Abstract
Aims/hypothesis
The genetic risk of type 1 diabetes has been extensively studied. However, the genetic determinants of age at diagnosis (AAD) of type 1 diabetes remain relatively unexplained. Identification of AAD genes and pathways could provide insight into the earliest events in the disease process.
Methods
Using ImmunoChip data from 15,696 cases, we aimed to identify regions in the genome associated with AAD.
Results
Two regions were convincingly associated with AAD (p < 5 × 10−8): the MHC on 6p21, and 6q22.33. Fine-mapping of 6q22.33 identified two AAD-associated haplotypes in the region nearest to the genes encoding protein tyrosine phosphatase receptor kappa (PTPRK) and thymocyte-expressed molecule involved in selection (THEMIS). We examined the susceptibility to type 1 diabetes at these SNPs by performing a meta-analysis including 19,510 control participants. Although these SNPs were not associated with type 1 diabetes overall (p > 0.001), the SNP most associated with AAD, rs72975913, was associated with susceptibility to type 1 diabetes in those individuals diagnosed at less than 5 years old (p = 2.3 × 10−9).
Conclusion/interpretation
PTPRK and its neighbour THEMIS are required for early development of the thymus, which we can assume influences the initiation of autoimmunity. Non-HLA genes may only be detectable as risk factors for the disease in individuals diagnosed under the age 5 years because, after that period of immune development, their role in disease susceptibility has become redundant.
Electronic supplementary material
The online version of this article (10.1007/s00125-017-4440-y) contains peer-reviewed but unedited supplementary material, which is available to authorised users.
Keywords: Age at diagnosis, Early diagnosis, Genetic risk, Type 1 diabetes
Introduction
Since the introduction of genome-wide association studies (GWAS), over 50 regions in the genome have been associated with susceptibility to type 1 diabetes [1–6], but less research has examined the genetic determinants of age at diagnosis (AAD) of type 1 diabetes. One study, limited to specific SNPs in regions associated with type 1 diabetes, identified that the MHC, IL-2 (IL2) and renalase (RNLS) gene regions showed evidence of association with AAD [7, 8]. However, the question has never been examined in a genome-wide fashion. Identification of genes associated with the initiation of the anti-islet autoimmunity, which is in most cases established by the age of 3 years [9], could help to establish the earliest events in the disease process.
Here we aimed to identify genetic determinants of AAD in a more powerful approach using data from an extensive SNP panel, the custom array ImmunoChip [4], by combining data from independent cases and affected sib-pairs (ASPs) to increase sample size and improve the genetic map through imputation.
Methods
We analysed data from six cohorts, independent cases from the UK Genetic Resource Investigating Diabetes (GRID) cohort [2], the Northern Irish GRID (NI) cohort (used here for the first time), and the Finnish IDDMGEN (Tyypin 1 Diabetekseen Sairastuneita Perheenjäsenineen) and T1DGEN (Tyypin 1 Diabeteksen Genetiikka) cohorts [10], in addition to ASPs from the Type 1 Diabetes Genetics Consortium (T1DGC) cohort [11] (from north America, Europe, Asia and the UK) and the UK Warren cohort [12]. The majority of individuals were diagnosed in childhood, with 92% diagnosed at less than 20 years of age. Genotyping (see electronic supplementary material [ESM] Genotyping) was performed on 16,015 affected individuals, 8683 (54%) independent participants and 7332 (46%) ASPs (Table 1). Quality control was performed prior to analysis to minimise the risk of reporting false-positive results (ESM Quality control, ESM Figs 1–3).
Table 1.
Baseline characteristics and inclusion in the primary AAD analysis after quality control
| Cohort | Country | Type | Genotyped | Included | AAD: median (IQR) | SNPs after QC |
|---|---|---|---|---|---|---|
| GRID | UK | Independent | 6799 | 6736 | 8 (4, 11) | 164,953 |
| IDDMGEN | Finland | Independent | 1111 | 1073 | 9 (5, 12) | 156,343 |
| NI | NI | Independent | 524 | 509 | 7 (4, 10) | 156,343 |
| T1DGEN | Finland | Independent | 249 | 249 | 16 (10, 24) | 156,343 |
| Warren | UK | ASP | 907 | 839 | 10 (5, 15) | 156,343 |
| T1DGC | Asia | ASP | 960 | 919 | 10 (5, 14) | 167,537 |
| T1DGC | Europe | ASP | 2521 | 2485 | 11 (6, 17) | 167,537 |
| T1DGC | USA | ASP | 2593 | 2544 | 8 (4, 13) | 167,537 |
| T1DGC | UK | ASP | 351 | 342 | 8 (4, 11) | 167,537 |
| Total | All | All | 16,015 | 15,696 | 9 (5, 12) | 150,381 |
The intersect of the SNPs that passed QC across genotype batch were included in the analysis. Total SNPs after QC refers to the common set of SNPs across all collections
QC, quality control
SNP imputation
Where mentioned, we used IMPUTE2 software (http://mathgen.stats.ox.ac.uk/impute/impute_v2.html) [13, 14], using 1000 Genomes Project data [15] (version III) as the reference dataset (https://mathgen.stats.ox.ac.uk/impute/1000GP_Phase3.html) to perform SNP imputation. We excluded SNPs with a minor allele frequency of less than 0.01 or an imputation information score less than 0.8 as a quality control measure (an imputation score of 0 indicates no certainty in the imputed genotype, whereas a score of 1 indicates no uncertainty in the imputed genotype).
Association discovery using ImmunoChip data
As we had a population that comprised related and independent cases, from a variety of cohorts containing individuals from multiple countries between and within cohorts, it was crucial to account for population structure, to avoid reporting spurious associations. We did this by performing an inverse-variance weighted meta-analysis [16], in which we stratified the samples by cohort and examined the effect of each SNP on loge AAD (assuming an additive mode of inheritance), adjusting for sex and the top five principal components within the cohort to account for population structure.
