Abstract
For organs to achieve their proper size, the processes of stem cell renewal and differentiation must be tightly regulated. We previously showed that in the developing kidney, Wnt9b regulates distinct β-catenin-dependent transcriptional programs in the renewing and differentiating populations of the nephron progenitor cells. How β-catenin stimulated these two distinct programs was unclear. Here, we show that β-catenin cooperates with the transcription factor Myc to activate the progenitor renewal program. Although in multiple contexts Myc is a target of β-catenin, our characterization of a cell type-specific enhancer for the Wnt9b/β-catenin target gene Fam19a5 shows that Myc and β-catenin cooperate to activate gene expression controlled by this element. This appears to be a more general phenomenon as we find that Myc is required for the expression of every Wnt9b/β-catenin progenitor renewal target assessed as well as for proper nephron endowment in vivo. This study suggests that, within the developing kidney, tissue-specific β-catenin activity is regulated by cooperation with cell type-specific transcription factors. This finding not only provides insight into the regulation of β-catenin target genes in the developing kidney, but will also advance our understanding of progenitor cell renewal in other cell types/organ systems in which Myc and β-catenin are co-expressed.
KEY WORDS: Wnt, Myc, β-Catenin, Nephron progenitors, Stem cells, Fam19a5, Mouse
Summary: Characterization of a cell type-specific enhancer for the Wnt9b/β-catenin target gene Fam19a5 shows that Myc is necessary for activation of the β-catenin nephron progenitor cell renewal program.
INTRODUCTION
The metanephric kidney consists of thousands of individual functional units known as nephrons. Nephron number (also known as nephron endowment) is established during embryonic development as the result of reciprocal interactions between two tissues: the ureteric bud (UB) and the metanephric mesenchyme (MM). In the mouse, this process begins around embryonic day (E) 10.5 when a caudal segment of the Wolffian duct invades the surrounding MM forming the UB. Signals produced by the MM promote reiterative branching morphogenesis of the UB, and signals from the UB support survival and self-renewal of nephron progenitor cells (NPCs) within the MM. In addition, the UB produces signals that induce a subset of the progenitors to undergo a mesenchymal-to-epithelial transition to form a structure known as a renal vesicle, which will eventually undergo morphogenesis and transform into the nephron (Little and McMahon, 2012).
We previously showed that within the cap mesenchyme of the embryonic kidney, Wnt9b plays dual roles in nephron progenitor renewal and differentiation (Karner et al., 2011). This is accomplished through activation of at least two distinct transcriptional programs within the responding cells. Both programs require β-catenin. Indeed, β-catenin plays multiple roles in distinct cellular lineages of the embryonic kidney, including differentiation of the stromal fibroblasts and collecting ducts (Stark et al., 1994; Yu et al., 2009; Marose et al., 2008; Carroll et al., 2005; Trowe et al., 2012). In addition to its normal role in development, dysregulated β-catenin activity has been implicated in numerous renal pathologies, including Wilms' tumors, polycystic kidney disease, diabetic nephropathy and renal fibrosis (Benzing et al., 2007; Tycko et al., 2007; Pulkkinen et al., 2008).
β-Catenin functions by interacting with members of the Lef/Tcf family of DNA-binding co-regulators. Under baseline conditions, free β-catenin is bound by a group of proteins known as the destruction complex. Proteins within the complex, including Gsk3β and casein kinase, phosphorylate serine and threonine residues on β-catenin, which leads to its degradation. Upon binding of ligands to their receptors, the destruction complex is inactivated and β-catenin is stabilized allowing it to move into the nucleus to interact with Lef/Tcfs to drive expression of target genes.
β-Catenin plays essential roles in the development of most, if not all, tissues in the mouse. Given the numerous and varied roles in normal and pathological biology, β-catenin must be considered a permissive factor, meaning it does not dictate a specific response within a cell, but instead acts together with other cell type-specific factors to regulate cellular identity. However, several β-catenin target genes have been identified as ‘universal’ targets, meaning they are expressed in all tissues where β-catenin is active. Examples include Axin2, Nkd1/2, cyclin D1 and Myc. How β-catenin might play pleiotropic roles, but also have universal target genes, is unclear. One possibility is that β-catenin (in cooperation with Lef/Tcfs) affects expression of a limited set of universal β-catenin targets and the combination of these targets along with other intracellular factors determines cell fate. Another possibility is that β-catenin has some universal targets that require only β-catenin/Lef/Tcf, and other targets that require additional cell type-specific transcription factors. This model is supported by the observation that Lef/Tcf-binding sites alone are sufficient to drive expression of reporter genes in numerous mouse tissues (DasGupta and Fuchs, 1999; Maretto et al., 2003; Currier et al., 2010; Ferrer-Vaquer et al., 2010). A final possibility is that β-catenin requires different tissue-specific transcription factors (in addition to the Lef/Tcfs) in all cellular contexts to activate target genes and that there are no truly universal targets.
In this study, we use the developing mouse kidney to gain insights into how β-catenin functions pleiotropically, by investigating the mechanisms underlying β-catenin regulation of specific gene programs in the nephron progenitors. Through characterization of a cell type-specific enhancer element for the Wnt9b/β-catenin target gene Fam19a5 (also known as Tafa5) that is sufficient to drive gene expression within the renewing NPCs, we find that c- and N-Myc (also known as Mycn), rather than being β-catenin targets, cooperate with β-catenin to activate the Wnt9b/β-catenin progenitor renewal program in vivo. Not only does this analysis shed light on the cell type-specific regulation of β-catenin activity within the developing kidney but it also provides insight into how β-catenin functions pleiotropically.
