SUMMARY
An atomic-detail model of the Marburg virus glycoprotein in complex with a neutralizing human monoclonal antibody designated MR78 was constructed using Phenix.Rosetta starting from a 3.6Å crystallographic density map. The Asp at T6 in the HCDR3’s bulged torso cannot form the canonical salt bridge as position T2 lacks an Arg or Lys residue. It instead engages in a hydrogen bond interaction with a Tyr contributed by the HCDR1 loop. This inter-CDR loop interaction stabilizes the bulged conformation needed for binding to the viral glycoprotein: a Tyr to Phe mutant displays a binding affinity reduced by a factor of at least 10. We found that 5% of a database of 465 million human antibody sequences has the same residues at T2 and T6 positions in HCDR3 and Tyr in HCDR1 that could potentially form this Asp-Tyr interaction and that this interaction might contribute to a non-canonical bulged torso conformation.
Keywords: Phenix.Rosetta Refinement, Complementarity Determining Region (CDR), Bulged Torso, Antibody Structure, Binding affinity
eTOC Blurb
Sangha et al. have discovered that the conformation of the human antibody MR78 HCDR3 loop that binds to Marburg virus glycoprotein is stabilized by a non-local hydrogen bond between an Asp at T6 position of HCDR3 and a Tyr in HCDR1.
INTRODUCTION
Marburg virus (MARV) belongs to the Filovirus family along with a cuevavirus (Lloviu virus) and five ebolaviruses (Ebola, Sudan, Reston, Bundibuygo and Tai Forest viruses). MARV was first discovered in 1967, and has since re-emerged multiple times to cause deadly outbreaks among humans. Recent outbreaks have been associated with up to ~90% lethality(MMWR, 2005), but no specific treatments are yet approved for MARV infection. Antibody therapy against filoviruses is an area of increasing interest, with many Ebola virus-specific therapies under development, including a compound in a clinical trial(Borio et al., 2015). The MARV surface glycoprotein (MARV GP) consists of a trimer of glycoprotein 1 (GP1) and glycoprotein 2 (GP2) and is the only known target of protective antibodies. Recently, a number of MARV-neutralizing monoclonal antibodies from a human survivor of MARV infection were isolated and shown to bind at GP1 epitopes and to potentially inhibit the binding of the NPC1 receptor(Flyak et al., 2015). The first structure of MARV GP, bound to one of these neutralizing antibodies, designated MR78, was determined using X-ray crystallography to 3.6Å resolution(Hashiguchi et al., 2015). These studies revealed the mechanism of inhibition of virus entry and paved the way for immunotherapeutic development against MARV disease.
Computational techniques(Marcatili et al., 2014, Maier and Labute, 2014, Yamashita et al., 2014, Shirai et al., 2014, Zhu et al., 2014, Sircar et al., 2009, BIOVIA, 2012, Messih et al., 2014) play an important role in constructing models for antibody/antigen complexes, as atomic-detail can be lacking from experimental density maps when determined at low resolution. Antibody modeling techniques use structure-based knowledge from high-resolution antibody structures available in the Protein Data Bank (PDB) to model the framework region and the six complementarity-determining regions (CDRs) of the heavy and light chains of antibody. Five out of the six CDRs typically assume canonical conformations that can be predicted from their amino acid sequence. CDR3 of the heavy chain (HCDR3), however, remains a challenge to the modeling techniques developed so far because of its variability in amino acid composition and length(Zhu et al., 2014). To overcome these limitations, rules have been proposed to classify the conformation of the base region of the HCDR3 loop, also termed the torso region(Shirai et al., 1996, Oliva et al., 1998, Morea et al., 1998, Shirai et al., 1999, Koliasnikov et al., 2006, North et al., 2011). The torso region comprises the first three residues on the N-terminal side of HCDR3 loop after the Cys residue of framework region 3, as well as the last four residues on the C-terminal side of HCDR3 loop before the Trp residue of framework region 4 (Figure 1). According to these rules, the presence of a basic Arg/Lys residue at position T2 plus Asp at position T6 should lead to a bulged (kinked) torso, while the absence of a basic residue at T2, but presence of Asp at T6 should lead to a non-bulged (extended) conformation in HCDR3 torso(Shirai et al., 1999). These rules have been extended to include the chemical nature of other residues in the stem part of the HCDR3 loop, as well as residues at positions 36, 46 and 49 in the light chain, in order to explain exceptions to the basic rules(Kuroda et al., 2008). However, the inter- and intra-chain CDR loop interactions in antibody structures have not yet been systematically investigated. Understanding the role of CDR loop interactions in antibody structural stability and in optimizing its binding to the target pathogen can potentially improve the antibody modeling and design of antibodies for biotherapeutic development.
Figure 1.
MR78 HCDR3 sequence. The seven residues comprising the HCDR3 torso in MR78 are shown in blue. Residues are numbered from T1 to T7 based on their position. Framework regions 3 (FR3) and 4 (FR4) are shown in green.
Weitzner et al. recently added another geometric parameter termed τ101, a pseudo bond angle of the Cα atoms of the three C-terminal residues in HCDR3 loop, along with a pseudo dihedral angle described previously as α101 (Shirai et al., 1999). The dihedral angle α101 positions the kink relative to the framework of the antibody and the bond angle τ101 measures the degree of the kink in the torso region. The study showed that the distribution of τ101 measured for antibody structures is centered at 39° and the distribution of α101 is centered at 101° (Weitzner et al., 2015).
Antibody MR78 lacks a basic residue in T2, but features an Asp in position T6 (Figure 1). The initial model of MR78 in complex with the MARV GP (PDB ID 3×2D) derived from a density map at a resolution of 3.6Å suggests an unusual conformation for HCDR3. Accordingly, we set out to use the Rosetta suite of programs(Kaufmann et al., 2010, Bender et al., 2016) to investigate this HCDR3 confirmation more closely. Specifically, we constructed an atomic detail model of MR78 in complex with the MARV GP that is consistent not only with the low-resolution density map but also biophysically realistic as evaluated with the Rosetta energy function. This method is an attractive approach as Rosetta provides techniques that optimize model coordinates to agree simultaneously with its energy function(DiMaio et al., 2015, Wang et al., 2015) and with experimental data, including scattering factors from X-ray crystallography or cryo-electron microscopy. This strategy can help improve experimental models lacking atomic details. Phenix.Rosetta refine(DiMaio et al., 2013) utilizes Rosetta’s conformational sampling technique and knowledge-based all-atom energy function during structure refinement in Phenix software(Adams et al., 2010). In the combined Phenix.Rosetta refinement approach, Phenix is used to perform a bulk solvent correction, calculate electron density maps and refine B factors, while Rosetta is used for force field, minimization and sampling to optimize model geometry.
In this paper, we report an atomic-detailed model of the MR78:MARV GP complex as calculated using Phenix.Rosetta refinement followed by loop modeling(Kaufmann et al., 2010) in Rosetta. The protocol resolved energetic frustrations in the conformations of HCDR3 and HCDR1 and also proposed additional interactions between the CDRs and GP1 of MARV that interestingly, were not observed in the experimental electron density maps(Hashiguchi et al., 2015).
