Abstract
Biodiversity varies from place to place due to environmental and historical factors. To improve our understanding of how history and the environment influence observed patterns, we need to address the limitations of the most commonly used biodiversity metric, species richness. Here, we show that scale-dependent dissections of species richness into components of total abundance, species relative abundances and spatial aggregations of species reveal that two well-known biogeographic reef fish species richness gradients emerge from very different underlying component patterns. Latitudinal richness is underpinned by scale-independent patterns of total and relative abundances, suggesting ecological constraints scale up to determine abundances within communities. In contrast, the longitudinal gradient of species richness typically attributed to historical biogeography only emerges at the largest scale and is accompanied by a similar pattern of relative abundances, suggesting that site-to-site compositional variation leading to species aggregation (i.e. a component of β-diversity) underlies this gradient. Examining relationships among the components that underpin biodiversity gradients reveals new patterns that can better identify processes influencing patterns of biodiversity.
Keywords: biodiversity, macroecology, biogeography
1. Introduction
The heterogeneous distribution of biodiversity on the planet has been a topic of keen interest for centuries [1,2]. For example, why do some areas of the world have very few species (e.g. boreal forests with only a few species in several thousand hectares), whereas the same basal area in other parts of the world can have a great many species (e.g. tropical forests that can have hundreds or thousands of tree species in only a few hectares)? We know that both contemporary factors, such as energy availability and temperature (e.g. [3]), and historical factors, such as evolutionary time and diversification rates [4], play central roles in driving this heterogeneity, and that they can interact [5]. However, the continued use of species richness as the most common indicator of biodiversity has constrained our descriptions of observed patterns of biodiversity and weakened tests of possible mechanisms underlying the observed patterns [6,7].
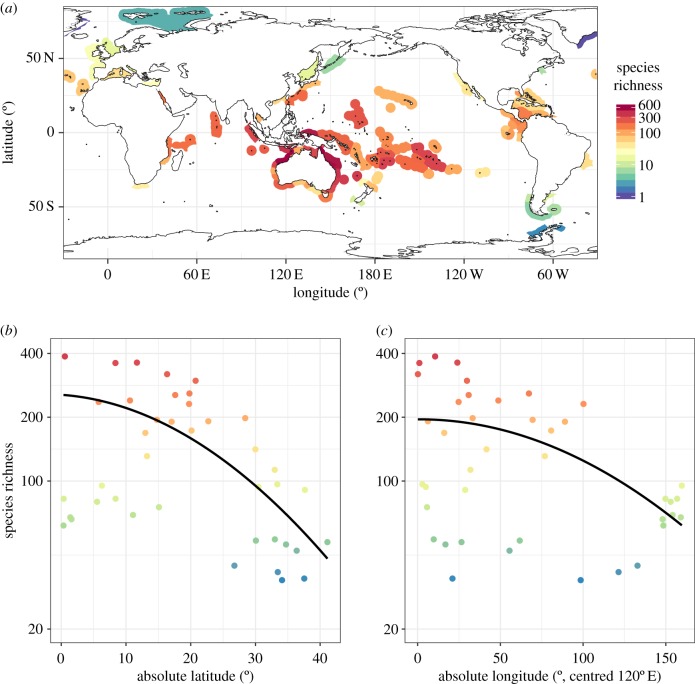
Species richness is simply the number of unique species in a sample. For reef fishes, when species richness is quantified at the scale of ecoregions (i.e. scales at which macroecological studies are usually conducted), two strong and well-known biogeographic gradients in species richness emerge (figure 1). First, there is a strong latitudinal gradient, which is often attributed to historical (e.g. time for speciation) as well as ecological drivers, such as energy availability [9,10]. Second, there is an equally strong longitudinal gradient centred in the tropics around the biodiversity hotspot in the Indo-Australian Archipelago (IAA), sometimes called the ‘coral triangle’ [11,12]. This pattern is typically explained as originating from historical processes such as high diversification rates and/or refugia during periods of environmental stress [9,12,13], and is not typically associated with contemporary gradients in energy or habitat. However, despite its ease of measurement and popularity, species richness is an extremely coarse metric, and such gradients can arise through changes in a number of components that determine species richness.
Figure 1.
Geographical gradients of reef fish species richness. (a) Map of total observed species richness within ecoregions [8]. (b) Declining species richness with increasing distance from the equator (i.e. absolute latitude). (c) Declining species richness with increasing distance from the centre of the coral triangle (absolute longitude centred on 120°E). Note that (a) shows the total species richness within ecoregions for the full dataset, whereas the points on panels (b) and (c) show only Indo-Pacific Ocean species richness within the 10 000 m2 ecoregion-scale grains used in all subsequent analyses; lines are predictions at the ecoregion scale from the best-fitting simultaneous autoregressive models. The colour scale is consistent for all panels.
