Abstract
The current investigation is trying to study the impact of the mixture of Chinese herbs Qi Wei Bai Zhu San (QWBZS) on bacterial lactase gene from antibiotics-induced diarrhea (AAD) mice, as the good curative effect of QWBZS on diarrhea. Mice (6 mice per group) were randomly selected as control, model and treatment groups. To induce diarrhea, mice in both model and treatment groups were intragastrically injected with mixture of gentamycin sulfate and cefradine (23.33 mL kg−1 day−1) twice per day and continuously for totally 5 days. After the success of establishing diarrhea model, the mice in treatment group were gavaged with QWBZS for 3 days. Intestinal contents in all three groups were then collected and DNA was extracted in aseptic environment for the following sequencing. The results showed that mice from QWBZS treatment group had obviously detectable levels of intestinal bacteria, such as Actinobacteria, Firmicutes and Proteobacteria, which produce phyla lactase specifically. In comparison with other groups, the mice in treatment group had more abundant expression of lactase gene from Acidovorax sp. KKs102, Stenotrophomonas sp. LMG11000, Pseudomonas oleovorans, Eggerthella and Burkholderia. Interestingly, the Shannon index decreased significantly after the treatment with QWBZS (P < 0.01 or P < 0.05). 63.1% of lactase genes detected in the mice in treatment group were unclassified, and 32.8% of them were non-homologous to any fragments in the gene bank, which means that most of lactase-producing bacteria are novel. Our results indicate that treatment with QWBZS did not increase the diversity of bacterial lactase gene. Its curative effect on diarrhea may be relevant to its role in facilitating the growth of novel or some key lactase-producing strains.
Keywords: Antibiotics-induced diarrhea, Lactase, Qi Wei Bai Zhu San, Intestine microbiota, High-throughput sequencing
Introduction
According to the theory of Chinese traditional medicine, people with malfunctions of spleen or stomach are easier to suffer from diarrhea than others do (Qiao 2014). Compound traditional Chinese medicine is the essence of traditional Chinese medicine, such as Qi Wei Bai Zhu San (QWBZS). With the stringent design, accurate and appropriate compatibility and the function of regulating intestinal microecology, QWBZS is a millennium ancient prescription to treat diarrhea, especially pediatric diarrhea (Cai et al. 2013; Wang et al. 2014). It is composed of Sijunzitang and is a kind of broth cooked with radix codonopsis, poria cocos, atractylodes macrocephala koidz, radix glycyrrhizae, radix aucklandiae, agastache rugosa and radix puerariae (Yang and Yang 2010; Wang et al. 2013). Previous studies demonstrated that QWBZS can adjust intestinal Lactobacillus and Bifidobacterrium, reverse the effect of antibiotics on activities of enzymes, such as lactase, xylanase, amylase and protease in mice and promote the growth of yeast and improve mucosa structure of small intestine in antibiotics-induced diarrhea (Guo et al. 2013; Guo et al. 2015a; Liu et al. 2014; Tan et al. 2012).
Diarrhea, a health condition with the symptom featured by over three times of watery stools daily, can be caused by malfunction of spleen or stomach and over-dose antibiotics. With the overuse of antibiotics, antibiotics-induced diarrhea (AAD) has become the most common iatrogenic diarrhea. There are several possible pathogenesis of antibiotics-induced diarrhea (Zhou 2004): breaking the balance of intestinal bacteria, interfering the glycometabolism and bile acid metabolism or decreasing the activity of disaccharidase, such as lactase. According to a previous study (Long et al. 2017a), a mix of antibiotics for diarrhea modeling in mice reduced the density of bacterial lactase gene and led to the decreased Chao1 and ACE indexes (P < 0.05) and the abundance of some bacteria genera producing lactase.
Lactase plays critical role in regulating intestinal function and is mainly produced by Proteobacteria, Actinobacteria or Firmicutes (Alexandre et al. 2013; Ojetti et al. 2010). Antibiotics, especially beta-lactam antibiotics such as cefradine used in our previous study, can destroy the intestinal lactase (Peng and Ren 2011). Due to the deficiency or low activity of lactase, the lactose cannot be absorbed but decomposed by intestinal microbiota, which causes intestinal water and osmotic pressure increase by producing a large number of short-chain fatty acids and hydrogen and eventually induce diarrhea (Hammer and Hammer 2012; Zhang et al. 2014).
16S rDNA sequencing has been widely used in the classification, identification and detection of bacteria, and research of intestinal microbiota has been a focus because of its crucial role in human health. However, there is few study on intestinal bacterial lactase gene while some studies on genetic xylanase and pectase diversity in rumen have been reported (Wang et al. 2011; Yuan 2012). As such, we designed and validated a pair of universal primers used for the amplification of lactase gene (Long et al. 2017b). The diversity of bacterial lactase gene were analyzed by high-throughput sequencing.
