Abstract
A new resazurin-based assay was evaluated and optimized using a microplate (384-well) format for high-throughput screening of antibacterial molecules against Klebsiella pneumoniae. Growth of the bacteria in 384-well plates was more effectively measured and had a > sixfold higher signal-to-background ratio using the resazurin-based assay compared with absorbance measurements at 600 nm. Determination of minimum inhibitory concentrations of the antibiotics revealed that the optimized assay quantitatively measured antibacterial activity of various antibiotics. An edge effect observed in the initial assay was significantly reduced using a 1-h incubation of the bacteria-containing plates at room temperature. There was an approximately 10% decrease in signal variability between the edge and the middle wells along with improvement in the assay robustness (Z′ = 0.99). This optimized resazurin-based assay is an efficient, inexpensive, and robust assay that can quantitatively measure antibacterial activity using a high-throughput screening system to assess a large number of compounds for discovery of new antibiotics against K. pneumoniae.
Keywords: Antibacterials, Edge effect, High-throughput screening, Klebsiella pneumoniae, Resazurin assay
Introduction
Klebsiella pneumoniae is a gram-negative bacterium. It is a major causative agent of nosocomial infections. Manifestations of Klebsiella infection range from uncomplicated skin and skin-structure infections to fatal bacteremia. Although antibiotics are routinely used to treat these infectious diseases, the emergence of drug-resistant K. pneumoniae (e.g., carbapenem-resistant K. pneumoniae) has limited the treatment options and contributed to the rapid increase in dependency on polymyxins and tigecycline, which are the last resort of the most cases of gram-negative bacterial infections (Petrosillo et al. 2008; Ruzin et al. 2005). This trend imposes a serious threat because polymyxin- and tigecycline-resistant K. pneumoniae cases have been reported (Bogdanovich et al. 2011; Elemam et al. 2009; Lee et al. 2009; Marchaim et al. 2011; Ruzin et al. 2005). Currently, drug candidates, such as combinations of a beta-lactam and a beta-lactamase inhibitor as well as existing classes of antibiotic derivatives, are in clinical development for treatment of nosocomial infections (Livermore et al. 2008; Stachyra et al. 2009). However, except for a new beta-lactamase inhibitor (Avibactam), no new classes of antibiotics have been approved for treatment of gram-negative bacterial infections since 2000 (Butler et al. 2016; Zetts 2016). For the discovery of first-in-class antibiotics, especially against gram-negative bacteria such as K. pneumoniae, it is crucial to develop an effective and efficient screening system that is specialized for use against a target pathogen.
High-throughput screening (HTS) is an approach widely used in drug discovery that has been also applied to antibiotic development. Assays for HTS need to be highly sensitive, rapid, and relatively simple so that reliable, robust results can be produced using automated-operation platforms. Fluorescence-based assays are the most likely candidates for HTS because they have high sensitivity and efficiency. A target-based fluorescence spectroscopic high-throughput screening (FLHTS) as well as gel microdroplet –fluorescence-activated cell sorting assay (GMD –FACS) using fluorescent dyes such as fluorescein maleimide and SYTOX orange, respectively, have been successfully implemented for HTS of antibacterial agents.(Dostal et al. 2015; Hanson et al. 2016; Nairn et al. 2017; Scanlon et al. 2014) Resazurin is a dye that detects metabolic activity of living cells through its conversion from the non-fluorescent form (blue, oxidized) to the fluorescent resorufin (pink, reduced). Resorufin fluorescence signals can be easily detected using a fluorescence spectrometer, and they provide quantitative measurements of cell proliferation. Resazurin has, therefore, been used extensively in cell proliferation, growth, and toxicity studies of cells types ranging from human to bacterial cells. Recent studies have revealed various applications for resazurin in the determination of antibiograms and detection of bacterial biofilm formation (Franzblau et al. 1998; Kirchner et al. 2012; Palomino and Portaels 1999; Van den Driessche et al. 2014). In this study, a resazurin-based phenotypic assay was developed for HTS of antibacterial molecules against K. pneumoniae. The developed assay was evaluated by determining the association of resazurin reduction with K. pneumoniae growth and the linearity of fluorescence signals against the viability of bacteria. Optimization of the assay for HTS was performed to maximize its sensitivity and stability as an automated-operation platform with reduced edge effects.