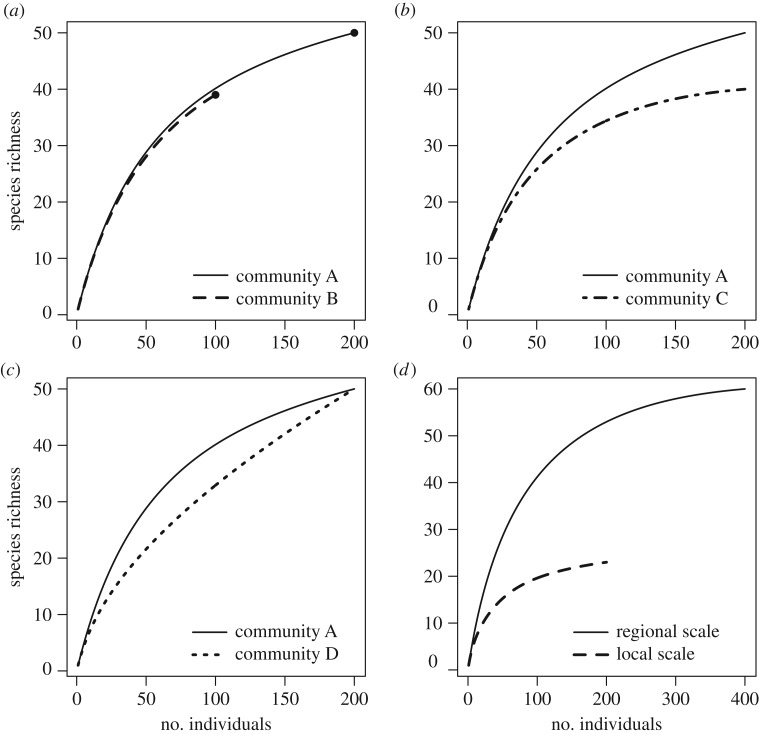
The components that underlie species richness include the numbers of individuals, as well as the relative abundances and spatial aggregations of species [7,14]. Through changes in these components, there are many pathways that can create variation in species richness (figure 2). For example, species richness can be higher in one community compared with another simply as a result of differences in the number of individuals and by sampling a high number of species, known as the ‘more individuals’ hypothesis (figure 2a) [16,17]. Species richness can also be higher in one community relative to another simply because of the presence of many rare species (figure 2b) [18]. Likewise, within a given sampling area, species richness will be higher when communities are more even (i.e. where no one species is overly dominant), because higher evenness leads to scale-dependent differences in species richness (figure 2c) [15]. Finally, changes in the spatial distribution of species (e.g. random versus aggregated) can result in changes to species richness (figure 2d) [7,15]. Because changes in any of these components may result in similar changes to species richness, traditional analyses would not differentiate them. Importantly, each of these pathways indicates a very different underlying structure of biodiversity, and examining them will provide new insights into the processes generating biodiversity gradients.
Figure 2.
Species richness as a function of the number of individuals (rarefaction curves). (a) Species in communities A and B have similar relative abundances (quantified in our analyses using the effective number of species conversion of the probability of interspecific encounter, ENSPIE), but different total numbers of species (black dots) due to more individuals in community A. (b) Communities A and C have similar total and relative abundances, but different species richness due to more rare species in community A. (c) Species in communities A and D have different relative abundances, leading to scale-dependent differences in species richness. (d) Individual-based rarefaction can also be used to infer within-species aggregation when relative abundances become more even with increasing scale [7,15].
In addition, species richness is a notoriously scale-sensitive metric that increases nonlinearly with sampling area (i.e. the ubiquitous species–area relationship). As a result of this nonlinear scaling, species richness is neither extensive nor intensive [19], meaning that species richness at large scales cannot simply be calculated as the sum of richness estimated at smaller scales (i.e. as an extensive variable), nor as a weighted average of small-scale richness estimates (i.e. as an intensive variable). This greatly limits traditional comparisons of species richness that are scale-agnostic, and that do not account for the components underlying species richness (e.g. total abundance of individuals, species relative abundances [7,19]).
Here we directly examine the components of species richness to gain deeper insight into the fundamental factors determining heterogeneity in biodiversity across broad biogeographic gradients. To do this, we used a unique dataset from a global survey of reef fishes on shallow hard substrate habitats from all major marine realms which contains individual-level, spatially explicit information on species abundances (i.e. the Reef Life Survey [20]). We dissected the latitudinal and longitudinal richness gradients into three main components: (i) changes to the total number of individuals; (ii) changes to the probability of interspecific encounter (a measure of the relative abundance of species, i.e. evenness [6]); and (iii) changes in richness. All components were calculated at four spatial scales to examine any scale dependencies. We show that these two well-known biogeographic gradients have divergent, scale-dependent changes in the components of species richness. Our results reveal that understanding the environmental and historical factors that promote patterns of biodiversity will be improved by scale-dependent quantification of the component changes that underpin large-scale biodiversity gradients.
2. Material and methods
(a). Marine biogeographic species richness gradients
We used the Reef Life Survey (RLS) data [20,21] to dissect the latitudinal and longitudinal biogeographic gradients of marine fish species richness into component parts: numbers of individuals, relative abundance of species and the spatial aggregation of species. The RLS data represent standardized quantitative estimates of reef fish abundance collected by trained recreational scuba divers on shallow hard-substrate habitats worldwide. Details of fish census methods, data quality and diver training are available in [20] and online at reeflifesurvey.com.
RLS data document the abundance of all individual fish along 500 m2 transects (2 × 250 m2 blocks). As we were interested in dissecting patterns of species richness, we discarded records where taxa were not recorded to species level. This resulted in a final dataset documenting 9 569 195 individuals from 2542 species observed along 8166 transects at 2804 sites worldwide, representing 89 ecoregions and 12 realms [8]. To simplify our analyses, we used absolute values of latitude (centred on the equator), and absolute longitude centred on 120°E. As (longitudinal) distance from the centre of the coral triangle (i.e. 120°E) may not be ecologically or evolutionarily relevant for fishes in the Atlantic Ocean, all models were fitted to data from the Indo-Pacific only (i.e. the Atlantic Ocean realms: the tropical Atlantic, temperate Northern Atlantic, temperate South America, and Arctic and Southern Ocean realms were removed before analysis). These Indo-Pacific data represent 8 946 214 individuals from 2203 species, observed along 7588 transects at 2492 sites within 66 ecoregions in 7 realms.