RESULTS
Identification of a Wnt9b/β-catenin-dependent nephron progenitor cell enhancer
We previously showed that Wnt9b/β-catenin regulates both renewal and differentiation of NPCs by activating expression of two distinct classes of target genes in spatially distinct cells (Karner et al., 2011). The mechanism regulating the expression of these target genes in distinct cell types is unknown. To determine whether Lef/Tcf-binding sites alone were sufficient to drive mRNA expression in either cell type, we analyzed the BAT-Gal reporter mouse in more detail. The BAT-Gal transgene consists of multimerized consensus Lef/Tcf-binding sites and a minimal promoter driving expression of a β-galactosidase cDNA. X-Gal staining of embryonic kidneys showed that BAT-Gal was expressed mosaically throughout the embryonic collecting duct system/ureteric bud. Little or no expression was detectable in the NPCs (Fig. S1A,B; data not shown).
As the BAT-Gal transgene is artificial, we next investigated the embryonic kidney expression of three so-called β-catenin universal reporter mouse lines: Axin2-LacZ, Nkd1-LacZ and Nkd2-LacZ. In each of these lines, the β-galactosidase gene lacZ has been knocked into the gene locus and thus expression is controlled by endogenous regulatory elements. Nkd1-LacZ and Nkd2-LacZ were both expressed exclusively in the medullary stroma (Fig. S1C,D; data not shown). Axin2-LacZ was active primarily in the tips of the ureteric bud and the medullary stroma although there was also low level expression in the cortical stroma (Fig. S1E,F; data not shown). Although expression of all four of these reporters is affected by loss of β-catenin activity, none shows β-catenin-dependent expression in the nephron progenitor cells/cap mesenchyme (the target of Wnt9b signaling) indicating either that additional factors cooperate with β-catenin to drive expression within this cell type or that Wnt9b signals to these cells in a β-catenin-independent manner.
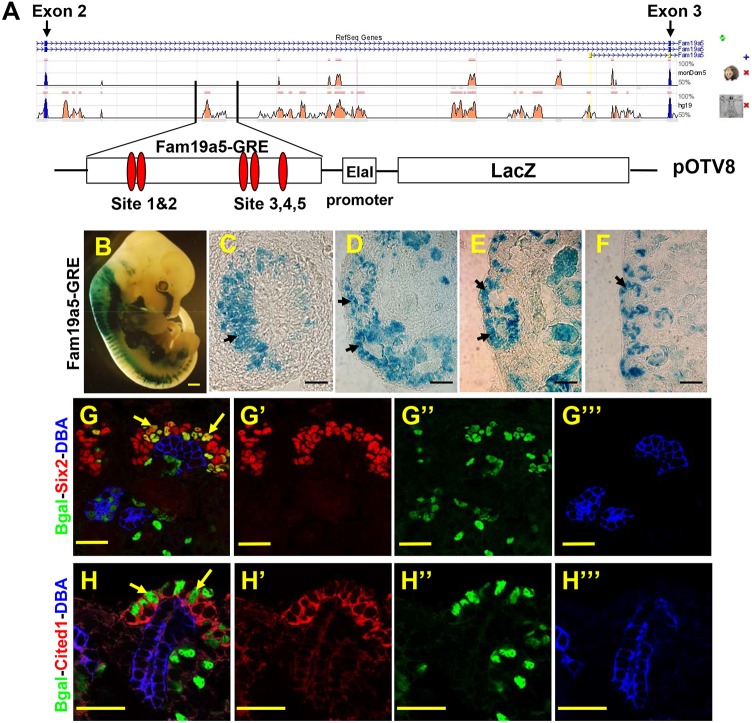
To address this possibility, we sought to understand more about the regulation of Wnt9b target gene expression. We and others have previously identified a region of DNA within intron 2 of the Wnt9b target gene Fam19a5 that contains five consensus Lef/Tcf-binding sites and has been shown to be physically associated with β-catenin (Fig. 1A; Karner et al., 2011; Park et al., 2012). To determine if this element could act as a cell type-specific enhancer, we cloned a 2.9 kb region of Fam19a5 intron 2 that was significantly conserved between mouse and human (hereafter referred to as Fam19a5-gene regulatory element or Fam19a5-GRE) into the pOTV8 vector, which consists of a minimal promoter from the human elastase gene (ELA1; CELA1) and a mouse codon-optimized lacZ cDNA (Fig. 1A). Transgenic mice carrying the Fam19a5-GRE-LacZ were generated by pronuclear injection. Of 18 G0 embryos carrying this transgene, five showed nearly identical β-galactosidase activity within the developing metanephric kidney, the brain, dorsal root ganglia, somites and lung (Fig. 1; data not shown). The remaining 13 showed no or sporadic β-galactosidase activity. The pattern of lacZ expression in the five positive transgenics shows extensive overlap with the expression pattern of Fam19a5 mRNA.
Fig. 1.
A 2.9-kb fragment in intron 2 of Fam19a5 activates transcription in the renewing nephron progenitor cell population. (A) Schematic of intron 2 of mouse Fam19a5. Peaks represent areas of homology between mouse, possum or human. The boxed area indicates the position of the fragment used to engineer the reporter. The 2.9-kb fragment was cloned into the pOTV8 plasmid containing a minimal promoter and a β-galactosidase cDNA. The relative positions of the five consensus Lef/Tcf-binding sites are indicated by red ovals. (B-F) Whole embryos (E12.5; B) and kidney sections from Fam19a5-GRE-stained 11.5 (C), E12.5 (D), E14.5 (E) and P1 (F) kidneys. For C-F, the cortical zone is to the left. (G-H‴) Sections of P1 Fam19a5-GRE kidneys stained with antibodies to Six2 (red in G,G′) or Cited1 (red in H,H′), β-gal (Bgal, green), the collecting duct marker DBA (blue in G,G‴,H,H‴). G′-G‴ and H′-H‴ are single-channel images; G and H are merged images. Arrows indicate the nephron progenitor cells. Scale bars: 500 μm (B); 50 μm (C-F); 30 μm (G-H‴).