Materials and methods
Bacterial strains
Klebsiella pneumoniae ATCC® 13883™ was purchased (American Type Culture Collection, Rockville, MD, USA) as a type strain. For routine maintenance, the bacteria were grown in Luria broth (LB) at 37 °C with a shaking. For longer storage, the culture was supplemented with 10% (v/v) glycerol and stored at − 70 °C. To prepare the bacterial culture, the frozen stock was thawed at room temperature, and then 10 µl of the stock was inoculated into a 50-ml tube containing 5 ml LB. The culture was incubated at 37 °C with a shaking. The next day, 1 ml of the 1:10 diluted grown bacteria was transferred to a disposable cuvette (Ratiolab, Dreieich, Germany) for measurement of optical density (OD600; ULTROSPEC10, Biochrom Ltd., Cambridge, England).
Chemicals and materials
The LB (Difco, Detroit, MI, USA) was prepared according to the manufacturer’s instructions. The reference antimicrobial agents, colistin, imipenem, and tetracycline were obtained (Sigma-Aldrich, Louis, MO, USA); the gentamicin solution (10 mg/ml) was purchased (Thermo-fisher, Waltham, MA, USA). The colistin, imipenem, and tetracycline were purchased in powder form, and were solubilized in the growth media (final concentration, 5 mg/ml). The resazurin powder was obtained from Sigma-Aldrich. A 10× solution (0.125 mg/ml) was prepared in water and filter-sterilized using a 0.22-µm membrane. The prepared resazurin solution was covered to prevent exposure to light and was stored at 4 °C. Sterile dimethyl sulfoxide (DMSO) solution, was purchased from Sigma-Aldrich. The Victor3 multi-label plate reader (Perkin and Elmer, Waltham, MA, USA) was used to measure optical density (OD600) and fluorescence (bottom-read) with 530 nm excitation and 590 nm emission. Black polystyrene microplates (384 wells per plate) with clear bottoms (Greiner-bio One, Frickenhausen, Germany) were used for optimization and validation procedures. Before the readings were taken, a plastic adhesive plate sealer was applied to each plate to prevent contamination of the reading equipment.
Assessment of bacterial growth kinetics
To determine the growth kinetics, the overnight bacterial culture was diluted in fresh media to prepare inoculation cultures with an OD600 of 0.02 and 0.0001 for a large culture in an Erlenmeyer flask and for a microplate culture, respectively. The large culture was incubated at 37 °C with a shaking. A 600 nm reading of a 1 ml sample of culture was collected each hour. For the microplate culture, a total of eight microplates were prepared, and bacteria were added to the designated wells. At the time of measurement, one plate was removed from the incubator, and 5 µl 10× resazurin was added to each well, followed by a 1-h incubation at room temperature. The fluorescence signals were measured with 530 nm excitation and 590 nm emission using a multi-label plate reader Victor 3 (Perkin and Elmer, Waltham, MA, USA). After the reading, 10 µl samples were removed from each well and were diluted in LB at 1:100, 1:1000, and 1:100,000 concentrations. Replicate 100 µl volumes were then plated onto an LB plate for every reading time point. The plates were incubated at 37 °C, and the colonies were counted the next day.
Assay validation experiment
For the assay validation experiment, signal variability was assessed using an interleaved 384-well plate and maximum and minimum signal conditions. Each interleaved plate was prepared by adding 5 µl 5% (v/v) DMSO or 200 µg/ml colistin to odd-numbered and even-numbered columns, respectively. The resazurin assay was performed by adding 45 µl bacteria to each interleaved plate (inoculation OD600 0.0001). After incubating the assay plate at 37 °C for 6 h, 5 µl 10× resazurin solution was added to each well and the plate was incubated for 1 h at room temperature. Duplicate plates were prepared for the edge effect reduction test. One plate was placed at room temperature for 1 h, before the 6-h incubation at 37 °C. The edge effect was analyzed using scatter-plot and heatmap analysis. Assay quality was characterized using Z′ factor values (Z′ = 1 − [3 × (σ p + σ n)/|μ p − μ n|], where the inputs were the mean (µ) and standard deviation (σ) values for the positive (p) and negative (n) controls.