To confirm the altered conformation of HCDR3, we obtained a high-resolution structure of the unbound MR78 and compared it to the Phenix.Rosetta model of the MR78 antibody component of this complex. An interaction between two complementarity-determining regions of heavy chain, HCDR3 (Asp116 at T6) and HCDR1 (Tyr37), observed in the refined model, the apo MR78 structure and the bound MR78 structure was noted as potentially important for antibody reactivity. To understand the role of this T6Asp116 -Tyr37 hydrogen bond, we compared the structure and binding affinity of the wild-type MR78 structure with MR78 containing a Tyr37Phe point mutation.
RESULTS
The Phenix.Rosetta model of the MR78:GP complex
The model discussed in the present work was constructed in a two-step procedure: the first step was a refinement of the original experimental coordinates (PDB ID 3×2D) with Phenix.Rosetta. The second step reconstructed the conformation of the HCDR1 loop to improve the geometry of Ser34 and Ser35. Changes to the experimental model (PDB ID 3×2D) upon Phenix.Rosetta refinement were observed mostly in the MR78 backbone. Further, several side-chain reorientations occurred in GP1 residues close to the interface. The quality of the model generated after Phenix.Rosetta refinement was measured by comparing the Molprobity score and Rfree factor to the published structure. The Rwork changed from 0.25 to 0.21 while the Rfree remained unaffected, suggesting that although the geometry was improved, we did not obtain a significantly better fit to experimental X-ray data. The refined structure had a Molprobity score of 1.67 compared to 2.08 of 3×2D, mainly due to improvements in the clashscore. The reconstruction of HCDR1 altered the geometry of Ser34 and Ser35 and improved the Molprobity score to 1.48, although a few more residues now were marked as Ramachandran outliers (Table 1).
Table 1.
Comparison of Molprobity score and Rfree for the experimental and the Phenix.Rosetta structures.
| Metric | Experimental | Phenix.Rosetta | HCDR1 modeling |
|---|---|---|---|
| R-free | 0.279 | 0.278 | - |
| R-work | 0.247 | 0.209 | - |
| Clashscore | 8.00 | 3.29 | 1.82 |
| Ramachandran Outliers | 0.80% | 1.55% | 2.19% |
| Sidechain Outliers | 1.0% | 0.0% | 0.0% |
| Molprobity Score | 2.08 | 1.67 | 1.48 |
Thr111A of MR78 interacts with the side chain of Arg73 of GP1
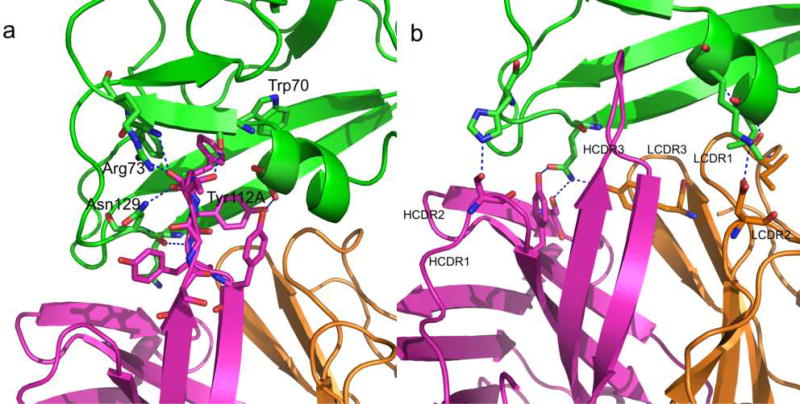
The original experimental structure and the subsequent Phenix.Rosetta model agree that the major interactions between MARV GP1 and MR78 occur via the HCDR3. Phe111B of MR78 HCDR3 packs between Trp70 and Phe72 of GP1 (Figure 2). Further, both the experimental and the refined model indicate that additional electrostatic interactions occur between the carbonyl oxygen of MR78 Phe111B and the side chain of GP1 Asn129 and the amide nitrogen of MR78 Tyr112A with the carbonyl oxygen of GP1 Gln128. Tyr112 of HCDR3 forms a hydrogen bond with Ser67 in α-helix 1 of GP1.
Figure 2.
Antibody:Antigen interactions in the Phenix.Rosetta model of MARV GP1 (green) and MR78 (heavy chain in magenta and light chain in orange). (a) MR78 interacts with MARV GP1 mainly through the HCDR3 via aromatic stacking and electrostatic interactions. (b) The remaining five CDRs also interact with GP1 via hydrophobic and electrostatic interactions.
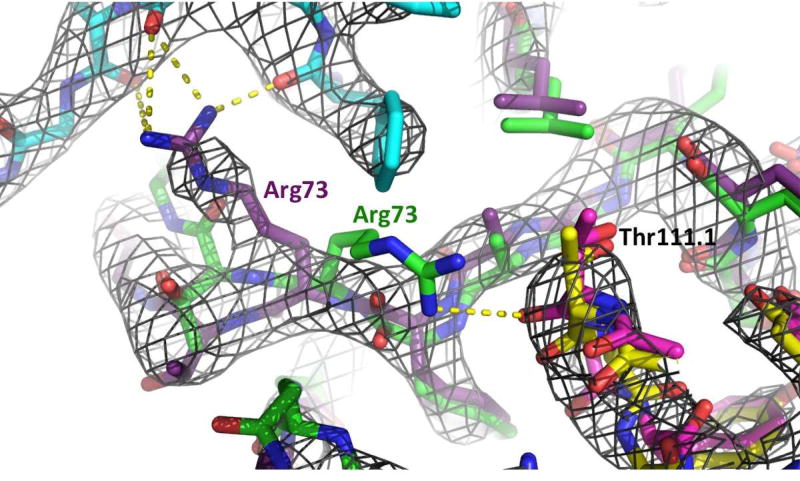
However, the Phenix.Rosetta model deviates from the experimental electron density map in the position of residue Arg73. In the Phenix.Rosetta model, the main chain carbonyl oxygen of Thr111A interacts with the side chain of Arg73 of GP1. In the experimental X-ray structure, Thr111A of HCDR3 does not interact with Arg73 of GP1 (Figure 3).
Figure 3.
Superimposition of the experimental X-ray structure (GP1; deep purple, GP2; cyan, MR78; Yellow) and the Phenix.Rosetta refined model (GP1; green, MR78; magenta) represented by stick models. In black is the 2Fo-Fc electron density map of the MARV GP – MR78 complex, created by Phenix and illustrated using PyMOL (www.pymol.org), contoured at 1.5σ. This experimental X-ray map indicates that Arg73 (purple) interacts with GP2 (cyan), while the energy-minimized model suggests that Arg73 (green) interacts with Thr111 of HCDR3 (magenta).
Residues of the other five CDRs of MR78 also interact with GP1: Ile29 and Asn37 of LCDR1 (QASQVISNYLN) interacts with the backbone nitrogen of Leu64 and Gln126 in GP1 α helix 1, respectively, Asp56 of LCDR2 (YDTSNLKT) interacts with the side chain of Lys68 in GP1 α helix 1, Tyr107 of LCDR3 (QQYENLQFT) interacts with the side chain of GP1 Gln128 in β strand 2 and Ser35 and Tyr38 of HCDR1 (TVSGGSISSSSYYWG) interact with His131 and Gln128 of β strand 2 of GP1. Ser66 of HCDR2 (SVYYSGGAS) interacts with Gln128 of β strand 2 of GP1 (Figure 2). These additional interactions led to an increase in the predicted binding affinity of MR78 to MARV GP1 by 4 Rosetta Energy Units (REU) after Phenix.Rosetta refinement.