The current research aimed to investigate pharmacological mechanisms of QWBZS’s effect on the activities of lactases at the genetic level and to provide experimental basis for the treatment of AAD with QWBZS.
Materials and methods
Reagents and medicine
Medicines and antibiotics were purchased from Suzhou Chung-Hwa Chemical & Pharmaceutical Industrial Co. Ltd, or Yichang Humanwell Pharmaceutical Co. Ltd. Protease K, acetone, lysozyme, tris saturated phenol–chloroform–isoamyl alcohol (25:24:1) and TE buffer were purchased from Beijing Dingguo Changsheng Biotechnology Co., Ltd. 10% SDS, 0.1 mol L−1 PBS buffer, 5 mol L−1 NaCl and CTAB/NaCl were prepared in the lab. QWBZS broth was made of Radix codonopsis, poria cocos, atractylodes macrocephala koidz, radix glycyrrhizae, radix aucklandiae, agastache rugosa and radix puerariae, which were purchased as prescription from The First Hospital of Hunan University of Chinese Medicine. The herbs were boiled for 1 h and the broth was collected and stored at 4 °C.
Animals and procedures
Adult mice were purchased from Hunan Slaccas Jingda Laboratory Animal Company with licence number SCKX (Xiang) 2013-0004. All experiments and procedures involving animals were performed according to the protocols approved by the Institutional Animal Care and Use Committee of Hunan University of Chinese Medicine. The experiments included equal number of male and female mice. Mice were randomly selected into three groups (6 mice per group): healthy control group (control), diarrhea model group (model) and diarrhea treatment group (treatment). Mice of control group were gavaged with 0.35 mL of sterile water twice a day for 5 days. To induce diarrhea, mice in both model group and treatment group were intragastrically injected with mixture of gentamycin sulfate and cefradine (23.33 mL kg−1 day−1) twice per day and continuously for totally 5 days by following the procedures described previously (Zeng et al. 2012). Once the diarrhea symptoms (specifically watery stool, reduced intake, erected coat and declined activity) were induced, the mice in treatment group were administered intragastrically with QWBZS broth at the dose of 0.16 g kg−1 day−1 for 3 days (Guo et al. 2015b). Mice were then euthanized and intestinal contents samples of each two mice (one male and one female) in every group were then collected for the following experiments (Zeng et al. 2012).
DNA extraction
Genomic DNAs from the intestinal contents were extracted by following the protocols (He et al. 2017; Wu et al. 2012). 2.0 g of intestinal feces was weighed in a sterile environment and was homogenized in 5 mL of 0.1 mol L−1 phosphate buffer solution (PBS), followed by centrifugation at 200 g for 2 min. After washing the sediments twice with PBS, the whole supernatants were transferred into new tubes and centrifuged for 8 min at 10,000g. The new sediments were then gathered, washed once with phosphate buffer solution, twice with acetone, and three times with phosphate buffer solution, then resuspended in 4 mL TE buffer. 500 μL of spreadhead were added with 45 μL TE buffer, 5 μL proteinase K and 20 μL lysozyme and homogenized in 1.5 mL germ-free EP tubes. Samples were incubated at 37 °C for 30 min, then mixed with 30 μL of 10% SDS, followed by incubation at 37 °C for 40 min with turning upside down once every 10 min. Afterwards, 100 μL of 5 mol L−1 NaCl and 80 μL of CTAB/NaCl were added and mixed well. The mixture was reacted at 65 °C for 10 min. An equal volume of Tris saturated phenol–chloroform–isoamyl alcohol (25:24:1) then were added to the sample, mixed well, and were centrifuged at 10,000g for 3 min. The supernatant were transferred to fresh sterile tubes, mixed with an equal volume of chloroform–isoamyl alcohol (24:1), centrifuged at 10,000g for 3 min. The supernatant was transferred into fresh sterile tubes and mixed with an equal volume of chloroform–isoamyl alcohol (24:1) again. After centrifugation at 10,000g for 3 min, the supernatant were transferred into new sterile tubes, added with 1/10 volume of 3 mol L−1 sodium acetate and double volume of absolute ethyl alcohol, and were precipitated at − 20 °C overnight. Samples were centrifuged at 10,000g for 3 min. The acquired sediment were washed with 70% ethanol, dried and eventually dissolved in 50 μL TE buffer.