To characterize further the kidney-specific activity of the Fam19a5-GRE element, we established a transgenic line and collected embryonic and postnatal kidneys from E11.5 to postnatal day (P) 1 offspring. Transgenic kidneys were subjected to X-Gal staining. Fam19a5-GRE drove expression of lacZ in kidneys at all stages examined (Fig. 1C-F). To determine in which cells the Fam19a5-GRE was active, we sectioned the transgenic kidneys and co-stained with antibodies to β-galactosidase and other cell type-specific markers. β-Galactosidase protein was mosaically expressed throughout the cortical zone of the kidney (Fig. 1G-H‴; data not shown). Co-staining showed β-galactosidase-positive cells within the Six2-positive cap mesenchyme and pre-tubular aggregates (PTAs) and the Lef1-positive renal vesicles and comma-shaped bodies (Fig. 1G,H; data not shown). With the exception of the comma-shaped bodies, these domains overlap with Fam19a5 mRNA expression. The ectopic expression within the comma-shaped bodies might be due to perdurance of the β-galactosidase protein or the absence of a repressor element within the GRE reporter.
Although we previously showed that Fam19a5 mRNA is expressed in the cap mesenchyme, it is not clear whether it is expressed within the renewing nephron progenitor populations or a subpopulation of the cap that has already been induced to differentiate. Cited1 protein currently reflects the most specific and restricted expression in the renewing nephron progenitor cell population (Boyle et al., 2008; Brown et al., 2015). To determine whether Fam19a5-GRE is expressed within the renewing NPCs, we co-stained sections of transgenic kidneys with antibodies to Cited1 and β-galactosidase. A subpopulation of the β-galactosidase-positive cells also expressed Cited1 indicating that the Fam19a5-GRE is active within the renewing NPC population (Fig. 1H).
Our previous work showed that expression of Fam19a5 mRNA required both Wnt9b and β-catenin (Karner et al., 2011). To determine whether the Fam19a5-GRE was regulated in a manner similar to endogenous Fam19a5, we crossed the Fam19a5-GRE line onto Wnt9b and β-catenin mutant backgrounds (Wnt9b−/− and Six2Cre;catnbflox/−, respectively) and assessed β-galactosidase activity. Similar to what is observed with Fam19a5 mRNA, the Fam19a5-GRE line requires both Wnt9b and β-catenin for its expression (Fig. 2). These data suggest that the Fam19a5-GRE represents an endogenous enhancer element for Fam19a5 and is the first β-catenin-responsive enhancer element that is sufficient to drive expression of a transgene in the renewing NPCs.
Fig. 2.
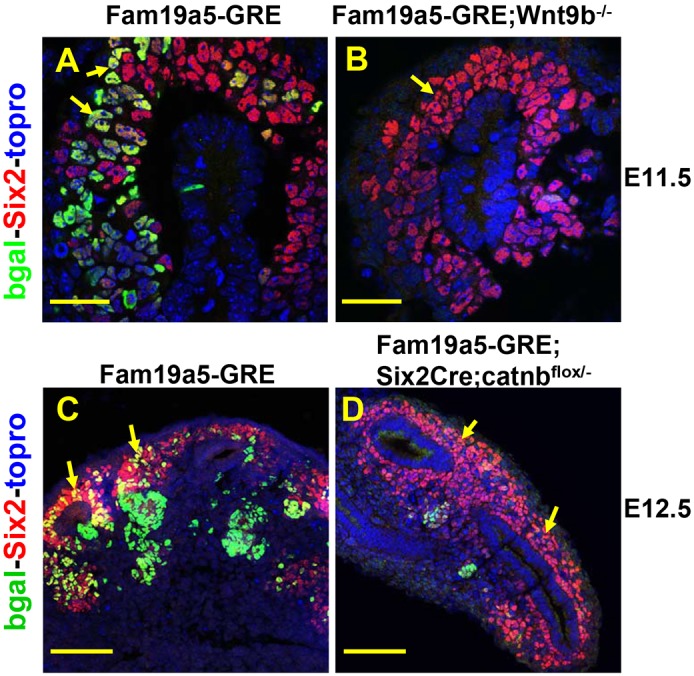
The Fam19a5-GRE is regulated by Wnt9b and β-catenin. (A-D) Sections of E11.5 (A,B) or E12.5 (C,D) Fam19a5-GRE kidneys from wild-type (A,C), Wnt9b−/− (B) and Six2Cre;catnbflox/− (D) kidneys stained with antibodies to β-gal (bgal, green), Six2 (red) and TO-PRO-3 (blue). Arrows indicate the nephron progenitor cells. Scale bars: 30 µm (A,B); 100 µm (C,D).