The six CDR conformations in the refined structure fall into canonical classes of L1-11-1, L2-8-1, L3-9-1, H1-15-1, H2-9-1 and H3-17-1, respectively. In the crystal structure, CDRs L1, L2, L3 and H3 were classified under different classes and H1 was not classifiable in the experimental structure(North et al., 2011).
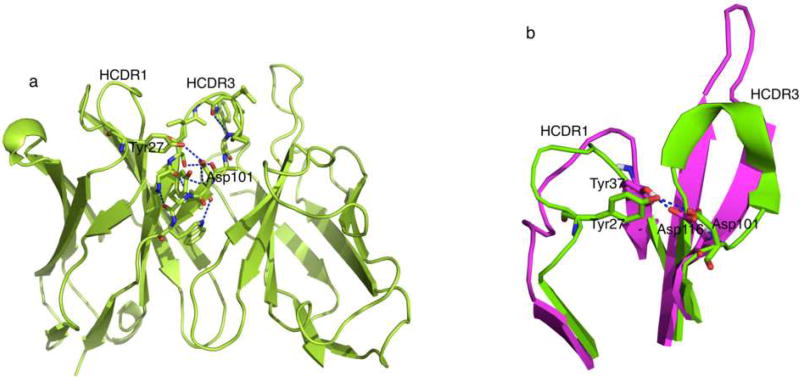
Bulged torso in MR78 HCDR3 is stabilized by a hydrogen bond between Asp116 at T6 and Tyr37 from HCDR1
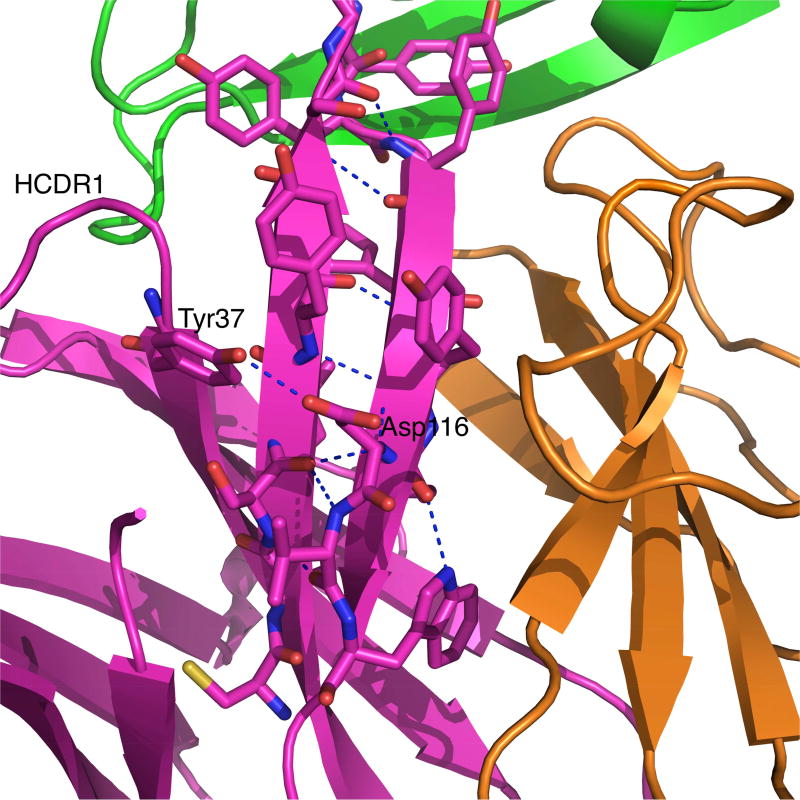
One major change upon Phenix.Rosetta refinement of the experimental model of the MR78:MARV GP complex occurred in the torso region of the HCDR3 loop. The first three residues on the N-terminal side and the last four residues on the C-terminal side of HCDR3 (ASIYGSGTFYYYFYMDV) constitute the torso region. These seven residues are named T1 to T7 starting from N-terminal of HCDR3 loop to the C-terminal such that Ala105 is at T1 and Val117 is at T7 position (Figure 1). The refined structure shows that antibody MR78 has a bulged torso. While the torso is also bulged in the original PDB structure (PDB ID 3×2D), the geometric parameters in the refined structure align much closer to the canonical values for antibody structures in Weitzner et al. Specifically, the τ101 is 100.3° and α101 is 28.2° for the refined structure. The canonical values are 101° and 39°. The values for τ101 and α101 in the four copies of MR78 in PDB ID 3×2D are 124.2, 115.1, 121.7, 110.6 and −7.6, 30.7, 11.5, −46.2 for antibody heavy chains D, H, L and P, respectively. An unusual interaction of the Asp116 at T6 of HCDR3 with Tyr37 in the HCDR1 (TVSGGSISSSSYYWG) loop was observed. One of the four interactions, a salt bridge between Asp at T6 and Arg/Lys at T2, usually seen in bulged torso is replaced by this inter HCDR loop T6Asp116-Tyr37 interaction in the absence of a basic residue at T2 in MR78. The other three typical interactions observed in bulged torsos, i.e., Trp118 with carbonyl oxygen of T5 (Met115 in MR78), a backbone-backbone hydrogen bond between T2 Ser106 and T6 Asp116 and a backbone-backbone hydrogen bond between Tyr108 and Tyr114, are preserved. Thus the refined MR78 bulged torso has all the four stabilizing hydrogen bonds present that could lead to ideal values for τ101 and α101.
A ladder of hydrogen bonds in the absence of a basic residue in the stem part between torso residues T3 and T4 can be observed in Figure 4. This HCDR3 loop structure follows the rule iii in Kuroda et al.(Kuroda et al., 2008, Shirai et al., 1999) which explains the effect of torso conformations on the conformation of the stem region of HCDR3 loop in antibody structure. A straight conformation of HCDR3 loop with H-bond ladder like in MR78 is infrequent in antibody structures as the majority of the long HCDR3 loops (with >14 amino acids) adopt non-straight (bent or broad) conformations(Tsuchiya and Mizuguchi, 2016). The position of Gly111 residue is more toward the N terminus of the HCDR3 loop, as is expected for loops with a bulged torso conformation(Weitzner et al., 2015).
Figure 4.
An unusual interaction of Asp116 at T6 with Tyr37 from HCDR1 loop in the MR78 torso region optimizes the bulge. The majority of the antibody structures with bulged torso have T6 Asp interacting with local HCDR3 residues, especially with Arg/Lys at T2 if present. A non-local interaction like T6Asp116-Ty37 between HCDR3 and HCDR1 loops seen in MR78 is extremely uncommon in known antibody structures.
The HCDR1 loop is stabilized in a H1-15-1 conformation
Another region with major conformational change from the experimental structure is the HCDR1 loop. The initial Phenix.Rosetta model maintained some of the energetic frustrations for Ser34 and Ser35 residues in HCDR1 loop stemming from the experimental coordinates (PDB ID 3×2D). Therefore, the geometry of these residues in the HCDR1 loop was reconstructed using Rosetta loop modeling. The optimized HCDR1 loop residues interact with GP1 via Ser35 to His131 and Phe38 to Gln128 in β-strand 2 of GP1 as mentioned above. In the original model (PDB ID 3×2D), Ser35 and His131 are 4.7 Å apart. Upon remodeling, the HCDR1 loop went from not being classified under any of the clusters to H1-15-1 cluster described in North et al.