We quantified the total number of individuals as the sum of the abundance of all individuals of all species observed at a given scale (see below). Relative abundance was quantified using the effective number of species (ENS [22]) conversion of the probability of interspecific encounter (PIE [6]) so that 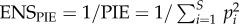 , where S is the number of species and pi is the proportion of the community represented by species i. The PIE represents the probability that two individuals randomly sampled from a community are different species [6], and is equal to the slope of the rarefaction curve at its base [23]. Our conversion to an effective number of species (or Hill number) means that it can be equivalently interpreted as the number of common species (i.e. ENSPIE is a diversity index of order q = 2 and the equivalent of the ENS conversion of Simpson's concentration [22]). While PIE (and therefore ENSPIE) are generally insensitive to sample grain and extent when individuals are randomly distributed through space, sample grain and extent can influence their values when individuals are spatially aggregated, resulting in scale-dependent estimates of PIE (and ENSPIE [7,23]). Accordingly, examining scale dependencies in ENSPIE along geographical gradients allows us to infer an effect of spatial aggregation on species richness patterns. Finally, species richness was quantified as the observed number of species at a given scale, and we additionally examined whether our results were sensitive to the number of undetected species using a non-parametric asymptotic richness estimator [24].
, where S is the number of species and pi is the proportion of the community represented by species i. The PIE represents the probability that two individuals randomly sampled from a community are different species [6], and is equal to the slope of the rarefaction curve at its base [23]. Our conversion to an effective number of species (or Hill number) means that it can be equivalently interpreted as the number of common species (i.e. ENSPIE is a diversity index of order q = 2 and the equivalent of the ENS conversion of Simpson's concentration [22]). While PIE (and therefore ENSPIE) are generally insensitive to sample grain and extent when individuals are randomly distributed through space, sample grain and extent can influence their values when individuals are spatially aggregated, resulting in scale-dependent estimates of PIE (and ENSPIE [7,23]). Accordingly, examining scale dependencies in ENSPIE along geographical gradients allows us to infer an effect of spatial aggregation on species richness patterns. Finally, species richness was quantified as the observed number of species at a given scale, and we additionally examined whether our results were sensitive to the number of undetected species using a non-parametric asymptotic richness estimator [24].
To examine scale dependencies of species richness patterns and of our dissected components, we aggregated transect-scale data to create samples at larger scales. Four scales were used for all analyses: 500 m2 (transect scale, no aggregation), 1000 m2 and 2000 m2 (aggregated within sites) and 10 000 m2 (aggregated within ecoregions). To control for the effects of bias associated with unequal sampling effort at the larger scales, we used sample-based rarefaction with 200 resamples at each of the site scales (1000 m2 and 2000 m2), and the ecoregion (10 000 m2) scale. To aggregate data at the site scales, the data were first reduced to sites with at least two (1000 m2) or four (2000 m2) unique transects. Then, either two or four transects were randomly resampled from these reduced data 200 times (without replacement), to create a new sample; species abundances were then aggregated at the new scale, and species richness and our dissection components (numbers of individuals, ENSPIE) were then calculated as the average over all of the resamples. Similarly, at the ecoregion-scale (10 000 m2), data were first reduced to ecoregions where at least 20 transects were sampled; 20 transects were then randomly resampled 200 times (without replacement), species abundances aggregated at the new larger scale, and diversity components calculated as the average over all of the resamples.
(b). Statistical analyses
As we were interested in comparing the biogeographic patterns of species richness with its component parts (total abundance, ENSPIE), we wanted all response variables to be on the same scale. Preliminary analyses showed log-transformed response variables to better meet the assumptions of our statistical models (particularly homoscedasticity) compared with untransformed response variables. Accordingly, we present results and analyses where all response variables were log-transformed before model fitting. Additionally, as our response variables were spatially auto-correlated, we used simultaneous autoregressive (SAR) models [25] to examine how they change with latitude, longitude and scale. We fitted SAR models that incorporate a spatially dependent error term that assumes the autoregressive process is found only in the error term. We used Aikaike's information criterion (AIC) to compare the fit of models with different spatial weights matrices constructed using different cut-off distances for determining neighbours; specifically, all neighbours within (i) the mean distance to nearest neighbour (3.75 km), (ii) 50 km, (iii) 100 km and (iv) 200 km. Preliminary analyses showed that all distances removed spatial autocorrelation in model residuals (electronic supplementary material, table S1), but that the 50 km cut-off distance for determining neighbours was strongly supported (AIC weight >99% support) as providing the best-fitting spatial weights matrix for all of our diversity components (electronic supplementary material, table S1).
To examine scale dependencies in biogeographic patterns of species richness and its component parts, we fitted SAR models with parameters for interactions between latitude and scale, and between longitude and scale. Preliminary analyses of linear models showed that there were nonlinear patterns remaining in the residuals, so we fitted second-order trend surfaces [26] with the additional scale-dependent parameters. This resulted in statistical models of the form
where the covariates x, y and s represent absolute longitude, absolute latitude and a categorical variable denoting scale (four levels: 500 m2, 1000 m2, 2000 m2, 10 000 m2), respectively; the β-values are estimated regression coefficients, λ is the estimated spatial autoregressive coefficient, W is the spatial weights matrix, u the spatially dependent error term and ε the (spatially) independent error term. To examine scale dependencies of geographical patterns (i.e. along longitudinal and latitudinal gradients), we set specific β coefficients to zero (e.g. setting β7 = 0 or β8 = 0 removes the scale dependency of the longitudinal and latitudinal gradients, respectively); as these models are nested within our full model, we used likelihood ratio tests assuming chi-square error and a p-value threshold of 0.05 to evaluate the significance of individual terms. Similarly, we used likelihood ratio tests to simplify our full model and to determine the simplest model for each response (see electronic supplementary material, tables S2–S4 for model selection statistics and parameter estimates from simplified models).
