AHA protein enrichment while the HILAQ protocol employs AHA peptide enrichment. Peptide enrichment has been demonstrated to be more sensitive than protein enrichment in biotin labeled protein identification (17). To illustrate the benefit of peptide enrichment for quantification of NSP, we developed QuaNCAT-pep, a modified QuaNCAT protocol which includes peptide enrichment rather than protein enrichment. (Supplementary Fig. 5). QuaNCAT-pep confidently identified twice as many NSPs than QuaNCAT (Supplementary Fig. 6) and NSP detection sensitivity was increased to 23.3% in QuaNCAT-pep from 6.19% in QuaNCAT (Supplementary Fig. 7). This analysis suggests that the improved performance of the HILAQ protocol over the QuaNCAT protocol is primarily due to their different AHA enrichment strategies rather than the use of different heavy molecules.
Figure 2.
Performance of HILAQ. (a) The number of NSP identified by HILAQ protocol was significantly increased over QuaNCAT. In the bottom table, confident NSP means the NSP overlapped in all three biological replicates. NSP sensitivity is the ratio of modified peptide/protein identifications over baseline. (**p<0.001; one tailed unpaired t-test, comparison with QuaNCAT) (b) Composition of identified protein by HILAQ and QuaNCAT. (c) Venn diagram analysis on quantification by HILAQ or QuaNCAT. Only proteins quantified confidently by all three replicates of HILAQ or QuaNCAT were counted and compared for this analysis. (d) Normal distribution of replicates of HILAQ and QuaNCAT. All datasets were not normalized. (e) Boxplot analysis on every replicate of HILAQ and QuaNCAT using Graphpad Prism (version 5.01). The boxplot was created with Turkey whiskers. (f) Proteins quantified by all three replicate of HILAQ or QuaNCAT were annotated using web based tool PANTHER (http://pantherdb.org/). There are twiceas many pathways enriched in HILAQ than QuaNCAT. (g) The quantification of NSP between HT22 and HEK293Tcell lines. The peptides ratios from two independent HILAQ experiments were compared. The AHA and HAHA labels were swapped in the two experiments.
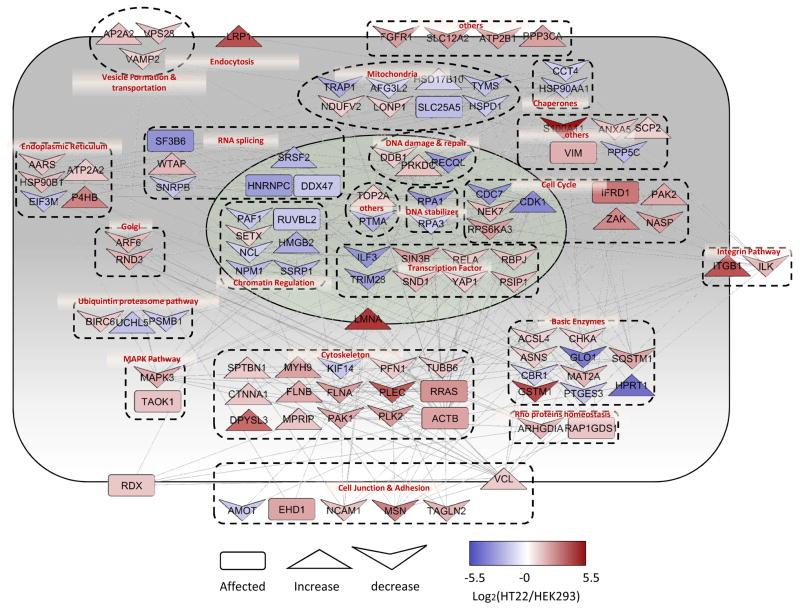
To demonstrate the usefulness of HILAQ, an in vitro model of oxytosis was analyzed. HILAQ analysis was employed to compare HT22 cells to HEK293T cells to determine the molecular vulnerability profile to oxytosis. HEK293T cells were pulsed with AHA and HT22 cells were pulsed with hAHA. In the second experiment labeling of the cells was reversed. A comparison of the swap experiments between two biological replicates showed sufficient reproducibility with a Pearson correlation coefficient of 0.79 (Fig. 2g). In total, 818 NSPs were quantified in the swap experiments. Pathway analysis was performed on 226 proteins that were observed to have at least a twofold change between the cell types with 136 up-regulated and 90 down-regulated in the HT22 cells. The most significantly (p value = 1.16 e −16) enriched pathway was cell death, with 108 proteins annotated (Fig. 3). Cell fate is dictated by the relative activity of a complex network pro- and anti-death proteins. 