Abstract
Casein kinase 2 (CK2) is a highly pleiotropic serine-threonine kinase, which catalyzed phosphorylation of more than 300 proteins that are implicated in regulation of many cellular functions, such as signal transduction, transcriptional control, apoptosis and the cell cycle. On the other hand, CK2 is abnormally elevated in a variety of tumors, and is considered as a promising therapeutic target. The currently available ATP-competitive CK2 inhibitors, however, lack selectivity, which has impeded their development in cancer therapy. Because allosteric inhibitors can avoid the shortcomings of conventional kinase inhibitors, this study was aimed to discover a new allosteric site in CK2α and to investigate the effects of mutations in this site on the activity of CK2α. Using Allosite based on protein dynamics and structural alignment, we predicted a new allosteric site that was partly located in the αC helix of CK2α. Five residues exposed on the surface of this site were mutated to validate the prediction. Kinetic analyses were performed using a luminescent ADP detection assay by varying the concentrations of a peptide substrate, and the results showed that the mutations I78C and I78W decreased CK2α activity, whereas V31R, K75E, I82C and P109C increased CK2α activity. Potential allosteric pathways were identified using the Monte Carlo path generation approach, and the results of these predicted allosteric pathways were consistent with the mutation analysis. Multiple sequence alignments of CK2α with the other kinases in the family were conducted using the ClustalX method, which revealed the diversity of the residues in the site. In conclusion, we identified a new allosteric site in CK2α that can be altered to modulate the activity of the kinase. Because of the high diversity of the residues in the site, the site can be targeted using rational drug design of specific CK2α inhibitors for biological relevance.
Keywords: CK2α, allosteric site, mutation, allosteric pathway
Introduction
Casein kinase 2 (CK2) is a highly pleiotropic serine-threonine kinase1 that is naturally endowed with constitutive activity and lacks gain-of-function mutants. It is often present as a heterotetramer composed of two catalytic α subunits (α or α′) and two regulatory β subunits in various combinations2,3. CK2 is a key regulator of various cellular events, and its phosphorylation targets include more than 300 proteins4 that are implicated in the regulation of many cellular functions, including signal transduction, transcriptional control, apoptosis, and the cell cycle5,6,7. On the other hand, CK2 is abnormally elevated in a wide variety of tumors6. Overexpression of CK2 was documented in kidney8, mammary gland9, lung10, head and neck11 and prostate cancers8. Thus, CK2 is currently considered to be a promising therapeutic target, and many CK2 inhibitors targeting the classical ATP substrate site have been developed12, such as emodin13, TBB14, (5-oxo-5,6-dihydroindolo[1,2-a]quinazolin-7-yl)aceticacid15, and CX-494516,17. Some CK2 inhibitors, such as 5,6-dichloro-1-b-D-ribofuranosylbenzimidazole (DRB)18, also block the interface between the CK2α and CK2β subunits. Nevertheless, the lack of selectivity of these known CK2 inhibitors has impeded their development in cancer therapy.
Allosteric inhibitors can avoid the shortcomings of most conventional ATP-competitive kinase inhibitors, such as poor selectivity, in vivo competition with a high ATP concentration, and reliance on highly conserved residues in the ATP binding site susceptible to mutation19. One way to discover new allosteric molecules is to identify a new allosteric pocket19,20. Inhibitors that bind these allosteric pockets exert control over the protein's function by perturbing the ensemble of structures that the protein adopts21,22,23. Such sites could have biological functions and serve as valuable targets for drug design. Unfortunately, the discovery of new allosteric pockets is challenging. Most known allosteric sites were serendipitously discovered from the results of protein-ligand structural complexes24,25. To date, only Jennifer Raaf has found an allosteric site in CK2α that can be alternatively filled by (DRB) or glycerol18 in the complex structure of CK2α and DRB. However, the specificity of the cavity is obviously low compared with that of other kinases, which is not good for developing selective inhibitors.
It is urgent to identify more allosteric pockets in CK2α for further inhibitor discovery. In the present study, computational and experimental experiments were performed to explore such allosteric sites for drug design in CK2α. Using Allosite prediction and point mutation analysis, we identified a new allosteric site in CK2α.
Materials and methods
Plasmid constructs and site-directed mutagenesis
The His-tagged CK2α construct was created by the direct insertion of the CK2α gene into the pET28a expression vector between the BamHI and HindIII restriction sites. The template of CK2α was amplified from pDB1-CK2α (Addgene, Cambridge, MA, USA). All DNA oligonucleotides were produced by Sangon Biotech Co, Ltd (Shanghai, China). Point mutations were created using the Site-Directed Mutagenesis Kit (Stratagene, La Jolla, California, USA) according to standard protocols. The sequences of the individual clones were verified.
