Abstract
The association between methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism and colorectal cancer (CRC) susceptibility has been researched in numerous studies. However, the results of these studies were controversial. Therefore, the objective of this meta-analysis was to offer a more convincible conclusion about such association with more included studies. Eligible studies published till May 1, 2017 were searched from PubMed, Embase, Web of Science, and CNKI database about such association. Pooled odds ratios (ORs) together with 95% confidence intervals (CIs) were calculated to evaluate such association. And the Begg’s funnel plot and Egger’s test were applied to assess the publication bias. This meta-analysis contained 37049 cases and 52444 controls from 87 publications with 91 eligible case–control studies. Because of lack of data for a particular genotype in several studies, all the included studies were analysed barely in the dominant model. Originally, there was no association between MTHFR C677T polymorphism and CRC susceptibility (OR =0.99, 95% CI =0.94–1.05). After excluding 13 studies according to their heterogeneity and publication bias, rs1801133 polymorphism was found to reduce the risks of CRC significantly (OR =0.96, 95% CI =0.94–0.99). In the subgroup analysis of ethnicity, there was a significant association in Asians (OR =0.94, 95% CI =0.89–1.00). Furthermore, when stratified by the source of controls and genotyping methods, the positive results were observed in population-based control group (OR =0.97, 95% CI =0.93–1.00) and PCR-restriction fragment length polymorphism (PCR-RFLP) method (OR =0.95, 95% CI =0.91–0.99. The results of the meta-analysis suggested that MTHFR C677T polymorphism was associated with CRC susceptibility, especially in Asian population.
Keywords: colorectal cancer, gene polymorphism, MTHFR, meta-analysis
Introduction
Colorectal cancer (CRC) is a critical public health problem, which is the third most commonly diagnosed cancer and the third common cause of cancer deaths in both males and females. There were 134490 new CRC cases and 49190 mortalities by estimation in the United States in 2016 [1]. The colorectal carcinogenesis is a complex multistep progress (a benign adenomatous polyp – an advanced adenoma with high-grade dysplasia – an invasive cancer) with altered expression of oncogenes, tumor suppressor genes and DNA repair genes [2]. However, the etiology of CRC is still unclear. It is known to all that CRC is a multifactorial and multigenic disease, and is influenced by environment conditions, diet habits, genetic mutations, and Escherichia coli infection [3,4]. With increasing numbers of studies, more gene polymorphisms were found to contribute to CRC [5]. These single nucleotide polymorphisms (SNPs) can be used as makers for improving cancer diagnosis and determination of treatment plans [6].
As a key enzyme and an important regulator for the metabolism of folate/vitamin B9, methylenetetrahydrofolate reductase (MTHFR) catalyzes the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate [7]. Simultaneously, the 5-methyltetrahydrofolate is the main circulatory form of folate in the body and provides a methyl group to convert the amino acid homocysteine into methionine, which is the precursor of S-adenosylmethionine (SAM). SAM is the major methyl donor in the cell and takes part in DNA methylation [8]. Therefore, MTHFR not only plays a role in making proteins and other important compounds, but also is an important factor in DNA methylation, synthesis, and repair [9]. The enzyme is encoded by the MTHFR gene located on the short arm of chromosome 1-1p36.3 [10]. Previously, several mutations of MTHFR gene have been found and MTHFR C677T (rs1801133) is the most common type amongst them. MTHFR C677T represents an alanine-to-valine substitution at nucleotide position 677 in exon 4 resulting in thermolability and concurrent decreased activity of the enzyme [11,12]. MTHFR gene mutations lead to MTHFR enzyme dificiency, low plasma folate levels, hyperhomocysteinemia [13,14] and certain diseases such as cardiovascular disease, pregnancy complications, neural defect, and several cancers including CRC [15–21]. With a growing number of studies conducted to explore such association, we hypothesized that rs1801133 was likely to relate to colorectal carcinogenesis.
Many researchers have carried out a large number of studies to examine the potential association between MTHFR C677T polymorphism and CRC susceptibility. But, the results are still inconclusive so far. Thus, the aim of this meta-analysis including all available case–control studies was to investigate a more reliable association.
Materials and methods
We searched several databases including PubMed, Embase, Web of Science, and CNKI database for published studies about exploring the association between MTHFR C677T polymorphism and CRC susceptibility till May 1, 2017. The search strategy included listed key words: ‘methylenetetrahydrofolate reductase’, ‘MTHFR polymorphism’, ‘C677T’, ‘rs1801133’, and ‘risk or susceptibility’ and ‘colorectal or colon or rectal cancer’. Furthermore, we manually searched the reference lists of clinical trials and former meta-analyses for more relevant studies. When duplicate data appeared in different publications, this meta-analysis only adopted the most recent study or the study with the most complete information. The meta-analysis was on the basis of the preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) [22]. The eligible studies needed to accord with the following inclusion criteria: (i) case–control studies; (ii) the language was not restricted to English; (iii) investigating the association between MTHFR C677T polymorphism and CRC susceptibility; (iv) offering enough raw data to calculate odds ratio (OR) with 95% confidence interval (CI). Additionally, exclusion criteria were as follows: (i) non-case–control studies; (ii) lack of sufficient data for calculating genotype frequency; (iii) case–control studies about examining the relationship between MTHFR C677T polymorphism and colorectal adenoma; (iv) duplicated publications.
Data extraction
In order to guarantee the accuracy of extracted information, two authors individually reviewed each publication and extracted useful data on the basis of the inclusion criteria listed above. When disagreements arose in the course of data extraction, discussion was carried out with other authors until the agreements were reached. The following information were extracted from each study to accomplish a standardized sheet: first author’s name, year of publication, ethnicity of population, source of controls (hospital based or population based), genotyping method, sample size of cases and controls, genotype frequency of rs1801133 in cases and controls, and the results of the Hardy–Weinberg equilibrium (HWE) test.
Statistical analysis
The relationship between MTHFR C677T polymorphism and CRC susceptibility was analyzed by using five models including the dominant model (CT + TT compared with CC), the recessive model (TT compared with CT + CC), the homozygous model (TT compared with CC), the heterozygous model (CT compared with CC), and the allele model (T compared with C). The goodness-of-fit χ2 test was conducted to evaluate the HWE in control groups and P<0.05 was regarded as significant disequilibrium [23]. Stratified analysis were performed by ethnicity, source of controls, and genotyping method. Besides, the pooled OR together with 95% CI were measured to bring out the strength of such association. The fixed effects model (Mantel–Haenszel method) and the random effects model (Dersimonian–Laird method) were selected to use based on heterogeneity in the meta-analysis. If there was no or little heterogeneity, the fixed effects model was used; otherwise, the random effects model was used. Due to only particular genotypes extracted in several studies, the dominant model analysis were carried out for all the included studies [84]. Galbraith graph was performed to explore the impossible cause of heterogeneity [24]. A sensitivity analysis was conducted to assess the stability of the results. Begg’s funnel plot was performed for potential publication bias and Egger’s linear regression test was executed to assess funnel plot asymmetry statistically. If P<0.05, publication bias existed [25]. All statistical data analyses were carried out by using Stata software (version 12.0, StataCorp LP, College Station, TX, U.S.A.).
Results
Characteristics of the studies
According to PRISMA-P, this meta-analysis contained 37049 cases and 52444 controls that were combined from 87 publications with 91 eligible case–control studies to examine the relationship between rs1801133 polymorphism and CRC risks [26–112]. The literature retrieval and selection process are shown in the flowchart in Figure 1. Detailed information of each study were listed in Table 1. The distribution of genotypes in controls was consistent with HWE except 15 studies [33–35,37,39,47,63,71,76,80,87,88,106,110,111]. In these studies, four ethnicities of population were included: Asian, Caucasian, African, and mixed ethnic group. Nine genotyping methods were applied: PCR-restriction fragment length polymorphism (PCR-RFLP), real-time PCR (RT-PCR), PCR-single strand conformation polymorphism (PCR-SSCP), methylation-specific PCR (MS-PCR), mutagenically separated PCR (MSP), MALDI-TOF-MS, Taqman, MassARRAY, and Sequenom. Depending on different sources of control, population-based and hospital-based control groups were distinguished in all the included studies.
Figure 1. Flowchart of literature search and selection process.
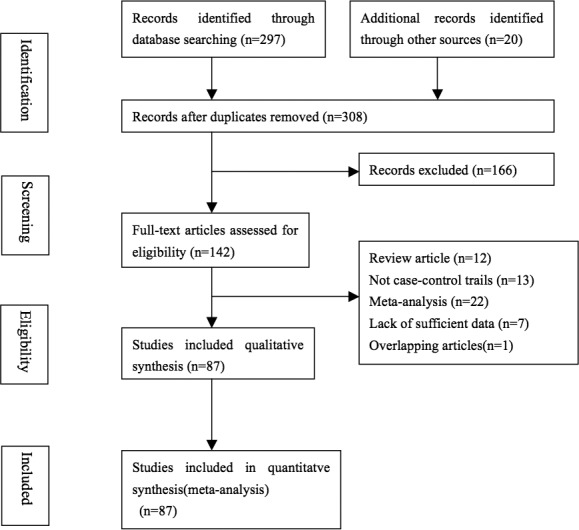
Table 1. Characteristics of individual studies included in the meta-analysis.