PCR amplification of intestinal bacterial lactase gene and sequencing
According to a previous report (Long et al. 2017b), the lactase genes were amplified by PCR with the forward primer 5′-TRRGCAACGAATACGGSTG-3′ and the reverse primer 5′-ACCATGAARTTSGTGGTSARCGG-3′. The PCR products were purified and applied for sequencing. Universal primers for amplifying bacteria beta-galactosidase gene were designed based on the sequence info from NCBI database and synthesized by Personal Biotechnology Co., Ltd. Shanghai, China. PCR mixture (25 μL) included 0.25 μL Q5 high-fidelity DNA polymerase, 5 μL 5× reaction buffer, 5 μL 5× high GC buffer, 0.5 μL 10 mmol L−1 dNTP, 1 μL 10 μmol L−1 forward primer, 1 μL 10 μmol L−1 reverse primer, 1 μL DNA template and 11.25 μL sterilized ddH2O. The PCR conditions were as follow: initial denaturation at 98 °C for 30 s, 32 cycles of 98 °C for 15 s, 46 °C for 30 s and 72 °C for 30 s, extension at 72 °C for 5 min and holding at 4 °C. PCR products were first examined by 2% Agarose gel electrophoresis. PCR products of bacteria lactase genes were purified, and were then sequenced by Illumina Miseq in Personal Biotechnology Co., Ltd.
Gene diversity analysis and statistical analysis
The sequencing results were blasted using software available online, including Qiime (http://qiime.org/) (Caporaso et al. 2010) and Mothur (http://www.mothur.org/). Lactase genes’ abundance and diversity were measured by Rank abundance curve and Alpha diversity index (Pitta et al. 2010, 2014; Shannon 1997; Mahaffee and Kloepper 1997). PCoA figure was generated according to UniFrac analysis and used for species evolution analysis (Lozupone and Knight 2005). The source and abundance of bacterial lactase genes at the specific taxonomic levels were presented by figure of Species evolution and abundance information. Species composition ratio was measured by pie charts. Differences among groups were considered significant when P-values less than 0.05.
Results
Quality of extracted DNA and statistics of sequences
Before sequencing, the quality of genomic DNAs in all samples was measured by OD260/OD280. All of the samples had OD260/OD280 above 1.8. There were totally 625,914 effective sequences and 614,414 high quality sequences acquired by sequencing. High quality sequences account for around 98% of effective sequences in each sample. The results were shown in Table 1. These results indicated that the samples were suitable for the following analyses.
Table 1.
Statistics of sequences intestinal bacteria DNA sequences
| Samples | Effective sequences | High quality sequences | Ratio (%) |
|---|---|---|---|
| tlcn1 | 63,052 | 61,821 | 98.05 |
| tlcn2 | 77,507 | 76,101 | 98.19 |
| tlcn3 | 75,377 | 73,418 | 97.40 |
| tlmn1 | 70,334 | 68,547 | 97.46 |
| tlmn2 | 65,217 | 63,929 | 98.03 |
| tlmn3 | 69,876 | 68,810 | 98.47 |
| tlqn1 | 73,310 | 72,454 | 98.83 |
| tlqn2 | 51,263 | 50,521 | 98.55 |
| tlqn3 | 79,978 | 78,813 | 98.54 |
| Total | 625,914 | 614,414 | 98.16 |
Effective sequence: index exactly matched sequence. High quality sequence: the effective sequences after filtering and removing chimeric. tlcn1-3, tlmn1-3 and tlqn1-3 are control group samples 1–3, model group samples 1–3 and treatment group samples 1–3, respectively
Richness and homogeneity of bacterial lactase gene
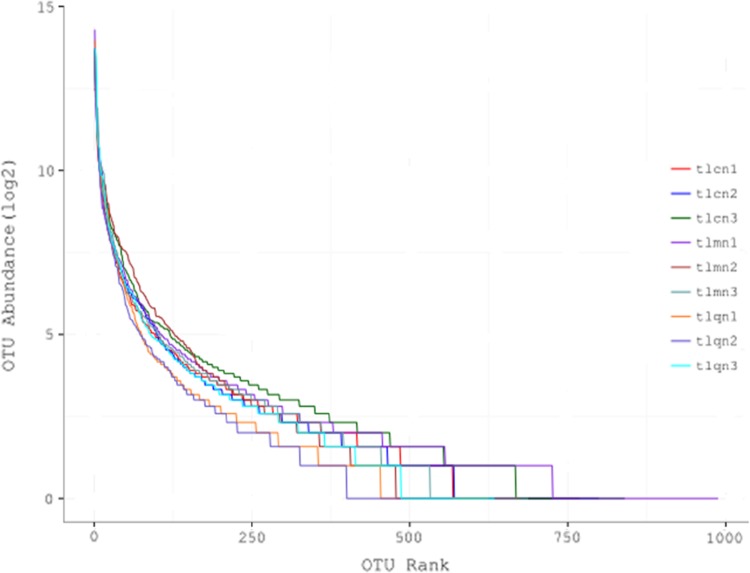
To investigate the impact of QWBZS treatment on diarrhea, the richness and homogeneity of the lactase genes were determined by Rank-abundance curve and Alpha index. As shown in Fig. 1, among three groups of mice, the treatment group had lowest abundance values. It indicates that the treatment decreased the richness of bacterial lactase genes in the intestines of mice. This is confirmed by the Alpha index result: the Chao 1 index was the highest in control group, followed by model group and treated group, while the difference had no significance (P > 0.05). However, the Shannon index of lactase genes was significantly lower in the QWBZS treatment group than those in model group and control group (P < 0.05 or P < 0.01) (Table 2).