Protein expression and purification
His-fusion vectors of CK2α and the indicated mutant vectors were transformed into BL21 Escherichia coli. All wild-type and mutated vectors were expressed and purified identically. Expression was induced with 0.5 mmol/L IPTG at 0.6–0.8 OD and proceeded for 12–16 h at 16 °C. Bacterial cells were harvested by centrifugation at 4000×g for 15 min, and the pellets were resuspended in lysis buffer containing 20 mmol/L TRIS-HCl at pH 7.5, 50 mmol/L NaCl, 1 mmol/L phenylmethylsulfonyl fluoride, and 20 mmol/L imidazole. The cells were lysed by sonication, and the lysates were cleared by centrifugation at 20 000×g for 40 min. The supernatants were incubated with 1 mL of previously washed Ni Sepharose Fast Flow (GE Healthcare, Chicago, Illinois, USA) per liter of bacterial culture and equilibrated with the lysis buffer. After 1 h at 4 °C, the beads were washed with lysis buffer and eluted with lysis buffer containing different concentrations of imidazole. After the Ni2+ affinity chromatography, CK2α and its mutants were purified by Superdex 200 gel filtration chromatography (GE Healthcare, Chicago, Illinois, USA) and equilibrated with a buffer containing 20 mmol/L TRIS-HCl at pH 7.5, 50 mmol/L NaCl, 1 mmol/L EDTA, and 2 mmol/L DTT. The peak fractions were combined and concentrated. Then, the purified proteins were aliquoted, snap-frozen in liquid nitrogen, and stored at −80 °C for subsequent use.
ADP-Glo™ CK2α kinase assay
Kinetic analyses of CK2α and the indicated mutants were performed using a luminescent ADP detection assay (Promega, Madison, WI, USA) by varying the concentration of the peptide substrate (RRRADDSDDDDD). The peptide substrate (>95% purity) was synthetized by Wuhan Holder Co, Ltd (Wuhan, China). White low-volume 384-well polystyrene plates (ProxiPlate-384 Plus, PerkinElmer, Waltham, MA, USA) were used for the kinase assay. In total, 2.5 μL of kinase were mixed with 2.5 μL of the kinase buffer (50 mmol/L TRIS at pH 7.6, 10 mmol/L MgCl2, 2 mmol/L DTT, varied concentrations of substrate, and 200 μmol/L ATP). The enzyme concentrations were chosen so that the reactions were linear during the 220-s incubation period, and all reactions were carried out in triplicate. Blank wells lacked the enzyme but included the kinase buffer, the substrate, and ATP. The plates were covered, and the reactions were carried out at 25 °C for up to 220 s. The reactions were stopped with the addition of 5 μL of ADP-GloTMReagent. After 40 min of incubation at room temperature, 10 μL of Kinase Detection Reagent were added, and the plates were incubated for another 60 min at room temperature. The plates were read on a Synergy H4 microplate reader (BioTek Instruments, Winooski, USA) with an integration time of 1 s per well. The apparent KM and Vmax values were obtained from a non-linear curve fitted to the Michaelis-Menten equation and analyzed using Prism 5.0 software (GraphPad, La Jolla, CA, USA). The kcat was calculated as Vmax divided by the valid enzyme concentration.
Prediction of the allosteric sites in CK2α
All 127 crystal structures of CK2α were downloaded from Protein Data Bank26. To cover potential sites as much as possible, all 127 structures with diverse conformations were used to identify allosteric sites in CK2α. These query structures were submitted into Allosite, and 1510 sites were found and merged into 31 primary pockets based on the overlay of the structures. First, 25 weighted descriptors (eg, Normalized maximum distance between two alpha spheres, normalized hydrophobicity density, flexibility score) in the Allosite model were chosen to characterize all primary pockets, and the feature score for each pocket was calculated. Second, normal mode analysis (NMA) was also performed to perturb each primary pocket with a dummy ligand. The extent of flexibility at each pocket was evaluated based on the difference between the B-factors before and after binding the ligand, and the dynamic score of each pocket was calculated using the flexibility. Then, the allosite score for each primary pocket was obtained according to the normalization of feature score and dynamic score. Finally, allosite scores larger than 0.5 were used for further validation.
Identification of the allosteric pathway between the allosteric site and the substrate site
The MCPath server (http://safir.prc.boun.edu.tr/clbet_server) is based on MC path generation runs with a PDB ID or an uploaded structure. Using the query structure from the server, the MCPath generation method was used to calculate all of the likely allosteric communication pathways by generating an ensemble of maximum probability paths, as well as an infinitely long path for plausible functional residues based on the graph centrality measures. Finally, the output of results included the top three populated pathways shown on the structure's ribbon diagram27.