| MTHFR rs1801133 | Case (n) | Control (n) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Year | Surname (References) | Ethnicity | SOC | Genotyping | Case | Control | CC | CT | TT | CC | CT | TT | HWE |
| 2016 | Haerian [26] | Asian | HB | Taqman | 1123 | 1298 | 607 | 421 | 95 | 667 | 523 | 108 | Y |
| 2015 | Kim [27] | Asian | PB | PCR-RFLP | 477 | 514 | 159 | 248 | 70 | 172 | 265 | 77 | Y |
| 2014 | Rai [28] | Asian | PB | PCR-RFLP | 155 | 294 | 137 | 17 | 1 | 261 | 31 | 2 | Y |
| 2014 | Ozen [29] | Caucasian | PB | RT-PCR | 86 | 212 | 36 | 32 | 18 | 207 | 5 | 0 | Y |
| 2013 | Ashmore [30] | Caucasian | PB | RT-PCR | 625 | 603 | 241 | 309 | 75 | 263 | 259 | 81 | Y |
| 2013 | Delgado- Plasencia [31] | Caucasian | HB | PCR-RFLP | 50 | 103 | 32 | 16 | 2 | 44 | 50 | 9 | Y |
| 2013 | Yousef [32] | Asian | PB | PCR-RFLP | 128 | 116 | 79 | 45 | 4 | 59 | 45 | 12 | Y |
| 2012 | Lee [33] | Caucasian | PB | Taqman | 531 | 1004 | 250 | 229 | 52 | 464 | 391 | 149 | N |
| 2012 | Promthet [34] | Asian | HB | PCR-RFLP | 112 | 242 | 93 | 18 | 1 | 185 | 49 | 8 | N |
| 2012 | Kim [35] | Asian | HB | Taqman | 787 | 656 | 265 | 393 | 129 | 205 | 289 | 162 | N |
| 2012 | Yin [36] | Asian | HB | RT-PCR | 370 | 370 | 124 | 167 | 79 | 139 | 178 | 53 | Y |
| 2011 | Sameer [37] | Asian | PB | PCR-RFLP | 86 | 160 | 59 | 18 | 9 | 121 | 27 | 12 | N |
| 2011 | Vossen [38] | Caucasian | PB | Taqman | 1762 | 1811 | 737 | 823 | 202 | 795 | 807 | 209 | Y |
| 2011 | Kang [39] | Asian | PB | PCR-RFLP | 255 | 448 | 87 | 134 | 34 | 145 | 238 | 65 | N |
| 2011 | Zhu [40] | Asian | PB | PCR-RFLP | 86 | 100 | 29 | 42 | 15 | 49 | 41 | 10 | Y |
| 2011 | Pardini [41] | Caucasian | HB | PCR-RFLP | 666 | 1376 | 317 | 307 | 42 | 613 | 627 | 136 | Y |
| 2011 | Kim [42] | Asian | HB | MSP | 67 | 53 | 30 | 30 | 7 | 15 | 21 | 17 | Y |
| 2011 | Prasad [43] | Asian | PB | PCR-RFLP | 110 | 241 | 97 | 12 | 1 | 228 | 12 | 1 | Y |
| 2011 | Li [44] | Asian | PB | PCR-RFLP | 137 | 145 | 68 | 54 | 15 | 55 | 64 | 26 | Y |
| 2011 | Jokic [45] | Caucasian | PB | Taqman | 300 | 300 | 139 | 130 | 31 | 142 | 130 | 28 | Y |
| 2011 | Guimaracs(a) [46] | Caucasian | HB | PCR-RFLP | 101 | 188 | 42 | 44 | 15 | 92 | 79 | 17 | Y |
| 2011 | Guimaracs(b) [46] | African | HB | PCR-RFLP | 12 | 188 | 6 | 6 | 0 | 92 | 79 | 17 | Y |
| 2010 | Komlosi [47] | Caucasian | PB | PCR-RFLP | 951 | 939 | 398 | 427 | 126 | 442 | 380 | 117 | N |
| 2010 | Karpinski [48] | Caucasian | HB | MSP | 186 | 140 | 74 | 97 | 15 | 71 | 55 | 14 | Y |
| 2010 | Cui [49] | Asian | PB | PCR-RFLP | 1829 | 1700 | 622 | 923 | 284 | 540 | 863 | 297 | Y |
| 2010 | Eussen [50] | Caucasian | PB | MALDI-TOF-MS | 1329 | 2366 | 567 | 608 | 154 | 1019 | 1076 | 271 | Y |
| 2010 | Chandy [51] | Asian | HB | PCR-RFLP | 100 | 86 | 74 | 25 | 1 | 66 | 19 | 1 | Y |
| 2010 | Naghibalhossaini [52] | Asian | PB | MS-PCR | 151 | 231 | 64 | 80 | 7 | 150 | 68 | 13 | Y |
| 2010 | Promthet [53] | Asian | HB | PCR-RFLP | 130 | 130 | 104 | 26 | 0 | 94 | 31 | 5 | Y |
| 2010 | Yang [54] | Asian | PB | Sequenom | 141 | 165 | 58 | 61 | 22 | 62 | 75 | 28 | Y |
| 2010 | Fernández - Peralta [55] | Caucasian | HB | PCR-RFLP | 143 | 103 | 89 | 52 | 2 | 44 | 50 | 9 | Y |
| 2010 | Zhu [56] | Asian | PB | PCR-RFLP | 216 | 111 | 88 | 102 | 26 | 50 | 53 | 8 | Y |
| 2009 | Vogel [57] | Caucasian | PB | RT-PCR | 689 | 1793 | 318 | 320 | 51 | 876 | 750 | 167 | Y |
| 2009 | Iacopetta [58] | Mixed | PB | PCR-SSCP | 850 | 958 | 382 | 386 | 82 | 428 | 429 | 101 | Y |
| 2009 | Arreola [59] | Caucasian | PB | PCR-RFLP | 369 | 170 | 124 | 126 | 119 | 59 | 79 | 32 | Y |
| 2009 | Reeves [60] | Caucasian | HB | Taqman | 206 | 211 | 105 | 83 | 18 | 101 | 91 | 19 | Y |
| 2009 | Awady [61] | African | HB | PCR-RFLP | 35 | 68 | 6 | 23 | 6 | 44 | 20 | 4 | Y |
| 2009 | Derwinger [62] | Caucasian | PB | Taqman | 544 | 299 | 273 | 216 | 55 | 167 | 107 | 25 | Y |
| 2008 | Haghighi [63] | Asian | HB | PCR/pyrosequencing | 234 | 257 | 117 | 68 | 49 | 94 | 80 | 83 | N |
| 2008 | Sharp [64] | Caucasian | PB | PCR-RFLP | 251 | 394 | 117 | 111 | 23 | 170 | 177 | 47 | Y |
| 2008 | Kury [65] | Caucasian | PB | Taqman | 1023 | 1121 | 435 | 452 | 136 | 457 | 515 | 149 | Y |
| 2008 | Mokarram [66] | Asian | HB | MSP | 151 | 81 | 64 | 80 | 7 | 40 | 31 | 10 | Y |
| 2008 | Cao [67] | Asian | PB | PCR-RFLP | 315 | 370 | 109 | 154 | 52 | 121 | 183 | 66 | Y |
| 2008 | Theodoratou [68] | Caucasian | PB | MassARRAY | 999 | 1010 | 447 | 441 | 111 | 439 | 455 | 116 | Y |
| 2008 | Ekolf [69] | Caucasian | PB | Taqman | 220 | 414 | 123 | 85 | 12 | 212 | 160 | 42 | Y |
| 2008 | Zhang [70] | Asian | HB | PCR-RFLP | 300 | 299 | 97 | 136 | 67 | 91 | 139 | 69 | Y |
| 2008 | Guerreiro [71] | Caucasian | HB | Taqman | 196 | 200 | 94 | 76 | 26 | 84 | 107 | 9 | N |
| 2007 | Osian [72] | Caucasian | HB | PCR-RFLP | 69 | 67 | 38 | 25 | 6 | 47 | 17 | 3 | Y |
| 2007 | Zeybek [73] | Asian | HB | PCR-RFLP | 52 | 144 | 18 | 27 | 7 | 64 | 65 | 15 | Y |
| 2007 | Lima(a) [74] | Caucasian | HB | PCR-RFLP | 90 | 300 | 36 | 40 | 14 | 143 | 127 | 30 | Y |
| 2007 | Lima(b) [74] | African | HB | PCR-RFLP | 10 | 300 | 4 | 5 | 1 | 143 | 127 | 30 | Y |
| 2007 | Chang [75] | Asian | HB | RT-PCR | 195 | 195 | 85 | 86 | 24 | 92 | 87 | 16 | Y |