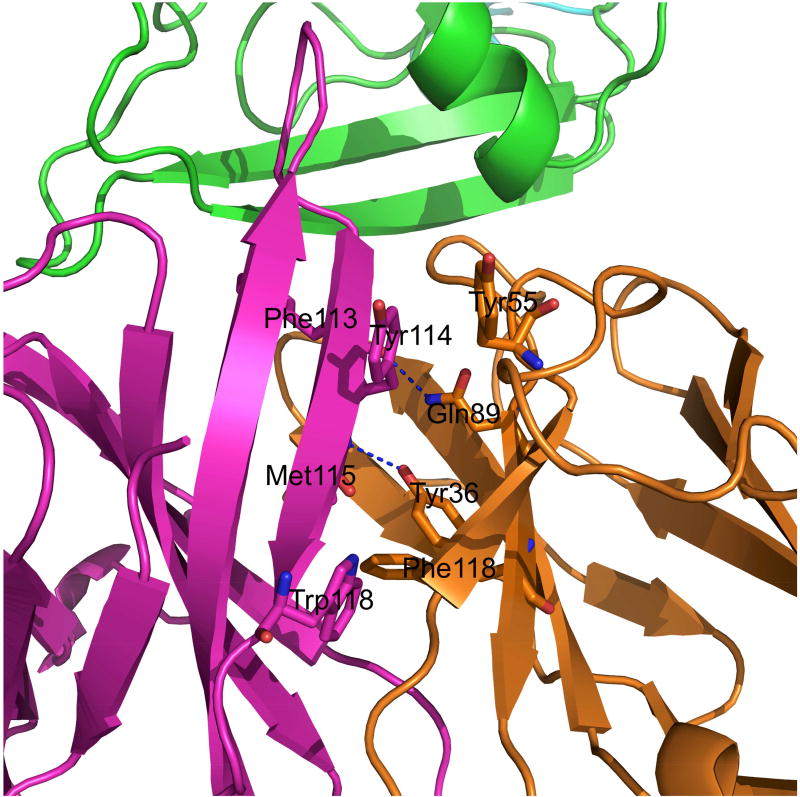
An interaction between Phe113 in the HCDR3 loop and Gln 89 of the MR78 light chain contribute to the relative orientation of heavy and light chain in MR78
The relative orientation of heavy and light chains plays an important role in the stability of the antibody structure and its interaction with the antigen at the interface as the HCDR3 loop stem is part of the heavy/light chain interface. Some interactions at the heavy and light chain interface are conserved among all antibody structures, e.g., the H-bond between L36 Tyr and residue at T5 position (Met115 in MR78). The Phenix.Rosetta model shows an additional interaction between Gln 89 of the MR78 light chain and the carbonyl oxygen of Phe113 in the HCDR3 loop of the heavy chain (Figure 5). Hydrophobic packing between two conserved pairs of aromatic residues in the heavy and light chain is also present: heavy chain Tyr114 - Tyr55 light chain, heavy chain Trp118 – Phe118 of light chain. The six AB angle parameters HL, HC1, LC1, HC2, LC2 and dc for MR78, as deduced from the ABangle program(Dunbar et al., 2013), are −58.3°, 70.1°, 122.8°, 117.7°, 81.3° and 16.8Å respectively.
Figure 5.
The heavy (magenta) and light chain (orange) interface packing in MR78. Along with all the interactions conserved in antibody structures, MR78 makes an additional interaction between Gln89 of light chain and Phe113 of heavy chain.
Comparison of MR79 with antibody structures from PDB reveals one other antibody with a similar interaction of the Asp at T6 with Tyr from HCDR1
Interactions of torso residues with distant residues from other CDR loops may affect the torso conformation. We wanted to investigate the role of the interaction between HCDR1 Tyr and HCDR3 T6 Asp in causing or supporting the bulge in MR78. Thus, we looked for antibody structures in the PDB with similar bulged torsos as in MR78. For this purpose, we collected a set of 345 antibody structures determined at a resolution better than 3Å, in which the T6 residue is Asp and T2 is neither Arg nor Lys. Only one antibody, termed 26-10 (PDB ID 1IGJ, resolution 2.5Å) had a bulged conformation with a similar interaction of Asp T6 with a Tyr from the HCDR1 loop (Figure 6(a)). Antibody 26-10 is a murine monoclonal antibody used to treat fatal digoxin (a steroid used in treatment of congestive heart failure) intoxication(Jeffrey et al., 1993). 26-10 has Asp101 at T6 and Gly94 at T2 like MR78. Although the position of these interacting Tyr residues is different in sequence between antibody 26-10 (fifth in a 13 residue long loop) and MR78 (third last in a 15 residue long loop), they are in close spatial proximity to Asp T6 in 3D space (Figure 6(b)). The τ101 and α101 values for 1IGJ are 101.1° and 49.8°, respectively. Along with T6 Asp101 to Tyr27 from HCDR1 interaction, there are only two out of three other hydrogen-bonds supporting a bulged torso present in antibody 2610: a) a hydrogen-bond between the T5 Met100B carbonyl oxygen and the Trp103 side chain and b) a backbone-backbone hydrogen bond between T2 Gly94 and T6 Asp101. The backbone-backbone hydrogen bond between T4 Ala100A and Ser96 on N terminus of HCDR3 loop is missing. The degree of kink (τ101 = 101.1°) with Asp-Tyr interaction present in antibody 26-10 is similar to that in refined MR78 structure and is the close to the median value for τ101 bond angle distributions in known antibody structures with bulged torso. The other copy of anti-digoxin antibody in 1IGJ (chain D), however, does not show the Asp-Tyr interaction between the two HCDR loops and the degree of kink in this chain deviates from the median value of 101° to 97.8° along with α101 = 50.2°.
Figure 6.
(a) In the absence of a basic residue at T2, the bulge in the torso region of HCDR3 loop in Antibody 26-10 (1IGJ_B.pdb) shows an interaction of Asp101 at T6 with Tyr27 from HCDR1 similar to the case of MR78. (b) Position of the Tyr residue interacting with Asp in HCDR3 torso is different in HCDR1 sequence in MR78 (magenta) and 1IGJ (green).
In the PDB, we identified 27 unique heavy chains with Tyr at fifth position of a 13 amino acid long HCDR1 and Asp at T6 without Arg/Lys at T2 in HCDR3 loop as in anti-digoxin antibody. None of these antibodies make the Asp-Tyr interaction seen in MR78 or antibody 26-10 (PDB 1IGJ, chain B). Three out of 27 have an extended torso (1UWE_H, 3C0_C and 4FQJ_L). Out of the remaining 24 antibody structures,11 maintain at least 3 out of the 4 hydrogen bond interactions that support the bulge and have τ101 closer to 101°, the median value for degree of kink in known antibody structures. In four of these 24 antibody structures, the Asp-Tyr interaction seen in 1IGJ is replaced by Asp-Arg (5I1L_B and 5L6Y_H) or Asp-Thr (5CEY_X and 5FYJ_H), residue immediately next to the T3 residue on N-terminal side, and the τ101 value is 100.1° (5L6Y_H), 101.7° (5I1L_B), 102° (5FYJ_H) and 100.8° (5CEY_X), respectively.