To further examine the role of species aggregation in driving the observed patterns of species richness, we quantified the log-ratio of species richness and ENSPIE estimated at increasing scales [7,23]. To calculate the ratios, we first calculated the mean values of species richness and ENSPIE at the smaller scales (500 m2, 1000 m2 and 2000 m2) within each ecoregion, and then calculated the ratio for each ecoregion where we had observations at every scale (30 ecoregions in 9 realms). Similar to previous analyses, data were reduced to Indo-Pacific ecoregions for the fitting of models with both longitude and latitude, resulting in an analysis of 26 ecoregions in 7 realms. We examined the geographical patterns of 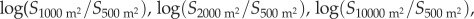
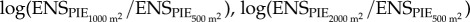 and
and 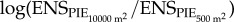 using linear models. Preliminary analyses showed that the residuals of linear models of were not spatially auto-correlated, but were heteroscedastic with respect to scale; additionally, the species richness model showed some residual non-linearity. Therefore, we fitted models with interactions between latitude and scale, longitude and scale, and latitude and longitude with a variance covariate for scale to deal with heteroscedasticity [27]; and the species richness log-ratio model additionally included second-order terms on latitude and longitude to address the non-linearity of the observed patterns. Models were fitted using maximum likelihood, and we assessed the significance of model terms using likelihood ratio tests (see electronic supplementary material, tables S5–S8 for model selection statistics and parameter estimates of simplified models).
using linear models. Preliminary analyses showed that the residuals of linear models of were not spatially auto-correlated, but were heteroscedastic with respect to scale; additionally, the species richness model showed some residual non-linearity. Therefore, we fitted models with interactions between latitude and scale, longitude and scale, and latitude and longitude with a variance covariate for scale to deal with heteroscedasticity [27]; and the species richness log-ratio model additionally included second-order terms on latitude and longitude to address the non-linearity of the observed patterns. Models were fitted using maximum likelihood, and we assessed the significance of model terms using likelihood ratio tests (see electronic supplementary material, tables S5–S8 for model selection statistics and parameter estimates of simplified models).
As we used a resampling process to generate our dissection metrics, differences in the total number of sites within the largest (ecoregion) scale meant that the geographical area (i.e. the extent) from which we resampled differed between ecoregions. To examine whether these differing extents influenced our results, we refitted all models with extent included as an additional covariate. Only sites in the RLS dataset have a unique geographical coordinate, so we could only calculate geographical extent at the ecoregion scale (where we resampled from multiple sites). At the ecoregion scale, extent was calculated as geographical area (i.e. area on an ellipsoid) within a convex hull that bounded all the sites from which we resampled within each ecoregion. Extent was log-transformed prior to model fitting. Including extent had no qualitative effect on our main results (electronic supplementary material, table S9).
All data manipulation and analyses used R 3.3.1 [28]. We used the dplyr package for data manipulation [29]; data analyses were conducted in spdep [30,31] and nlme [32]; and plots were generated using AICcmodavg [33], ggplot2 [34] and meowR packages [35].
3. Results
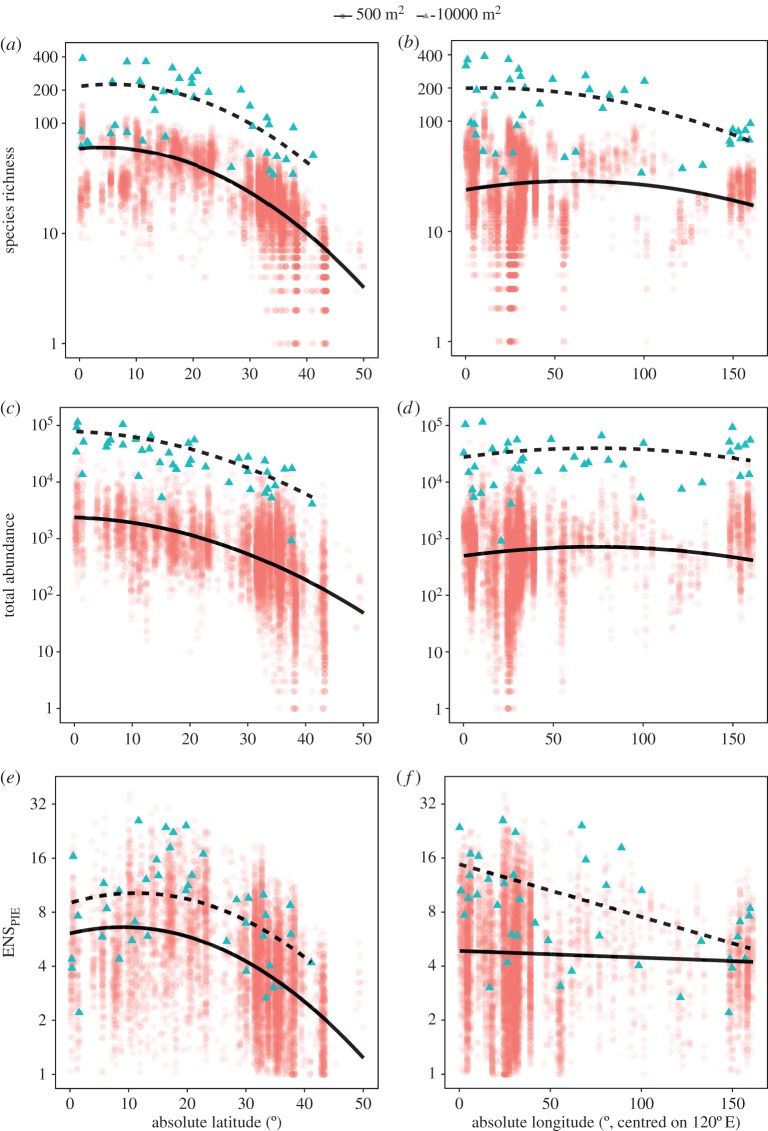
Across latitudes, we find the expected decrease in species richness (figure 3a) accompanied by a similar pattern in the total numbers of individuals (figure 3c). Latitudinal decreases in both the total numbers of individuals and species richness are scale-independent (i.e. the latitude × scale interaction was not significant: likelihood ratio test; individuals: χ2 = 4.91, d.f. = 3, p = 0.18; species richness: χ2 = 1.18, d.f. = 3, p = 0.76), meaning that across all scales a sampling effect (i.e. the more individuals hypothesis) is an important component of the observed latitudinal gradient in marine fishes species richness. In contrast, total numbers of individuals do not change across longitudes or with scale (figure 3d; longitude × scale interaction: χ2 = 1.01, d.f. = 3, p = 0.8), confirming that the longitudinal gradient in richness is not formed by gradients in the total numbers of individuals, and is not likely to be associated with changes in energy or habitat availability (electronic supplementary material, figures S2–S3).