| 2007 | Murtaugh [76] | Mixed | PB | PCR-RFLP | 742 | 970 | 357 | 301 | 84 | 466 | 392 | 112 | N |
| 2007 | Jin [77] | Asian | PB | Taqman | 449 | 672 | 182 | 211 | 56 | 211 | 325 | 136 | Y |
| 2007 | Curtin [78] | Mixed | PB | PCR-RFLP | 916 | 1972 | 432 | 402 | 82 | 887 | 858 | 227 | Y |
| 2007 | Hubner [79] | Caucasian | PB | Taqman | 1685 | 2691 | 743 | 759 | 183 | 1173 | 1192 | 326 | Y |
| 2006 | Koushik [80] | Caucasian | PB | Taqman | 349 | 794 | 166 | 145 | 38 | 355 | 327 | 112 | N |
| 2006 | Battistelli [81] | Caucasian | HB | PCR-RFLP | 93 | 100 | 32 | 40 | 21 | 30 | 51 | 19 | Y |
| 2006 | Van Guelpen [82] | Caucasian | PB | Taqman | 220 | 415 | 123 | 85 | 12 | 212 | 161 | 42 | Y |
| 2006 | Wang [83] | Asian | PB | PCR-RFLP | 302 | 291 | 257 | 43 | 2 | 255 | 36 | 0 | Y |
| 2006 | Chen [84] | Asian | PB | PCR-RFLP | 138 | 340 | 52 | 86 | 133 | 207 | - | ||
| 2005 | Matsuo [85] | Asian | HB | PCR-RFLP | 256 | 771 | 106 | 114 | 36 | 289 | 348 | 134 | Y |
| 2005 | Landi [86] | Caucasian | HB | RT-PCR | 350 | 309 | 128 | 158 | 64 | 109 | 139 | 61 | Y |
| 2005 | Marchand [87] | Mixed | PB | PCR-RFLP | 817 | 2021 | 394 | 336 | 87 | 987 | 779 | 255 | N |
| 2005 | Jiang [88] | Asian | PB | PCR-RFLP | 125 | 339 | 51 | 59 | 15 | 134 | 143 | 62 | N |
| 2005 | Otani [89] | Asian | HB | MassARRAY | 106 | 222 | 32 | 49 | 25 | 51 | 114 | 57 | Y |
| 2005 | Miao [90] | Asian | PB | PCR-RFLP | 198 | 420 | 53 | 87 | 58 | 133 | 201 | 86 | Y |
| 2004 | Kim [91] | Asian | HB | PCR-RFLP | 243 | 225 | 86 | 122 | 35 | 83 | 109 | 33 | Y |
| 2004 | Ulvik [92] | Caucasian | PB | Taqman | 2159 | 2190 | 1103 | 899 | 157 | 1092 | 886 | 212 | Y |
| 2004 | Yin [93] | Asian | PB | PCR-RFLP | 685 | 778 | 270 | 330 | 85 | 278 | 367 | 133 | Y |
| 2004 | Curtin [94] | Mixed | HB | PCR-RFLP | 1608 | 1972 | 734 | 724 | 150 | 887 | 858 | 227 | Y |
| 2003 | Pufulete [95] | Caucasian | HB | PCR-RFLP | 28 | 76 | 16 | 6 | 6 | 41 | 29 | 6 | Y |
| 2003 | Plaschke [96] | Caucasian | PB | PCR-RFLP | 287 | 346 | 133 | 120 | 34 | 149 | 159 | 38 | Y |
| 2003 | Toffoli [97] | Caucasian | PB | PCR-RFLP | 276 | 279 | 93 | 145 | 38 | 83 | 140 | 56 | Y |
| 2003 | Heijmans [98] | Caucasian | PB | PCR-RFLP | 18 | 793 | 7 | 7 | 4 | 399 | 329 | 65 | Y |
| 2003 | Huang [99] | Asian | HB | PCR-RFLP | 82 | 82 | 36 | 40 | 6 | 40 | 33 | 9 | Y |
| 2003 | Barna [100] | Caucasian | PB | PCR-RFLP | 101 | 196 | 46 | 48 | 7 | 84 | 97 | 15 | Y |
| 2002 | Keku(a) [101] | Caucasian | PB | Taqman/PCR-PFLP | 308 | 539 | 144 | 140 | 24 | 265 | 223 | 51 | Y |
| 2002 | Keku(b) [101] | African | PB | Taqman/PCR-PFLP | 244 | 329 | 198 | 43 | 3 | 264 | 59 | 6 | Y |
| 2002 | Marchand(a) [102] | Caucasian | PB | PCR-RFLP | 149 | 171 | 66 | 64 | 19 | 66 | 81 | 24 | Y |
| 2002 | Marchand(b) [102] | Asian | PB | PCR-RFLP | 399 | 485 | 170 | 180 | 49 | 191 | 214 | 80 | Y |
| 2002 | Shannon [103] | Caucasian | PB | PCR-SSCP/RFLP | 501 | 1207 | 249 | 197 | 55 | 533 | 560 | 114 | Y |
| 2002 | Matsuo [104] | Asian | HB | PCR-RFLP | 142 | 241 | 39 | 81 | 22 | 81 | 124 | 36 | Y |
| 2002 | Sachse [105] | Caucasian | PB | PCR-RFLP | 490 | 592 | 238 | 199 | 53 | 271 | 272 | 49 | Y |
| 2002 | Chen [106] | Caucasian | PB | PCR-RFLP | 202 | 326 | 92 | 92 | 18 | 145 | 132 | 49 | N |
| 2001 | Ryan | Caucasian | PB | PCR-RFLP | 136 | 848 | 49 | 73 | 14 | 439 | 326 | 83 | Y |
| 2000 | Slattery [108] | Caucasian | PB | PCR-RFLP | 232 | 164 | 106 | 107 | 19 | 73 | 71 | 20 | Y |
| 1999 | Slattery [109] | Mixed | PB | PCR-RFLP | 1467 | 1821 | 673 | 655 | 139 | 827 | 787 | 207 | Y |
| 1999 | Park [110] | Asian | PB | PCR-RFLP | 200 | 460 | 65 | 107 | 28 | 140 | 246 | 74 | N |
| 1997 | Ma [111] | Caucasian | PB | PCR-RFLP | 202 | 326 | 92 | 92 | 18 | 145 | 132 | 49 | N |
| 1996 | Chen [112] | Caucasian | PB | PCR-RFLP | 144 | 627 | 67 | 64 | 13 | 280 | 263 | 84 | Y |
These 13 studies in bold were removed afterward because of its heterogeneity and publication bias. Abbreviations: HB: hospital-based control; PB, population-based control; SOC, source of control.
Results of quantitative synthesis
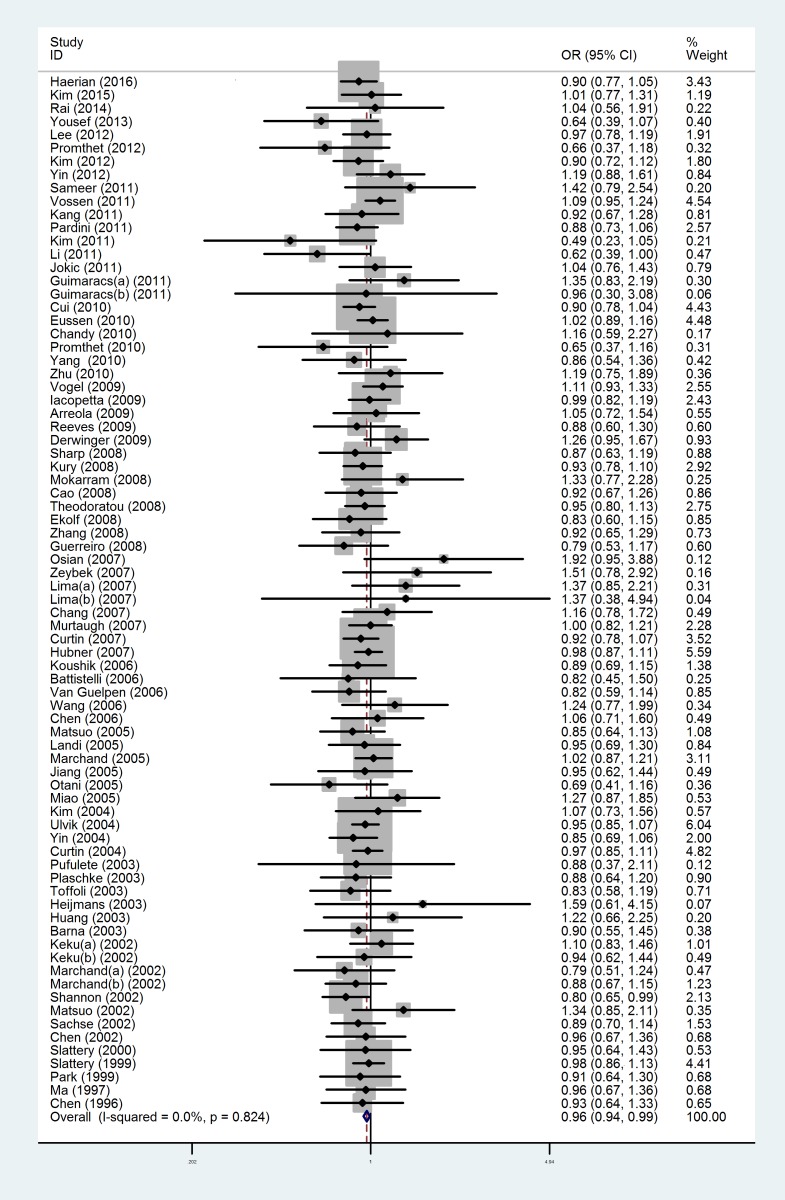
Initially, there was no association between MTHFR C677T polymorphism and CRC susceptibility in the dominant model (OR =0.99, 95% CI =0.94–1.05). 0.94–1.05). Nevertheless, for the sake of looking for possible reasons that might lead to such result, we performed heterogeneity analysis and tested publication bias. According to these results, 13 studies were excluded [29–31,40,43,47,48,52,55,61,63,77,107], the P-value was estimated to be 0.824, and the fixed effect model was applied. Ultimately, the results demonstrated that the rs1801133 polymorphism was significantly correlated with the risk of CRC (Figure 2) (dominant model: OR =0.96, 95% CI =0.94–0.99; recessive model: OR =0.90, 95% CI =0.83–0.96; homozygous model: OR =0.88, 95% CI =0.82–0.95; allele model: OR =0.95, 95% CI =0.93–0.98). All detailed results in the present meta-analysis are shown in Table 2.
Figure 2. Forest plots of the association between MTHFR C677T polymorphism and CRC susceptibility in dominant model after omitting these 13 studies with heterogeneity and publication bias.
Table 2. Meta-analysis results for the included studies of the association between MTHFR rs1801133 polymorphism and risk of CRC.