Dose–response curve and determination of susceptibility
To generate the dose–response curve, each reference agent (colistin, imipenem, tetracycline, gentamicin) (1 mg/ml) was diluted using a 1:2 ratio (15 total points); 5 µl of each dilution was transferred to an assay plate. The bacterial culture was diluted to 0.0001 OD600, 45 µl was added to each well containing an antibiotic. After addition of the bacteria, the assay plate was incubated at room temperature for 1 h, and was transferred to 37 °C incubation conditions for 6 h. A total of 5 µl 10 × resazurin was added to each well and the plate was incubated for 1 h at room temperature, followed by the fluorescence measurements. The values obtained from nonlinear regression of the read-out values were analyzed against concentration (logarithmic scale; Graphpad Prism 6.07) to determine the values for MIC.
Results and discussion
Klebsiella pneumoniae growth in 384-well microplates and efficiency comparison of optical density and resazurin-based viability measurements as an assay method
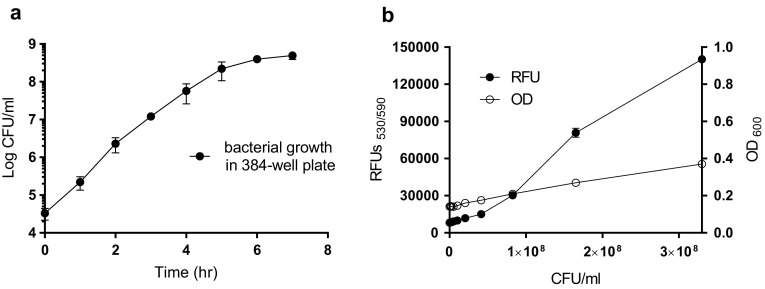
Bacterial growth kinetics in a 384-well microplate were measured to optimize the assay conditions for HTS. Exponential growth phase K. pneumoniae ATCC 13883 were diluted in fresh LB medium at a 0.0001 optical density (OD; 3 × 104 colony forming units [CFU]/ml); 50 µl of the diluted bacteria were transferred to each well of a 384-well plate, followed by incubation at 37 °C without agitation. The bacterial growth was measured (CFU/ml) using samples collected each hour. The growth in the microplate was exponential; the number of bacteria doubled every 22 min during the 6-h period, until the total CFU concentration was 4 × 108 cells per ml (Fig. 1a). This growth kinetics were comparable to the growth kinetics seen with a 20-ml culture in an Erlenmeyer flask incubated at 37 °C with agitation (data not shown). Based on the results, an OD600 = 0.0001 (~ 1 × 103 CFU/ml) and a 6-h incubation period were chosen as the initial bacterial concentration and the total culture time, respectively, in the 384-well plates. Turbidometry (OD600) is a conventional phenotypic screening method used to detect bacterial growth, but the signal-to-background (S/B) ratio was only 2.6 ± 0.04 when the bacteria had increased from 0 to 3.3 × 108 CFU/ml. The bacterial viability measurement of the resazurin samples revealed a > sixfold increase in the S/B ratio (17.2 ± 0.28); this result suggested a higher sensitivity, especially during HTS (Fig. 1b).
Fig. 1.
a Klebsiella pneumoniae ATCC 13883 growth in 384-well microplates (filled circle), b comparison of optical density (OD600) (open circle) and resazurin-based relative fluorescence units (RFU) (filled circle) viability measurements. Mean ± SD values (n = 4)
Optimization of a resazurin applying condition
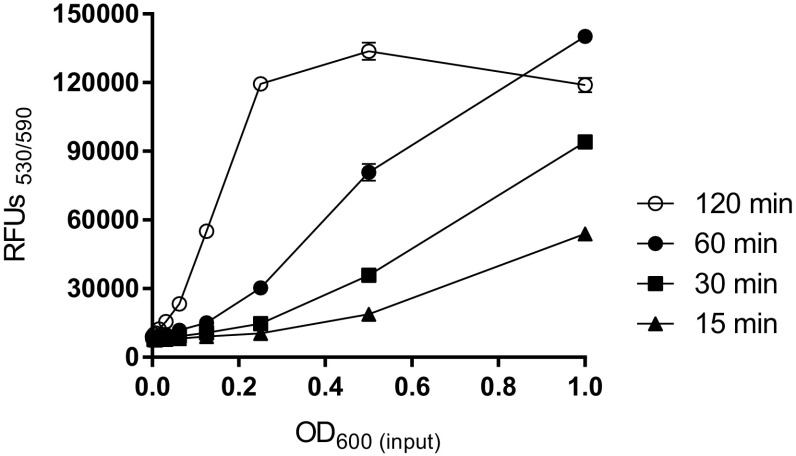
Applying conditions of resazurin can be varied based on experimental purposes (Franzblau et al. 1998; Palomino and Portaels 1999; Van den Driessche et al. 2014). To find an optimal condition of the resazurin-based assay for automated large-scale drug screenings against K. pneumoniae, the effect of incubation temperature and time with resazurin were accessed. Higher fluorescence signals were observed after incubation at room temperature compared to 37 °C in given time (data not shown). We found that a 1-h room-temperature incubation resulted in the best linearity (R 2 = 0.99) of fluorescence signal to the amount of input bacteria (Fig. 2). Less than 30 min of incubation resulted in decreased signals, which indicated that resazurin had not fully reduced and lower S/B ratios were obtained compared with the 60-min incubation. In contrast, a prolonged (2-h) incubation resulted in saturated signals, plateaus of medium to high numbers of bacteria, and low linearity (R 2 = 0.65) (Fig. 2). Slight signal reduction occurred with high numbers of input bacteria (OD600 = 1.0) and a prolonged incubation time (Fig. 2). Therefore, 60 min of resazurin incubation at room temperature was used for subsequent optimization experiments.