Characterization of the Fam19a5-GRE identifies multiple conserved transcription factor-binding sites
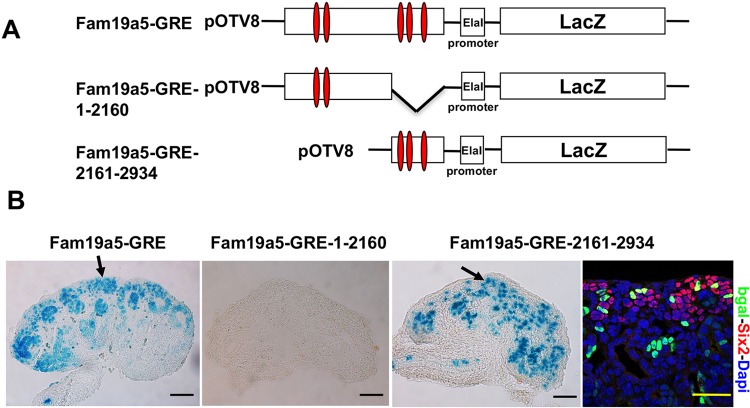
As indicated by the expression of the various reporter genes outlined above and β-catenin target genes in distinct and frequently non-overlapping cell types in the kidney, cell type-specific factors must cooperate with β-catenin to drive expression of target genes in the kidney. To help identify such factors, we first sought to narrow down the region of DNA that is sufficient to drive expression within the NPCs. As mentioned, there are five conserved consensus Lef/Tcf-binding sites spread throughout the 2934-bp construct and β-catenin was shown to be physically associated with at least two of them (Karner et al., 2011; Park et al., 2012). To determine which, if any, of these elements was necessary to drive expression within the NPCs, we generated two additional β-galactosidase reporter lines: one containing 2160 bp with two Lef/Tcf-binding sites at the 5′ end of the Fam19a5-GRE (Fam19a5-GRE-1-2160) and the other 774 bp with three Lef/Tcf-binding sites at the 3′ end (Fam19a5-GRE-2161-2934) (Fig. 3A). Whereas the 774-bp fragment at the 3′ end of the Fam19a5-GRE was sufficient to drive expression of lacZ within the NPCs (although expression was in general more mosaic than that observed with the 2934-bp element), none of the 65 embryos carrying the 5′ 2160-bp fragment showed β-galactosidase activity in the kidney (Fig. 3B). These findings suggest that sites contained within this 774-bp region of DNA are necessary and sufficient for expression within the NPCs.
Fig. 3.
A poorly conserved 774-bp element is sufficient to drive expression within the NPCs. (A) Schematic of three different transgenic constructs used to generate transgenic lines. Red ovals represent consensus Lef/Tcf-binding sites. (B) Sections of stained kidneys from embryos injected with each construct whose DNA was scored as positive for carrying the lacZ transgene. The right-hand image shows a P1 Fam19a5-GRE-2161-2934 section stained with β-gal (bgal, green), Six2 (red) and DAPI (blue). Arrows indicate the nephron progenitor cells. Scale bars: 50 μm.
To facilitate further investigation of the regulation of Fam19a5-GRE, we generated a luciferase reporter under the control of Fam19a5-GRE (Fig. 4A). We first characterized activity of this construct in the presence or absence of the β-catenin pathway agonist LiCl in HEK293T cells. LiCl treatment resulted in mild (relative to Top-Flash) but consistent activation of the reporter (Fig. 4B; data not shown). LiCl is thought to primarily function by inhibiting Gsk3β but there are other known targets. To determine if this effect was specific to repression of Gsk3β activity, we used an independent Gsk3β antagonist CHIR. Similar to LiCl, we observed a weak but significant stimulation of luciferase with CHIR (Fig. S3A), suggesting this was a Gsk3β-specific defect.
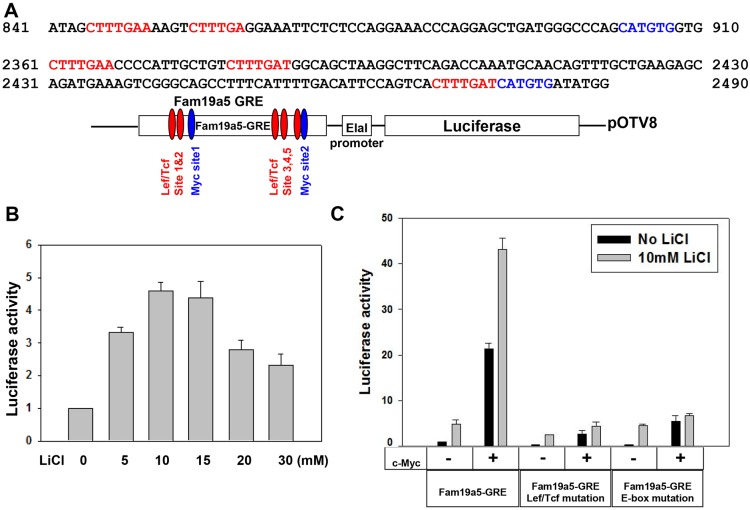
Fig. 4.
c-Myc promotes Fam19a5-GRE reporter activity in vitro. (A) Sequence fragments from the Fam19a5-GRE highlighting the consensus Lef/Tcf (red)- and Myc (blue)-binding sites. (B) Graph of luciferase reporter activity from HEK 293 cells transfected with a Fam19a5-GRE luciferase reporter and treated with LiCl at different concentrations. (C) Graph of luciferase activity from HEK 293 cells transfected with wild-type or various mutant Fam19a5-GRE reporters. Some cells were co-transfected with a full length c-Myc expression plasmid as indicated. The media was replaced with fresh media (black bars) or media supplemented with 10 mM LiCl (gray bars) 24 h after transfection and luciferase activity was measured 18 h later. Data are represented as mean+s.e.m.
LiCl has previously been shown to affect Fam19a5 expression through activation of β-catenin (Karner et al., 2011). There are five consensus Lef/Tcf-binding sites in the Fam19a5-GRE that are conserved in the human FAM19A5 intron 2. To determine whether enhancer activity was dependent on them, we mutated all five sites within this element (Fig. S2B). In the absence of these putative binding sites, LiCl did not activate reporter expression over background (Fig. 4C). Although these data show that Lef/Tcf-binding sites are necessary for response of this element to LiCl, the relatively mild expression of luciferase suggests that other factors are required in addition to β-catenin for robust activity of the Fam19a5-GRE enhancer.