| Variables | Number of studies | Dominant model | Recessive model | Homozygous model | Heterozygous model | Allele model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P-values | I-squared (%) | OR (95% CI) | P-values | I-squared (%) | OR (95% CI) | P-values | I-squared (%) | OR (95% CI) | P-values | I-squared (%) | OR (95% CI) | P-values | I-squared (%) | ||
| rs1801133C>T | (CT + TT) compared with CC | TT compared with (CT + CC) | TT compared with CC | CT compared with CC | T compared with C | |||||||||||
| All | 78 | 0.96 (0.94–0.99) | 0.824 | 0.0 | 0.90 (0.83–0.96) | <0.001 | 49.9 | 0.88 (0.82–0.95) | <0.001 | 42.5 | 0.99 (0.96–1.02) | 0.950 | 0.0 | 0.95 (0.93–0.98) | 0.006 | 31.2 |
| Ethnicity | ||||||||||||||||
| Asian | 33 | 0.94 (0.89–1.00) | 0.418 | 3.0 | 0.88 (0.77–1.00) | 0.001 | 51.2 | 0.86 (0.75–1.00) | 0.001 | 49.2 | 0.96 (0.91–1.02) | 0.933 | 0.0 | 0.94 (0.88–1.00) | 0.002 | 47.9 |
| Caucasian | 36 | 0.97 (0.93–1.01) | 0.711 | 0.0 | 0.93 (0.83–1.04) | <0.001 | 57.8 | 0.91 (0.82–1.01) | 0.001 | 47.7 | 0.99 (0.95–1.03) | 0.505 | 0.0 | 0.96 (0.93–1.00) | 0.079 | 26.2 |
| African | 3 | 0.98 (0.67–1.42) | 0.866 | 0.0 | 0.69 (0.24–2.03) | 0.873 | 0.0 | 0.72 (0.24–2.15) | 0.837 | 0.0 | 1.02 (0.69–1.51) | 0.852 | 0.0 | 0.93 (0.67–1.30) | 0.816 | 0.0 |
| Mixed | 6 | 0.98 (0.92–1.04) | 0.959 | 0.0 | 0.83 (0.75–0.92) | 0.829 | 0.0 | 0.84 (0.75–0.93) | 0.830 | 0.0 | 1.02 (0.95–1.09) | 0.967 | 0.0 | 0.95 (0.90–0.99) | 0.908 | 0.0 |
| Source of control | ||||||||||||||||
| HB | 28 | 0.96 (0.90–1.03) | 0.357 | 7.2 | 0.97 (0.81–1.16) | <0.001 | 59.6 | 0.96 (0.80–1.15) | <0.001 | 54.4 | 0.98 (0.92–1.04) | 0.550 | 0.0 | 0.97 (0.90–1.05) | 0.007 | 44.4 |
| PB | 50 | 0.97 (0.93–1.00) | 0.911 | 0.0 | 0.88 (0.81–0.95) | 0.001 | 43.3 | 0.87 (0.80–0.93) | 0.012 | 34.1 | 0.99 (0.96–1.03) | 0.970 | 0.0 | 0.95 (0.92–0.98) | 0.087 | 22.4 |
| Geotyping | ||||||||||||||||
| Taqman | 14 | 0.96 (0.92–1.01) | 0.568 | 0.0 | 0.86 (0.73–1.00) | <0.001 | 65.0 | 0.85 (0.74–0.99) | 0.004 | 57.3 | 0.99 (0.94–1.05) | 0.460 | 0.0 | 0.94 (0.89–0.99) | 0.085 | 36.4 |
| PCR-RFLP | 50 | 0.95 (0.91–0.99) | 0.886 | 0.0 | 0.90 (0.81–0.99) | 0.001 | 43.6 | 0.88 (0.79–0.97) | 0.005 | 37.5 | 0.98 (0.94–1.03) | 0.992 | 0.0 | 0.95 (0.91–0.99) | 0.027 | 30.0 |
| RT-PCR | 4 | 1.10 (0.97–1.26) | 0.746 | 0.0 | 1.12 (0.76–1.64) | 0.017 | 70.4 | 1.15 (0.79–1.66) | 0.042 | 63.4 | 1.11 (0.96–1.27) | 0.771 | 0.0 | 1.08 (0.95–1.22) | 0.207 | 34.2 |
These 13 studies by Ozen et al., Ashmore et al., Delgado-Plasencia et al., Zhu et al., Prasad et al., Komlosi et al., Karpinski et al., Naghibalhossaini et al., Fernández-Peralta et al., Awady et al., Haghighi et al., Jin et al., Ryan et al. were removed [29, 30, 31, 40, 43, 47, 48, 52, 55, 61, 63, 77, 107].
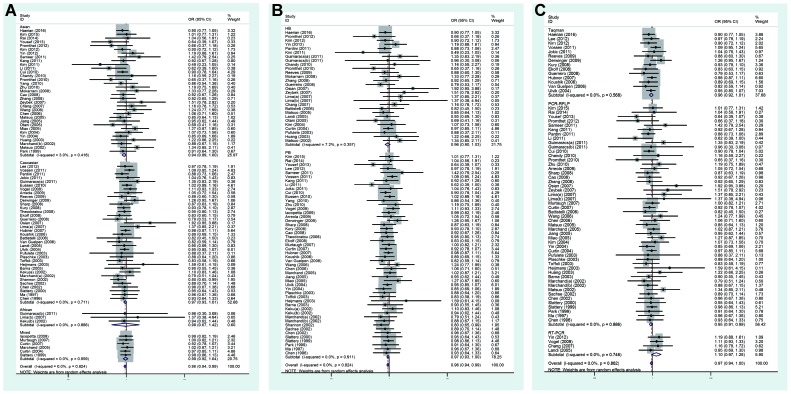
In the subgroup analysis of ethnicity, MTHFR C677T polymorphism was found to reduce CRC susceptibility in Asians significantly (dominant model: OR =0.94, 95% CI =0.89–1.00 (Figure 3A); recessive model: OR =0.88, 95% CI =0.77–1.00; homozygous model: OR =0.86, 95% CI =0.75–1.00; allele model: OR =0.92, 95% CI =0.88–1.00). Simultaneously, significantly reduced risks were also found in mixed group (recessive model: OR =0.83, 95% CI =0.75–0.92; homozygous model: OR =0.84, 95% CI =0.75–0.93; allele model: OR =0.95, 95% CI =0.90–0.99). Amongst Caucasians, yet significantly reduced risks were only observed in the allele model (OR =0.96, 95% CI =0.93–1.00). Nevertheless, no significant associations were detected in Africans for all genetic models. When stratified by the source of controls, the positive results were observed in population-based control group (dominant model: OR =0.97, 95% CI =0.93–1.00 (Figure 3B); recessive model: OR =0.88, 95% CI =0.81–0.95; homozygous model: OR =0.87, 95% CI =0.80–0.93; allele model: OR =0.95, 95% CI =0.92–0.98). The similar significant associations were absent from hospital-based group for all the genetic models. The stratified analysis by genotyping methods showed that PCR-RFLP method (dominant model: OR =0.95, 95% CI =0.91–0.99 (Figure 3C); recessive model: OR =0.90, 95% CI =0.81–0.99; homozygous model: OR =0.88, 95% CI =0.79–0.97; allele model: OR =0.95, 95% CI =0.91–0.99) and Taqman method (recessive model: OR =0.86, 95% CI =0.73–1.00; homozygous model: OR =0.85, 95% CI =0.74–0.99; allele model: OR =0.94, 95% CI =0.89–0.99) were significantly correlated with risks of decreased CRC. However, RT-PCR method was not relevant to significant associations for all genetic models. In conclusion, the present meta-analysis suggested that MTHFR C677T polymorphism was connected with CRC susceptibility.
Figure 3. Forest plots of subgroup analysis of the association between MTHFR C677T polymorphism and CRC susceptibility in dominant model.
(A) Stratified by ethnicity; (B) stratified by source of controls; (C) stratified by genotyping method.
Test of heterogeneity
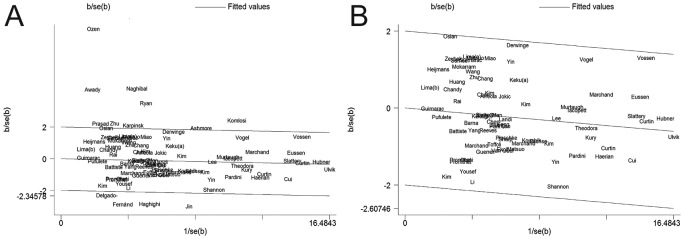
Heterogeneity analysis was performed in this meta-analysis, and heterogeneity was significantly observed between all the included studies in the dominant model (I2 =62.0%, P<0.001; Figure 4A). In addition, the Galbraith radial plot illustrated heterogeneity obviously. Meanwhile, it specifically pointed out 13 studies that might have led to the obvious heterogeneity and insignificant results of the meta-analysis [27–29,38,41,45,46,50,53,59,61,75,105]. After excluding 13 studies, the heterogeneity decreased significantly (I2 =0.0%, P=0.789; Figure 4B) in the present meta-analysis.
Figure 4. Galbraith plot of the association between MTHFR C677T polymorphism and CRC susceptibility in dominant model.
(A) Before removing these 13 studies. (B) After the exclusion of these studies.
Publication bias
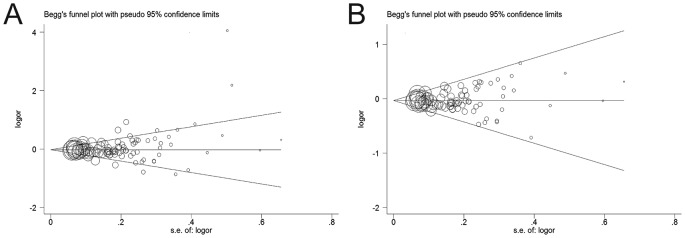
The Begg’s funnel plot and Egger’s test were performed to assess the publication bias. Initially, the Begg’s funnel plot was asymmetrical obviously with all the included studies and it suggested a potential publication bias (Begg’s test: P=0.103; Egger’s test: P=0.058; Figure 5A). After the removal of 13 studies mentioned above [27–29,38,41,45,46,50,53,59,61,75,105], the plots seemed to have a symmetrical distribution in the funnel plot and then Egger’s test was used to provide statistical evidence (Begg’s test: P=0.369; Egger’s test: P=0.136; Figure 5B). No significant publication bias was observed in the present studies.
Figure 5. Begg’s funnel plot of publication bias test.
(A) Before omitting these 13 studies. (B) After the exclusion of these studies.
Sensitivity analysis
In order to distinguish the impact of each study on the pooled ORs, we conducted one-way sensitivity analysis. Each time one study was omitted, meta-analysis was repeated and the statistical significance of the results was not changed. Therefore, the results confirmed that the present meta-analysis was relatively stable and reliable.
Discussion
MTHFR is a key enzyme in the folate metabolism and may play a role in the CRC carcinogenesis. It is an essential enzyme in the catalytic reaction that converts 5,10-methylenetetrahydrofolate into 5-methyltetrahydrofolate. On one hand, 5,10-methylenetetrahydrofolate takes part in the thymidylate synthesis. On the other hand, 5-methyltetrahydrofolate promotes methionine synthesis and SAM-mediated methylations. In brief, MTHFR has an influence on DNA synthesis, methylation, and repair [113]. The MTHFR polymorphisms result in the decreased enzyme activity and then low levels of plasma folate and high homocysteine come to light. Folate is one of water-soluble B vitamins that takes part in various biochemical reactions with its activity to provide or accept one-carbon units [13]. Folate deficiency is likely to contribute to the development of CRC, and several mechanisms may explain how it leads to CRC, including DNA strand breaks, abnormal DNA methylation, and impaired DNA repair [114].