Fig. 1.
Rank-abundance curve of each sample. The abscissa represents the ordinal of the OTU, and the ordinate represents the abundance of the OTU. The larger the curve span is, the richer the composition of the species is. The more flat the curve, the higher the uniformity of the species composition. tlcn1-3, tlmn1-3 and tlqn1-3 are control group samples 1–3, model group samples 1–3 and treatment group samples 1–3, respectively
Table 2.
Alpha biodiversity fares
| Group | Chao1 | ACE | Shannon |
|---|---|---|---|
| Control group | 532.00 ± 48.14 | 570.22 ± 77.44 | 5.30 ± 0.24 |
| Model group | 517.00 ± 116.25 | 585.54 ± 148.49 | 5.12 ± 0.33 |
| Treatment group | 401.00 ± 19.67 | 479.00 ± 41.04 | 4.46 ± 0.14aB |
a: P < 0.01 in comparison with the control group. B: P < 0.05 in comparison with compared with the model group
Difference analysis of bacterial community producing lactase
The method PCoA was applied for difference analysis of bacterial community. The results show that the clusters from treatment group were distinctly separated from others in control and model groups, which suggests that the diversity of bacteria in QWBZS-treated mice differed from the mice in other two groups. There was no significant difference, however, between the control group and model group. The percentages attributing to the variations of PC1, PC2 and PC3 were 44.82, 25.72 and 10.9%, respectively (Fig. 2).
Fig. 2.
PCoA analysis of the samples showing 3D sort graph based on Weighted UniFrac. Each point represents a sample. The points with the same colors belong to the same group. The closer the distance between the two points, the smaller difference between the two samples of microbial community. tlcn1-3, tlmn1-3 and tlqn1-3 are control group samples 1–3, model group samples 1–3 and treatment group samples 1–3, respectively
Community structure of lactase-producing microbiota and lactase gene abundance
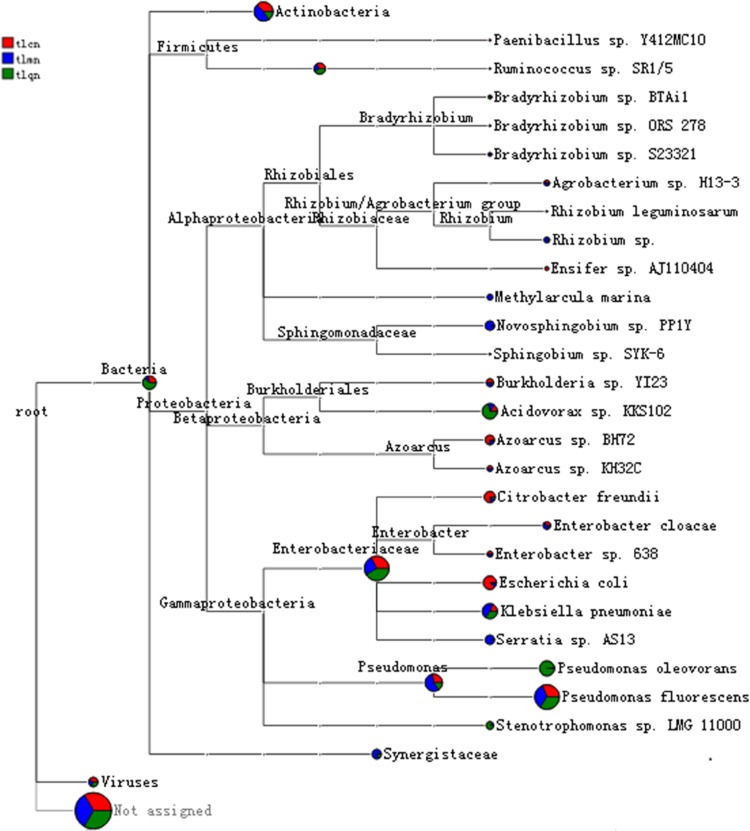
The evolution of lactase genes was investigated by evolution tree which show the similarity of the genes. As shown Fig. 3, intestinal lactase was mainly produced by Actinobacteria, Firmicutes and Proteobacteria, in particular Proteobacteria, in which Enterobacteriaceae and Pseudomonas were major species for lactase gene. In comparison with control group and model group, QWBZS treatment increased the abundance of lactase gene originated from Acidovorax sp. KKs102, Stenotrophomonas sp. LMG11000 and Pseudomonas oleovorans (Fig. 3).