68 of enriched proteins have been reported to decrease cell death, suggesting that they have a protective function, while 27 have been reported to increase cell death, suggesting a pro-death function or that they are required for cell death to proceed. The remaining thirteen proteins were reported to be involved in cell death without direct evidence for halting or activating the process. Many of these altered proteins have previously been connected to cell death in brain tissue and disease models. For example, BIRC6 (Baculoviral IAP repeat-containing protein 6) is part of a protein family that inhibits cell death through the inactivation of caspases and was up-regulated in HT22 cells. It has been reported that the hippocampus down-regulated this protein in response to glutamate toxicity(29). In order for cell death to proceed, cells degrade BIRC6 through the proteasome (30). Interestingly, it has been reported that proteasome inhibitors can prevent oxytosis in HT22 cells (31, 32). TRAP1 (Tumor necrosis factor type 1 receptor-associated protein) is mitochondrial molecular chaperone and was down-regulated in HT22 cells. It has been reported that PINK1 (PTEN-induced putative kinase protein 1) phosphorylates TRAP1, which is essential for PINK1 to protect cells from cell death induced by oxidative stress (33). Mutations in PINK1 can cause Parkinson’s disease(PD), which is characterized by neurodegeneration of dopamine neurons induced by oxidative stress(34, 35). The major source of ROS in oxytosis is generated from the mitochondrial complex I and complex I dysfunction is implicated in PD pathogenesis (36, 37). The core subunit of complex I, NDUFV2, was up-regulated in HT22 cells. Finally, p62/SQSTM1 (Sequestosome-1) was up-regulated in HT22 cells. This protein has been localized to protein aggregates that accumulate in multiple neurodegenerative diseases (38–40). It is an autophagy receptor that binds poly-ubiquitinated proteins to direct them to lysosomes for degradation. It was reported that SQSTM1 can protect cells from cell death induced by mutant huntingtin, which causes the neurodegenerative disease, Huntington’s Disease (41). In summary, this HILAQ analysis demonstrates the validity of HT22 cells as a tool to study the molecular details of cell death involved in neurodegenerative diseases.
Figure 3.
HILAQ analysis of oxytosis cell model. Interaction network analysis on 108 cell death enriched proteins with 2 fold change after 1 h labeling in HEK293T and HT22 cells. The protein network and localization were analyzed by String and visualized by Cytoscape. The log ratio used in visualization was the average ratio of replicates. The red color represents proteins up-regulated in HT22 while the blue color indicates down-regulation. Different shapes used in the figure represents the effects of the protein on cell apoptosis according to previous reports.
Conclusion
We have described a novel strategy, HILAQ, to quantify the newly synthesized proteome. Since the HILAQ strategy employs two modified AA while the QuaNCAT method employs five modified AA, HILIQ simplifies the workflow and bioinformatics analysis. While QuaNCAT employs AHA solely for enrichment of NSP and relies on the SILAC AA for confirmation and quantification of a NSP, the HILAQ method exploits the AHA molecule for enrichment, confirmation and quantification of the NSP. We also demonstrated that the quantification of NSP is greatly improved when peptide AHA enrichment is employed rather than protein AHA enrichment. Overall, the high sensitivity of this approach allows it to be used to monitor proteomic dynamics using short-time pulse labeling instead of traditional whole-proteome labeling.
Supplementary Material
Acknowledgments
Funding from the following National Institute of Health grants: P41 GM103533, R01 MH067880, R01 MH100175 to Yates laboratory.