Several polymorphisms have been reported about the MTHFR gene coding relevant enzyme, and MTHFR C677T polymorphism is the most common one. Heretofore, various studies conducted to detect such association and obtained inconsistent results. Chen et al. [112], first reported that MTHFR variant homozygous (TT) genotype was closely linked to reduced incidence of CRC with low consumption of alcohol. In the next few years, similar results were replicated by several other studies [109–111]. However, another study of a homogeneous northern European population obtained different conclusions that MTHFR CT heterozygote had a significantly increased risk of developing CRC and no increased cancer risk was observed in TT homozygotes [107]. In addition, a hospital-based case–control study conducted by Matsuo et al. [104] found no significant relativity between MTHFR C677T and the risks of CRC. Owing to the difference in study design and the sample size, the different ethnicity, and the diverse stratification, these controversial results were found in published studies. Hence, meta-analysis is essential to be carried out by combining all studies that meet the requirements to get more precise conclusions.
In recent years, there were several meta-analyses performed to elucidate the association of MTHFR C677T polymorphism and the susceptibility to CRC before [26,115–118]. Compared with them, this meta-analysis included the most eligible reported studies with the largest sample size and had no restrictions in ethnicity. Since the quality of included documents were disequilibrium, our initial analysis achieved no significant results with all eligible studies. In order to obtain more reliable results, the final conclusion were obtained excluding 13 studies in accordance with the analysis of heterogeneity and publication bias. In this meta-analysis, the pooled conclusions revealed that rs1801133 polymorphism significantly reduced the risk of CRC in the dominant model. The findings agreed with the overwhelming majority results reported by the published studies.
When stratified by ethnicity, there was a significant association with reduced risks of CRC in Asians. The result was consistent with the two previous meta-analysis based on the Asians [116,117]. Zhong et al. [118], carried out a meta-analysis obtaining similar results in East Asians and further subgroup analyses by country identified such association in Korea and Japan. Nevertheless, the recent meta-analysis failed to identify that rs1801133 polymorphism was connected with CRC susceptibility in Iranian population [26]. By means of stratified analysis based on the source of controls and genotyping methods, the positive results were observed in population-based control group and PCR-RFLP method. In general, the source of controls included healthy individuals and patients without CRC. Since the risks of CRC varies amongst individuals over a few years, it might have an impact on the results of relevant studies and make them unreliable. Therefore, inclusion criteria should be improved and studies with large sample sizes should be accepted. In the subgroup of genotyping method, there were nine methods applied for genotyping such as PCR-RFLP, RT-PCR, PCR-SSCP, MS-PCR, MSP, MALDI-TOF-MS, Taqman, MassARRAY, and Sequenom in the including studies. Specific methods and steps were described in each article. Amongst these 87 studies, the majority method was PCR-RFLP. Different methods have their own merits, and when all included studies used the same method, the final results would be more reliable.
In the present meta-analysis, we had obtained weak associations significantly with a large sample size. However, the potential limitations of the meta-analysis should be acknowledged. First, this meta-analysis was based on unadjusted effect estimates and 95% CI, and the influence of multiple cofactors such as age, gender, diet habits including intake of alcohol and consumption of cigarette, the level of folate, and the other environmental factors should be taken into consideration. Second, because of incomplete data of some genotypes, only the dominant model was analyzed in all the included studies. Third, we did not perform stratification analysis by serum folate levels, locations of the tumor and so on, which might result in confounding bias. In addition, after excluding 13 studies according to the analysis of heterogeneity and publication bias, the heterogeneity decreased significantly and the publication bias seemed to disappear. However, the selection bias existed because all the studies were published. Furthermore, the gene–gene and gene–environment interactions were not mentioned in this meta-analysis. In addition, the potential roles of the gene polymorphism which were hidden or magnified by other interactions were omitted.
Conclusion
In summary, the present meta-analysis revealed that there was a significant association between MTHFR C677T polymorphism and susceptibility to CRC. Simultaneously, the TT genotype of MTHFR C677T polymorphism could reduce the risk of CRC. In addition, the associated risk of CRC was also reduced in Asians and those studies with population-based controls and used the PCR-RFLP method. Therefore, detection of the MTHFR C677T polymorphism might be used as markers for CRC prediction and treatment selection.
Abbreviations
- CI
confidence interval
- CRC
colorectal cancer
- HWE
Hardy–Weinberg equilibrium
- MSP
mutagenically separated PCR
- MS-PCR
methylation-specific PCR
- MTHFR
methylenetetrahydrofolate reductase
- OR
odds ratio
- PCR-RFLP
PCR-restriction fragment length polymorphism
- PCR-SSCP
PCR-single strand conformation polymorphism
- PRISMA-P
preferred reporting items for systematic review and meta-analysis protocol
- RT-PCR
real-time PCR
- SAM
S-adenosylmethionine
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work has been supported by the Natural Science Funding of Jiangsu Province [grant number BK20141492]; and the ‘333 Project’ of Jiangsu Province [grant number BRA2016517].
Author contribution
Y.G., H.Y., and Z.Q. were responsible for conception and design. Y.G., H.Y., and F.W. provided the administrative support. S.S., Z.Q., and L.L. were responsible for the collection and assembly of data. P.L., X.H., and X.C. were responsible for data analysis and interpretation. L.X., Z.Q., and F.W. were responsible for manuscript writing. All the authors approved the final manuscript.
References
- 1.Siegel R.L., Miller K.D. and Jemal A. (2016) Cancer statistics, 2016. CA Cancer J. Clin. 66, 7–30 [DOI] [PubMed] [Google Scholar]
- 2.Markowitz S.D. and Bertagnolli M.M. (2009) Molecular basis of colorectal cancer. N. Engl. J. Med. 361, 2449–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baroudi O. and Benammar-elgaaied A. (2016) Involvement of genetic factors and lifestyle on the occurrence of colorectal and gastric cancer. Crit. Rev. Oncol. Hemat. 107, 72–81 [DOI] [PubMed] [Google Scholar]
- 4.Khan A.A., Khan Z., Malik A., Kalam A.M., Cash P., Ashraf T.M. et al. (2017) Colorectal cancer-inflammatory bowel disease nexus and felony of Escherichia coli. Life Sci. 180, 60–67 [DOI] [PubMed] [Google Scholar]
- 5.Nassiri M., Kooshyar M.M., Roudbar Z., Mahdavi M. and Doosti M. (2013) Genes and SNPs associated with non-hereditary and hereditary colorectal cancer. Asian Pac. J. Cancer Prev. 14, 5609–5614 [DOI] [PubMed] [Google Scholar]
- 6.Noci S., Dugo M., Bertola F., Melotti F. and Vannelli A. (2016) A subset of genetic susceptibility variants for colorectal cancer also has prognostic value. Pharmacogenomics J. 16, 173–179 [DOI] [PubMed] [Google Scholar]
- 7.Guo X.P., Wang Y., Zhao H., Song S.D., Zhou J. and Han Y. (2014) Association of MTHFR C677T polymorphisms and colorectal cancer risk in Asians: evidence of 12,255 subjects. Clin. Transl. Oncol. 16, 623–629 [DOI] [PubMed] [Google Scholar]
- 8.Fang X., Xu W., Huang Q., Yang X.K., Liu Y.Y., Leng R.X. et al. (2014) 5,10-Methylenetetrahydrofolate reductase polymorphisms and colon cancer risk: a meta-analysis. Asian Pac. J. Cancer Prev. 15, 8245–8250 [DOI] [PubMed] [Google Scholar]
- 9.Ueland P.M., Hustad S., Schneede J., Refsum H. and Vollset S.E. (2001) Biological and clinical implications of the MTHFR C677T polymorphism. Trends Pharmacol. Sci. 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 10.Goyette P., Pai A., Milos R., Frosst P., Tran P., Chen Z.T. et al. (1998) Gene structure of human and mouse methylenetetrahydrofolate reductase (MTHFR). Mamm. Genome 9, 652–656 [DOI] [PubMed] [Google Scholar]
- 11.Rozen R. (1997) Genetic predisposition to hyperhomocysteinemia: deficiency of methylenetetrahydrofolate reductase (MTHFR). Thromb. Haemost. 78, 523–526 [PubMed] [Google Scholar]
- 12.Frosst P., Milos R., Goyette P., Sheppard C.A., Matthews R.G., Boers G.J.H. et al. (1995) A candidate genetic risk factor for vascular disease:a common mutation in methylenetetrahydrofolate reductase. Nature 10, 111–113 [DOI] [PubMed] [Google Scholar]
- 13.Duthie S.J. (1999) Folic acid deficiency and cancer: mechanisms of DNA instability. Br. Med. Bull. 55, 578–592 [DOI] [PubMed] [Google Scholar]