However, in cohorts with related individuals, standard principal components analysis may not correctly identify the population structure, as the population-level clusters are confounded by the relatedness between individuals. Therefore, in these cohorts, we used principal components analysis in related-samples, PC-AiR [17], which estimates principal components by identifying a subset of genetically dissimilar individuals and performs principal components analysis on this subset, before using these principal components to estimate the ancestry of the remaining individuals in the cohort. This was performed using the GENESIS R package (https://www.bioconductor.org/packages/devel/bioc/html/GENESIS.html, version 2.2.7) [18]. We applied a variance-components model using the GenABEL R package (http://www.genabel.org/packages/GenABEL, version 1.8-0, Grammar-Gamma method) [19] to analyse the effect of each SNP in cohorts of related individuals, which takes into account relatedness between individuals. ESM Fig. 4 provides a schematic overview of the association discovery meta-analysis procedure.
We also performed the association discovery analysis using an alternative approach. First, we fitted a linear mixed model [20] with the loge AAD as the outcome, adjusting for sex as a fixed effect and including random effects for cohort, country and family identifier. We then used the residuals from the linear mixed model as the outcome variable and tested the association of each SNP using a linear regression model. We called this approach the ‘residual-based model’, and it has been proposed by Aulchenko et al [21] to make genome-wide analysis for related individuals possible. Since our aim was eventually to fine-map statistically significant regions, the advantage of the residual-based model is that the residuals can also be used as the outcome variable in a fine-mapping analysis for continuous traits. ESM Fig. 5 illustrates the steps of this second approach used for variants discovery.
SNPs were declared to be associated with AAD if the p value was less than a genome-wide significance threshold of 5 × 10−8. We also highlight regions associated with a false discovery rate (FDR) [22] of less than 0.05, to identify the regions next most likely to influence AAD. In this analysis, we used a stringent definition, removing the MHC region before calculating the FDR, as including all the highly associated SNPs from the MHC can inflate the threshold below which SNPs are declared to be associated, increasing the probability of reporting false-positive results.
To add further evidence to the detected associated SNPs, we combined cases with 19,510 control individuals (ESM Table 1) and performed an inverse-variance weighted meta-analysis across cohorts, examining the effect of the SNPs on risk of type 1 diabetes overall and in those who were diagnosed at less than 5 years of age. Cohorts of independent individuals were analysed by fitting a logistic regression model adjusting for the top five principal components and examining the effect of the SNP of interest on risk of type 1 diabetes. Cohorts of related individuals were analysed using a generalised linear mixed model association test [23], using the GMMAT R package (https://www.hsph.harvard.edu/han-chen/software/, version 0.7-1), adjusting for the top five principal components as fixed effects, and using a kinship matrix to define the covariance structure of the random effect included in the model. We present ORs for the SNPs associated with AAD for their association with type 1 diabetes overall and for those diagnosed at less than 5 years of age, to compare the direction of effect between analyses, with a consistent direction of effect adding further evidence that the association was genuine.
Association discovery: imputed data
A subset of 1768 cases from the GRID cohort had data that had been genotyped using the Affymetrix GeneChip Mapping 500K, and 3833 had been genotyped using the Illumina 550K Infinium microarray platform [3], both of which are GWAS chips and cover a broader spectrum across the genome than the ImmunoChip. We stratified the GRID cohort into strata, one containing cases who had been genotyped using Affymetrix technology, and the other containing cases who had been genotyped using Illumina. We imputed in 1 Mb blocks across the entire genome, and then tested the association of each SNP with loge AAD for both strata using SNPTEST software (version II, using the frequentist option, https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html) [24], before combining results by an inverse-variance weighted, fixed-effects meta-analysis. Owing to an uncertain genotype call, we used the expectation maximisation option to estimate the association of each SNP while performing the imputation of the missing genotypes.
Fine-mapping
In AAD-associated regions (p < 5 × 10−8), we imputed SNPs to obtain the densest SNP set possible in that region, and performed fine-mapping to identify groups of candidate causal SNPs. We used GUESSFM (https://github.com/chr1swallace/GUESSFM version 1.0.1), [25] a fine-mapping algorithm [26, 27] that allows more than one causal SNP in the region to be identified. Briefly, GUESSFM identifies models, that is, combinations of SNPs, with high posterior support given the likelihood and the priors, by carrying out a stochastic search. Models are ranked by the frequency with which they appear in the search. The SNPs included in the most visited models are most likely to be causally associated with AAD. In contrast to other fine-mapping methods that use summary statistics, GUESSFM makes use of raw genotype data. One output from GUESSFM is the group marginal posterior probability of inclusion (gMPPI), which groups SNPs in tight linkage disequilibrium (LD) and can be thought of as the posterior support that exactly one of the SNPs in the group is causal.
The outcome variable was the same set of residuals used in the association discovery analysis residual-based model, so population and family structure had been accounted for. Our primary analyses used the default prior for the expected number of causal variants in the region to be three. For reproducibility of the results, we examined also the effect of changing the expected number of causal SNPs to two or six. For comparison, we fitted stepwise linear regression models in each region of interest (with the same set of residuals as the outcome), initially including the most significant SNP in the region, followed by the most associated SNP conditional on the initial SNP. We repeated this process until one of the SNPs in the model fell below a commonly used [25, 28] significance threshold of p = 5 × 10−6.