Myc synergizes with β-catenin to promote Fam19a5 enhancer activity in vitro
To identify additional transcription factors that cooperate with β-catenin to drive expression of Fam19a5 within the NPCs, we took two different approaches. First, microarray data comparing isolated wild-type and Wnt9b mutant E11.5 metanephric mesenchyme and E15.5 wild-type and Wnt9b hypomorphic mutant (Wnt9bcneo/cneo) whole kidneys (Karner et al., 2011) was re-analyzed. We determined whether specific transcription factor targets were enriched among our significantly downregulated genes. For both time points, Myc target genes were significantly over-represented in genes listed as being significantly downregulated in mutants (P=2.6×10−5 for E11.5 and 4.16×10−4 for E15.5).
We also examined the 2934-bp Fam19a5 nephron progenitor enhancer fragment using Transfac software to identify additional consensus transcription factor-binding sites. Conserved binding sites for several transcriptional regulators were present surrounding both the 5′ and 3′ Lef/Tcf sites (Fig. S2A), including STAT1- and STAT3-binding sites around the 5′ sites and two phylogenetically conserved E-box sites adjacent to the 5′ and 3′ Lef/Tcf-binding sites (Fig. 4A). E-boxes bind basic helix-loop-helix transcription factors including members of the Myc family. As both of the E-boxes were close to the conserved Lef/Tcf-binding sites, Wnt9b target mRNAs are enriched for Myc targets and as Mycs have been implicated in progenitor cell survival and/or renewal in multiple tissues including the kidney (Couillard and Trudel, 2009; Bates et al., 2000), we decided to investigate the role of Myc in regulating Wnt9b target gene expression.
To determine whether Myc could regulate the activity of the Fam19a5-GRE, we assessed luciferase activity in response to c-Myc. Transfection with a c-Myc cDNA resulted in luciferase activity that was nearly five times the levels stimulated by LiCl alone. To determine whether the E-box sites were necessary for this activation, we mutated the two conserved sights, which resulted in a drastic reduction in luciferase expression in response to c-Myc transfection (Fig. 4C).
To determine whether β-catenin could cooperate with c-Myc, we co-treated c-Myc-transfected cells with LiCl. Co-treatment increased luciferase activity more than 10-fold over LiCl alone and it was nearly double that of c-Myc alone (Fig. 4C). Nearly identical results were observed with the specific Gsk3β inhibitor CHIR (Fig. S3B).
In addition to de-stabilizing β-catenin, Gsk3β has previously been shown to de-stabilize Myc (Yeh et al., 2004). Thus, it is possible that LiCl treatment only serves to stabilize Myc. If this is the case, then the presence of the Lef/Tcf-binding sites in the Fam19a5-GRE should not impact Myc's ability to activate this element. To determine whether Lef/Tcf co-factors were required for Myc activity, we assessed the effect of c-Myc treatment on the Lef/Tcf mutant enhancers. Although mutation of the E-boxes had no effect on LiCl-stimulated activity, c-Myc was not able to stimulate luciferase activity over background levels when the Lef/Tcf binding sites were mutated (Fig. 4C). These data suggest that c-Myc and β-catenin functionally interact to activate expression of Fam19a5.
c-Myc expression in the nephron progenitor cells is Wnt9b independent
The findings that c-Myc cooperates with β-catenin to activate gene expression are somewhat unexpected. Previous studies have suggested that c- and N-Myc (Mycn) expression is directly regulated by β-catenin. Direct regulation of c-Myc expression by Wnt9b/β-catenin would be inconsistent with cooperativity of these factors in nephron progenitor renewal. To determine whether c- or N-Myc were targets of Wnt9b activity, we performed a detailed analysis of the expression of both genes in wild-type and Wnt9b mutant embryonic kidneys.
At E11.5, both c- and N-Myc are expressed in the NPCs, the target cells for Wnt9b (Fig. S4A,E). At E15.5, c-Myc continues to show strong expression in the NPCs whereas N-Myc expression becomes restricted to the pre-tubular aggregates and/or renal vesicles (Fig. S4C,G). At P1, c-Myc is expressed in the NPCs and N-Myc is expressed in the renal vesicles (Fig. S4D,H).
To determine if expression of either gene was regulated by Wnt9b, we examined their expression in E11.5 Wnt9b mutants. c-Myc expression appears to be unaffected by loss of Wnt9b whereas N-Myc expression is significantly downregulated (Fig. S4B,F). These data suggest that although N-Myc may be a direct target (although it is important to note that the N-Myc-expressing PTAs do not form in Wnt9b mutants), c-Myc expression is independent of Wnt9b and presumably β-catenin activity.