- 14.Zhu X.L., Liu Z.Z., Yan S.X., Wang W., Chang R.X., Zhang Y.C. et al. (2016) Association between the MTHFR A1298C polymorphism and risk of cancer: evidence from 265 case-control studies. Mol. Genet. Genomics 291, 51–63 [DOI] [PubMed] [Google Scholar]
- 15.Long S. and Goldblatt J. (2016) MTHFR genetic testing: controversy and clinical implications. Aust. Fam. Physician 45, 237–240 [PubMed] [Google Scholar]
- 16.Liew S. and Gupta E.D. (2015) Methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism: Epidemiology, metabolism and the associated diseases. Eur. J. Med. Genet. 58, 1–10 [DOI] [PubMed] [Google Scholar]
- 17.Shi H., Yang S.W., Liu Y., Huang P., Lin N., Sun X.R. et al. (2015) Study on environmental causes and SNPs of MTHFR, MS and CBS genes related to congenital heart disease. PLoS ONE 10, e128646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sohda S., Arinami T., Hamada H., Yamada N. and Hamaguchi H. (1997) Methylenetetrahydrofolate reductase polymorphism and pre-eclampsia. J. Med. Genet. 34, 525–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stonek F., et al. (2007) Methylenetetrahydrofolate reductase C677T polymorphism and pregnancy complications. Obstet. Gynecol. 110, 363–368 [DOI] [PubMed] [Google Scholar]
- 20.van der Put N.M.J., Gabreels F., Stevens E.M.B., Smeitink J.A.M., Trijbels F.J., Eskes T.K.A.B. et al. (1998) A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am. J. Hum. Genet. 62, 1044–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiao S.P.K. and Yu C.H. (2016) Meta-prediction of MTHFR gene polymorphism mutations and associated risk for colorectal cancer. Biol. Res. Nurs. 18, 357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shamseer L., Moher D., Clarke M., Ghersi D., Liberati A., Petticrew M. et al. (2015) Preferred reporting items for systematic review and meta-analysis protocols(PRISMA-P)2015:elaboration and explanation. BMJ 349, g7647. [DOI] [PubMed] [Google Scholar]
- 23.Wei G.S. and Thompson E.A. (1992) Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48, 361–372 [PubMed] [Google Scholar]
- 24.Anzures-Cabrera J. and Higgins J.P. (2010) Graphical displays for meta-analysis: an overview with suggestions for practice. Res. Synth. Methods 1, 66–80 [DOI] [PubMed] [Google Scholar]
- 25.Hayashino Y., Noguchi Y. and Fukui T. (2005) Systematic evaluation and comparison of statistical tests for publication bias. J. Epidemiol. 15, 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haerian M.S., Haerian B.S., Molanaei S., Kosari F., Sabeti S., Bidari-Zerepoosh F. et al. (2016) MTHFR rs1801133 polymorphism and susceptibility to colorectal cancer in Iranian population: evidence of a case-control study and meta-analysis. Pharmacogenomics 17, 1957–1965 [DOI] [PubMed] [Google Scholar]
- 27.Kim J.W., Jeon Y.J., Jang M.J., Kim J.O. and Chong S.Y. (2015) Association between folate metabolism-related polymorphisms and colorectal cancer risk. Mol. Clin. Oncol. 3, 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rai P.S., Pai G.C., Alvares J.F., Bellampalli R., Gopinath P.M. and Satyamoorthy K. (2014) Intraindividual somatic variations in MTHFR gene polymorphisms in relation to colon cancer. Pharmacogenomics 15, 349–359 [DOI] [PubMed] [Google Scholar]
- 29.Ozen F., Sen M. and Ozdemir O. (2014) Methylenetetrahydrofolate reductase gene germ-line C677T and A1298C SNPs are associated with colorectal cancer risk in the Turkish population. Asian Pac. J. Cancer Prev. 15, 7731–7735 [DOI] [PubMed] [Google Scholar]
- 30.Ashmore J.H., Lesko S.M., Muscat J.E., Gallagher C.J., Berg A.S., Miller P.E. et al. (2013) Association of dietary and supplemental folate intake and polymorphisms in three FOCM pathway genes with colorectal cancer in a population-based case-control study. Gene. Chromosome Canc. 52, 945–953 [DOI] [PubMed] [Google Scholar]
- 31.Delgado-Plasencia L., Medina-Arana V., Bravo-Gutiérrez A., Perez-Palma J., Alvarez-Arguelles H., Salido-Ruiz E. et al. (2013) Impact of the MTHFR C677T polymorphism on colorectal cancer in a population with low genetic variability. Int. J. Colorectal Dis. 28, 1187–1193 [DOI] [PubMed] [Google Scholar]
- 32.Yousef A., Shomaf M., Berger S., Ababneh N., Bobali Y., Ali D. et al. (2013) Allele and genotype frequencies of the polymorphic methylenetetrahydrofolate reductase and colorectal cancer among Jordanian population. Asian Pac. J. Cancer Prev. 14, 4559–4565 [DOI] [PubMed] [Google Scholar]
- 33.Lee J.E., Wei E.K., Fuchs C.S., Hunter D.J., Lee I.M., Selhub J. et al. (2012) Plasma folate, methylenetetrahydrofolate reductase (MTHFR), and colorectal cancer risk in three large nested case-control studies. Cancer Cause Control 23, 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Promthet S., Pientong C., Ekalaksananan T., Songserm N., Poomphakwaen K., Chopjitt P. et al. (2012) Risk factors for rectal cancer and methylenetetrahydrofolate reductase polymorphisms in a population in northeast Thailand. Asian Pac. J. Cancer Prev. 13, 4017–4023 [DOI] [PubMed] [Google Scholar]
- 35.Kim J., Cho Y.A., Kim D.H., Lee B.H., Hwang D.Y., Jeong J. et al. (2012) Dietary intake of folate and alcohol, MTHFR C677T polymorphism, and colorectal cancer risk in Korea. Am. J. Clin. Nutr. 95, 405–412 [DOI] [PubMed] [Google Scholar]
- 36.Yin G., Ming H., Zheng X., Xuan Y., Liang J. and Jin X. (2012) Methylenetetrahydrofolate reductase C677T gene polymorphism and colorectal cancer risk: a case-control study. Oncol. Lett. 4, 365–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sameer A.S., Shah Z.A., Nissar S., Mudassar S. and Siddiqi M.A. (2011) Risk of colorectal cancer associated with the methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism in the Kashmiri population. Genet. Mol. Res. 10, 1200–1210 [DOI] [PubMed] [Google Scholar]
- 38.Vossen C.Y., Hoffmeister M., Chang-Claude J.C., Rosendaal F.R. and Brenner H. (2011) Clotting factor gene polymorphisms and colorectal cancer risk. J. Clin. Oncol. 29, 1722–1727 [DOI] [PubMed] [Google Scholar]
- 39.Kang B.S., Ahn D.H., Kim N.K. and Kim J.W. (2011) Relationship between metabolic syndrome and MTHFR polymorphism in colorectal cancer. J. Korean Soc. Coloproctol. 27, 78–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Q., Jin Z., Yuan Y., Lu Q., Ge D. and Zong M. (2011) Impact of MTHFR gene C677T polymorphism on Bcl-2 gene methylation and protein expression in colorectal cancer. Scand. J. Gastroenterol. 46, 436–445 [DOI] [PubMed] [Google Scholar]
- 41.Pardini B., Kumar R., Naccarati A., Prasad R.B., Forsti A., Polakova V. et al. (2011) MTHFR and MTRR genotype and haplotype analysis and colorectal cancer susceptibility in a case-control study from the Czech Republic. Mutat. Res. 721, 74–80 [DOI] [PubMed] [Google Scholar]
- 42.Kim J.W., Park H.M., Choi Y.K., Chong S.Y. and Oh D. (2011) Polymorphisms in genes involved in folate metabolism and plasma DNA methylation in colorectal cancer patients. Oncol. Rep. 25, 167–172 [PubMed] [Google Scholar]
- 43.Prasad V.V.T.S. and Wilkhoo H. (2011) Association of the functional polymorphism C677T in the methylenetetrahydrofolate reductase gene with colorectal, thyroid, breast, ovarian, and cervical cancers. Onkologie 34, 422–426 [DOI] [PubMed] [Google Scholar]
- 44.Li H., Xu W.L., Shen H.L., Chen Q.Y., Hui L.L., Long L.L. et al. (2011) Methylenetetrahydrofolate reductase genotypes and haplotypes associated with susceptibility to colorectal cancer in an eastern Chinese Han population. Genet. Mol. Res. 10, 3738. [DOI] [PubMed] [Google Scholar]
- 45.Jokić M., Brčić-Kostić K., Stefulj J., Ivkovic ´ T.C., Bozˇo L., Gamulin M. et al. (2011) Association of MTHFR, MTR, MTRR, RFC1, and DHFR gene polymorphisms with susceptibility to sporadic colon cancer. DNA Cell Biol. 30, 771–776 [DOI] [PubMed] [Google Scholar]
- 46.Guimarães J.L.M., Ayrizono M.D.L., Coy C.S.R. and Lima C.S.P. (2011) Gene polymorphisms involved in folate and methionine metabolism and increased risk of sporadic colorectal adenocarcinoma. Tumor Biol. 32, 853–861 [DOI] [PubMed] [Google Scholar]
- 47.Komlósi V., Hitre E., Pap E., Adleff V., Réti A., Székely E. et al. (2010) SHMT1 1420 and MTHFR 677 variants are associated with rectal but not colon cancer. BMC Cancer 10, 1471–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karpinski P., Myszka A., Ramsey D., Misiak B., Gil J., Laczmanska I. et al. (2010) Polymorphisms in methyl-group metabolism genes and risk of sporadic colorectal cancer with relation to the CpG island methylator phenotype. Cancer Epidemiol. 34, 338–344 [DOI] [PubMed] [Google Scholar]
- 49.Cui L., Shin M., Kweon S., Kim H.N., Song H., Piao J. et al. (2010) Methylenetetrahydrofolate reductase C677T polymorphism in patients with gastric and colorectal cancer in a Korean population. BMC Cancer 10, 1471–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eussen S.J.P.M., Vollset S.E., Igland J., Meyer K., Fredriksen A., Ueland P.M. et al. (2010) Plasma folate, related genetic variants, and colorectal cancer risk in EPIC. Cancer Epidemiol. Biomarkers Prev. 19, 1328–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chandy S., Adiga M.N.S., Ramachandra N., Krishnamoorthy S., Ramaswamy G., Savithri H.S. et al. (2010) Association of methylenetetrahydrofolate reducíase gene polymorphisms & colorectal cancer in India. Indian J. Med. Res. 131, 659–664 [PubMed] [Google Scholar]