Phenix.Rosetta model of MR78 agrees with subsequently determined high-resolution structure of the apo MR78 Fab
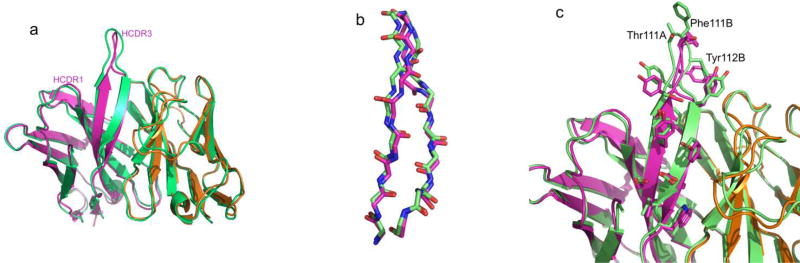
A structure of the unbound MR78 Fab was determined at 1.9Å resolution to test the conformation of the HCDR3 torso region and the HCDR1 in the apo state. This unbound, apo MR78 structure also shows a bulged torso with T6Asp116-Tyr37 interaction maintained and a conformation of the HCDR1 loop similar to that of the Phenix.Rosetta model (Figure 7). The interaction between Ser35 of HCDR1 and Tyr57 of HCDR2 seen in the experimentally determined complex was not observed in the unbound MR78 structure or the optimized model. Overall the Cα-RMSD between the Fv region of the unbound structure and the Phenix.Rosetta bound model of MR78 was 0.73Å.
Figure 7.
(a) Comparison of MR78 as observed in the Phenix.Rosetta-optimized GP complex (magenta and orange) and the high-resolution apo (unbound) structure (green). (b) The backbone conformation and (c) residue side chain orientation differs at the apex of HCDR3 loop in apo MR78.
Many antibodies use a pre-formed structure apo structure with minimal changes upon antigen binding to minimize the entropic cost of binding(Sela-Culang et al., 2012). Only a few antibodies are known to undergo major conformational changes including a transformation from a non-bulged to bulged torso (BV04-01: 1cbv and 1nbv)(Herron et al., 1991). For MR78, the apex of the HCDR3 loop undergoes backbone conformational change upon binding with some side-chain reorientations for residues Thr111A, Phe111B and Tyr112B (Figure7b and 7c). The CG-CB-CA-N dihedral angle for the three residues differ by 100° or more between apo and bound form of MR78.
An MR78 Tyr37Phe mutant antibody displays slightly increased B-factors in the HCDR regions while maintaining the MR78 wild-type apo conformation
To understand the effect of the non-canonical inter-HCDR T6Asp116 -Tyr37 interaction seen in MR78, we determined the apo structure of MR78 Tyr37Phe mutant at 2.0 Å. While this structure maintains the bulge in the absence of Asp-Tyr interaction, the normalized B-factors in the HCDR3 loop show a small increase of 0.87 in the apex residues and 0.66 in the torso residues. From the unbound wild-type to the unbound mutant MR78 structure, the τ101 and α101 values change from 107.3° and 38.1° to 113.3° and 21.1°, respectively. As a result, the degree of kink in the loop and the position of the kink relative to the framework of the antibody are altered. In the absence of this inter HCDR interaction in mutant MR78, the other three hydrogen bonds supporting the bulge in the torso region are maintained, i.e., Trp118 side chain to T5 Met115 carbonyl oxygen, a backbone-backbone H-bond between T4 Tyr114 and Tyr108, and a backbone-backbone H-bond between T2 Ser106 and T6 Asp116. Changes in the degree of the kink in the loop, however affect the pseudobond angles and torsional angles between C-alpha atoms of the stem residues in HCDR3 loop in the mutant MR78, i.e. HCDR3 is in a slightly different conformation and slightly more flexible in the MR78 Tyr37Phe mutant.
T6Asp116-Tyr37 inter CDR loop interaction affects binding affinity of MR78
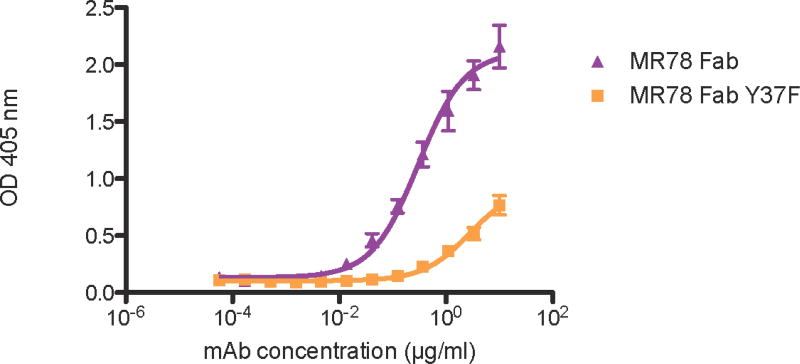
We hypothesize that the role of the Asp-Tyr interaction, in the absence of Arg/Lys at T2, is to stabilize the bulge in the conformation necessary for interaction with MARV GP thereby reducing the entropic cost of binding. To test this hypothesis, the binding affinity of the MR78 Tyr37Phe mutant was determined and compared with MR78 using ELISA. The shift in the concentration response curves shows that wild-type MR78 binds to the MARV GP approximately ten times tighter than the MR78 Tyr37Phe mutant, supporting our hypothesis (Figure 8).
Figure 8.
Binding of MR78 Fab and MR78 Tyr37Phe Fab to MARV GP. MR78 Fab binds MARV GP with an EC50 value of 0.30 µg/mL (95% confidence interval: 0.22–0.42 µg/mL) and MR78 Tyr37Phe Fab binds MARV GP with an EC50 value of 2.71 µg/mL (95% confidence interval: 1.77–4.15 µg/mL), resulting in a 10-fold reduction in binding to MARV GP. EC50 values of MR78 and MR78 Tyr37Phe Fab to MARV GP were obtained via ELISA. Technical triplicates of each were performed. Plates were coated with 1 µg/mL of MARV GP. MR78 and MR78 Tyr37Phe Fab molecules were serially diluted 3-fold from 10 µg/mL to 56.5 ng/mL. EC50 values and 95% confidence intervals were calculated using a non-linear regression analysis of the curves generated in Prism v.5. (GraphPad Software).
DISCUSSION
The Phenix.Rosetta optimization method provides an invaluable aid to crystallographic structure determination tools when low-resolution data challenges model building. Although it can add atomic detail not present in the experimental data with high confidence, there is also the possibility of occasional prediction of positions and contacts that are not in agreement with the electron density maps. This does however not exclude the possibility that some of these contacts occur, perhaps transiently, in the biological environment.