N- and c-Myc cooperate with Wnt9b/β-catenin to activate target genes and nephron progenitor renewal in vivo
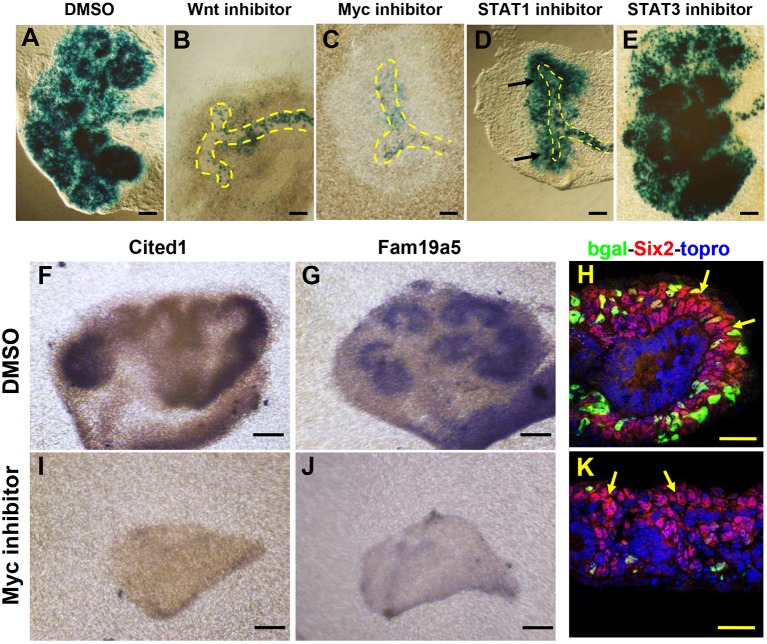
To determine if Myc cooperated with β-catenin in vivo, we first utilized an ex vivo organ culture system. E11.5 Fam19a5-GRE kidneys were isolated and cultured in the presence of the Myc inhibitor 10058-F4 for 48 h. The porcupine inhibitor IWP2, which we previously showed was capable of efficiently blocking Wnt9b activity (Karner et al., 2011), was used as a positive control. Treatment of cultured kidneys with the Myc inhibitor blocked kidney development and repressed the expression of Fam19a5-GRE-LacZ to a similar degree as IWP2 (Fig. 5A-C). To determine the specificity of the Myc inhibitor to the Wnt/β-catenin pathway, treated kidneys were assessed for the expression of additional Wnt9b/β-catenin-dependent target mRNAs (Cited1, Pla2g7 and Fam19a5 itself) as well as the Wnt9b/β-catenin-independent NPC marker Six2. Although the expression of Cited1, Pla2g7 and Fam19a5 was undetectable, the expression of Six2 was maintained at normal levels in 10058-F4-treated kidneys (Fig. 5F-K; data not shown) suggesting that the effect of Myc inhibition was specific to Wnt9b NPC targets.
Fig. 5.
Myc is necessary for Fam19a5 reporter activity and Wnt9b target gene expression in organ culture. E11.5 Fam19a5-GRE kidneys were dissected out and cultured for 48 h in media supplemented with DMSO (A,F-H), the Wnt production inhibitor IWP2 (B), a Myc inhibitor (C,I-K), a STAT1 inhibitor (D) or a STAT3 inhibitor (E). Cultured kidneys were fixed and stained with X-gal (A-E), antisense mRNA for Cited1 (F,I) or Fam19a5 (G,J), or sectioned and stained with antibodies to β-gal (bgal, green), Six2 (red) and TO-PRO-3 (blue) (H,K). Arrows indicate the nephron progenitor cells and ureteric bud is outlined. Scale bars: 30 µm.
As a further test of specificity, we also assessed the effect of STAT activity on the Fam19a5-GRE as there are several STAT1 and STAT3 sites present within the 5′ 2160-bp region of the Fam19a5-GRE that is neither necessary nor sufficient for expression of lacZ. Inhibitors of neither STAT1 nor STAT3 had any effect on Fam19a5-GRE expression even though the STAT1 inhibitor clearly repressed kidney development indicating it was active in this setting (Fig. 5D,E).
To determine the role of c- and/or N-Myc in the NPCs in vivo, we ablated both genes individually or in tandem using floxed alleles and the Six2Cre transgene (Six2Cre;c-Mycflox/flox;N-Mycflox/flox). Six2Cre is active specifically within the nephron progenitors and thus should ablate the expression of these genes specifically within this lineage (Kobayashi et al., 2008). In situ hybridization with probes to c- and N-Myc mRNAs showed that levels of both genes were significantly reduced by E12.5 and neither gene was detectable in respective single or double mutant kidneys at E13.5 (Fig. S5).
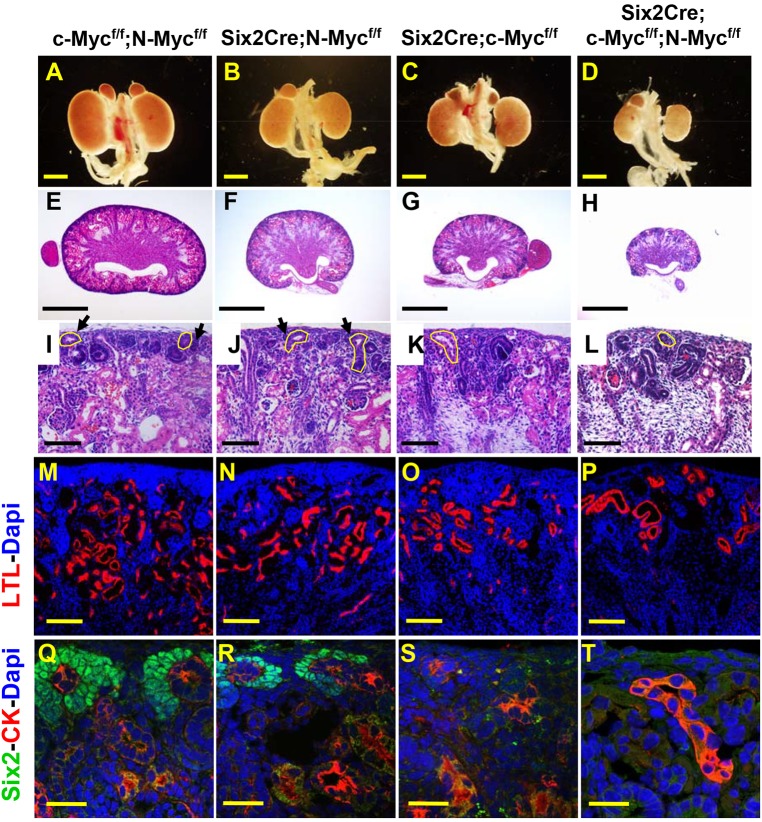
Pups of all genotypes were present at the expected ratio at E18.5 and there was no significant difference in body sizes amongst the different genotypes. c- and N-Myc mutants survived at least until one month of age although c-Myc mutant kidneys were extremely cystic (not shown). No Six2Cre;c-Mycflox/flox;N-Mycflox/flox double mutants pups survived beyond P1, consistent with a severe defect in renal function.