Haplotype and diplotype analyses
The output from GUESSFM identifies groups of candidate causal SNPs in LD that are most likely to be associated with the phenotype. We examined these groups detected by GUESSFM in a haplotype analysis to highlight transmission patterns and to visualise haplotype membership of the selected SNPs. We phased SNPs using SNPHAP software (https://github.com/chr1swallace/snphap, version 1.3), which generates the posterior probability of each haplotype for each individual. To capture the uncertainty of the haplotype phase, we simulated ten haplotype datasets, where, in each dataset, the haplotype for every individual was sampled from their haplotype posterior distribution. The effect size of each haplotype relative to the most common was calculated in each dataset using a linear mixed model, with loge AAD as the outcome, adjusting for haplotype and sex as fixed effects, and family identifier, cohort and country as random effects. Results were pooled across the ten datasets using the mice R package (http://www.jstatsoft.org/v45/i03/, version 2.9) for combining results from multiply imputed datasets [29]. Once haplotypes had been estimated, we extended the analysis to examine diplotypes by combining the haplotypes from the two chromosomes and using the methods described above to estimate the effect of each diplotype.
All analyses were carried out using R version 3.3.2.
All samples were collected after approval from the relevant research ethics committees, and written informed consent was obtained from the participants.
Results
Association discovery ImmunoChip analysis
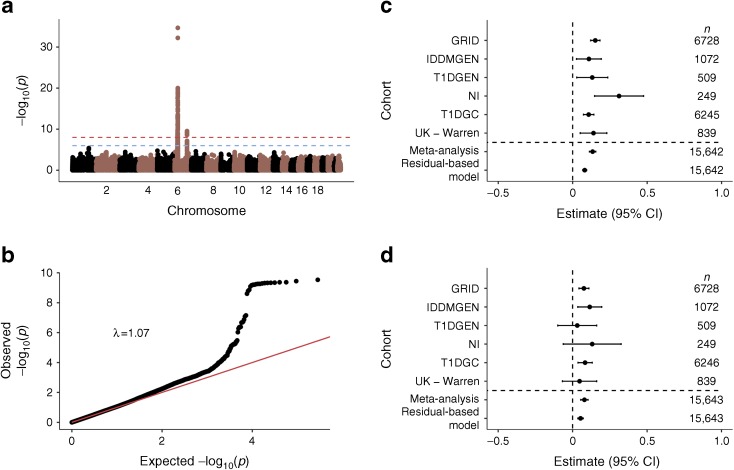
Results from the meta-analysis identified two regions that were associated at p < 5 × 10−8, the MHC region and the 6q22.33 region, the latter of which contains the genes encoding protein tyrosine phosphatase receptor kappa (PTPRK) and thymocyte-expressed molecule involved in selection (THEMIS) (Fig. 1). The index SNP in the MHC was rs9273363 (p = 2.16 × 10−35), which has previously been shown to be associated with type 1 diabetes [30], to tag the HLA DQB1*03:02 genotype [31] and to be located in a potential enhancer region of the major type 1 diabetes gene, HLA-DQB1 [32].
Fig. 1.
(a) Manhattan plot from the association discovery meta-analysis for AAD of type 1 diabetes. (b) Quantile–quantile plot (excluding the MHC region). (c) Forest plot for the lead SNP in the MHC region, rs9273363. (d) Forest plot for the lead SNP in the 6q22.33 region, rs72975913
In the chromosome 6q22.33 region, the lead SNP was rs72975913 (p = 2.94 × 10−10), which is in LD (r 2 = 0.99) with a key SNP associated with coeliac disease (rs72975916) [33]. Results from the residual-based model were similar, with the same two regions, and no others, reaching genome-wide significance. Manhattan and quantile–quantile plots for the residual-based model can be found in ESM Fig. 6, and residual plots for this analysis in ESM Figs 7 and 8.
The results from the residual-based model estimate that the addition of an A allele (the minor allele in controls) at the lead MHC SNP, rs9273363, is associated with an 11.8% decrease in AAD, which translates to a decrease of 1.11 years (13 months). The addition of the major C allele at the lead 6q22.33 SNP, rs72975913, is associated with a younger AAD by 0.7 years (8 months). If an individual is homozygous for the AAD risk allele at the lead SNP in both regions, they are estimated to be diagnosed 4.12 years younger than those who are homozygous for the non-risk allele at both loci.
Although there were just two regions that reached genome-wide significance, Table 2 shows that region 1q24.3, which contains the Fas ligand (FASLG) gene, contains at least one SNP with some evidence (FDR <0.05) of association with AAD. It adds weight to the evidence that this SNP might be truly related to AAD since the type 1 diabetes risk effect direction in those diagnosed at less than 5 years of age is the same as the AAD effect direction for each SNP, that is, the risk allele for type 1 diabetes is associated with younger AAD.
Table 2.
Regions with evidence (FDR <0.05) of association with AAD of type 1 diabetes
| Region | Lead SNP | Risk < major allele | Nearest gene(s) | Estimated % change in AAD (95% CI) from linear mixed model | Meta-analysis, p value | Residual-based, p value | Region associated with autoimmune diseases? | Overall type 1 diabetes OR (95% CI) | Type 1 diabetes under age 5 years, OR (95% CI) |
|---|---|---|---|---|---|---|---|---|---|
| 1q24.3 | rs10912265 | T < C | FASLG | 0.06 (0.04, 0.08) | 4.0 × 10−6 | 1.2 × 10−6 | CEL, CRO, T1D | 0.95 (0.90, 0.99) | 0.82 (0.76, 0.88) |
| 6q22.33 | rs802719 | C < T | PTPRK, THEMIS | −0.06 (−0.08, −0.04) | 7.0 × 10−8 | 5.6 × 10−9 | CEL, CRO, MS | 1.01 (0.98, 1.05) | 1.14 (1.07, 1.20) |
| 6q22.33 | rs72975913 | A < C | PTPRK, THEMIS | 0.08 (0.05, 0.10) | 2.9 × 10−10 | 7.4 × 10−9 | CEL, CRO, MS | 0.92 (0.88, 0.97) | 0.78 (0.72, 0.85) |
| 6p21.32 | rs9273363 | A < C | HLA-DRB1, HLA-DQB1 | −0.12 (−0.10, −0.14) | 2.2 × 10−35 | 2.09 × 10−32 | Multiple | 3.52 (3.37, 3.67) | 5.03 (4.67, 5.42) |
Estimates are for addition of a minor allele. The 6q22.33 region contains two associations: the lead SNP from the residual-based model and the lead SNP from the meta-analysis
CEL, coeliac disease; CRO, Crohn’s disease; MS, multiple sclerosis; T1D, type 1 diabetes
Association discovery analysis—imputed data
In total, 7,476,246 SNPs were examined for their association with AAD. Just one region reached genome-wide significance, the MHC region (ESM Fig. 9), which indicates that there was not sufficient power to detect associated regions outside the ImmunoChip due to a smaller sample size in this analysis (5601 vs 15,696 in the ImmunoChip analysis).