Figure 3.
Scale-dependent dissection of geographical gradients of reef fish species richness into component parts: latitudinal gradients of (a) species richness, (c) total numbers of individuals and (e) ENSPIE (effective number of species conversion of the probability of interspecific encounter), and longitudinal gradients of (b) species richness, (d) total numbers of individuals and (f) ENSPIE. Points depict data from the Indo-Pacific only, lines depict predictions of the best-fitting simultaneous autoregressive models at the transect and ecoregion scales; predictions for latitudinal gradients were made with longitude set to its mean value for each scale; similarly, longitudinal predictions were generated with latitude set to its mean calculated for each scale. Panels depict two scales for clarity; however, all models were fitted to four scales: 500 m2 (transect), 1000 m2 (site), 2000 m2 (site) and 10 000 m2 (ecoregion) (see electronic supplementary material, figure S1). (Online version in colour.)
At local scales, community composition and evenness are thought to reflect the outcome of environmental (abiotic) and biotic filters, which act to determine which members of the regional species pool occupy a given community, and in what proportions [36]. Higher values of ENSPIE indicate more even communities, and changes in the ENSPIE at small scales indicate that the outcomes of species coexistence are changing to affect evenness. Here, we find contrasting patterns across latitudes (figure 3e) and longitudes (figure 3f). Across latitudes, patterns of the ENSPIE closely resemble the latitudinal gradients of the total numbers of individuals and species richness (figure 3a,c,e), and are scale-independent (χ2 = 4.94, d.f. = 3, p = 0.18). Combined, this lack of strong scale-dependence suggests that latitudinal gradients are most likely to be driven by constraints imposed by contemporary ecological factors that act similarly across scales to determine the size of, and relative abundances within, communities. In contrast, there is no change in the ENSPIE at local scales across longitudes (figure 3f). The similar longitudinal patterns of our dissection components imply that fish communities observed along single transects in the IAA biodiversity hotspot and, for example, in the comparatively species poor French Polynesia may be remarkably similar in terms of total abundance, relative species abundances and species richness.
Across longitudes both species richness (figure 3b) and the ENSPIE (figure 3f) show marked scale dependence (species richness: χ2 = 14.25, d.f. = 3, p = 0.003; ENSPIE: χ2 = 16.42, d.f. = 3, p = 0.001). Such scale-dependent patterns in species richness, whereby gradients only emerge at large scales, are due to changes to one or both of two of the underlying diversity components: rare species being sampled with increases in scale and/or large-scale within-species aggregation. Increasing values of the ENSPIE with increasing sample grain means that new, relatively common species are being sampled as scale increases. Species contributing to such a scale-dependent pattern of the ENSPIE are probably aggregated in space [7,23]. Here, we found ENSPIE and species richness show similar, scale-dependent patterns across longitudes (i.e. only decreasing with increasing distance from the IAA at the largest scale), suggesting that both within-species aggregation and rare species are contributing to the longitudinal diversity gradient.
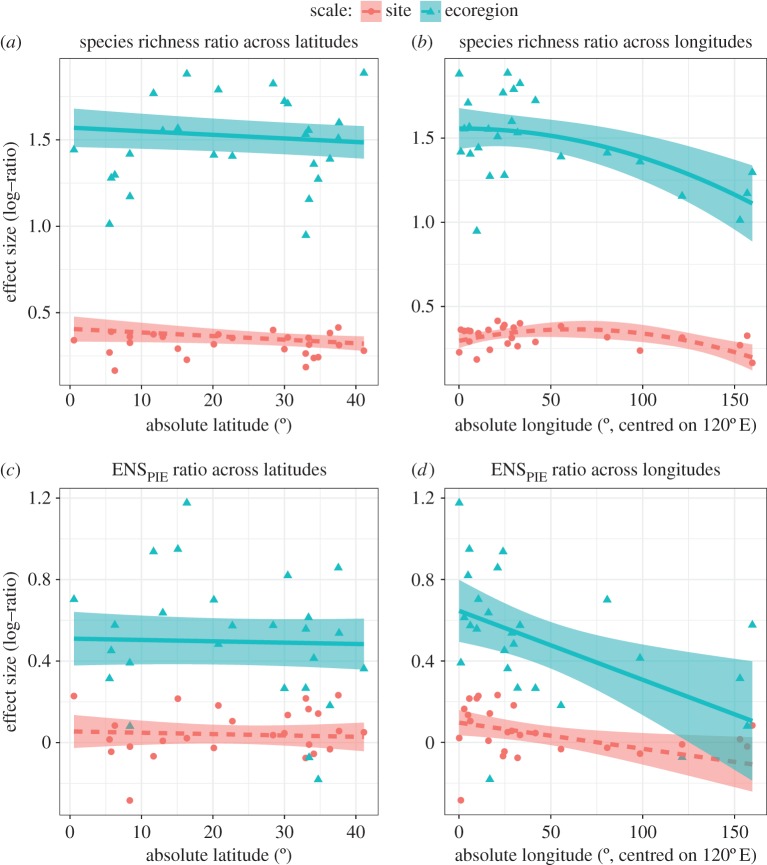
We further visualized how the spatial aggregation of common and rare species factors into species richness gradients by quantifying the ratios of species richness (figure 4a,b) and ENSPIE (figure 4c,d) at increasing scales. The species richness ratio is multiplicative β-diversity (i.e. the ratio of γ/α), and is more sensitive to rare species than the ENSPIE ratio [22]; positive values mean that new, relatively rare species are being sampled as the scale of the sample increases. The ENSPIE ratio measures how evenness is changing as the scale of the sample grain increases (termed ‘beta-evenness’ by Olszewski [23], and conceptually related to the beta-diversity of common species [22]). Positive values of the ENSPIE ratio indicate that new, relatively common species are being sampled as the scale of the sample increases. We find that both ratios are more strongly scale-dependent across longitudes than latitudes (figure 4; species richness: longitude × scale: χ2 = 9.2, d.f. = 2, p = 0.01; latitude × scale: χ2 = 0.58, d.f. = 2, p = 0.75; ENSPIE: longitude × scale: χ2 = 7.3, d.f. = 2, p = 0.03; latitude × scale: χ2 = 5.84, d.f. = 2, p = 0.05). Additionally, we find that the rate of decay with increasing longitudinal distance from the coral triangle is greater for the ENSPIE ratio (figure 4d) compared with the species richness ratio (figure 4b). The steep gradient of the ENSPIE ratio suggests that within the IAA biodiversity hotspot, each site may contain different species with relatively high abundances that contribute to the high spatial turnover in community structure, while sites at more remote pacific islands (e.g. Easter Island) may all be dominated by the same few species. Moreover, these analyses show that spatial turnover of both common and rare species contributes to the longitudinal decline in species richness, and emphasizes the contributions of regional-scale aggregation to the longitudinal, but not the latitudinal, diversity gradient.