Molecular graphics
All of the protein structure figures were generated with PyMOL Molecular Graphics System (Version 1.8; Schrödinger, LLC). The amino acid sequences of human CK2α (P68400), CK2α'(P19784), CDKL2(Q92772), CDKL3(Q82VW4), CDKL4(Q5MAI5), CDK1(P06493), CDK2(P24941), CDK5(Q00535), CDK7(P50613), CDK8(P49336), CDK9(P50750), CDK10(Q15131), CDK11B(P21127), CDK12(Q9NYV4), CDK14(O94921), CDK16(Q00536), CDK20(Q8IZL9), MK01(P28482), MK03(P27361), MK04(P31152), MK06(Q16659), MK07(Q13164), MK08(P45983), MK09(P45984), MK10(P53779), MK11(Q15759), MK12(P53778), MK13(O15264), MK14(Q16539), CLK1(P49759), CLK3(P49761), CLK4(Q9HAZ1), GSK3A(P49840) and GSK3B(P49841) were aligned using Clustal X28, and the figures were generated with ESPript (http://espript.ibcp.fr/ESPript/ESPript/)
Results
Computational detection of the known hidden allosteric site in CK2α
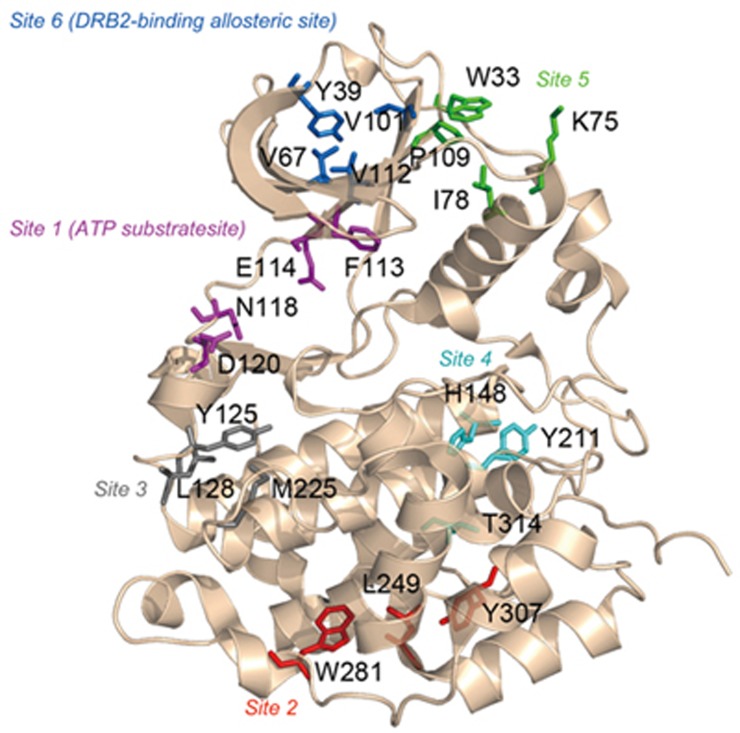
Using our algorithm Allosite24,25, we computationally identified the potential allosteric sites in CK2α. The Allosite results showed that there are 6 predicted allosteric sites distributed around the surface of CK2α (Figure 1 and Table 1). Interestingly, site 1 corresponded to the ATP substrate site18, whereas the DRB2-binding allosteric site was identified as site 6. These results could be considered an internal validation of the Allosite methodology.
Figure 1.
Predicted allosteric sites using the Allosite method. The structure of CK2α is represented by a cartoon, and the residues at the predicted sites are shown in sticks. Residues at site 1 (ATP substrate site) are colored in purple, residues at site 2 are colored in red, residues at site 3 are colored in gray, residues at site 4 are colored in cyan, residues at site 5 are colored in green, and residues at site 6 (DRB2-binding allosteric site) are colored in blue.
Table 1. The predicted allosteric sites in the CK2α proteins.
| Rank | Representative residues | Feature score | Perturbation score | Allosite score |
|---|---|---|---|---|
| Site 1 | F113, E114, N118, D120 | 0.985 | 0.649 | 0.918 |
| Site 2 | L249, W281, Y307 | 0.610 | 0.131 | 0.514 |
| Site 3 | M225, Y125, L128 | 0.967 | 0.155 | 0.805 |
| Site 4 | Y211, H148, T314 | 0.820 | 0.165 | 0.689 |
| Site 5 | W33, K75, I78, P109 | 0.819 | 0.209 | 0.697 |
| Site 6 | Y39, V67, V112, V101 | 0.573 | 0.310 | 0.520 |
In addition to the known sites, a new potential allosteric site at rank 5 (referred to as site 5 in Table 1) was predicted around the loop linking the β3 strand and the αC helix (Figure 1). The conformation of the αC helix has been reported to correlate with kinase activity in another kinase, PDK129, leading to the identification of an allosteric site in that kinase30. Then, further analysis of site 5 showed that the structural conservation of the pocket located on the surface of the αC helix was low, 27.5%, compared with other kinases. In addition, the pocket is present even in the absence of CK2α substrate. This type of pocket may be more suitable for an allosteric pocket20. Therefore, we expected that perturbations at this site could modulate the ensemble of CK2α conformations and regulate the protein's activity.