The conformation of short antibody CDRs, typically HCDR1,2 and LCDR1,2,3, can often be predicted from their sequence alone. HCDR3 is an exception as it is often between 12 and 20 residues long, sometimes longer. HCDR3s adopt kinked/bulged or extended structures. The bulge observed in the MR78 HCDR3 torso cannot be classified by the rules by Shirai(Shirai et al., 1999), Kuroda(Kuroda et al., 2008) or North(North et al., 2011) as shown recently that these rules no longer hold for a large number of antibody structures. Exceptions to the H3-rule 1b have been discussed due to the interaction of Asp at T6 to other basic residues either in the HCDR3 loop itself or from light chain (position 46 and 49)(Kuroda et al., 2008). Interactions of torso residues with distant residues seem to play role in optimizing the bulge for better binding as shown here in the case of MR78 antibody. Such interactions already exist in antibody 26-10 (PDB ID 1IGJ) and can be expected in new antibody structures based on our analysis of antibody sequence database. Therefore, inter CDR loop interactions should be taken into account while classifying and modeling torso regions in the HCDR3 loop of antibody structures.
The H3-rules website (http:/www.protein.osaka-u.ac.jp/rcsfp/pi/H3-rules/)(Shirai et al., 1999, Kuroda et al., 2008) predicts an extended torso for MR78 based on a predicted interaction between T6 Asp and Trp118, the first residue after the C terminus of HCDR3. A class A hairpin was predicted for HCDR3 without a meaningful feature such as an H-bond ladder. These results show that classification software fails to recognize the bulge in the HCDR3 torso. Effects of the surrounding environment on the torso conformation such as in the 26-10 and MR78 antibody structures can be used to improve predictions in antibody modeling software.
A database containing 465 million human antibody sequences was analyzed to find the number of sequences with similar residues in HCDR3 torso, i.e., absence of Arg/Lys at T2 and presence of Asp at T6 position, that could potentially lead to an interaction with Tyr in HCDR1 loop. 56.5 million antibody sequences were found to have Asp (D) at T6 and not Arg/Lys at T2. Out of these 56.5 million sequences, 23.7 million sequences (5% of all human antibodies) have Tyr at third to last position in HCDR1 loop like in MR78 and 11.8 million sequences (2.5% of all human sequences) have Tyr at fifth position in HCDR1 loop as in the 12-10 antibody. This analysis shows that this interaction between HCDR3 and HCDR1 might contribute to the torso conformation in the structure of many human antibodies.
CONCLUSIONS
Here we present the crystal structures of the unbound MR78 Fab fragment and a point mutant of this antibody, as well as a Phenix.Rosetta refined model of the MR78-GP1 complex. The HCDR3 of MR78, as well as the previously determined 26-10 antibody, forms a bulged torso structure with an unusual interaction of Asp at the T6 position of HCDR3 with a Tyr residue in the HCDR1. 5% of all human antibodies share sequence features consistent with this bulge conformation. The non-local interactions from HCDR1 can complement for the absence of Arg/Lys at T2 and support the bulge for better binding of the antibody.
The HCDR3 loop in MR78 is not flexible due to its H-bond ladder structure and does not undergo large backbone conformational changes upon binding to MARV GP. The tip, however, adjusts to maximize the aromatic stacking interactions.
Energy minimization in Phenix.Rosetta allows improvement of model geometry in the moderate resolution X-ray structure, especially in the HCDR3 torso. Energy minimization, however, predicts a few additional contacts between MR78 and GP1 that are contradicted by electron density maps and do not appear in any of the four independent copies of the MR78-GP1 complex in the asymmetric unit. Formation of these contacts would improve the energetics of binding. It is striking then, that the experimental electron density maps do not support all the possible contacts that could be made. The crystals were grown at pH 6.5, which itself likely would not significantly affect binding. It may be that the biological complex is not energetically “ideal”. Alternatively, the additional contacts between HCDR1 and GP1 may be transient, and not captured in the crystal structure. Nonetheless, all the structures and models indicate that the HCDR3 possesses an atypical bulge containing particular contacts of the HCDR3 to HCDR1. This non-local interaction defines the structural determinants of the interaction of the HCDR3 loop with the antigen and is therefore key for antibody efficacy. These models provide a starting point for computational design of antibodies against MARV disease. Antibody modeling software can include new rules to take into account the long distant interactions of torso residues resulting in better antibody models.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jens Meiler (jens@meilerlab.org).
Experimental Model and Subject Details
Cells
The human hybridoma cell line expressing MR78 mAb was grown in post-fusion medium ((ClonaCell-HY Medium E, STEMCELL Technologies #03805), and was then expanded in serum free medium (Hybridoma-SFM, GIBCO #12045-076) at 37°C with 7% CO2. MR78 mutant Fab was produced with Expi293F cells (Thermo Fisher Scientific #A14527) grown in Epi293 Expression Medium (TheromFisher Scientific #A1435104) at 37°C with 7% CO2.
Method Details
Phenix.Rosetta refinement of 3×2D
Phenix software is used to determine protein structures from X-ray scattering data, although modeling some regions using moderate-to low-resolution scattering data still remains a challenge. The combined Phenix.Rosetta refinement approach(DiMaio et al., 2013) has been shown to improve model geometry (with better Rfree factor and Molprobity score) for structures determined in the resolution range of 3.0 – 4.5Å. For the 3.6Å structure of MARV GP in complex with antibody MR78 (PDB ID 3×2D), we used the low_resolution_refinement script (available at Rosetta/source/src/apps/public/crystal_refinement/) with symmetry. A weight of 20 was used for the elec_dens_fast option to reweight scoring function with electron density data. The XML script and options used for refinement are explained in Dimaio et al. (Nature Methods, 2013). The resulting model then was dual-space relaxed to generate 100 additional models. The lowest energy model was chosen as the optimized model for the MR78:MARV GP complex.
HCDR1 loop modeling
The HCDR1 loop residues still showed energetic frustrations after Phenix.Rosetta refinement and dual-space relaxation. The loop modeling technique(Kaufmann et al., 2010) in Rosetta was used to rebuild the 10 residue-long HCDR1. A thousand models were generated that grouped into five clusters. A representative structure from the largest cluster (size 263) was chosen as the final model. The HCDR1 then was grafted onto the MR78:MARV GP structure and the complex was relaxed to generate 100 additional models. The lowest scoring model was chosen as the final model to analyze antibody-antigen interactions.
MR78 Fab production and purification
The human hybridoma cell line expressing MR78 mAb was grown in post-fusion medium, as previously described(Flyak et al., 2015). HiTrap MabSelectSure columns were used to purify MR78 from filtered hybridoma supernates. The purified MR78 mAb was cleaved with papain to obtain the MR78 Fab fragment (Pierce Fab Preparation Kit, ThermoFisher Scientific), and the MR78 Fab was purified further with IgG-CH1 affinity chromatography (CaptureSelect, Thermofisher Scientific).
Crystallization, data collection, and structure determination
MR78 Fab and MR78 mutant (12 mg/mL in 20 mM Tris, 7.5, 50 mM NaCl) was crystalized in 4.1 ~ 4.5 M NaCl, 0.1 M HEPES, pH 7.5 with protein to precipitant volume ratio of 1.5:1. Crystals were flash frozen in liquid nitrogen using Parabar 10312 oil as cryoprotectant. Diffraction data were collected at the beamline 21-ID-F at the Advanced Photon Source. The diffraction data were processed with imosflm, XDS and CCP4 suite(Winn et al., 2011). The crystal structure was solved by molecular replacement using the MR78 structure in the MR78 and the MARV GP complex (PDB ID 3×2D) with the program Phaser(Mccoy et al., 2007). The structure was refined and rebuilt manually with Phenix(Adams et al., 2010) and Coot(Emsley and Cowtan, 2004), respectively. The final statistics of final structures of wild type and mutant MR78 Fabs are shown in Table 2. The models have been deposited into the Protein Data Bank (PDB ID 5JRP and 5WEQ).