Figure 4.
Scale-dependent geographical patterns of the species richness and ENSPIE ratios: the species richness ratio as a function of (a) absolute latitude and (b) absolute longitude, and the ENSPIE ratio as a function of (c) absolute latitude and (d) absolute longitude. The site and ecoregion scales are the log ratios estimated at site (1000 m2) and ecoregion (10 000 m2) over the transect-scale (500 m2) estimate, respectively. We show only two scales for clarity (see Material and methods). (Online version in colour.)
4. Discussion
Our dissection of richness into components of the numbers of individuals and species relative abundances shows that at the scales examined here, the latitudinal richness gradient is underpinned by scale-insensitive component patterns. This means that processes affecting the total numbers of individuals and species relative abundances are changing similarly across scales. In particular, the pattern of richness increasing with the total number of individuals emphasizes a role for contemporary ecological factors, such as available energy [16,37,38], and higher evenness at low latitudes is associated with processes that allow more species to coexist at small scales. For example, resource partitioning and ecological specialization are often thought to be greater in the tropics (e.g. [39]), though empirical evidence for this is mixed and there have been very few tests of this hypothesis in marine systems [40]. The strong complementary patterns between species richness and both evenness and the number of individuals make alternative, historical hypotheses for latitudinal gradients in marine fishes species richness (such as differences in evolutionary time or diversification rates [4]) less plausible. Nevertheless, the latitudinal patterns of total abundance and evenness observed here could also result from higher diversification rates at low latitudes that allow for finer niche partitioning by constituent species, and hence more individuals overall.
When gradients of species richness are scale-dependent and emerge only at larger spatial scales, we can infer that they are caused by some combination of rare species and/or within-species aggregation. Teasing apart such changes in the components of richness is essential for increasing our understanding of contemporary biodiversity patterns. In particular, different processes probably drive increased numbers of rare species versus within-species aggregations. Increased numbers of rare species at low latitudes have been hypothesized to be associated with a greater availability of niches, increased specialization, and temperature-dependence of ecological and evolutionary rates [37,38]. In contrast, large-scale aggregation is associated with very different processes, such as habitat heterogeneity, spatial frequency dependence, allopatric speciation and/or dispersal limitation [41,42]. Here, our finding of scale-dependent longitudinal patterns of species richness and ENSPIE, and their ratios, means that new species, both rare and common, are sampled with increases in scale near the IAA biodiversity hotspot. Hence, both common and rare species are more spatially aggregated near the IAA biodiversity hotspot, and both within-species aggregation and rare species are contributing to the longitudinal diversity gradient of marine fish.
Our scale-dependent dissections of species richness allowed us to show that similar large-scale richness gradients of reef fishes are underpinned by very different component patterns. Importantly, the identification of different scale-dependent changes in the components indicates that divergent underlying processes probably drive the latitudinal and longitudinal gradients in reef fish species richness. While it is accepted that historical processes of diversification dynamics as well as range expansion from refugia cause longitudinal diversity gradients [9,13], the scale dependence of the ENSPIE shows that for reef fishes, these processes mostly promote coexistence regionally through a component of β-diversity, quantified as within-species aggregation (e.g. via spatial niche partitioning, dispersal limitation, allopatric speciation), and do not trickle down to influence local-scale patterns. In contrast, latitudinal gradients are largely insensitive to scale, and imply that bottom-up constraints on total and relative abundance (e.g. available energy) scale up to shape the species pool.