References
- 1.McClatchy DB, Ma Y, Liu C, Stein BD, Martínez-Bartolomé S, Vasquez D, Hellberg K, Shaw RJ, Yates JR. Pulsed Azidohomoalanine Labeling in Mammals (PALM) Detects Changes in Liver-Specific LKB1 Knockout Mice. Journal of Proteome Research. 2015;14(11):4815–4822. doi: 10.1021/acs.jproteome.5b00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolb HC, Finn MG, Sharpless KB. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem Int Ed Engl. 2001;40(11):2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 3.Eichelbaum K, Winter M, Diaz MB, Herzig S, Krijgsveld J. Selective enrichment of newly synthesized proteins for quantitative secretome analysis. Nat Biotech. 2012;30(10):984–990. doi: 10.1038/nbt.2356. [DOI] [PubMed] [Google Scholar]
- 4.Howden AJM, Geoghegan V, Katsch K, Efstathiou G, Bhushan B, Boutureira O, Thomas B, Trudgian DC, Kessler BM, Dieterich DC, Davis BG, Acuto O. QuaNCAT: quantitating proteome dynamics in primary cells. Nat Meth. 2013;10(4):343–346. doi: 10.1038/nmeth.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Y, Bao J, Zhao X, Shen H, Lv J, Ma S, Zhang X, Li Z, Wang S, Wang Q, Ji J. Activated Cyclin-Dependent Kinase 5 Promotes Microglial Phagocytosis of Fibrillar β-Amyloid by Up-regulating Lipoprotein Lipase Expression. Molecular & Cellular Proteomics : MCP. 2013;12(10):2833–2844. doi: 10.1074/mcp.M112.026864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheldon AL, Robinson MB. The role of glutamate transporters in neurodegenerative diseases and potential opportunities for intervention. Neurochem Int. 2007;51(6–7):333–55. doi: 10.1016/j.neuint.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olney JW. Brain lesions, obesity, and other disturbances in mice treated with monosodium glutamate. Science. 1969;164(3880):719–21. doi: 10.1126/science.164.3880.719. [DOI] [PubMed] [Google Scholar]
- 8.Choi DW. Glutamate neurotoxicity and diseases of the nervous system. Neuron. 1988;1(8):623–34. doi: 10.1016/0896-6273(88)90162-6. [DOI] [PubMed] [Google Scholar]
- 9.Tan S, Wood M, Maher P. Oxidative stress induces a form of programmed cell death with characteristics of both apoptosis and necrosis in neuronal cells. J Neurochem. 1998;71(1):95–105. doi: 10.1046/j.1471-4159.1998.71010095.x. [DOI] [PubMed] [Google Scholar]
- 10.Vanden Berghe T, Linkermann A, Jouan-Lanhouet S, Walczak H, Vandenabeele P. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat Rev Mol Cell Biol. 2014;15(2):135–47. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 11.Peter C, Braidy N, Zarka M, Welch J, Bridge W. Therapeutic approaches to modulating glutathione levels as a pharmacological strategy in Alzheimer’s disease. Curr Alzheimer Res. 2015;12(4):298–313. doi: 10.2174/1567205012666150302160308. [DOI] [PubMed] [Google Scholar]
- 12.Jiang T, Sun Q, Chen S. Oxidative stress: A major pathogenesis and potential therapeutic target of antioxidative agents in Parkinson’s disease and Alzheimer’s disease. Prog Neurobiol. 2016;147:1–19. doi: 10.1016/j.pneurobio.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Davis JB, Maher P. Protein kinase C activation inhibits glutamate-induced cytotoxicity in a neuronal cell line. Brain Res. 1994;652(1):169–73. doi: 10.1016/0006-8993(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 14.Soucek T, Cumming R, Dargusch R, Maher P, Schubert D. The regulation of glucose metabolism by HIF-1 mediates a neuroprotective response to amyloid beta peptide. Neuron. 2003;39(1):43–56. doi: 10.1016/s0896-6273(03)00367-2. [DOI] [PubMed] [Google Scholar]
- 15.Seiler A, Schneider M, Forster H, Roth S, Wirth EK, Culmsee C, Plesnila N, Kremmer E, Radmark O, Wurst W, Bornkamm GW, Schweizer U, Conrad M. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab. 2008;8(3):237–48. doi: 10.1016/j.cmet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Speers AE, Cravatt BF. Activity-Based Protein Profiling (ABPP) and Click Chemistry (CC)-ABPP by MudPIT Mass Spectrometry. Curr Protoc Chem Biol. 2009;1:29–41. doi: 10.1002/9780470559277.ch090138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiapparelli LM, McClatchy DB, Liu HH, Sharma P, Yates JR, 3rd, Cline HT. Direct detection of biotinylated proteins by mass spectrometry. J Proteome Res. 2014;13(9):3966–78. doi: 10.1021/pr5002862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Washburn MP, Wolters D, Yates JR. 3rd, Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 19.McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, Yates JR. 3rd, MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun Mass Spectrom. 2004;18(18):2162–8. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 20.Xu T, Venable JD, Park SK, Cociorva D, Lu B, Liao L, Wohlschlegel J, Hewel J, Yates JR. ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Molecular & Cellular Proteomics. 2006;5(10):S174–S174. [Google Scholar]