Fig. 2.
Comparison of signal (RFU) linearity at different reaction time with resazurin. 1:2 serial dilutions (1.0–0.0004) of bacteria were prepared and added to a 384-well plate. Five microliters 10× resazurin solution was added to each well containing bacteria and media (50 µl total volume per well). The resazurin reaction took place at room temperature for 15 (filled triangle), 30 min (filled square), 60 min (filled circle), and 120 min (open circle) min. Mean ± SD values (n = 4)
Validation of the resazurin-based assay using signal variability assessment
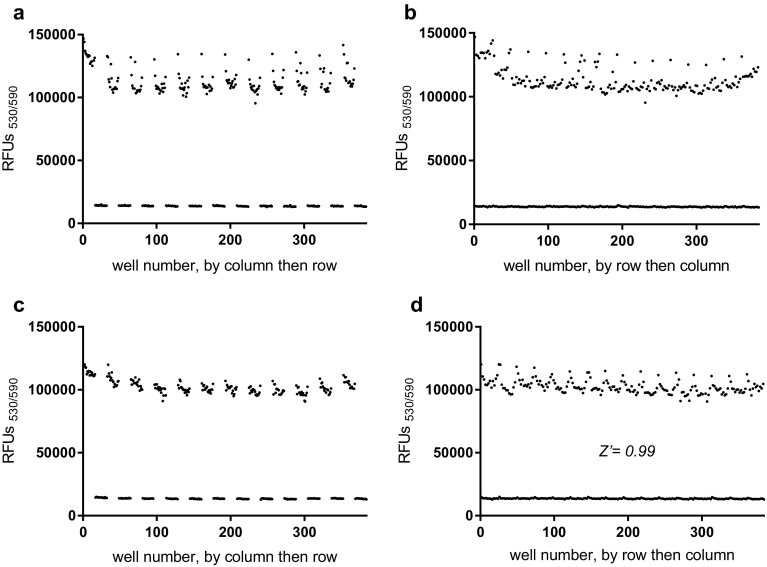
To validate the developed resazurin-based assay as a HTS system, fluorescence signal variability was assessed in 384-well plates. The assessment was performed by comparing fluorescence signals from different wells containing bacteria in 20 µg/ml colistin (inhibited signals, n = 192) or 0.5% (v/v) DMSO (maximum signals, n = 192). The heat map (data not shown) and dot-graph analysis results revealed the presence of significant edge effects in the plates (Fig. 3a, b). The maximum signal values for wells located at the edges of the plates (columns 1, 2, 23, and 24 and rows A, B, O, and P) were almost 14% higher than signals from the middle wells (i.e., average signals of 123,700 ± 9990 and 108,700 ± 4900, respectively). Edge effects are issues because of increased noise and variability they contribute during HTS. Previously, the pre-incubation approach was used to decrease an edge effect by reducing a thermal gradient in a microplate (Lundholt et al. 2003). Significant reduction of the edge effect in our resazurin-based assay was also achieved when the plates containing the bacteria were left at room temperature for 1 h before the 6-h incubation at 37 °C (Fig. 3c, d). Compared with plates with no room-temperature pre-incubation, pre-incubation resulted in a 4.7% signal difference between the edge and the middle wells (Fig. 3). Other changes in method, such as post-incubation at room temperature after the 6-h incubation, did not significantly decrease the edge effect (data not shown).