Fine-mapping
It is beyond the scope of the current study to consider fine-mapping of the MHC region, and it is well established that the HLA-DQB1, HLA-DRB1, HLA-A and HLA-B genes are the primary determinants of the MHC risk in type 1 diabetes [30].
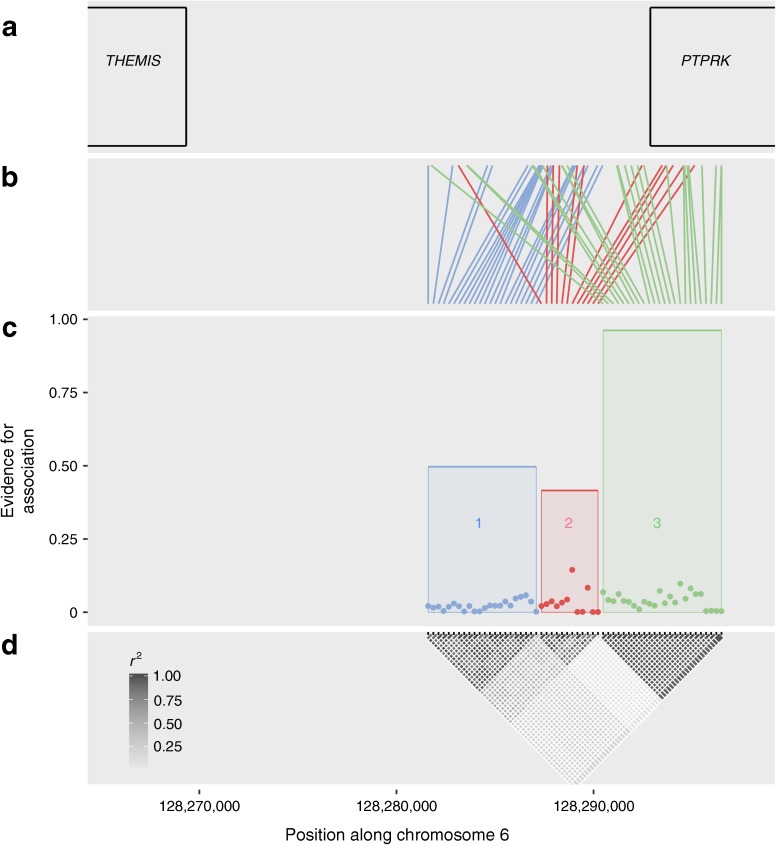
In the 6q22.33 region (positions 127,952,182 to 128,340,790 on chromosome 6, NCBI build 37, https://www.ncbi.nlm.nih.gov), we performed SNP imputation with a concordance of 97.9% (i.e. for each SNP of known genotype, the imputed genotype matched the known genotype at least 97.9% of the time). In total, 786 SNPs were analysed in this region, including 319 imputed SNPs. The GUESSFM analysis highlighted three potential signals in the region (Fig. 2, ESM Table 2). Group 1 comprised 22 SNPs, had a gMPPI of 0.50 and was in LD (r 2 = 0.49) with group 2, which contained 12 SNPs and had a gMPPI of 0.42; this group contains rs802719, the SNP with the highest posterior support (SNP marginal posterior probability of inclusion = 0.14). Finally, group 3, with the strongest signal, contained 24 SNPs and had a gMPPI of 0.96, including the index SNP from the association discovery analysis, rs72975913. However, the LD between groups 1 and 2 implies that these groups are probably the same signal; in addition, the fact that the SNP with the highest posterior probability is contained in group 2 rather than group 1 implies that the true signal is more likely to be from group 2. The results were similar when changing the prior number of SNPs expected in the model to two or six, both showing three signals in the region. Results from the stepwise linear regression approach indicated that there were two signals: the lead SNP once imputed data was included was SNP rs11753289, which is contained in group 3 from the GUESSFM analysis, with a borderline second signal, from SNP rs802719 (p = 7.5 × 10−6), which is contained in group 2.
Fig. 2.