Effects of distal mutations in the predicted site on CK2α catalysis activity
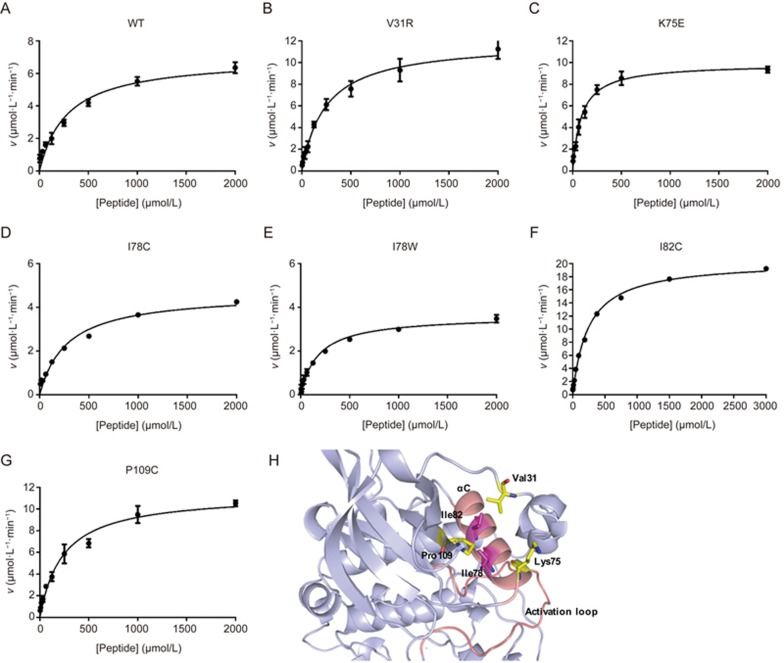
The predicted site 5 was tested experimentally using site-directed mutagenesis, a method that has been useful for the determination of allosteric sites and key residues that play a role in signaling31,32. We selected residues exposed on the surface of site 5 for experimental validation after inspecting the CK2α structure using PyMOL. These mutations include I78, I82 and P109, which form the bottom of the pocket, and V31 and K75, which are on the flank of the pocket (Figure 2H). Among these residues, K75 and I78 are part of the αC helix, V31 is located in the N-terminal segment loop, and P109 is in the β5 strand. First, we chose residue I78, which was close to a basic center related to substrate recognition33. Our computational model also predicted that it could form a bump when the residue was mutated to tryptophan, and we hypothesized that this mutation minimized potential perturbations to the protein but changed the conformation of the pocket performing the reaction. Thus, we introduced a mutation into this region by replacing isoleucine with tryptophan or cysteine and performed an enzyme activity assay to determine the effect on CK2α activity. The kinase activities and catalytic rate of CK2α were measured with the ADP-Glo™ kinase assay34 using the peptide substrate RRRDDDSDDD35 and ATP. This mutation resulted in a decrease in the catalytic rate kcat from 290.5 min−1 for wild-type CK2α to 150.2 min−1 for the I78W mutant and 193.9 min−1 for the I78C mutant. The I78W and I78C mutants catalyzed the reaction at 52% and 66.7% of the rate of wild-type CK2α, respectively. The KM of I78C was not changed relative to wild-type CK2α, but the KM of I78W was reduced from 285.2 μmol/L to 179.9 μmol/L (Figure 2A, 2D, 2E and Table 2). The mutation of I78 to hydrophilic cysteine or hydrophobic tryptophan had a different influence on the activity of CK2α, which indicated that I78 is an important residue in maintaining the activity of CK2α.
Figure 2.
Michaelis–Menten plot of CK2α and its mutants with a peptide substrate. (A) Wild-type CK2α, (B) CK2α-V31R, (C) CK2α-K75E, (D) CK2α-I78C, (E) CK2α-I78W, (F) CK2α-I82C, and (G) CK2α-P109C. The reaction velocities for CK2α and its mutants were assayed in the presence of 3.9–4000 μmol/L of RRRDDDSDDD, the peptide substrate, and the results were fitted to the Michaelis–Menten equation. (H) Residues selected for mutation in the predicted pocket. Ribbon diagram of CK2α highlighting the residues on the bottom of the predicted allosteric pocket (I78 and I82 colored in magenta) and the flank of the predicted allosteric pocket (V31, K75 and P109 colored in yellow). The amino acids are shown in stick diagram. The αC helix and the activation loop are colored in light pink.
Table 2. The effect of mutations in the CK2α proteins on the kinetic constants for CK2-catalyzed substrate phosphorylationa.
| CK2α | Km (μmol/L) | kcat (min−1) | kcat/Km (μmol·L−1·min−1) |
|---|---|---|---|
| WT | 285.2±35.27 | 290.54±16.43 | 1.02±0.27 |
| V31R | 245.4±43.92 | 397.66±45.46 | 1.62±0.60 |
| I82C | 242.8±12.2 | 848.33±25.6 | 3.49±0.22 |
| P109C | 236.1±26.6 | 475.41±8.13 | 2.01±0.22 |
| I78C | 278.4±13.2 | 193.87±12.3 | 0.70±0.02 |
| I78W | 179.9±19.6 | 150.20±1.02 | 0.83±0.04 |
| K75E | 88.5±28.55 | 411.45±34.19 | 4.65±0.85 |
aThe kinase activity experiments were run with CK2α or its mutants as described in the methods section. All data were the mean of at least three separate determinations. The values are expressed as the mean±SEM.
We also mutated four other important residues in the pocket. The kinetic constants of all of the CK2α mutants with the peptide substrate were determined and compared with wild-type CK2α. The kcat and KM values and the overall catalytic efficiencies expressed by the kcat/KM are summarized in Figure 2 and Table 2. Surprisingly, the mutants V31R, K75E, I82C and P109C displayed significantly increased catalytic efficiencies relative to that of wild-type CK2α, with each showing a 1.6-fold, 4.6-fold, 3.5-fold, and 2-fold increase in the value of kcat/KM, respectively. A detailed examination of the data in Table 2 allowed a rough subdivision of the mutants into the following two categories: (i) mutants with a KM for the substrate peptide that was only slightly decreased, whereas the kcat was actually increased or decreased relative to that of wild-type CK2α including V31R, I82C, P109C, and I78C; (ii) mutants with a KM and kcat for the peptide that were altered in a sharply different manner compared with wild-type CK2, including K75E and I78W. K75 and I78 are located in the αC helix and may have a stronger influence than the other four residues on CK2α activity. In fact, lysine mutated to glutamic acid has also been implicated in activating mutations in encephalocraniocutaneous lipomatosis36. At the same time, the large increase in the catalytic efficiencies of K75E, I82C and P109C resulted from significantly and reproducibly higher kcat values and lower KM values. A possible interpretation was that these mutants might also stimulate CK2α activity by interacting with V116, an important residue in the binding of ATP to CK217. These results showed that mutations in this site could both activate and inhibit the activity of CK2α.