Table 2.
Unbound MR78 wild type and Tyr37Phe mutant data collection and refinement statistics
| MR78 Wild type | MR78 Tyr37Phe mutant | |
|---|---|---|
| Wavelength, Å | 0.97872 | 0.97856 |
| Space group | P3121 | P3121 |
| Unit cell dimensions | a=b=117.57, c=91.80, α=β=90°, γ=120° | a=b=117.18, c=92.10, α=β=90°, γ=120° |
| Resolution, Å | 50.91 – 2.00 | 50.00 – 2.00 |
| Highest resolution shell, Å | 2.11 – 2.00 | 2.11 – 2.00 |
| Rsym,* % | 11.2 (43.9) | 7.7 (74.2) |
| Mean I/σ(I) | 13.6 (4.9) | 9.8 (1.7) |
| Total no. reflections | 557, 044 (79, 594) | 278, 149 (50000) |
| Completeness, % | 100 | 99.2 |
| Refinement statistics | ||
| Rwork/Rfree, # % | 17.1/19.4 | 18.6/21.5 |
| Average B factor, Å2 | 33.7 | 52.3 |
| Total no. atoms | 3815 | 3505 |
| Water molecules | 544 | 211 |
| Bond angles, ° | 0.846 | 0.890 |
| Bond length, Å | 0.0034 | 0.0076 |
| Ramachandran: favored/allowed (%) | 98.77/100 | 97.147/100 |
Rsym = Σ Σ |Ihkl − Ihkl(j)|/Σ Ihkl, where Ihkl(j is the observed intensity and Ihkl is the final average intensity.
Rwork = Σ ‖Fobs| − |Fcalc‖/Σ |Fobs| and Rfree = Σ ‖Fobs| − |Fcalc‖/Σ |Fobs|, where Rfree and Rwork are calculated using a randomly selected test set of 5% of the data and all reflections excluding the 5% test set, respectively. Numbers in parentheses are for the highest resolution shell.
Half-maximal effective concentration (EC50) binding ELISA analysis
The soluble form of the full-length extracellular domain of MARV GP (1 µg/µL) was diluted in 1× D-PBS to coat 384-well ELISA plates (Thermo Scientific #265202) at 25 µL/well and incubated at 4 °C overnight. The plates were washed 3× with D-PBS-T (1× DPBS + 0.05% Tween 20) and blocked for 1 hour at room temperature with blocking solution (1% non-fat dry milk (Blotting Grade Blocker Bio-rad #170-6404), 1% goat serum (Gibco 16210-072) in D-PBS-T). After blocking, the plates were then washed 3× with D-PBS-T and 25 µL/well of 3-fold serially diluted purified Fab MR78 or Fab MR78 Tyr37Phe (10 µg/mL – 56.5 ng/mL) in blocking solution was added. Plates were incubated for 2 hours at room temperature and then washed 3× with D-PBS-T. Secondary antibody (goat anti-human kappa-alkaline phosphatase conjugated; Southern Biotech 2060-04) at a 1:4,000 dilution in blocking solution was added at 25 µL/well for 1 hour at room temperature. Alkaline phosphatase substrate solution (phosphatase substrate tablets (Sigma #S0942) in AP substrate buffer (1M Tris aminomethane (Fisher #BP152-5), 0.3 mM MgCl2 (Sigma #M1028)) was added at 25 µL/well following plate washing 4× with D-PBS-T. Plates were incubated at room temperature in the dark for 2 hours then read at an optical density of 405 nm with a Biotek plate reader. EC50 values and 95% confidence intervals were calculated using a non-linear regression analysis of the curves generated in Prism v.5. (GraphPad Software).
Quantification and Statistical Analysis
Graphpad Prism software was used to determine average values, standard errors, and standard deviations for Figure 8.
Data and Software availability
The structure factors and experimental model have been deposited in the RCSB Protein Data Bank under ID code 5JRP, 5WEQ and 5UQY. Rosetta software is available at https://www.rosettacommons.org.
Supplementary Material
HIGHLIGHTS.
Phenix.Rosetta refinement optimizes the model of human antibody MR78
An unusual inter CDR loop interaction supports the bulged torso conformation of HCDR3
Removal of inter CDR loop H-bond lowers the MR78 binding affinity by ten fold
Acknowledgments
Author A.K.S. is grateful to Dr. Frank DiMaio for technical discussions on Phenix.Rosetta refinement technique. A.K.S. is grateful to Jessica Ann Finn, Alexander Sevy and Alberto Cisneros for helpful discussions. The work was supported through NIH grants R21 AI121799 (Meiler), U19 AI117905 (Crowe and Meiler), and U19 AI109711 (Crowe) and DTRA grant HDTRA1-13-1-0034 (Crowe). Work in the Saphire laboratory is supported through NIH U19AI1097652 and R01 AI089498. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract Number DE-AC02-06CH11357. Updated coordinates for the MR78-MARV GP complex, including additional insights from separate work (King et al., 2017)have been deposited in the Protein Data Bank 5UQY.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
A.K.S and J.M. planned research. A.K.S, J.D. and L.W. performed research. A.K.S, J.D., L.W. and J.M. analyzed the data. A.K.S, J.D., L.W., T.H., E.O.S, J.E.C. and J. M. wrote the paper.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystalallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender BJ, Cisneros A, Duran AM, Finn JA, Fu D, Lokits AD, Mueller BK, Sangha AK, Sauer MF, Sevy AM, Sliwoski G, Sheehan JH, DiMaio F, Meiler J, Moretti R. Protocols for molecular modeling with Rosetta3 and RosettaScripts. Biochem. 2016;55(34):4748–4763. doi: 10.1021/acs.biochem.6b00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIOVIA. Discovery studio modeling environment. Accelrys software (3.5) 2012 [Google Scholar]
- Borio L, Cox E, Lurie N. Combating Emerging Threats - Accelerating the Availability of Medical Therapies. New Engl. J. Med. 2015;373(11):993–995. doi: 10.1056/NEJMp1508708. [DOI] [PubMed] [Google Scholar]
- DiMaio F, Echols N, Headd JJ, Terwilliger TC, Adams PD, Baker D. Improved low-resolution crystallographic refinement with Phenix and Rosetta. Nature Methods. 2013;10(11):1102–1104. doi: 10.1038/nmeth.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio F, Song Y, Li X, Brunner MJ, Xu C, Conticello V, Egelman E, Marlovits TC, Cheng Y, Baker D. Atomic-accuracy models from 4.5-A cryo-electron microscopy data with density-guided iterative local refinement. Nature Methods. 2015;12(4):361–5. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar J, Fuchs A, Shi J, Deane CM. ABangle: Characterising the VH-VL orientation in antibodies. Protein Eng. Des. Sel. 2013;26(10):611–620. doi: 10.1093/protein/gzt020. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Flyak AI, Ilinykh PA, Murin CD, Garron T, Shen XL, Fusco ML, Hashiguchi T, Bornholdt ZA, Slaughter JC, Sapparapu G, Klages C, Ksiazek TG, Ward AB, Saphire EO, Bukreyev A, Crowe JE. Mechanism of human antibody-mediated neutralization of Marburg virus. Cell. 2015;160(5):893–903. doi: 10.1016/j.cell.2015.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi T, Fusco ML, Bornholdt ZA, Lee JE, Flyak AI, Matsuoka R, Kohda D, Yanagi Y, Hammel M, Crowe JE, Saphire EO. Structural basis for Marburg virus neutralization by a cross-reactive human antibody. Cell. 2015;160(5):904–912. doi: 10.1016/j.cell.2015.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron JN, He XM, Ballard DW, Blier PR, Pace PE, Bothwell AL, Voss EW, Jr, Edmundson AB. An autoantibody to single-stranded DNA: Comparison of the three-dimensional structures of the unliganded Fab and a Deoxynucleotide-Fab complex. Proteins. 1991;11(3):159–75. doi: 10.1002/prot.340110302. [DOI] [PubMed] [Google Scholar]