There are three main sources of patchiness in our data. The first is that the number of observations (transects) is different in the different regions. This was controlled for with the resampling process used to generate the larger scales, which is the equivalent of sample-based rarefaction, and effectively standardizes sampling within scales. The second source of patchiness is the location of transects within regions. For example, for a given sampling effort, regions that are larger may sample more of the environmental heterogeneity and hence contain higher spatial turnover and aggregation. We examined this effect of patchiness at the largest (ecoregion) scale (where geographical extent of the region from which we resample may differ) by refitting the models including the different extents from which the data were resampled. Model selection presented in the supplementary material suggests that the differing extents do not qualitatively alter our main findings (electronic supplementary material, table S9). The third source of patchiness is the large gaps, where we have no data from entire regions. For example, we do not have data from Borneo and the Philippines. While it is undeniable including data from these regions would be desirable, we have no reason to believe that adding these missing locations would have changed the results.
In all, we have demonstrated that understanding variation in biodiversity is a more complex endeavour than simply measuring and comparing patterns of species richness at a single spatial scale. Our results illustrate that examining how species-relative abundances and spatial aggregations change will be required to truly understand how species richness varies across the planet, as well as to open a window onto why those values are changing. Knowing which components underpin variation in species richness is vital for improving how we conserve and manage biodiversity, as well as for understanding its potential response in the face of ongoing environmental change. For example, if regional species richness is largely maintained by rare species or intraspecific aggregations, protected areas will need to be larger relative to situations where species richness is simply a function of more individuals randomly dispersed in space. Quantifying patterns in the components of richness provides important information for distinguishing among competing hypothesized processes driving biodiversity gradients, and promises to improve our understanding of the relative roles of contemporary and historical factors in shaping heterogeneous distributions of biodiversity.
Supplementary Material
Acknowledgements
We thank the many Reef Life Survey (RLS) divers who collected the data, and acknowledge Graham Edgar and Rick Stuart-Smith for leading the collection and organization of the data, and thank them for making it available from reeflifesurvey.com.
Ethics
Fieldwork was conducted according to local legislation.
Data accessibility
Data are available from reeflifesurvey.com.
Authors' contributions
S.A.B., J.B. and J.M.C. conceived the research; S.A.B. analysed the data; all authors wrote the manuscript.
Competing interests
The authors declare no competing interests.
Funding
This research was supported by a grant from the German-Israeli Foundation (GIF) for Scientific Research and Development (project no. I-2373-203.13/2014), and the Israel Science Foundation (ISF) grant no. 1356/15 to J.B. J.M.C. and S.A.B. gratefully acknowledge the support of the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, funded by the German Research Foundation (FZT 118).
References
- 1.Darwin C. 1859. On the origin of species. London, UK: John Murray. [Google Scholar]
- 2.Gaston KJ. 2000. Global patterns in biodiversity. Nature 405, 220–227. ( 10.1038/35012228) [DOI] [PubMed] [Google Scholar]
- 3.Wang Z, Brown JH, Tang Z, Fang J. 2009. Temperature dependence, spatial scale, and tree species diversity in eastern Asia and North America. Proc. Natl Acad. Sci. USA 106, 13 388–13 392. ( 10.1073/pnas.0905030106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mittelbach GG, et al. 2007. Evolution and the latitudinal diversity gradient: speciation, extinction and biogeography. Ecol. Lett. 10, 315–331. ( 10.1111/j.1461-0248.2007.01020.x) [DOI] [PubMed] [Google Scholar]
- 5.Ricklefs RE. 2015. Intrinsic dynamics of the regional community. Ecol. Lett. 18, 497–503. ( 10.1111/ele.12431) [DOI] [PubMed] [Google Scholar]
- 6.Hurlbert SH. 1971. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52, 577–586. ( 10.2307/1934145) [DOI] [PubMed] [Google Scholar]
- 7.Chase JM, Knight TM. 2013. Scale-dependent effect sizes of ecological drivers on biodiversity: why standardised sampling is not enough. Ecol. Lett. 16, 17–26. ( 10.1111/ele.12112) [DOI] [PubMed] [Google Scholar]
- 8.Spalding MD, et al. 2007. Marine ecoregions of the world: a bioregionalization of coastal and shelf areas. Bioscience 57, 573–583. ( 10.1641/B570707) [DOI] [Google Scholar]
- 9.Mora C, Chittaro PM, Sale PF, Kritzer JP, Ludsin SA. 2003. Patterns and processes in reef fish diversity. Nature 421, 933–936. ( 10.1038/nature01393) [DOI] [PubMed] [Google Scholar]
- 10.Bellwood D, Hughes T, Connolly SR, Tanner J. 2005. Environmental and geometric constraints on Indo-Pacific coral reef biodiversity. Ecol. Lett. 8, 643–651. ( 10.1111/j.1461-0248.2005.00763.x) [DOI] [Google Scholar]
- 11.Bellwood DR, Hughes TP. 2001. Regional-scale assembly rules and biodiversity of coral reefs. Science 292, 1532–1535. ( 10.1126/science.1058635) [DOI] [PubMed] [Google Scholar]
- 12.Renema W, et al. 2008. Hopping hotspots: global shifts in marine biodiversity. Science 321, 654–657. ( 10.1126/science.1155674) [DOI] [PubMed] [Google Scholar]
- 13.Pellissier L, et al. 2014. Quaternary coral reef refugia preserved fish diversity. Science 344, 1016–1019. ( 10.1126/science.1249853) [DOI] [PubMed] [Google Scholar]
- 14.Gotelli NJ, Colwell RK. 2001. Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol. Lett. 4, 379–391. ( 10.1046/j.1461-0248.2001.00230.x) [DOI] [Google Scholar]
- 15.He F, Legendre P. 2002. Species diversity patterns derived from species–area models. Ecology 83, 1185–1198. [Google Scholar]
- 16.Wright DH. 1983. Species-energy theory: an extension of species–area theory. Oikos 41, 496–506. ( 10.2307/3544109) [DOI] [Google Scholar]
- 17.Srivastava DS, Lawton JH. 1998. Why more productive sites have more species: an experimental test of theory using tree-hole communities. Am. Nat. 152, 510–529. [DOI] [PubMed] [Google Scholar]
- 18.McGill BJ, et al. 2007. Species abundance distributions: moving beyond single prediction theories to integration within an ecological framework. Ecol. Lett. 10, 995–1015. ( 10.1111/j.1461-0248.2007.01094.x) [DOI] [PubMed] [Google Scholar]
- 19.McGill BJ. 2011. Linking biodiversity patterns by autocorrelated random sampling. Am. J. Bot. 98, 481–502. ( 10.3732/ajb.1000509) [DOI] [PubMed] [Google Scholar]
- 20.Edgar GJ, Stuart-Smith RD. 2014. Systematic global assessment of reef fish communities by the Reef Life Survey program. Sci. Data 1, 140007 ( 10.1038/sdata.2014.7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar GJ, Stuart-Smith RD. 2016. Reef Life Survey (RLS): global reef fish dataset. See http://catalogue-rls.imas.utas.edu.au/geonetwork/srv/en/metadata.show?uuid=9c766140-9e72-4bfb-8f04-d51038355c5.