Fig. 3.
Species evolution and abundance information. The number of sequences of different samples on a branch is presented by a pie chart with different colors: the larger the area of the pie chart, the more the number of sequences at the branch. The different colors represent the different samples. The larger the fan-shaped area of a color indicates that the number of sequences corresponding to the sample on the branch is greater than that of the other samples. tlcn: control group; tlmn: model group; tlqn: treatment group
Overall, thirty species of bacteria were detected at genus level. Rhodococcus, Paenibacillus and Ensifer were detected only in control group, while bacteria of Ruminococcus and Methylarcula were exclusively identified in model group mice. In addition, bacteria of Mycobacterium, Frankia, Sphingobium and Serratia were undetectable in treatment group mice. Except for Eggerthella, Burkholderia and Azoarcus, the abundance of lactase genes in most of the originated genus in all three groups showed no significant difference. The abundance of lactase genes in Eggerthella and Burkholderia were seen to be increased while those in Azoarcus were shown to be decreased in treatment group mice compared to the control group and model group (Table 3).
Table 3.
The abundance of bacterial lactase genes from known species at genus level
| Genus | Control group | Model group | Treatment group |
|---|---|---|---|
| Corynebacterium | 9.63E−04 ± 9.59E−05 | 9.78E−04 ± 6.91E−05 | 4.35E−04 ± 2.61E−05 |
| Mycobacterium | 8.53E−04 ± 2.80E−04 | 3.98E−04 ± 6.46E−04 | - |
| Rhodococcus | 1.28E−04 ± 2.22E−04 | - | - |
| Frankia | 1.03E−04 ± 4.7E−05 | 4.5E−05 ± 6.4E−05 | - |
| Modestobacter | 2.29E−04 ± 3.29E−04 | 2.8E−05 ± 4.8E−05 | 1.2E−05 ± 1.1E−05 |
| Microbacterium | 1.57E−04 ± 2.71E−04 | 1.842E−03 ± 3.132E−03 | 4.2E−05 ± 4.9E−05 |
| Arthrobacter | 8.28E−04 ± 6.00E−04 | 7.23E−04 ± 3.14E−04 | 5.24E−04 ± 2.41E−04 |
| Micromonospora | - | 4.9E−05 ± 4.2E−05 | 0.5E−05 ± 0.8E−05 |
| Streptomyces | 3.99E−04 ± 2.84E−04 | 4.13E−04 ± 3.40E−04 | 0.7E−05 ± 1.2E−05 |
| Eggerthella | 4.1E−05 ± 2.3E−05 | 2.2E−05 ± 0.9E−05A | 4.32E−04 ± 3.98E−04AB |
| Paenibacillus | 0.8E−05 ± 1.3E−05 | – | – |
| Ruminococcus | – | 0.6E−05 ± 1.0E−05 | – |
| Bradyrhizobium | 2.5E−05 ± 1.0E−05 | 2.7E−05 ± 2.5E−05 | 4.6E−05 ± 2.8E−05 |
| Agrobacterium | 4.2E−05 ± 1.6E−05 | 3.9E−05 ± 3.9E−05 | 1.0E−05 ± 0.8E−05 |
| Ensifer | 4.6E−05 ± 8.0E−05 | – | – |
| Rhizobium | – | 0.80E−04 ± 1.39E−04 | 1.4E−05 ± 1.4E−05 |
| Methylarcula | – | 0.67E−04 ± 1.16E−04 | – |
| Novosphingobium | – | 4.07E−04 ± 4.47E−04 | 1.2E−05 ± 1.1E−05 |
| Burkholderia | 8.7E−05 ± 1.9E−05 | 5.5E−05 ± 3.9E−05 | 0.15E−04 ± 1.5E−04A |
| Acidovorax | 2.49E−04 ± 0.74E−04 | 2.18E−04 ± 2.03E−04 | 1.063E−03 ± 0.745E−05 |
| Azoarcus | 2.95E−04 ± 1.54E−04 | 1.01E−04 ± 1.33E−04 | 0.26E−04 ± 3.3E−04A |
| Citrobacter | 3.56E−04 ± 3.32E−04 | 5.6E−05 ± 8.3E−05 | 3.7E−05 ± 4.8E−05 |
| Enterobacter | 1.27E−04 ± 0.56E−04 | 1.28E−04 ± 0.69E−04 | 3.0E−05 ± 2.2E−05 |
| Escherichia | 0.804E−03 ± 1.378E−03 | 0.99E−04 ± 1.01E−04 | 0.5E−05 ± 0.9E−05 |
| Klebsiella | 4.14E−04 ± 2.73E−04 | 8.96E−04 ± 6.41E−04 | 6.91E−04 ± 5.00E−04 |
| Serratia | 1.3E−05 ± 1.2E−05 | 2.63E−04 ± 4.56E−04 | – |
| Pseudomonas | 1.446E−02 ± 0.809E−02 | 1.768E−02 ± 1.062E−02 | 1.95E−02 ± 1.206E−02 |
| Stenotrophomonas | – | 2.2E−05 ± 2.6E−05 | 1.06E−04 ± 0.94E−04 |
| Fretibacterium | 0.6E−05 ± 1.0E−05 | 3.18E−04 ± 2.62E−04A | 5.3E−05 ± 6.1E−05 |
A: P < 0.05 in comparison with the control group; B: P < 0.05 in comparison with the model group
Besides those of known bacteria, other lactase genes in unclassified bacteria or bacteria without blast hit were found to be in the level of phylum and genus. The percentage of unclassified and no blast hit bacteria were 63.1 and 32.8%, respectively (Fig. 4).