The predicted allosteric pathway between the allosteric site and the active site
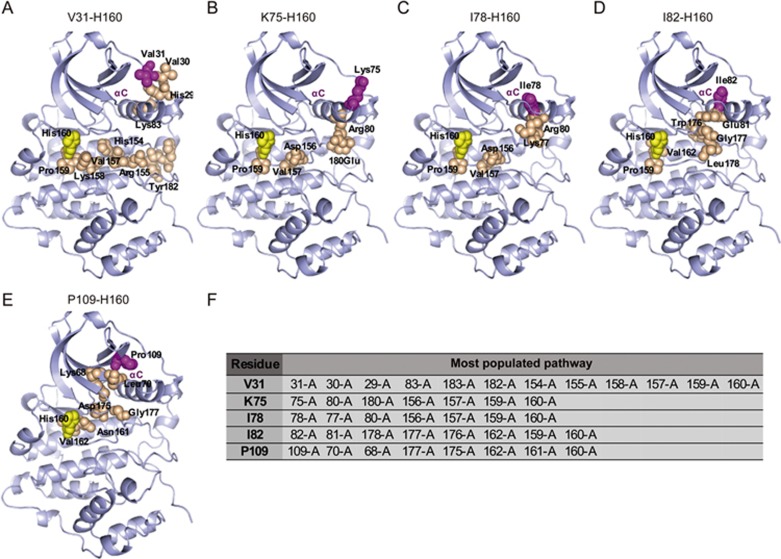
The MCPath generation approach is an effective tool for the prediction of key residues that mediate allosteric communication in an ensemble of functionally pathways27. Therefore, we employed the MCPath approach to investigate the potential communication between the predicted pockets and the active site, as indicated by correlations in the ensemble of structures that CK2α adopts. Residue H160 is homologous to PKA E170, which is responsible for the binding of the basic residue in the peptide substrate of CK2α33,37,38. H160 has been recently shown to contribute to the recognition of an acidic residue whenever it is present in the peptide substrates of CK239. The mutation of H160 caused the enzyme to be nearly inactive, hampering the calculation of kinetic parameters, which indicates the importance of H160 in the recognition of substrate39. Therefore, the intra-molecular communication pathways and functional sites between H160 and mutant residues were analyzed by the MCPath server. The apo-CK2α protein (PDB code 3AT2) was selected for analysis. The most popular pathways between the active site H160 and allosteric site using the MCPath method were mapped on the structure, as shown in Figure 3. Perturbation of the pathways could propagate, as visualized with spheres. The allosteric pathways of K75 and I78 to H160 are relatively short pathways, both passing through R80, D156, V157, and P159 (Figure 3B, 3C and 3F). The differences between the two pathways are the two residues with opposite charges, E180 in the K75-H160 allosteric pathway and K77 in the I82-H160 allosteric pathway. These results may correspond to the change in both the KM and kcat and may have the opposite influence on catalytic efficiencies. The allosteric pathways of I82 and P109 both generated 6 steps to H160 with two of the same residues, V162 and G177 (Figure 3D, 3E and 3F). The I82 and P109 mutants have nearly the same KM but different kcat values, which may be caused by the other four residues in the allosteric pathways. V31 generated an ensemble of pathways (10-steps-long) to H160 (Figure 3A and 3F), which the interaction transmits through the activation loop and then into the core of the domain. The whole pathway belongs to a loop that has good flexibility, which may exert the least influence after mutation. The results of these predicted allosteric pathways were consistent with the mutation analysis.
Figure 3.
The possible communication pathway between the active site His160 (colored in yellow) and the allosteric sites (colored in magenta) from CK2α (PDB code: 3AT2). The most populated pathways are shown in the sphere. The residues in the allosteric pathway are colored in orange. (A) The pathway from an ensemble of 12-step paths generated starting from V31. (B) The pathway from an ensemble of 7-step paths generated starting from K75. (C) The pathway from an ensemble of 7-step paths generated starting from I78. (D) The pathway from an ensemble of 8-step paths generated starting from I82. (E) The pathway from an ensemble of 8-step paths generated starting from P109. (F) The tables list the most populated pathway.