Output from GUESSFM fine-mapping the 6q22.33 region. (a) Location of the PTPRK and THEMIS genes, the closest genes to the candidate causal SNPs. (b) Map of the candidate causal SNPs to their physical location along chromosome 6. (c) The dots depict the strength of association (marginal posterior probability of inclusion) for each SNP, while the height of the shaded region is the gMPPI, the probability that one of the SNPs in the group is causal for AAD. It shows three signals in the region, termed groups 1 (blue), 2 (red) and 3 (green). (d) LD between SNPs
Haplotype and diplotype analyses
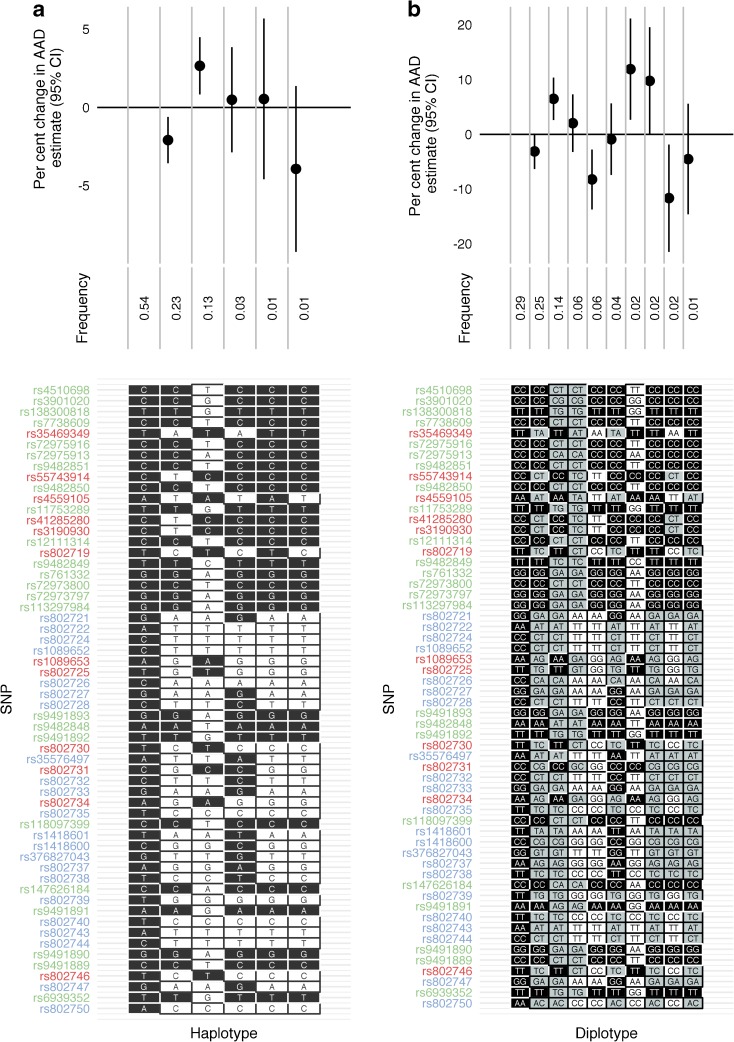
In the 6q22.33 region, we examined the 58 SNPs contained in groups 1, 2 and 3 from the GUESSFM analysis and estimated the effect of each haplotype relative to the most common, which was the major allele at each of the 58 SNPs. The minor allele haplotype from the group 2 SNPs is associated with younger AAD, while the minor allele haplotype from the group 3 SNPs is associated with an older AAD (Fig. 3). The LD between groups 1 and 2 can be observed, and the two main signals in the region appear to be from groups 2 and 3. Extending the analysis to examine diplotypes showed that being heterozygous at group 2 SNPs leads to an estimated 3.1% decrease in AAD relative to the most common diplotype, which translates to an estimated younger AAD of 3.1 months. Having two copies of the minor group 2 SNPs leads to an estimated younger AAD of 8.3%, which translates to 8.1 months. In contrast, individuals with one copy of the minor group 3 haplotype have an estimated AAD 6.4% (6.8 months) older than those with the most common diplotype, which increases to 11.9% (12.9 months) if they have two copies of the minor group 3 haplotype (Fig. 3). These results show there are two main risk haplotypes that predispose to younger AAD in the region: the minor allele haplotype for group 2 and the major allele haplotype for group 3.
Fig. 3.
(a) Haplotype analysis of the 6q22.33 region with respect to the AAD of type 1 diabetes using SNPs highlighted from the GUESSFM analysis. SNPs are colour-coded according to GUESSFM group 1 (blue), 2 (red) and 3 (green). Black, major alleles; white, minor alleles. (b) Diplotype analysis of the same region. Black, major homozygotes; grey, heterozygotes; white, minor homozygotes
Type 1 diabetes risk analysis
We examined one SNP from each of groups 2 and 3 (rs802719 and rs72975913, respectively) from the fine-mapping analysis and examined the effect of these SNPs on type 1 diabetes risk in a meta-analysis including the control participants from ESM Table 1. As expected from the lack of previously reported associations in this region, neither of the SNPs was strongly associated with type 1 diabetes overall (p > 0.001) (forest plots are shown in ESM Fig. 10).
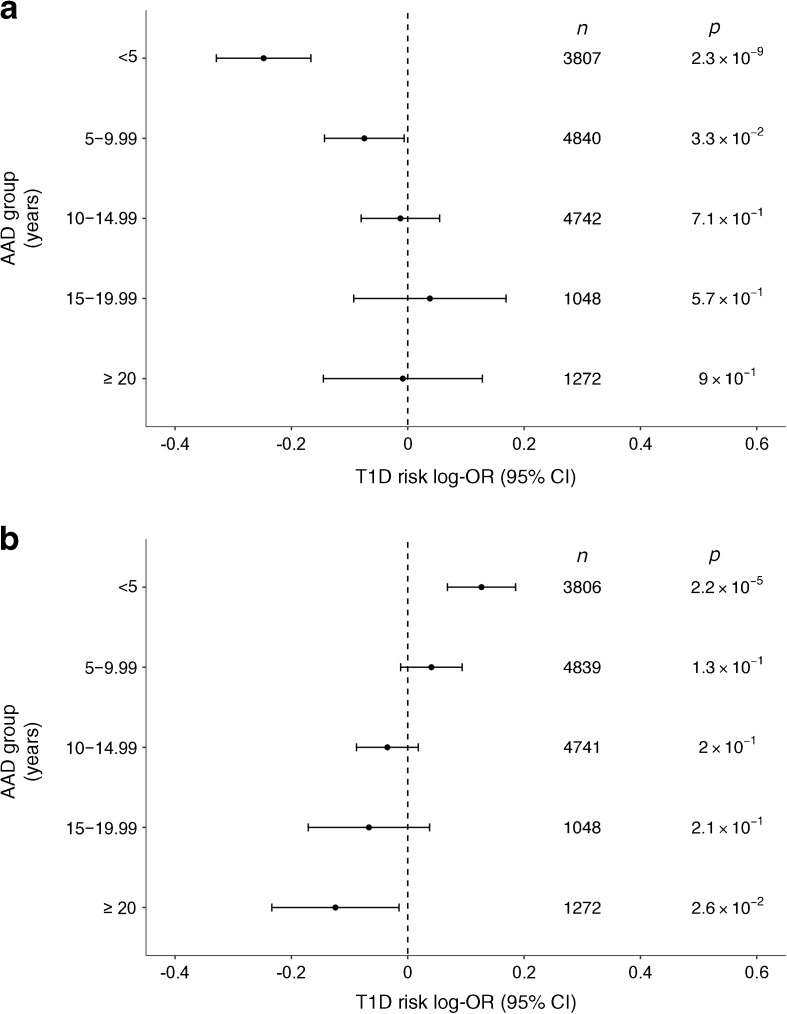
However, owing to the association with AAD, we stratified each cohort into age group at diagnosis (0–4.99, 5–9.99, 10–14.99, 15–19.99 and ≥20 years) and assessed the association of both SNPs with risk of type 1 diabetes within strata. A strong association was observed in the youngest AAD category at rs72975913 (p = 2.3 × 10−9, OR 0.78, n = 3807) and evidence of an association at rs802719 (p = 2.2 × 10−5, OR 1.14, n = 3806), with the forest plots shown in ESM Fig. 11. The effect estimate was in the same direction as the AAD analysis, with the minor A allele associated with decreased disease susceptibility in those diagnosed at less than 5 years old at rs72975913, which is associated with an older AAD, and for rs802719 the minor C allele increases disease susceptibility and decreases AAD. To examine the sensitivity of our results to the cut-off for which to define the youngest AAD strata, we repeated the analysis, defining the youngest age category first as <4 years and then as <6 years. The results did not change significantly, with rs72975913 remaining associated on a genome-wide basis regardless of cut-off and with a similar effect size observed at rs802719 (ESM Table 3). There is a trend showing how the effect size on disease susceptibility decreases at both SNPs as the age group at diagnosis increases, indicating that the genetic effects in this region act in those diagnosed at a young age (Fig. 4, ESM Figs 12 and 13 for minor allele frequencies). This may explain why this region has not previously been associated with type 1 diabetes as, typically, GWAS datasets include cases who have been diagnosed across a range of ages.