Fig. 4.
Percentage of different lactase-producing bacteria. a In the level of phylum; b in the level of genus
Discussion
In China, it is very common to treat diarrhea with Chinese traditional medicines (herbs). As reported, their effects were partly due to the regulation of lactase activities (Zhang et al. 2009, 2010). Our previous study showed that the lactase activity was decreased in mice with antibiotics-induced diarrhea. Therefore, our current study aimed to figure out whether QWBZS, a common medicine used for treating diarrhea, affects the lactase activity and further prove that the diarrhea is strongly associated with change in lactase activity.
Our results showed that as with control group and model group, the QWBZS treatment group had no significant difference in Chao index and ACE index of intestinal bacteria lactase genes, whereas Shannon index in the QWBZS treatment group was decreased significantly (P < 0.01 or P < 0.05). As shown in PCoA figure, samples in treatment group were clearly distinguished from those of control group and model group. These results indicated that QWBZS treatment decreased the richness of lactase genes of intestinal bacterial contents and changed the composition of bacteria community for producing lactase.
Bacterial lactase activities in the intestine can be affected by the quantity and different species of bacteria producing lactase, as well as the variation (including mutations) of lactase gene. The species producing lactase can be divided into three types on the basis of lactase-producing capability: high lactase-producing strains, low lactase-producing strains and inactive lactase-producing strains (Tan and Shi 2008; Jiang et al. 2014). Our current results showed that while the majority of species expressing lactase genes were unclassified, Actinobacteria, Firmicutes and Proteobacteria were the major known phyla for producing lactases in gut. Among the known lactase-producing genus, Mycobacterium, Frankia, Sphingobium and Serratia could not be detected in the treatment group. These results suggested that most of the identified bacteria expressing lactase genes in our study are novel and that Mycobacterium, Frankia, Sphingobium and Serratia make little contribution to lactase activity. Elguezabal et al. also reported no association was found between Mycobacterium avium subsp. paratuberculosis infection and lactase gene expression (Elguezabal et al. 2012).
It is known that lactase activities can be influenced by not only bacteria species but also their gene abundance. At the genus level, the abundance of lactase genes was shown to be higher in Corynebacterium, Arthrobacter, Eggerthella, Acidovorax, Klebsiella and Pseudomonas. Treatment with QWBZS increased the abundance of lactase gene originated from Acidovorax sp. KKs102, Stenotrophomonas sp. LMG11000, Pseudomonas oleovorans, Eggerthella and Burkholderia, but decreased that from Azoarcus. A recent study by Wang et al. (2017) reported that the metabolite by Eggerthella showed an inhibitory effect on growth of human colon cancer line. In addition, the relative abundance of Eggerthella was reduced in Rett syndrome subjects as compared to healthy controls (Strati et al. 2016). These results suggested that Eggerthella is one of the major strains with high level of lactase gene expression and lactase production.
In summary, our study suggests that the probable mechanism of QWBZS improving lactase activity would be the increase in the abundance of lactase genes by some key lactase-producing species rather than an increase in the lactase gene diversity. In addition, a large number of novel bacteria expressing lactase genes were found in our study and they are worth being explored in future studies.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81573951).
Compliance with ethical standards
Conflict of interest
No conflict of interest was declared.
Contributor Information
Huaying Hui, Phone: (+86)13487575642, Email: huihuaying2003@126.com.
Zhoujin Tan, Phone: (+86) 13974954942, Email: tanzhjin@sohu.com.