The conservation of the allosteric pocket on the CK2α surface in the CMGC kinase family
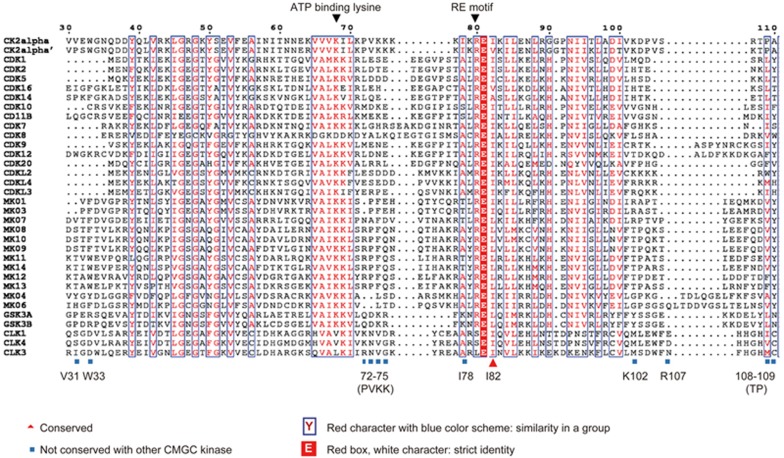
CK2α is a member of the CMGC family of eukaryotic protein kinases40. To determine the conservation of the residues in the newly discovered allosteric pocket of the whole CMGC kinase family, we aligned the protein sequences of 9 members of the CMGC kinase family, including cyclin dependent kinases (CDK), CDK-like kinases (CDKL), casein kinase 2 (CK2), glycogen synthase kinase(GSK), mitogen-activated protein kinase(MAPK), SR-rich protein kinase (SRPK), CDK-like kinase(CLK), dual specificity tyrosine regulated kinase (DYRK) and RCK41. A global score was calculated for all sequences. The similarity global score was defined as 0.7. The sequence alignment results of the ClustalX analysis of the CMGC family are shown in Figure 4. The predicted pocket consisted of 12 residues, with 3 residues (I78, I82 and P109) located on the bottom of the pockets and 9 residues (V31, W33, P72, V73, K74, K75, K102, R107 and T108) located on the side of the pockets. Thus, we assessed the conservation level of the amino acid residues in the predicted pocket of CK2α. The inspection of the amino acid sequences within this site showed that almost all residues were diverse, except for W33 and I82. V31, P72, V73, K74, K75, I78, K102, R107, T108 and P109 were not conserved throughout the whole CMGC family. Residue I82 showed considerable conservation in the CDK family, CDKL family and CLK family, and residue W33 showed considerable conservation in the MAPK family. In addition, none of these 12 residues showed conservation between CK2α and SRPK, DYRK and RCK kinases (data not shown). These unique residues could be good determinants for the design of specific inhibitors.
Figure 4.
Multiple sequence alignment of CK2α with the other CMGC kinases, CDK, CDKL, GSK, CLK and MAPK. The sequences were aligned using the ClustalX method. The figure was created with ESpript. The sequence of 30-110 of CK2α is shown. Residues with a conservation of greater than 70% are color-coded. Mutations are indicated at the corresponding positions. Residues that were not conserved (V31, W33, P72, V73, K74, K75, I78, K102, R107, T108 and P109) are shown in gray. A red character with a blue color scheme indicates similarity in a group (I82), and a red box with a white character indicates strict identity.
Discussion
There is currently increasing interest in identifying more specific next-generation CK2 inhibitors that do not directly compete with ATP and that exhibit different mechanisms of action. However, there are few known allosteric inhibitors of CK2. Allosteric pockets are being increasingly exploited for finding therapeutic molecules19. One way to find allosteric inhibitors is to locate a new allosteric pocket for which structure-based drug design can be used. We found a new allosteric pocket in CK2α by combining bioinformatics and biochemistry methods. Mutation of this pocket both activated and inhibited the activity of CK2α. Both the binding and the extent of activation or inhibition were highly dependent on the residues chosen.
It is intriguing that the mutation of a single allosteric site on a kinase, such as in pocket 2 on CK2α, can generate a continuum of agonistic and antagonistic effects. Previous studies have shown that even changing a single atom42 or a small group43 may result in opposite effects or large differences in efficacy. Furthermore, even the same allosteric ligand bound at the same site can lead to opposing effects in different environments44. As shown using MCPath, generating the predicted allosteric pathways may interrupt the opposing effects. Further experiments should be conducted to explain the results.
The pocket is not conserved among the whole CMGC kinase family, which suggests the potential for designing specific inhibitors. We should also avoid activating residues in this pocket, such as V31, K75, I82 and P109, when designing allosteric inhibitors based on this pocket.
Although we have not discovered new molecules that bind to this allosteric site, we note that the allosteric inhibitor discovered by Horn et al45. demonstrates that it is possible for small molecules to bind such hidden allosteric sites strongly enough to stabilize the open form of a pocket and to alter an enzyme's activity. In fact, Bowman et al found that almost any binding pocket could also serve as an allosteric site because of the coupling of many residues to different portions of the active site46. However, the drugability of different pockets is different. Given the central location of these residues within the allosteric pocket, new allosteric sites can be targeted with rational drug design or further studied to determine their biological relevance. In summary, this study yielded a new allosteric site in CK2α, which we believe will foster efforts to investigate its potential as a candidate pocket for subsequent drug design.
Acknowledgments
We sincerely thank Prof Ai-wu ZHOU at the Shanghai Jiao-Tong University for help with the protein expression and purification. This work was supported in part by grants from the National Basic Research Program of China (No 973 Program) (No 2015CB910403), the National Natural Science Foundation of China (No 81322046, 81473137, 81302698, and 81473232), the Research Program of Shanghai Municipal Commission of Health and Family Planning (No 201440576), and the Science Foundation of Shanghai Jiaotong University School of Medicine (No 14XJ10002).