Fig. 4.
(a) Risk of type 1 diabetes (T1D) at SNPs contained in group 3 (GUESSFM analysis) rs72975913, stratified by AAD group. (b) Risk of type 1 diabetes at the SNPs contained in group 2, rs802719, stratified by AAD group. The effect size is for addition of a minor allele at the loci, assuming an additive mode of inheritance on the log-odds scale
Discussion
In the first ImmunoChip analysis examining the AAD of type 1 diabetes, we found, as expected, that the MHC was the major genetic influence, while the 6q22.33 region was a second associated region. We performed fine-mapping across the 6q22.33 region, which contains the adjacent PTPRK and THEMIS genes, and has never previously been associated with AAD [7, 8] or type 1 diabetes, but has been reported to be associated with susceptibility to other autoimmune diseases [33–35].
We identified two haplotypes in the region that were associated with younger AAD. These haplotypes showed evidence of influencing type 1 diabetes susceptibility in those diagnosed at less than 5 years of age but not for other age groups. This implies that the region impacts on risk of type 1 diabetes at a young age but not once the immune system is more fully developed. In an era of increasing sample sizes, this study highlights the benefit of refining a phenotype in order to identify SNPs associated with a subset of individuals who develop a disease. In this case, we highlight a region associated with early-diagnosed type 1 diabetes, but this approach can be applied to heterogeneous diseases to more accurately identify the main genetic determinants in a particular subset. Analyses of the immunology and pancreas histology of type 1 diabetes do reveal distinct autoimmune features in children diagnosed under age 5 years [36]. Genetic findings such as ours will help to identify the key cells and tissues involved, pointing, in this case, to the thymus being particularly important in early, aggressive disease.
PTPRK and THEMIS are both important for transition of double-positive (CD4+, CD8+) thymocytes to single-positive thymocytes [37, 38], and there is a reduction in number of mature CD4+ T cells in mice that have both genes knocked out, over and above the effect that each gene has independently, indicating that they are both vital to thymopoiesis [39]. Chromosome conformation capture analyses have identified the PTPRK promoter as a target of disease-associated sequences [40], supporting its candidacy as the causal gene for AAD.
The 6q22.33 region has been associated with other autoimmune diseases: the index SNP for coeliac disease is rs55743914 [33], which is contained in group 2 in our analysis, and the minor allele is associated with increased risk of coeliac disease and also younger AAD for type 1 diabetes. The secondary signal for coeliac disease (rs72975916) is contained in group 3, and the minor allele is associated with reduced risk of coeliac disease and older AAD for type 1 diabetes, so the directions of effect between AAD of type 1 diabetes and coeliac disease are consistent.
However, the lead SNP for multiple sclerosis, rs802734 [35], which is contained in group 2 in this analysis, has the opposite direction of effect (the minor allele being protective against multiple sclerosis). This signal in multiple sclerosis was not replicated in a larger ImmunoChip analysis [41], and hence the risk of multiple sclerosis at this SNP may also depend on another factor, for example age at onset. Just one signal was detected in the region as associated with Crohn’s disease, rs9491891 [42], which is contained in group 3, with a direction of effect also opposite to that for type 1 diabetes and coeliac disease. The age at onset of multiple sclerosis and Crohn’s disease in the cohorts in these analyses is older than the AAD of the type 1 diabetic individuals in our dataset, while there can be a long delay in coeliac disease between onset and diagnosis [43], so it may be that the difference in effect direction could be to do with a changing immune system with age.