- 21.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2(1):43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 22.Sadygov RG, Eng J, Durr E, Saraf A, McDonald H, MacCoss MJ, Yates JR. 3rd, Code developments to improve the efficiency of automated MS/MS spectra interpretation. J Proteome Res. 2002;1(3):211–5. doi: 10.1021/pr015514r. [DOI] [PubMed] [Google Scholar]
- 23.Cociorva D, DLT, Yates JR. Validation of tandem mass spectrometry database search results using DTASelect. Curr Protoc Bioinformatics. 2007;Chapter 13(Unit 13):4. doi: 10.1002/0471250953.bi1304s16. [DOI] [PubMed] [Google Scholar]
- 24.Tabb DL, McDonald WH, Yates JR. 3rd, DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1(1):21–6. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C, Song CQ, Yuan ZF, Fu Y, Chi H, Wang LH, Fan SB, Zhang K, Zeng WF, He SM, Dong MQ, Sun RX. pQuant improves quantitation by keeping out interfering signals and evaluating the accuracy of calculated ratios. Anal Chem. 2014;86(11):5286–94. doi: 10.1021/ac404246w. [DOI] [PubMed] [Google Scholar]
- 26.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF, Res P Inflamm; Host Response to Injury Large Scale Collab. A network-based analysis of systemic inflammation in humans. Nature. 2005;437(7061):1032–7. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 27.Eng J, McCormack A, Yates J. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 28.Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK, Cociorva D, Lu B, Liao L, Hewel J, Han X, Wong CC, Fonslow B, Delahunty C, Gao Y, Shah H, Yates JR., 3rd ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J Proteomics. 2015;129:16–24. doi: 10.1016/j.jprot.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sokka AL, Mudo G, Aaltonen J, Belluardo N, Lindholm D, Korhonen L. Bruce/apollon promotes hippocampal neuron survival and is downregulated by kainic acid. Biochem Biophys Res Commun. 2005;338(2):729–35. doi: 10.1016/j.bbrc.2005.09.197. [DOI] [PubMed] [Google Scholar]
- 30.Qiu XB, Markant SL, Yuan J, Goldberg AL. Nrdp1-mediated degradation of the gigantic IAP, BRUCE, is a novel pathway for triggering apoptosis. EMBO J. 2004;23(4):800–10. doi: 10.1038/sj.emboj.7600075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Leyen K, Siddiq A, Ratan RR, Lo EH. Proteasome inhibition protects HT22 neuronal cells from oxidative glutamate toxicity. J Neurochem. 2005;92(4):824–30. doi: 10.1111/j.1471-4159.2004.02915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maher P. Proteasome inhibitors prevent oxidative stress-induced nerve cell death by a novel mechanism. Biochem Pharmacol. 2008;75(10):1994–2006. doi: 10.1016/j.bcp.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pridgeon JW, Olzmann JA, Chin LS, Li L. PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol. 2007;5(7):e172. doi: 10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatano Y, Li Y, Sato K, Asakawa S, Yamamura Y, Tomiyama H, Yoshino H, Asahina M, Kobayashi S, Hassin-Baer S, Lu CS, Ng AR, Rosales RL, Shimizu N, Toda T, Mizuno Y, Hattori N. Novel PINK1 mutations in early-onset parkinsonism. Ann Neurol. 2004;56(3):424–7. doi: 10.1002/ana.20251. [DOI] [PubMed] [Google Scholar]
- 35.Tan S, Sagara Y, Liu Y, Maher P, Schubert D. The regulation of reactive oxygen species production during programmed cell death. J Cell Biol. 1998;141(6):1423–32. doi: 10.1083/jcb.141.6.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schapira AH, Cooper JM, Dexter D, Jenner P, Clark JB, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. Lancet. 1989;1(8649):1269. doi: 10.1016/s0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- 37.Betarbet R, Sherer TB, Greenamyre JT. Animal models of Parkinson’s disease. Bioessays. 2002;24(4):308–18. doi: 10.1002/bies.10067. [DOI] [PubMed] [Google Scholar]
- 38.Kuusisto E, Salminen A, Alafuzoff I. Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport. 2001;12(10):2085–90. doi: 10.1097/00001756-200107200-00009. [DOI] [PubMed] [Google Scholar]
- 39.Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M, Aguzzi A, Denk H. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol. 2002;160(1):255–63. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuusisto E, Salminen A, Alafuzoff I. Early accumulation of p62 in neurofibrillary tangles in Alzheimer’s disease: possible role in tangle formation. Neuropathol Appl Neurobiol. 2002;28(3):228–37. doi: 10.1046/j.1365-2990.2002.00394.x. [DOI] [PubMed] [Google Scholar]
- 41.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.