Analysis of P1 pups revealed that c/N-Myc double mutant kidneys were significantly smaller than controls (Fig. 6A-D). N- and c-Myc single mutant kidneys were also smaller than controls although the difference was less pronounced than double mutants. Examination of Hematoxylin and Eosin (H&E)-stained sections and staining with the proximal tubule marker lotus tetragonolobus lectin (LTL) showed a significantly reduced nephrogenic zone and reduced numbers of nephrons in both single mutants and the double mutant kidneys with the double mutants showing the most severe reduction (Fig. 6E-P). Staining with an antibody to Six2 revealed a complete loss of NPCs in c-Myc and c/N-Myc double mutants at P1 (Fig. 6Q,S). N-Myc mutants had a reduced population of nephron progenitors at P1, consistent with the observation that after the earliest stages of development, N-Myc is primarily expressed in the PTAs/renal vesicles (Fig. 6Q,R) and may play only a minor role in NPC renewal.
Fig. 6.
c- and N-Myc play partially redundant roles in nephron progenitor renewal and nephron endowment. Whole-mount images (A-D) or sections through P1 urogenital systems/kidneys of wild-type (c-Mycflox/flox;N-Mycflox/flox; A,E,I,M,Q), N-Myc (Six2Cre;N-Mycflox/flox; B,F,J,N,R), c-Myc (Six2Cre;c-Mycflox/flox; C,G,K,O,S) or c/N-Myc double mutant (Six2Cre;c-Mycflox/flox;N-Mycflox/flox; D,H,L,P,T) kidneys. E-L show low (E-H) or high (I-L) magnification images of H&E-stained kidneys. M-T show sectioned P1 kidneys stained with antibodies to the proximal tubule marker lotus tetragonolobus lectin (LTL, red) and DAPI (blue) (M-P) or Six2 (green), the collecting duct marker cytokeratin (CK, red) and DAPI (blue) (Q-T). Arrows indicate cap mesenchyme and ureteric bud is outlined. Scale bars: 1 mm (A-H); 100 μm (I-L); 200 µm (M-P); 30 µm (Q-T).
We previously showed that Wnt9b played roles in both NPC proliferation and survival (Karner et al., 2011). To determine whether c-/N-Myc double mutants were defective in either or both of these processes, we assessed the number of E15.5 Six2-positive cells expressing cleaved caspase or phospho-histone H3 (pHH3). At E15.5, double mutant kidneys already had significantly reduced numbers of Six2-positive cells. Although we did not detect differences in the percentage of cells positive for pHH3, there were significantly more cells expressing cleaved caspase in double mutants (Fig. S6) suggesting that increased rates of cell death contributed to the loss of the NPC population in double mutants.
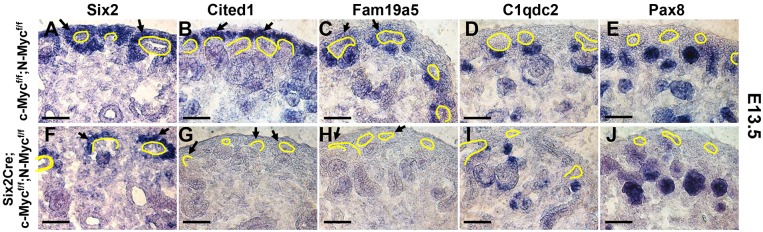
To determine whether ablation of c- and N-Myc specifically affected the expression of Wnt9b/β-catenin targets, we analyzed expression of the Wnt9b-dependent NPC renewal genes Cited1 and Fam19a5 as well as the Wnt9b-independent NPC marker Six2 in E13.5 Six2Cre;c-Mycf/f;N-mycf/f mutant kidneys. This represents a time point at which both N- and c-Myc mRNAs were greatly reduced/absent in mutants (Fig. S5). Although a Six2-positive population of cells was still present at E13.5 (Fig. 7A,B) Cited1, Pla2g7 and Fam19a5 were undetectable in c/N-Myc double mutant kidneys (Fig. 7C-F; data not shown). To determine whether Wnt/β-catenin differentiation target genes were affected, we examined the expression of the Wnt9b-dependent PTA markers C1qdc2 (C1qtnf12) and Pax8. Both genes showed normal expression on the medullary side of the collecting ducts at E13.5 and 15.5 (Fig. 7G-J; data not shown). However, by E17.5, PTA markers were absent from double mutants. These data suggest that Myc activity is necessary for NPC renewal but not differentiation. Loss of PTA markers at later stages is most likely due to the loss of nephron progenitor cells (not shown).
Fig. 7.
c-Myc and N-Myc are necessary for expression of Wnt/β-catenin progenitor renewal targets. Sections of E13.5 wild-type (c-Mycflox/flox;N-Mycflox/flox; A-E) and mutant (Six2Cre;c-Mycflox/flox;N-Mycflox/flox; F-J) kidneys hybridized with antisense in situ probes for the Wnt9b-independent nephron progenitor marker Six2 (A,F), the Wnt9b-dependent progenitor renewal targets Cited1 (B,G) and Fam19a5 (C,H), or the Wnt9b-dependent nephron progenitor differentiation targets C1qdc2 (D,I) and Pax8 (E,J). Arrows indicate the nephron progenitor cells and ureteric bud is outlined. Scale bars: 50 μm.