- 52.Naghibalhossaini F., Mokarram P., Khalili I., Vasei M., Hosseini S.V., Ashktorab H. et al. (2010) MTHFR C677T and A1298C variant genotypes and the risk of microsatellite instability among Iranian colorectal cancer patients. Cancer Genet. Cytogenet. 197, 142–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Promthet S.S., Pientong C., Ekalaksananan T., Wiangnon S., Poomphakwaen K., Songserm N. et al. (2010) Risk factors for colon cancer in northeastern Thailand: interaction of MTHFR codon 677 and 1298 genotypes with environmental factors. J. Epidemiol. 20, 329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang X.X., Li F.X., Yi J.P., Li X., Sun J.Z. and Hu N.Y. (2010) Impact of methylenetetrahydrofolate reductase C677T polymorphism on the risk of gastric cancer, colorectal cancer and lung cancer. Guangdong Med. 31, 2375–2378 [Google Scholar]
- 55.Fernández-Peralta A.M., Daimiel L., Nejda N., Iglesias D., Medina Arana V. and González-Aguilera J.J. (2010) Association of polymorphisms MTHFR C677T and A1298C with risk of colorectal cancer, genetic and epigenetic characteristic of tumors, and response to chemotherapy. Int. J. Colorectal Dis. 25, 141–151 [DOI] [PubMed] [Google Scholar]
- 56.Zhu F., Wang Y.-m. and ZhangQY Q.-y. (2010) A case-control study of plasma homocysteine, serum folate, the polymorphism of methylenetetrahydrofolate reductase in colorectal cancer. J. Southeast Univ. Med. Sci. Edi. 29, 88–92 [Google Scholar]
- 57.de Vogel S., Wouters K.A.D., Gottschalk R.W.H., van Schooten F.J., de Goeij A.F.P.M., de Bruïne A.P. et al. (2009) Genetic variants of methyl metabolizing enzymes and epigenetic regulators: associations with promoter CpG island hypermethylation in colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 18, 3086–3096 [DOI] [PubMed] [Google Scholar]
- 58.Iacopetta B., Heyworth J., Girschik J., Grieu F., Clayforth C. and Fritschi L. (2009) The MTHFR C677T and ΔDNMT3B C-149T polymorphisms confer different risks for right- and left-sided colorectal cancer. Int. J. Cancer 125, 84–90 [DOI] [PubMed] [Google Scholar]
- 59.Gallegos-Arreola M.P., Garcia-Ortiz J.E., Figuera L.E., Puebla-Perez A.M., Morgan-Villela G., Zuniga-Gonzalez G.M. et al. (2009) Association of the 677C→T polymorphism in the MTHFR Gene with Colorectal cancer in Mexican patients. Cancer Genome Proteomics 6, 183–188 [PubMed] [Google Scholar]
- 60.Reeves S.G., Meldrum C., Groombridge C., Spigelman A.D., Suchy J., Kurzawski G. et al. (2009) MTHFR 677 C>T and 1298 A>C polymorphisms and the age of onset of colorectal cancer in hereditary nonpolyposis colorectal cancer. Eur. J. Hum. Genet. 17, 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.El Awady M.K., Karim A.M., Hanna L.S., El Husseiny L.A., El Sahar M., Abdel Menem H.A. et al. (2009) Methylenetetrahydrofolate reductase gene polymorphisms and the risk of colorectal carcinoma in a sample of Egyptian individuals. Cancer Biomark. 5, 233–240 [DOI] [PubMed] [Google Scholar]
- 62.Derwinger K., Wettergren Y., Odin E., Carlsson G. and Gustavsson B. (2009) A study of the MTHFR gene polymorphism C677T in colorectal cancer. Clin. Colorectal Cancer 8, 43–48 [DOI] [PubMed] [Google Scholar]
- 63.Haghighi M.M., Mohebbi S.R., Khatami F., Ghiasi S., Derakhshan F., Atarian H. et al. (2008) Reverse association between MTHFR polymorphism (C677T) with sporadic colorectal cancer. Gastroenterol. Hepatol. 1, 57–63 [Google Scholar]
- 64.Sharp L., Little J., Brockton N.T., Cotton S.C., Masson L.F., Haites N.E. et al. (2008) Polymorphisms in the methylenetetrahydrofolate reductase (MTHFR) gene, intakes of folate and related B vitamins and colorectal cancer: a case-control study in a population with relatively low folate intake. Br. J. Nutr. 99, 379–389 [DOI] [PubMed] [Google Scholar]
- 65.Kury S., Buecher B., Robiou-du-Pont S., Scoul C., Colman H., Neel T.L. et al. (2008) Low-penetrance alleles predisposing to sporadic colorectal cancers: a French case-controlled genetic association study. BMC Cancer 8, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mokarram P., Naghibalhossaini F., Firoozi M.S., Hosseini S.V., Izadpanah A., Salahi H. et al. (2008) Methylenetetrahydrofolate reductase C677T genotype affects promoter methylation of tumor-specific genes in sporadic colorectal cancer through an interaction with folate/vitamin B12 status. World J. Gastroenterol. 14, 3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cao H., Gao C., Takezaki T., Wu J., Ding J., Liu Y. et al. (2008) Genetic polymorphisms of methylenetetrahydrofolate reductase and susceptibility to colorectal cancer. Asian Pac. J. Cancer Prev. 9, 203–208 [PubMed] [Google Scholar]
- 68.Theodoratou E., Farrington S.M., Tenesa A., McNeill G., Cetnarskyj R., Barnetson R.A. et al. (2008) Dietary vitamin B6 intake and the risk of colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 17, 171–182 [DOI] [PubMed] [Google Scholar]
- 69.Eklöf V., Van Guelpen B., Hultdin J., Johansson I., Hallmans G. and Palmqvist R. (2009) The reduced folate carrier (RFC1) 80G>A and folate hydrolase 1 (FOLH1) 1561C>T polymorphisms and the risk of colorectal cancer: a nested case-referent study. Scand. J. Clin. Lab. Inv. 68, 393–401 [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y.L., Yuan X.Y., Zhang C., Yang Y., Pan Y.M., Zhou Z.Y. et al. (2008) Relationship between polymorphisms of thymidylate synthase and methylenetetrahydrofolate reductase and susceptibility in Liaoning Benxi colorectal cancer patients. Cancer J. Clin. 13, 769–773 [Google Scholar]
- 71.Guerreiro C.S., Carmona B., Gonçalves S., Carolino E., Fidalgo P., Brito M. et al. (2008) Risk of colorectal cancer associated with the C677T polymorphism in 5,10-methylenetetrahydrofolate reductase in Portuguese patients depends on the intake of methyl-donor nutrients. Am. J. Clin. Nutr. 88, 1413–1418 [DOI] [PubMed] [Google Scholar]
- 72.Osian G., Procopciuc L. and Vlad L. (2007) MTHFR polymorphisms as prognostic factors in sporadic colorectal cancer. J. Gastrointestin. Liver Dis. 16, 251–256 [PubMed] [Google Scholar]
- 73.Zeybek U., Yaylim I., Yilmaz H., Agachan B., Ergen A., Arikan S. et al. (2007) Methylenetetrahydrofolate reductase C677T polymorphism in patients with gastric and colorectal cancer. Cell Biochem. Funct. 25, 419–422 [DOI] [PubMed] [Google Scholar]
- 74.Lima C.S.P., Nascimento H., Bonadia L.C., Teori M.T., Coy C.S.R., Goes J.R.N. et al. (2007) Polymorphisms in methylenetetrahydrofolate reductase gene (MTHFR) and the age of onset of sporadic colorectal adenocarcinoma. Int. J. Colorectal Dis. 22, 757–763 [DOI] [PubMed] [Google Scholar]
- 75.Chang S., Lin P., Lin J., Yang S., Wang H. and Li A. (2007) Role of MTHFR polymorphisms and folate levels in different phenotypes of sporadic colorectal cancers. Int. J. Colorectal Dis. 22, 483–489 [DOI] [PubMed] [Google Scholar]
- 76.Murtaugh M.A., Curtin K., Sweeney C., Wolff R.K., Holubkov R. and Caan B.J. (2007) Dietary intake of folate and co-factors in folate metabolism, MTHFR polymorphisms, and reduced rectal cancer. Cancer Cause Control 18, 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jin X.X., Zhu Z.Z., Wang A.Z. and Jia H.R. (2007) Association of methylenetetrahydrofoIate reductase C677T polymorphism with genetic susceptibility to colorectal cancer. World Chin. J. Dig. 15, 2754–2757 [Google Scholar]
- 78.Curtin K., Slattery M.L., Ulrich C.M., Bigler J., Levin T.R., Wolff R.K. et al. (2007) Genetic polymorphisms in one-carbon metabolism: associations with CpG island methylator phenotype (CIMP) in colon cancer and the modifying effects of diet. Carcinogenesis 28, 1672–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hubner R.A., Lubbe S., Chandler I. and Houlston R.S. (2007) MTHFR C677T has differential influence on risk of MSI and MSS colorectal cancer. Hum. Mol. Genet. 16, 1072–1077 [DOI] [PubMed] [Google Scholar]
- 80.Koushik A., Kraft P., Fuchs C.S., Hankinson S.E., Willett W.C., Giovannucci E.L. et al. (2006) Nonsynonymous polymorphisms in genes in the one-carbon metabolism pathway and associations with colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 15, 2408–2417 [DOI] [PubMed] [Google Scholar]
- 81.Battistelli S., Vittoria A., Stefanoni M., Bing C. and Roviello F. (2006) Total plasma homocysteine and methylenetetrahydrofolate reductase C677T polymorphism in patients with colorectal carcinoma. World J. Gastroenterol. 12, 6128–6132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Van Guelpen B., Hultdin J., Johansson I., Hallmans G., Stenling R., Riboli E. et al. (2006) Low folate levels may protect against colorectal cancer. Gut 55, 1461–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang J., Gajalakshmi V., Jiang J., Kuriki K., Suzuki S., Nagaya T. et al. (2006) Associations between 5,10-methylenetetrahydrofolate reductase codon 677 and 1298 genetic polymorphisms and environmental factors with reference to susceptibility to colorectal cancer: a case-control study in an Indian population. Int. J. Cancer 118, 991–997 [DOI] [PubMed] [Google Scholar]