Another possibility is that the SNPs affecting early-diagnosed type 1 diabetes are affecting a different pathway, tissue or cell type from the same SNPs that have the opposite effect in multiple sclerosis and Crohn’s disease; that is, an increased level of a protein in one cell type might increase the risk of type 1 diabetes, while the increase in that same protein in a different cell type might protect against multiple sclerosis or Crohn’s disease. There is no evidence that the 6q22.33 region is associated with age at onset of multiple sclerosis or Crohn’s disease, although there are very few individuals in these analyses who were diagnosed in childhood [35, 42] and it is difficult to assess in coeliac disease given the time between onset and diagnosis [43]. We hypothesise that there is co-localisation in the region between AAD of type 1 diabetes and coeliac disease, given the similar genetic risk variants and also the fact that individuals diagnosed with type 1 diabetes at a young age are more likely to have coeliac disease [44]. Our analysis offers a genetic explanation for this phenomenon.
There was some evidence (FDR <0.05) of an association with AAD at one other region, 1q24.3. This region contains the FASLG gene and has been shown to be associated with type 1 diabetes itself [5]; therefore it might be involved in a pathway that acts early in the disease course of type 1 diabetes, leading to the anti-islet autoimmunity that we now know is established in most cases by the age of 3 years [9, 45].
A potential limitation of our study is that the majority (92%) of individuals with type 1 diabetes were diagnosed at less than 20 years old, and it is unlikely that we have identified all the variants associated with the AAD of type 1 diabetes. However, there is scope to perform a similar analysis in a population with more individuals diagnosed at over 18 years of age when data are generated in the future. Finally, some caution should be taken when interpreting the association at 1q24.3 (near FASLG), as the association did not reach a stringent genome-wide significance.
In conclusion, we have identified a novel AAD region at 6q22.33, as well as confirmed the well-established association of the MHC. The two risk haplotypes at 6q22.33 show evidence of association with type 1 diabetes in individuals diagnosed at less than 5 years of age and might thus guide therapeutic strategies in those with early-diagnosed type 1 diabetes.
Electronic supplementary material
(PDF 7898 kb)
Acknowledgements
We gratefully acknowledge all individuals who provided biological samples or data for this study. This research uses resources provided by the T1DGC, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), the National Institute of Allergy and Infectious Diseases (NIAID), the National Human Genome Research Institute (NHGRI), the National Institute of Child Health and Human Development (NICHD) and the JDRF, and supported by grant U01 DK062418 from the US National Institutes of Health. Further support was provided by grants from the JDRF (9-2011-253 and 5-SRA-2015-130-A-N) and the Wellcome Trust (091157 and 107212) to the Diabetes and Inflammation Laboratory at Oxford and Cambridge Universities.
We obtained DNA samples from the British 1958 Birth Cohort collection, funded by the UK Medical Research Council and the Wellcome Trust. This study makes use of data generated by the Wellcome Trust Case Control Consortium, funded by Wellcome Trust award 076113; a full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk/. We thank the University of Virginia for their support in genotyping samples from the Northern Irish GRID cohort, the IDDMGEN and T1DGEN cohorts from Finland and the Warren cohort using the T1DGC grant (U01 DK062418) and JDRF grant (JDRF 9-2011-530).
We thank N. J. Cooper for allowing us to use the imputation map generated to examine AAD variants across the entire genome.
Abbreviations
- AAD
Age at diagnosis
- ASP
Affected sib-pair
- FDR
False discovery rate
- gMPPI
Group marginal posterior probability of inclusion
- GRID
Genetic Resource Investigating Diabetes
- GWAS
Genome-wide association study
- IDDMGEN
Tyypin 1 Diabetekseen Sairastuneita Perheenjäsenineen
- LD
Linkage disequilibrium
- NI
Northern Ireland
- T1DGC
Type 1 Diabetes Genetics Consortium
- T1DGEN
Tyypin 1 Diabeteksen Genetiikka
Data availability
The data analysed during the current study are available from the corresponding author on reasonable request.
Funding
CW is funded by the Wellcome Trust (WT107881) and the Medical Research Council (MC_UP_1302/5). LB was supported by the Alan Turing Institute under the EPSRC grant EP/N510129/1.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
JRJI carried out statistical analyses, wrote the manuscript and approved the final version. NMW helped with acquisition of the data and preparation of the data for analysis, revised the article critically and approved the final version of the manuscript. CW provided assistance implementing GUESSFM, input into the design of the study, provided code to carry out the haplotype analysis, and revised the article critically and approved the final version of the manuscript. LB provided statistical support throughout the project to JRJI and support with interpretation of data, revised the article critically and approved the final version. JAT directed research, provided biological expertise, helped with data acquisition and study design, revised the article critically and approved the final version of the manuscript. JAT is the guarantor of this work.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s00125-017-4440-y) contains peer-reviewed but unedited supplementary material, which is available to authorised users.