Fig. 3.
Validation of resazurin-based assay using signal variability assessment in 384-well plates: without pre-incubation at room temperature for 1 h (a, b) and with pre-incubation (c, d). For the scatter plots, the well numbers are arranged by column then row (a, c) or by row then column (b, d). Max signal: DMSO 0.5% (v/v) (n = 192), Inhibited signal: colistin 20 µM (n = 192)
Dose responses of various antibiotics against K. pneumoniae measured using the resazurin-based assay
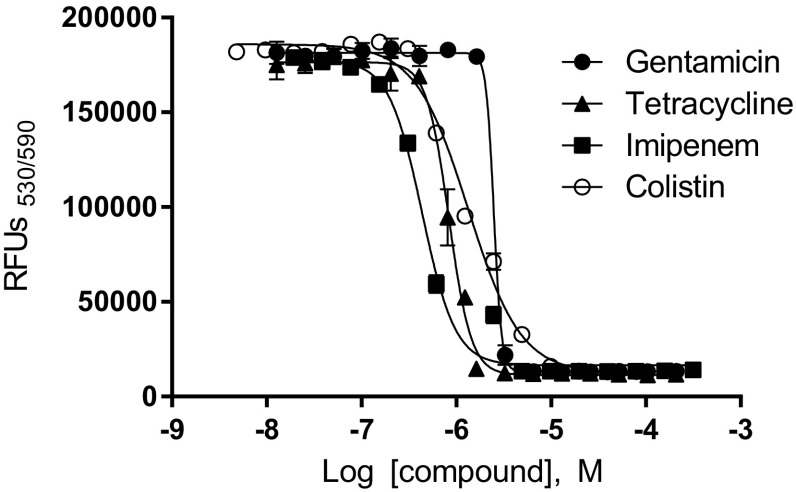
To investigate whether the use of the optimized HTS assay was valid for quantitative assessment of antibacterial activity of compounds, the MIC values for several antibiotics (gentamicin, colistin, imipenem, and tetracycline) were determined for K. pneumoniae by dose–response curves. The resulting MIC values for gentamicin, colistin, imipenem, and tetracycline were 3.0 (± 0.45), 5.1 (± 0.51), 1.9 (± 0.43), and 1.2 (± 0.16) μM, respectively. These results suggested that during automated large-scale screening, the optimized assay was able to quantitatively measure the antibacterial activity of compounds against K. pneumoniae (Fig. 4).
Fig. 4.
Dose responses of gentamicin (filled circle), tetracycline (filled triangle), imipenem (filled square), and colistin (open square) against Klebsiella pneumoniae measured using the resazurin-based assay. Mean ± SD values (n = 4), Nonlinear regression curve fit
Conclusion
The simple principle behind the resazurin-based assay is the measurement of antibacterial activity of compounds through the detection of living K. pneumoniae. Therefore, this assay can easily be used for various large-scale screenings to discover biofilm inhibitors or drug combinations, even against multidrug resistant K. pneumoniae. The resazurin-based assay optimized in this study was efficient, sensitive, and robust and can be used effectively for HTS of drugs against K. pneumoniae. We expect that this assay will facilitate discovery of new antibiotics.
Acknowledgements
We thank Kevin Pethe for invaluable advice on this research. This work was supported by National Research Foundation of Korea (NRF) grants funded by the Korean government (MSIP) (2012-00011) (NRF-2014K1A4A7A01074645), Gyeonggi-do, and KISTI.
Compliance with ethical standards
Conflict of interest
No conflict of interest declared.