- Jeffrey PD, Strong RK, Sieker LC, Chang CYY, Campbell RL, Petsko GA, Haber E, Margolies MN, Sheriff S. 26-10 Fab-Digoxin complex - Affinity and specificity due to surface complementarity. Proc. Natl. Acad. Sci. USA. 1993;90(21):10310–10314. doi: 10.1073/pnas.90.21.10310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann KW, Lemmon GH, DeLuca SL, Sheehan JH, Meiler J. Practically Useful: What the ROSETTA protein modeling suite can do for you. Biochem. 2010;49(14):2987–2998. doi: 10.1021/bi902153g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King LB, Fusco ML, Flyak AI, Ilinykh P, Huang K, Gunn B, KIrchdoerfer RN, Hastie KM, Sangha AK, Meiler J, Alter G, Bukreyev A, Crowe JE, Saphire EO. Therapeutic human monoclonal antibody MR191 against Marburg virus targets a conserved site to block receptor binding. 2017 doi: 10.1016/j.chom.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koliasnikov OV, Kiral MO, Grigorenko VG, Egorov AM. Antibody CDR H3 modeling rules: Extension for the case of absence of Arg H94 and Asp H101. J. Bioinform. Comput. Biol. 2006;4(2):415–24. doi: 10.1142/s0219720006001874. [DOI] [PubMed] [Google Scholar]
- Kuroda D, Shirai H, Kobori M, Nakamura H. Structural classification of CDR-H3 revisited: A lesson in antibody modeling. Proteins: Struct. Funct. Bioinf. 2008;73(3):608–620. doi: 10.1002/prot.22087. [DOI] [PubMed] [Google Scholar]
- Maier JK, Labute P. Assessment of fully automated antibody homology modeling protocols in molecular operating environment. Proteins. 2014;82(8):1599–610. doi: 10.1002/prot.24576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcatili P, Olimpieri PP, Chailyan A, Tramontano A. Antibody structural modeling with prediction of immunoglobulin structure (PIGS. Nat. Protoc. 2014;9(12):2771–2783. doi: 10.1038/nprot.2014.189. [DOI] [PubMed] [Google Scholar]
- Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystal. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messih MA, Lepore R, Marcatili P, Tramontano A. Improving the accuracy of the structure prediction of the third hypervariable loop of the heavy chains of antibodies. Bioinformatics. 2014;30(19):2733–2740. doi: 10.1093/bioinformatics/btu194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MMWR. Brief report: Outbreak of Marburg virus hemorrhagic fever - Angola, October 1, 2004–March 29, 2005 (Reprinted from MMWR, vol 54, pg 308–309, 2005) J. Am. Med. Assoc. 2005:2336–2336. [PubMed] [Google Scholar]
- Morea V, Tramontano A, Rustici M, Chothia C, Lesk AM. Conformations of the third hypervariable region in the VH domain of immunoglobulins. J. Mol. Biol. 1998;275(2):269–294. doi: 10.1006/jmbi.1997.1442. [DOI] [PubMed] [Google Scholar]
- North B, Lehmann A, Dunbrack RL. A new clustering of antibody CDR loop conformations. J. Mol. Biol. 2011;406(2):228–256. doi: 10.1016/j.jmb.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva B, Bates PA, Querol E, Aviles FX, Sternberg MJE. Automated classification of antibody complementarity determining region 3 of the heavy chain (H3) loops into canonical forms and its application to protein structure prediction. J. Mol. Biol. 1998;279(5):1193–1210. doi: 10.1006/jmbi.1998.1847. [DOI] [PubMed] [Google Scholar]
- Sela-Culang I, Alon S, Ofran Y. A systematic comparison of free and bound antibodies reveals binding-related conformational changes. J. Immunol. 2012;189(10):4890–4899. doi: 10.4049/jimmunol.1201493. [DOI] [PubMed] [Google Scholar]
- Shirai H, Ikeda K, Yamashita K, Tsuchiya Y, Sarmiento J, Liang SD, Morokata T, Mizuguchi K, Higo J, Standley DM, Nakamura H. High-resolution modeling of antibody structures by a combination of bioinformatics, expert knowledge, and molecular simulations. Proteins Struct. Funct. Bioinf. 2014;82(8):1624–1635. doi: 10.1002/prot.24591. [DOI] [PubMed] [Google Scholar]
- Shirai H, Kidera A, Nakamura H. Structural classification of CDR-H3 in antibodies. FEBS Lett. 1996;399(1–2):1–8. doi: 10.1016/s0014-5793(96)01252-5. [DOI] [PubMed] [Google Scholar]
- Shirai H, Kidera A, Nakamura N. H3-rules: Identification of CDR-H3 structures in antibodies. FEBS Lett. 1999;455(1–2):188–197. doi: 10.1016/s0014-5793(99)00821-2. [DOI] [PubMed] [Google Scholar]
- Sircar A, Kim ET, Gray JJ. RosettaAntibody: Antibody variable region homology modeling server. Nucleic Acids Res. 2009;37(Web Server issue):W474–9. doi: 10.1093/nar/gkp387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Mizuguchi K. The diversity of H3 loops determines the antigen-binding tendencies of antibody CDR loops. Protein Science. 2016;25(4):815–825. doi: 10.1002/pro.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RY, Kudryashev M, Li X, Egelman EH, Basler M, Cheng Y, Baker D, DiMaio F. De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nature Methods. 2015;12(4):335–8. doi: 10.1038/nmeth.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzner BD, Dunbrack RL, Gray JJ. The origin of CDR H3 structural diversity. Structure. 2015;23(2):302–311. doi: 10.1016/j.str.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita K, Ikeda K, Amada K, Liang SD, Tsuchiya Y, Nakamura H, Shirai H, Standley DM. Kotai Antibody Builder: Automated high-resolution structural modeling of antibodies. Bioinformatics. 2014;30(22):3279–3280. doi: 10.1093/bioinformatics/btu510. [DOI] [PubMed] [Google Scholar]
- Zhu K, Day T, Warshaviak D, Murrett C, Friesner R, Pearlman D. Antibody structure determination using a combination of homology modeling, energy-based refinement, and loop prediction. Proteins: Struct. Funct. Bioinf. 2014;82(8):1646–1655. doi: 10.1002/prot.24551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.