Contributor Information
Jamie R. J. Inshaw, Email: jinshaw@well.ox.ac.uk
John A. Todd, Email: jatodd@well.ox.ac.uk
References
- 1.The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Todd JA, Walker NM, Cooper JD, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett JC, Clayton D, Concannon P, et al. Genome-wide association study and meta-analysis finds over 40 loci affect risk of type 1 diabetes. Nat Genet. 2010;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Onengut-Gumuscu S, Chen W-M, Burren O, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fortune MD, Guo H, Burren O, et al. Statistical colocalization of genetic risk variants for related autoimmune diseases in the context of common controls. Nat Genet. 2015;47:839–846. doi: 10.1038/ng.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evangelou M, Smyth DJ, Fortune MD, et al. A method for gene-based pathway analysis using genomewide association study summary statistics reveals nine new type 1 diabetes associations. Genet Epidemiol. 2014;38:661–670. doi: 10.1002/gepi.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howson JMM, Cooper JD, Smyth DJ, et al. Evidence of gene-gene interaction and age-at-diagnosis effects in type 1 diabetes. Diabetes. 2012;61:3012–3017. doi: 10.2337/db11-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howson JMM, Rosinger S, Smyth DJ, et al. Genetic analysis of adult-onset autoimmune diabetes. Diabetes. 2011;60:2645–2653. doi: 10.2337/db11-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ziegler AG, Rewers M, Simell O, et al. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA. 2013;309:2473–2479. doi: 10.1001/jama.2013.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiao Q, Osterholm A-M, He B, et al. A genome-wide scan for type 1 diabetes susceptibility genes in nuclear families with multiple affected siblings in Finland. BMC Genet. 2007;8:84. doi: 10.1186/1471-2156-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rich SS, Akolkar B, Concannon P, et al. Overview of the type I diabetes genetics consortium. Genes Immun. 2009;10:S1–S4. doi: 10.1038/gene.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bain SC, Todd JA, Barnett AH. The British Diabetic Association—Warren Repository. Autoimmunity. 1990;7:83–85. doi: 10.3109/08916939008993380. [DOI] [PubMed] [Google Scholar]
- 13.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3. 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conomos MP, Reiner AP, Weir BS, Thornton TA. Model-free estimation of recent genetic relatedness. Am J Hum Genet. 2016;98:127–148. doi: 10.1016/j.ajhg.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conomos MP, Thornton T (2016) GENESIS: GENetic EStimation and Inference in Structured samples (GENESIS): statistical methods for analyzing genetic data from samples with population structure and/or relatedness. R package, version 2.2.7
- 19.Svishcheva GR, Axenovich TI, Belonogova NM, et al. Rapid variance components–based method for whole-genome association analysis. Nat Genet. 2012;44:1166–1170. doi: 10.1038/ng.2410. [DOI] [PubMed] [Google Scholar]
- 20.Pinheiro JC, Bates DM. Mixed-effects models in S and S-PLUS. Heidelberg: Springer; 2000. [Google Scholar]
- 21.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 22.Banjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 23.Chen H, Wang C, Conomos MP, et al. Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am J Hum Genet. 2016;98:653–666. doi: 10.1016/j.ajhg.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchini J, Howie BN, Myers S, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 25.Wallace C, Cutler AJ, Pontikos N, et al. Dissection of a complex disease susceptibility region using a Bayesian stochastic search approach to fine mapping. PLoS Genet. 2015;11:e1005272. doi: 10.1371/journal.pgen.1005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bottolo L, Richardson S. Evolutionary stochastic search for bayesian model exploration. Bayesian Anal. 2010;5:583–618. doi: 10.1214/10-BA523. [DOI] [Google Scholar]
- 27.Bottolo L, Chadeau-Hyam M, Hastie DI, et al. GUESS-ing polygenic associations with multiple phenotypes using a GPU-based evolutionary stochastic search algorithm. PLoS Genet. 2013;9:31003657. doi: 10.1371/journal.pgen.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Ferreira T, Morris AP, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stef van Buuren KG-O. Mice: multivariate imputation by chained equations in R. J Stat Softw. 2011;45:1–67. [Google Scholar]
- 30.Nejentsev S, Howson JM, Walker NM, et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450:887–892. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen C, Varney MD, Harrison LC, Morahan G. Definition of high-risk type 1 diabetes HLA-DR and HLA-DQ types using only three single nucleotide polymorphisms. Diabetes. 2013;62:2135–2140. doi: 10.2337/db12-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miao F, Chen Z, Zhang L, et al. Profiles of epigenetic histone post-translational modifications at type 1 diabetes susceptible genes. J Biol Chem. 2012;287:16335–16345. doi: 10.1074/jbc.M111.330373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trynka G, Hunt KA, Bockett NA, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawcer S, Hellenthal G, Pirinen M, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan NG. Bringing the human pancreas into focus: new paradigms for the understanding of type 1 diabetes. Diabet Med. 2017;34:879–886. doi: 10.1111/dme.13365. [DOI] [PubMed] [Google Scholar]
- 37.Johnson AL, Aravind L, Shulzhenko N, et al. Themis is a member of a new metazoan gene family and is required for the completion of thymocyte positive selection. Nat Immunol. 2009;10:831–839. doi: 10.1038/ni.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asano A, Tsubomatsu K, Jung C-G, et al. A deletion mutation of the protein tyrosine phosphatase kappa (Ptprk) gene is responsible for T-helper immunodeficiency (thid) in the LEC rat. Mamm Genome. 2007;18:779–786. doi: 10.1007/s00335-007-9062-0. [DOI] [PubMed] [Google Scholar]
- 39.Iwata R, Sasaki N, Agui T. Contiguous gene deletion of Ptprk and Themis causes T-helper immunodeficiency (thid) in the LEC rat. Biomed Res. 2010;31:83–87. doi: 10.2220/biomedres.31.83. [DOI] [PubMed] [Google Scholar]
- 40.Javierre BM, Burren OS, Wilder SP, et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016;167:1369–1384. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beecham AH, Patsopoulos NA, Xifara DK, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang H, Fang M, Jostins L, et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nat Publ Gr. 2017;547:173–178. doi: 10.1038/nature22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fasano A. Clinical presentation of celiac disease in the pediatric population. Gastroenterology. 2005;128:68–73. doi: 10.1053/j.gastro.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 44.Cerutti F, Bruno G, Chiarelli F, et al. Younger age at onset and sex predict celiac disease in children and adolescents with type 1 diabetes: an Italian multicenter study. Diabetes Care. 2004;27:1294–1298. doi: 10.2337/diacare.27.6.1294. [DOI] [PubMed] [Google Scholar]
- 45.Bonifacio E, Mathieu C, Nepom GT, et al. Rebranding asymptomatic type 1 diabetes: the case for autoimmune beta cell disorder as a pathological and diagnostic entity. Diabetologia. 2016;60:6–9. doi: 10.1007/s00125-016-4144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 7898 kb)
Data Availability Statement
The data analysed during the current study are available from the corresponding author on reasonable request.