- 22.Jost L. 2006. Entropy and diversity. Oikos 113, 363–375. ( 10.1111/j.2006.0030-1299.14714.x) [DOI] [Google Scholar]
- 23.Olszewski TD. 2004. A unified mathematical framework for the measurement of richness and evenness within and among multiple communities. Oikos 104, 377–387. ( 10.1111/j.0030-1299.2004.12519.x) [DOI] [Google Scholar]
- 24.Chao A. 1984. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11, 265–270. [Google Scholar]
- 25.Kissling WD, Carl G. 2008. Spatial autocorrelation and the selection of simultaneous autoregressive models. Glob. Ecol. Biogeogr. 17, 59–71. ( 10.1111/j.1466-8238.2007.00379.x) [DOI] [Google Scholar]
- 26.Legendre P, Legendre L. 1998. Numerical ecology, 2nd edn Amsterdam, the Netherlands: Elsevier. [Google Scholar]
- 27.Zuur AF, Ieno EN, Walker NJ, Saveliev AA, Smith GM. 2009. Mixed effects models and extensions in ecology with R. Berlin, Germany: Springer. [Google Scholar]
- 28.R Development Core Team. 2016. R: a language and environment for statistical computing. Vienna, Austria: R Development Core Team. [Google Scholar]
- 29.Wickham H, Francois R. 2016. Dplyr: a grammar of data manipulation, 0.5.0 edn. See https://cran.r-project.org/web/packages/dplyr/index.html . [Google Scholar]
- 30.Bivand R, Hauke J, Kossowski T. 2013. Computing the Jacobian in Gaussian spatial autoregressive models: an illustrated comparison of available methods. Geogr. Anal. 45, 150–179. ( 10.1111/gean.12008) [DOI] [Google Scholar]
- 31.Bivand R, Piras G. 2015. Comparing implementations of estimation methods for spatial econometrics. J. Stat. Softw. 63, 1–36. ( 10.18637/jss.v063.i18) [DOI] [Google Scholar]
- 32.Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team. 2016 Nlme: linear and nonlinear mixed effects models, 3.1-128 edn. See https://cran.r-project.org/web/packages/nlme/nlme.pdf. [Google Scholar]
- 33.Mazerolle MJ. 2016. AICcmodavg: model selection and multimodel inference based on (Q)AIC(c), 2.0-4 edn. See https://cran.r-project.org/web/packages/AICcmodavg/index.html . [Google Scholar]
- 34.Wickham H. 2009. ggplot2: elegant graphics for data analysis. New York, NY: Springer. [Google Scholar]
- 35.Byrnes J. 2016 meowR: marine ecoregions of the world in R, 0.6.2 edn. See https://github.com/jebyrnes/meowR . [Google Scholar]
- 36.HilleRisLambers J, Adler P, Harpole W, Levine J, Mayfield M. 2012. Rethinking community assembly through the lens of coexistence theory. Annu. Rev. Ecol. Evol. Syst. 43, 227 ( 10.1146/annurev-ecolsys-110411-160411) [DOI] [Google Scholar]
- 37.Rohde K. 1992. Latitudinal gradients in species diversity: the search for the primary cause. Oikos 65, 514–527. ( 10.2307/3545569) [DOI] [Google Scholar]
- 38.Brown JH. 2014. Why are there so many species in the tropics? J. Biogeogr. 41, 8–22. ( 10.1111/jbi.12228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacArthur RH. 1972. Geographical ecology: patterns in the distribution of species. Princeton, NJ: Princeton University Press. [Google Scholar]
- 40.Moles AT, Ollerton J. 2016. Is the Notion that species interactions are stronger and more specialized in the tropics a zombie idea? Biotropica 48, 141–145. ( 10.1111/btp.12281) [DOI] [Google Scholar]
- 41.Tuomisto H, Ruokolainen K, Yli-Halla M. 2003. Dispersal, environment, and floristic variation of western Amazonian forests. Science 299, 241–244. ( 10.1126/science.1078037) [DOI] [PubMed] [Google Scholar]
- 42.Myers JA, Chase JM, Jiménez I, Jørgensen PM, Araujo-Murakami A, Paniagua-Zambrana N, Seidel R. 2013. Beta-diversity in temperate and tropical forests reflects dissimilar mechanisms of community assembly. Ecol. Lett. 16, 151–157. ( 10.1111/ele.12021) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from reeflifesurvey.com.