References
- Alexandre V, Even PC, Larue-Achagiotis C, Blouin JM, Blachier F, Benamouzig R, Tomé D, Davila AM. Lactose malabsorption and colonic fermentations alter host metabolism in rats. Br J Nutr. 2013;110:625–631. doi: 10.1017/S0007114512005557. [DOI] [PubMed] [Google Scholar]
- Cai GX, Zeng A, Xiao NQ, Zhou SN, Guo KX, Tan ZJ. Effects of Jianwei Qiweibaizhusan on the intestinal microorganisms and enzyme activities. J Pharm Technol Drug Res. 2013;2(1):6. doi: 10.7243/2050-120X-2-6. [DOI] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elguezabal N, Chamorro S, Molina E, Garrido JM, Izeta A, Rodrigo L, Juste RA. Lactase persistence, NOD2 status and Mycobacterium avium subsp. paratuberculosis infection associations to Inflammatory Bowel Disease. Gut Pathog. 2012;4(1):6. doi: 10.1186/1757-4749-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo KX, Zhou SN, Tan ZJ, Cai Y, She Y, Cai GX. Study on inhibition and boosting action of Qiweibaishusan to intestinal yeast. Prog Mod Biomed. 2013;13(27):5259–5263. [Google Scholar]
- Guo KX, Xiao XY, Liu YJ, Long CX, Tan ZJ. Effect of Qiweibaizhusan on the intestinal lactobacilli diversity in dysbacteria diarrheal mice. Chin J Appl Environ Biol. 2015;21:1071–1075. [Google Scholar]
- Guo KX, Tan ZJ, Xie MZ, She Y, Wang XH. The synergic effect of ultra-micro powder Qiweibaizhusan combined with yeast on dysbacteriotic diarrhea mice. Chin J Appl Environ Biol. 2015;21:61–67. [Google Scholar]
- Hammer HF, Hammer J. Diarrhea caused by carbohydrate malabsorption. Gastroenterol Clin N Am. 2012;41(3):611–627. doi: 10.1016/j.gtc.2012.06.003. [DOI] [PubMed] [Google Scholar]
- He L, Long CX, Liu YJ, Guo YF, Xiao NQ, Tan ZJ. Effects of Debaryomyces hansenii treatment on intestinal microorganisms in mice with antibiotics-induced diarrhea. 3 Biotech. 2017;7:347. doi: 10.1007/s13205-017-0953-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang XD, Guo GG, Zhang J. Association of genetic diversity for Amy6-4 gene activity in germplasm of barley. Acta Agron Sin. 2014;40(2):205–213. doi: 10.3724/SP.J.1006.2014.00205. [DOI] [Google Scholar]
- Liu QS, Xu XH, Liu H, Liu YF, Tan ZJ. Effect on intestinal villi and crypts in flora diarrhea mice by Qiweibaizhu powder. Chin Med Mod Distance Educ China. 2014;12(23):154–155. [Google Scholar]
- Long CX, He L, Guo YF, Liu YW, Xiao NQ, Tan ZJ. Diversity of bacterial lactase genes in intestinal contents of mice with antibiotics-induced diarrhea. World J Gastroenterol. 2017;23(42):7584–7593. doi: 10.3748/wjg.v23.i42.7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CX, He L, Liu YW, Hui HY, Tan ZJ, Li DD. Universal primer for analysis of the diversity of intestinal bacterial lactase gene. Chin J Appl Environ Biol. 2017;23(4):758–763. [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaffee WF, Kloepper JW. Temporal changes in the bacterial communities of soil, rhizosphere, and endorhiza associated with Field-Grown Cucumber (Cucumis sativus L) Microb Ecol. 1997;34(3):210–223. doi: 10.1007/s002489900050. [DOI] [PubMed] [Google Scholar]
- Ojetti V, Gigante G, Gabrielli M, Ainora ME, Mannocci A, Lauritano EC, Gasbarrini G, Gasbarrini A. The effect of oral supplementation with Lactobacillus reuteri or tilactase in lactose intolerant patients: randomized trial. Eur Rev Med Pharmacol Sci. 2010;14(3):163–170. [PubMed] [Google Scholar]
- Peng HZ, Ren LH. Relationship between antibiotic associated diarrhea and lactose intolerance. Chin Gen Pract. 2011;14(26):2999–3000. [Google Scholar]
- Pitta DW, Pinchak E, Dowd SE, Osterstock J, Gontcharova V, Youn E, Dorton K, Yoon I, Min BR, Fulford JD, Wickersham TA, Malinowski DP. Rumen bacterial diversity dynamics associated with changing from bermudagrass hay to grazed winter wheat diets. Microb Ecol. 2010;59(3):511–512. doi: 10.1007/s00248-009-9609-6. [DOI] [PubMed] [Google Scholar]