References
- Bogdanovich T, Adams-Haduch JM, Tian GB, Nguyen MH, Kwak EJ, Muto CA, Doi Y. Colistin-resistant, Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae belonging to the international epidemic clone ST258. Clin Infect Dis Off Publ Infect Dis Soc Am. 2011;53:373–376. doi: 10.1093/cid/cir401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MS, Blaskovich MA, Cooper MA. Antibiotics in the clinical pipeline at the end of 2015. J Antibiot. 2016 doi: 10.1038/ja.2016.72. [DOI] [PubMed] [Google Scholar]
- Dostal SM, Fang Y, Guerrette JC, Scanlon TC, Griswold KE. Genetically enhanced lysozyme evades a pathogen derived inhibitory protein. ACS Chem Biol. 2015;10:1110–1117. doi: 10.1021/cb500976y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemam A, Rahimian J, Mandell W. Infection with panresistant Klebsiella pneumoniae: a report of 2 cases and a brief review of the literature. Clin Infect Dis Off Publ Infect Dis Soc Am. 2009;49:271–274. doi: 10.1086/600042. [DOI] [PubMed] [Google Scholar]
- Franzblau SG, et al. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J Clin Microbiol. 1998;36:362–366. doi: 10.1128/jcm.36.2.362-366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson M, Jordan LD, Shipelskiy Y, Newton SM, Klebba PE. High-throughput screening assay for inhibitors of TonB-dependent iron transport. J Biomol Screen. 2016;21:316–322. doi: 10.1177/1087057115613788. [DOI] [PubMed] [Google Scholar]
- Kirchner S, Fothergill JL, Wright EA, James CE, Mowat E, Winstanley C. Use of artificial sputum medium to test antibiotic efficacy against Pseudomonas aeruginosa in conditions more relevant to the cystic fibrosis lung Journal of visualized experiments. JoVE. 2012 doi: 10.3791/3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Patel G, Huprikar S, Calfee DP, Jenkins SG. Decreased susceptibility to polymyxin B during treatment for carbapenem-resistant Klebsiella pneumoniae infection. J Clin Microbiol. 2009;47:1611–1612. doi: 10.1128/JCM.02466-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore DM, Mushtaq S, Warner M, Miossec C, Woodford N. NXL104 combinations versus Enterobacteriaceae with CTX-M extended-spectrum beta-lactamases and carbapenemases. J Antimicrob Chemother. 2008;62:1053–1056. doi: 10.1093/jac/dkn320. [DOI] [PubMed] [Google Scholar]
- Lundholt BK, Scudder KM, Pagliaro L. A simple technique for reducing edge effect in cell-based assays. J Biomol Screen. 2003;8:566–570. doi: 10.1177/1087057103256465. [DOI] [PubMed] [Google Scholar]
- Marchaim D, et al. Outbreak of colistin-resistant, carbapenem-resistant Klebsiella pneumoniae in metropolitan Detroit. Mich Antimicrob Agents Chemother. 2011;55:593–599. doi: 10.1128/AAC.01020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairn BL, et al. Fluorescence high-throughput screening for inhibitors of TonB action. J Bacteriol. 2017 doi: 10.1128/JB.00889-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomino JC, Portaels F. Simple procedure for drug susceptibility testing of Mycobacterium tuberculosis using a commercial colorimetic assay. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. 1999;18:380–383. doi: 10.1007/PL00015025. [DOI] [PubMed] [Google Scholar]
- Petrosillo N, Ioannidou E, Falagas ME. Colistin monotherapy vs. combination therapy: evidence from microbiological, animal and clinical studies. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2008;14:816–827. doi: 10.1111/j.1469-0691.2008.02061.x. [DOI] [PubMed] [Google Scholar]
- Ruzin A, Visalli MA, Keeney D, Bradford PA. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2005;49:1017–1022. doi: 10.1128/AAC.49.3.1017-1022.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon TC, Dostal SM, Griswold KE. A high-throughput screen for antibiotic drug discovery. Biotechnol Bioeng. 2014;111:232–243. doi: 10.1002/bit.25019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachyra T, Levasseur P, Pechereau MC, Girard AM, Claudon M, Miossec C, Black MT. In vitro activity of the {beta}-lactamase inhibitor NXL104 against KPC-2 carbapenemase and Enterobacteriaceae expressing KPC carbapenemases. J Antimicrob Chemother. 2009;64:326–329. doi: 10.1093/jac/dkp197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Driessche F, Rigole P, Brackman G, Coenye T. Optimization of resazurin-based viability staining for quantification of microbial biofilms. J Microbiol Methods. 2014;98:31–34. doi: 10.1016/j.mimet.2013.12.011. [DOI] [PubMed] [Google Scholar]
- Zetts R (2016) Antibiotics currently in clinical development. The Pew Charitable Trusts. http://www.pewtrusts.org/en/multimedia/data-visualizations/2014/antibiotics-currently-in-clinical-development