References
- Ruzzene M, Pinna LA. Addiction to protein kinase CK2: a common denominator of diverse cancer cells? Biochim Biophys Acta 2010; 1804: 499–504. [DOI] [PubMed] [Google Scholar]
- Litchfield DW. Protein kinase CK2:structure regulation and role in cellular decisions of life and death. Biochem J 2003; 369: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filhol O, Martiel JL, Cochet C. Protein kinase CK2: a new view of an old molecular complex. EMBO Rep 2004; 5: 351–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J 2003; 17: 349–68. [DOI] [PubMed] [Google Scholar]
- Filhol O, Cochet C. Protein kinase CK2 in health and disease: Cellular functions of protein kinase CK2: a dynamic affair. Cell Mol Life Sci 2009; 66: 1830–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trembley JH, Wang G, Unger G, Slaton J, Ahmed K. Protein kinase CK2 in health and disease: CK2: a key player in cancer biology. Cell Mol Life Sci 2009; 66: 1858–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loizou JI, El-Khamisy SF, Zlatanou A, Moore DJ, Chan DW, Qin J, et al. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell 2004; 117: 17–28. [DOI] [PubMed] [Google Scholar]
- Hao Y, Wang C, Cao B, Hirsch BM, Song J, Markowitz SD, et al. Gain of interaction with IRS1 by p110α-helical domain mutants is crucial for their oncogenic functions. Cancer Cell 2013; 23: 589–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landesman-Bollag E, Romieu-Mourez R, Song DH, Sonenshein GE, Cardiff RD, Seldin DC. Protein kinase CK2 in mammary gland tumorigenesis. Oncogene 2001; 20: 3247–57. [DOI] [PubMed] [Google Scholar]
- Daya-Makin M, Sanghera JS, Mogentale TL, Lipp M, Parchomchuk J, Hogg JC, et al. Activation of a tumor-associated protein kinase (p40TAK) and casein kinase 2 in human squamous cell carcinomas and adenocarcinomas of the lung. Cancer Res 1994; 54: 2262–8. [PubMed] [Google Scholar]
- Tornatore L, Sandomenico A, Raimondo D, Low C, Rocci A, Tralau-Stewart C, et al. Cancer-selective targeting of the NF-kappaB survival pathway with GADD45beta/MKK7 inhibitors. Cancer Cell 2014; 26: 495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JS, Litchfield DW. Too much of a good thing: the role of protein kinase CK2 in tumorigenesis and prospects for therapeutic inhibition of CK2. Biochim Biophys Acta 2008; 1784: 33–47. [DOI] [PubMed] [Google Scholar]
- Battistutta R, Sarno S, De Moliner E, Papinutto E, Zanotti G, Pinna LA. The replacement of ATP by the competitive inhibitor emodin induces conformational modifications in the catalytic site of protein kinase CK2. J Biol Chem 2000; 275: 29618–22. [DOI] [PubMed] [Google Scholar]
- Szyszka R1, Grankowski N, Felczak K, Shugar D. Halogenated benzimidazoles and benzotriazoles as selective inhibitors of protein kinases CK I and CK II from Saccharomyces cerevisiae and other sources. Biochem Biophys Res Commun 1995; 208: 418–24. [DOI] [PubMed] [Google Scholar]
- Vangrevelinghe E, Zimmermann K, Schoepfer J, Portmann R, Fabbro D, Furet P. Discovery of a potent and selective protein kinase CK2 inhibitor by high-throughput docking. J Med Chem 2003; 46: 2656–62. [DOI] [PubMed] [Google Scholar]
- Battistutta R, Cozza G, Pierre F, Papinutto E, Lolli G, Sarno S, et al. Unprecedented selectivity and structural determinants of a new class of protein kinase CK2 inhibitors in clinical trials for the treatment of cancer. Biochemistry 2011; 50: 8478–88. [DOI] [PubMed] [Google Scholar]
- Pierre F, Chua PC, O'Brien SE, Siddiqui-Jain A, Bourbon P, Haddach M, et al. Discovery and SAR of 5-(3-chlorophenylamino)benzo[c][2,6]naphthyridine-8-carboxylic acid (CX-4945), the first clinical stage inhibitor of protein kinase CK2 for the treatment of cancer. J Med Chem 2011; 54: 635–54. [DOI] [PubMed] [Google Scholar]
- Raaf J, Brunstein E, Issinger OG, Niefind K. The CK2 alpha/CK2 beta interface of human protein kinase CK2 harbors a binding pocket for small molecules. Chem Biol 2008; 15: 111–7. [DOI] [PubMed] [Google Scholar]
- Lu S, Li S, Zhang J. Harnessing allostery: a novel approach to drug discovery. Med Res Rev 2014; 34: 1242–85. [DOI] [PubMed] [Google Scholar]
- Bowman GR, Bolin ER, Hart KM, Maguire BC, Marqusee S. Discovery of multiple hidden allosteric sites by combining Markov state models and experiments. Proc Natl Acad Sci U S A 2015; 112: 2734–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins 2004; 57: 433–43. [DOI] [PubMed] [Google Scholar]
- Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature 2014; 508: 331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Cheng F, Song H, Lu W, Zhao J, An X, et al. Proteome-scale investigation of protein allosteric regulation perturbed by somatic mutations in 7,000 cancer genomes. Am J Hum Genet 2017; 100: 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Wang G, Li S, Liu X, Lu S, Chen Z, et al. ASD v3.0: unraveling allosteric regulation with structural mechanisms and biological networks. Nucleic Acids Res 2016; 44: D527–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Lu S, Huang Z, Liu X, Mou L, Luo Y, et al. Allosite: a method for predicting allosteric sites. Bioinformatics 2013; 29: 2357–9. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res 2000; 28: 235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya C, Armutlulu A, Ekesan S, Haliloglu T. MCPath: Monte Carlo path generation approach to predict likely allosteric pathways and functional residues. Nucleic Acids Res 2013; 41: W249–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23: 2947–8. [DOI] [PubMed] [Google Scholar]
- Palmieri L, Rastelli G. alphaC helix displacement as a general approach for allosteric modulation of protein kinases. Drug Discov Today 2013; 18: 407–14. [DOI] [PubMed] [Google Scholar]
- Hindie V, Stroba A, Zhang H, Lopez-Garcia LA, Idrissova L, Zeuzem S, et al. Structure and allosteric effects of low-molecular-weight activators on the protein kinase PDK1. Nat Chem Biol 2009; 5: 758–64. [DOI] [PubMed] [Google Scholar]
- Matsui H, Lazareno S, Birdsall NJ. Probing of the location of the allosteric site on m1 muscarinic receptors by site-directed mutagenesis. Mol Pharmacol 1995; 47: 88–98. [PubMed] [Google Scholar]
- Valentini G, Chiarelli L, Fortin R, Speranza ML, Galizzi A, Mattevi A. The allosteric regulation of pyruvate kinase. J Biol Chem 2000; 275: 18145–52. [DOI] [PubMed] [Google Scholar]
- Sarno S, Boldyreff B, Marin O, Guerra B, Meggio F, Issinger OG, et al. Mapping the residues of protein kinase CK2 implicated in substrate recognition: mutagenesis of conserved basic residues in the alpha-subunit. Biochem Biophys Res Commun 1995; 206: 171–9. [DOI] [PubMed] [Google Scholar]
- Liu S, Hsieh D, Yang YL, Xu Z, Peto C, Jablons DM, et al. Coumestrol from the national cancer Institute's natural product library is a novel inhibitor of protein kinase CK2. BMC Pharmacol Toxicol 2013; 14: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiganis T, House CM, Kemp BE. Protein kinase CK2: biphasic kinetics with peptide substrates. Arch Biochem Biophys 1996; 325: 289–94. [DOI] [PubMed] [Google Scholar]
- Bennett JT, Tan TY, Alcantara D, Tetrault M, Timms AE, Jensen D, et al. Mosaic activating mutations in FGFR1 cause encephalocraniocutaneous lipomatosis. Am J Hum Genet 2016; 98: 579–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Sekiguchi Y, Fukada H, Nakaniwa T, Tada T, Nakamura S, et al. A detailed thermodynamic profile of cyclopentyl and isopropyl derivatives binding to CK2 kinase. Mol Cell Biochem 2011; 356: 97–105. [DOI] [PubMed] [Google Scholar]
- Ferguson AD, Sheth PR, Basso AD, Paliwal S, Gray K, Fischmann TO, et al. Structural basis of CX-4945 binding to human protein kinase CK2. FEBS Lett 2011; 585: 104–10. [DOI] [PubMed] [Google Scholar]
- Dobrowolska G, Meggio F, Marin O, Lozeman FJ, Li D, Pinna LA, et al. Substrate recognition by casein kinase-II: the role of histidine-160. FEBS Lett 1994; 355: 237–41. [DOI] [PubMed] [Google Scholar]
- Niefind K, Yde CW, Ermakova I, Issinger OG. Evolved to be active: sulfate ions define substrate recognition sites of CK2alpha and emphasise its exceptional role within the CMGC family of eukaryotic protein kinases. J Mol Biol 2007; 370: 427–38. [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Keskitalo S, Van Drogen A, Nurkkala H, Vichalkovski A, Aebersold R, et al. The protein interaction landscape of the human CMGC kinase group. Cell Rep 2013; 3: 1306–20. [DOI] [PubMed] [Google Scholar]
- Badireddy S, Yunfeng G, Ritchie M, Akamine P, Wu J, Kim CW, et al. Cyclic AMP analog blocks kinase activation by stabilizing inactive conformation: conformational selection highlights a new concept in allosteric inhibitor design. Mol Cell Proteomics 2011; 10: M110.004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowsky JD, Burlingame MA, Wolan DW, McClendon CL, Jacobson MP, Wells JA. Turning a protein kinase on or off from a single allosteric site via disulfide trapping. Proc Natl Acad Sci U S A 2011; 108: 6056–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R, Tsai CJ. Unraveling structural mechanisms of allosteric drug action. Trends Pharmacol Sci 2014; 35: 256–64. [DOI] [PubMed] [Google Scholar]
- Horn JR, Shoichet BK. Allosteric inhibition through core disruption. J Mol Biol 2004; 336: 1283–91. [DOI] [PubMed] [Google Scholar]
- Bowman GR, Geissler PL. Equilibrium fluctuations of a single folded protein reveal a multitude of potential cryptic allosteric sites. Proc Natl Acad Sci U S A 2012; 109: 11681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]