- Pitta DW, Parmar N, Patel AK, Indugu N, Kumar S, Prajapathi KB, Patel AB, Reddy B, Joshi C. Bacterial diversity dynamics associated with different diets and different primer pairs in the rumen of Kankrej cattle. PLoS ONE. 2014;9(11):e111710. doi: 10.1371/journal.pone.0111710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao WH. 30 Cases of dysbacteriosis-related diarrhea treated with flavored Shenling Baizhu powder. Shanxi J Tradit Chin Med. 2014;35(1):17–19. [Google Scholar]
- Shannon CE. The mathematical theory of communication. 1963. MD Comput. 1997;14:306–317. [PubMed] [Google Scholar]
- Strati F, Cavalieri D, Albanese D, De Felice C, Donati C, Hayek J, Jousson O, Leoncini S, Pindo M, Renzi D, Rizzetto L, Stefanini I, Calabrò A, De Filippo C. Altered gut microbiota in Rett syndrome. Microbiome. 2016;4(1):41. doi: 10.1186/s40168-016-0185-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CC, Shi L. Diversity of Helicobacter pylori clinical isolates by the method of polymerase chain reaction-restriction fragment length polymorphism. Mod Dig Interv. 2008;13(1):18–21. [Google Scholar]
- Tan ZJ, Wu H, Liu FL, Cai Y, Cai GX, Zhang HL, Zeng A. Effect of ultra-micro powder Qiweibaizhusan on the intestinal microbiota and enzyme activities in mice. Acta Ecol Sin. 2012;32(21):6856–6863. doi: 10.5846/stxb201109271422. [DOI] [Google Scholar]
- Wang G, Luo H, Meng K, Wang Y, Huang H, Shi P, Pan X, Yang P, Diao Q, Zhang H, Yao B. High genetic diversity and different distributions of glycosyl hydrolase family 10 and 11 xylanases in the goat rumen. PLoS ONE. 2011;6(2):e16731. doi: 10.1371/journal.pone.0016731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yu LK, Han XH, Yuan LY, Cui Y, Ma N. Curative effect of Qiweibaizhusan on antibiotics-associated diarrhea. Lishizhen Med Mater Medica Res. 2013;24(10):2451–2452. [Google Scholar]
- Wang H, Zeng A, Cao R, Guo ZH, He YS, Tan ZJ. The material base of Qiweibaizhusan regulating intestinal microecology. World Chin J Digestol. 2014;22(13):1773–1777. doi: 10.11569/wcjd.v22.i13.1773. [DOI] [Google Scholar]
- Wang Y, Yu F, Liu MY, Zhao YK, Wang DM, Hao QH, Wang XL. Isolation and characterization of a human intestinal bacterium Eggerthella sp. AUH-JLD49s for the conversion of (−)-3′-desmethylarctigenin. J Agric Food Chem. 2017;65(20):4051–4056. doi: 10.1021/acs.jafc.7b00114. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhou SN, Guo C, Tan ZJ, Cai GX, Zeng A, Zhang HL. A metagenome DNA extracting method of intestinal flora in mice for molecular diversity analysis based on PCR technology. Chin J Microecol. 2012;24(7):648–651. [Google Scholar]
- Yang ZH, Yang WH. 23 Cases of infantile spleen deficiency diarrhea treated with Qiweibaizhusan. Gansu J TCM. 2010;23(12):36–37. [Google Scholar]
- Yuan P (2012) Gene cloning and expression of pectinases and their diversity analysis in the rumen of Small Tail Han sheep. Dissertation, Chinese Academy of Agricultural Sciences
- Zeng A, Zhang HL, Tan ZJ, Cai Y, Cai GX, Zhou SN. The construction of mice diarrhea model due to dysbacteriosis and curative effect of ultra-micro Qiweibaizhusan. Microbiol China. 2012;39(9):1341–1348. [Google Scholar]
- Zhang XL, Xu QQ, Wang LY, Huo XQ, Cao LZ, Chen LG, Ren YH, Dong SS (2009) The influence of Sijunzi decoction on the lactase activity in diarrheal mice intestinal mucosa. The 16th Veterinary Pathology Branch in Chinese Association of Animal Science and Veterinary Medicine and the 15th Seminar of Animal Pathophysiology Professional Committee in Chinese Association of Pathophysiology
- Zhang XL, Wang YC, Xu QQ, Wang LY, Huo XQ, Cao LZ, Zhang YH, Dong SS. The influence of decoction of compound Radix pulsatillae on the lactase activity in diarrheal mice intestinal mucosa. J Agric Univ Hebei. 2010;33(2):99–102. [Google Scholar]
- Zhang YY, Zhou LL, Yang SF. The basic research of primary lactase deficiency. Int J Pediatr. 2014;41(3):302–304. [Google Scholar]
- Zhou XY. Pathogenesis of antibiotics-induced diarrhea. Chin J Microbiol. 2004;16(6):376–377. [Google Scholar]