- 84.Chen K., Song L., Jin M.J., Fang C.H., Jiang X.D. and Yu W.P. (2006) Associations between folate metabolism enzyme gene polymorphisms and colorectal susceptibility. Chin. J. Oncol. 28, 429–432 [PubMed] [Google Scholar]
- 85.Matsuo K., Ito H., Wakai K., Hirose K., Saito T., Suzuki T. et al. (2005) One-carbon metabolism related gene polymorphisms interact with alcohol drinking to influence the risk of colorectal cancer in Japan. Carcinogenesis 26, 2164–2171 [DOI] [PubMed] [Google Scholar]
- 86.Landi S., Gemignani F., Moreno V., Gioia-Patricola L., Chabrier A., Guino E. et al. (2005) A comprehensive analysis of phase I and phase II metabolism gene polymorphisms and risk of colorectal cancer. Pharmacogenet. Genomics 15, 535–546 [DOI] [PubMed] [Google Scholar]
- 87.Le Marchand L., Wilkens L.R., Kolonel L.N. and Henderson B.E. (2005) The MTHFR C677T polymorphism and colorectal cancer: the multiethnic cohort study. Cancer Epidemiol. Biomarkers Prev. 14, 1198–1203 [DOI] [PubMed] [Google Scholar]
- 88.Jiang Q., Chen K., Ma X.Y., Yao K.Y., Yu W.P., Li L.Y. et al. (2005) Diets, polymorphisms of methylenetetrahydrofolate reductase, and the susceptibility of colon cancer and rectal cancer. Cancer Detect. Prev. 29, 146–154 [DOI] [PubMed] [Google Scholar]
- 89.Otani T., Iwasaki M., Hanaoka T., Kobayashi M., Ishihara J., Natsukawa S. et al. (2005) Folate, vitamin B6, vitamin B12, and vitamin B2 intake, genetic polymorphisms of related enzymes, and risk of colorectal cancer in a hospital-based case-control study in Japan. Nutr. Cancer 53, 42–50 [DOI] [PubMed] [Google Scholar]
- 90.Miao X.P., Yang S., Tan W., Zhang X.M., Ye Y.J., Lin Y.J. et al. (2005) Association between genetic variations in methylenetetrahydrofolate reductase and risk of colorectal cancer in a Chinese population. Chin. Prev. Med. 39, 409–411 [PubMed] [Google Scholar]
- 91.Kim D., Ahn Y., Lee B., Tsuji E., Kiyohara C. and Kono S. (2004) Methylenetetrahydrofolate reductase polymorphism, alcohol intake, and risks of colon and rectal cancers in Korea. Cancer Lett. 216, 199–205 [DOI] [PubMed] [Google Scholar]
- 92.Ulvik A., Vollset S.E., Hansen S., Gislefoss R., Jellum E. and Ueland P.M. (2004) Colorectal cancer and the methylenetetrahydrofolate reductase 677C→T and methionine synthase 2756A→G polymorphisms: a study of 2,168 case-control pairs from the JANUS cohort. Cancer Epidemiol. Biomarkers Prev. 13, 2175–2180 [PubMed] [Google Scholar]
- 93.Yin G., Kono S., Toyomura K., Hagiwara T., Nagano J., Mizoue T. et al. (2004) Methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and colorectal cancer: the Fukuoka Colorectal Cancer Study. Cancer Sci. 95, 908–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Curtin K., Bigler J., Slattery M.L., Caan B., Potter J.D. and Ulrich C.M. (2003) MTHFR C677T and A1298C polymorphisms: diet, estrogen, and risk of colon cancer. Cancer Epidemiol. Biomarkers Prev. 13, 285–292 [DOI] [PubMed] [Google Scholar]
- 95.Pufulete M., Al-Ghnaniem R., Leather A.J.M., Appleby P., Gout S., Terry C. et al. (2003) Folate status, genomic DNA hypomethylation, and risk of colorectal adenoma and cancer: a case control study. Gastroenterology 124, 1240–1248 [DOI] [PubMed] [Google Scholar]
- 96.Plaschke J., Schwanebeck U., Pistorius S., Saeger H.D. and Schackert H.K. (2003) Methylenetetrahydrofolate reductase polymorphisms and risk of sporadic and hereditary colorectal cancer with or without microsatellite instability. Cancer Lett. 191, 179–185 [DOI] [PubMed] [Google Scholar]
- 97.Toffoli G., Gafà R., Russo A., Lanza G., Dolcetti R., Sartor F. et al. (2003) Methylenetetrahydrofolate reductase 677 C → T polymorphism and risk of proximal colon cancer in north Italy. Clin. Cancer Res. 9, 743–748 [PubMed] [Google Scholar]
- 98.Heijmans B.T., Boer J.M.A., Suchiman H.E.D., Cornelisse C.J., Westendorp R.G.J., Kromhout D. et al. (2003) A common variant of the methylenetetrahydrofolate reductase gene (1p36) is associated with an increased risk of cancer. Cancer Res. 63, 1249–1253 [PubMed] [Google Scholar]
- 99.Huang P., Zhou Z.Y., Ma H.T., Liu J.Y., Zhou Y.H., Cao J. et al. (2003) MTHFR polymorphisms and colorectal cancer susceptibility in Chongqing people. Acta Acad. Med. Mil. Tert. 25, 1704–1710 [Google Scholar]
- 100.Barna B., Erika H., Vilmos A., Ferenc C., Fruzsina G., Istvan L. et al. (2004) A metiléntetrahidrofolát-reduktáz (MTHFR) C677T polimorfizmus klinikai jelentôsége a metasztatikus colorectalis daganatok 5-fluoropirimidin-alapú kezelésében. Magyar Onkológia 48, 253–257 [PubMed] [Google Scholar]
- 101.Keku T., Millikan R., Worley K., Winkel S., Eaton A., Biscocho L. et al. (2002) 5,10-Methylenetetrahydrofolate reductase codon 677 and 1298 polymorphisms and colon cancer in African and Whites. Cancer Epidemiol. Biomarkers Prev. 11, 1611–1621 [PubMed] [Google Scholar]
- 102.Le Marchand L., Donlon T., Hankin J.H., Kolonel L.N., Wilkens L.R. and Seifried A. (2002) B-vitamin intake, metabolic genes, and colorectal cancer risk (United States). Cancer Cause Control 13, 239–248 [DOI] [PubMed] [Google Scholar]
- 103.Shannon B., Gnanasampanthan S., Beilby J. and Iacopetta B. (2002) A polymorphism in the methylenetetrahydrofolate reductase gene predisposes to colorectal cancers with microsatellite instability. Gut 50, 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Matsuo K., Hamajima N., Hirai T., Kato T. and Inoue M. (2002) Methionine synthase reductase gene A66G polymorphism is associated with risk of colorectal cancer. Asian Pac. J. Cancer Prev. 3, 353–359 [PubMed] [Google Scholar]
- 105.Sachse C., Smith G., Wilkie M., Barrett J.H. and Waxman R. (2002) A pharmacogenetic study to investigate the role of dietary carcinogens in the etiology of colorectal cancer. Carcinogenesis 23, 1839–1849 [DOI] [PubMed] [Google Scholar]
- 106.Chen J., Ma J., Stampfer M.J., Palomeque C., Selhub J. and Hunter D.J. (2002) Linkage disequilibrium between the 677C>T and 1298A>C polymorphisms in human methylenetetrahydrofolate reductase gene and their contributions to risk of colorectal cancer. Pharmacogenetics 12, 339–342 [DOI] [PubMed] [Google Scholar]
- 107.Ryan B.M., Molloy A.M., McManus R., Arfin Q., Kelleher D., Scott J.M. et al. (2001) The methylenetetrahydrofolate reductase (MTHFR) gene in colorectal cancer. Int. J. Gastrointestin. Cancer 30, 105–111 [DOI] [PubMed] [Google Scholar]
- 108.Slattery M.L., Edwards S.L., Samowitz W. and Potter J. (2000) Associations between family history of cancer and genes coding for metabolizing enzymes (United States). Cancer Cause Control 11, 799–803 [DOI] [PubMed] [Google Scholar]
- 109.Slattery M.L., Potter J.D. and Samowitz W. (1999) Methylenetetrahydrofolate reductase, diet, and risk of colon cancer. Cancer Epidemiol. Biomarkers Prev. 8, 513–518 [PubMed] [Google Scholar]
- 110.Park K.S., Mok J.W. and Kim J.C. (1999) The 677C >T mutation in 5,10-methylenetetrahydrofolate reducíase and colorectal cancer risk. Genet. Test 3, 233–236 [DOI] [PubMed] [Google Scholar]
- 111.Jing M., Stampfer M.J. and Giovannucci E. (1997) Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res. 57, 1098–1102 [PubMed] [Google Scholar]
- 112.Chen J., Giovannucci E. and Kelsey K. (1996) A methylenetetrahydrofolate reductase polymorphism and the risk of colorectal cancer. Cancer Res. 56, 4862–4864 [PubMed] [Google Scholar]
- 113.Zhou D., Mei Q., Luo H., Tang B. and Yu P. (2012) The polymorphisms in methylenetetrahydrofolate reductase, methionine synthase, methionine synthase reductase, and the risk of colorectal cancer. Int. J. Biol. Sci. 8, 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kennedy D.A., Stern S.J., Matok I., Moretti M.E., Sarker M., Adams-Webber T. et al. (2012) Folate intake, MTHFR polymorphisms, and the risk of colorectal cancer: a systematic review and meta-analysis. J. Cancer Epidemiol. 2012, 952508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Haerian B.S. and Haerian M.S. (2015) Evaluation of association studies and meta-analyses of MTHFR gene polymorphisms in colorectal cancer. Pharmacogenomics 16, 413–425 [DOI] [PubMed] [Google Scholar]
- 116.Yang Z., Zhang X., Liu H., Hao Y. and Zhao C. (2012) MTHFR C677T polymorphism and colorectal cancer risk in asians, a meta-analysis of 21 Studies. Asian Pac. J. Cancer Prev. 13, 1203–1208 [DOI] [PubMed] [Google Scholar]
- 117.Guo X.P., Wang Y., Zhao H., Song S.D., Zhou J. and Han Y. (2014) Association of MTHFR C677T polymorphisms and colorectal cancer risk in Asians: evidence of 12,255 subjects. Clin. Transl. Oncol. 16, 623–629 [DOI] [PubMed] [Google Scholar]
- 118.Zhong S., Yang J., Liu K., Jiao B.H. and Chang Z. (2012) Quantitative assessment of the association between MTHFR C677T polymorphism and colorectal cancer risk in East Asians. Tumor Biol. 33, 2041–2051 [DOI] [PubMed] [Google Scholar]