DISCUSSION
Proper balance of nephron progenitor renewal and differentiation is essential for proper kidney development and function. Failure to achieve proper nephron number results in renal insufficiency in the most severe cases and may predispose individuals to hypertension and susceptibility to kidney injury in relatively minor cases. We previously showed that Wnt9b/β-catenin signaling was necessary for both progenitor renewal and differentiation (Karner et al., 2011). How β-catenin regulated these two distinct and contradictory processes was unknown. Here, we have identified and characterized an enhancer element for the Wnt9b/β-catenin target gene Fam19a5 that is sufficient to drive gene expression in the renewing NPCs. Wnt9b and β-catenin are required but not sufficient for robust activity of this element suggesting that other cell type-specific transcription factors are involved. Analysis of Wnt9b target genes as well as identification of conserved transcription factor-binding sites within the Fam19a5 enhancer implicated Myc activity in this process. Ex vivo analysis shows that Myc function is necessary for activity of this element and sufficient to activate transcription downstream of this element in vitro. Further, we found that c-Myc was necessary for the Wnt9b/β-catenin renewal (but not differentiation) program in vivo. Based on these findings, we propose that Wnt9b/β-catenin signaling cooperates with c- and N-Myc to activate a molecular program (which includes direct activation of Fam19a5 transcription) within the renewing NPC population that is necessary for NPC proliferation and/or survival.
Although it has previously been suggested that c-Myc interacts with proximal promoter elements to stimulate transcriptional elongation, we think this is unlikely to be the mechanism utilized here. Mutation of putative Myc-binding sites within the Fam19a5-GRE abrogates both Myc-induced and β-catenin-induced reporter expression. Further, stimulation of the Fam19a5 reporter by treatment with both Myc and β-catenin is more than additive over individual treatment. Thus, we propose that β-catenin and Myc cooperate to activate Fam19a5 at the NPC renewal enhancer. We were able to identify conserved Myc-binding sites adjacent to other Wnt9b/β-catenin NPC renewal targets and Myc was necessary for the expression of all renewal targets analyzed in vivo whereas it did not appear to be necessary for differentiation targets (Fig. 6). Thus, we feel that cooperation between Myc and β-catenin might be a more general mechanism for activation of renewal targets. However, we were unable to confirm physical interaction between c-Myc protein and the Fam19a5 enhancer by chromatin immunoprecipitation (data not shown). This could in part be a technical issue as the Myc antibody tested did not appear to be suitable for chromatin immunoprecipitation (not shown). However, without this information we cannot conclusively state that Myc directly promotes transcription from this or other renewal target enhancers. It is interesting to note that Myc has previously been shown to form a complex with Six2 and Six2 has been shown to be necessary for activation of Fam19a5 (Xu et al., 2014; Karner et al., 2011). Further, Park recently showed that Six2 physically interacts with Tcf3 within the NPCs (Xu et al., 2014; Park et al., 2012). Thus, it is tempting to speculate that Myc is part of a transcription factor complex consisting of Six2 and Tcf3 that cooperate to promote nephron progenitor renewal.
In many tissues, N- and c-Myc appear to be direct transcriptional targets of β-catenin. In fact, the Mycs are frequently referred to as universal β-catenin targets. This does not appear to be the case in the developing kidney as ablation of Wnt9b does not affect c-Myc expression at E12.5. Although we cannot rule out the possibility that there is some Wnt9b-independent β-catenin activity in the NPCs, ablation of Wnt9b largely mimics β-catenin ablation in these cells, making this unlikely. Although N-Myc expression is significantly reduced in Wnt9b mutants, it is important to note that N-Myc is normally expressed in the pre-tubular aggregates/renal vesicles and these structures do not form in Wnt9b mutant kidneys, thus the loss of expression may be indirect. Either way, our data suggest that Wnt9b/β-catenin activity is not required for c-Myc expression in early stage NPCs and thus caution should be taken before assuming that this gene is a universal β-catenin target.
It is of note that, similar to our observations in the developing kidney, Myc signaling has also been shown to promote Wnt/β-catenin activity in breast cancers. In this case, Myc was shown to repress the expression of Dkk1 thus indirectly leading to Wnt pathway activation (Cowling et al., 2007; Cowling and Cole, 2007). Although a similar mechanism does not appear to be in place in the nephron progenitors, these data further support the idea that the interaction between β-catenin and Myc is not simply linear.
Our results contrast slightly with a previous analysis of Bmp7Cre;c-Mycflox/flox mutants (Couillard and Trudel, 2009). First, Couillard and Trudel reported that the NPC population was never lost in these mutants. This is most likely explained by the fact that the Six2Cre deleter appears to have more uniform activity than the Bmp7Cre as supported by the fact that they did not observe efficient deletion of Myc until E15.5 and never observed a complete loss of Myc-expressing cells. Another difference is that Couillard and Trudel did not observe changes in the rates of apoptosis but they did observe decreased levels of Ki67 within mutant NPCs. This difference once again could be attributed to differences in the efficiency of ablation. The increased apoptosis we observe could be the result of a loss of survival cues, such as Fgf8, normally provided by the renal vesicles. However, the difference in the rate of Ki67- and pHH3-positive NPCs cannot be explained by differences in Cre activity. A possible explanation is that the two antibodies mark different stages of mitosis, with Ki67 labeling cells in interphase and pHH3 labeling cells in late metaphase and early anaphase. It is possible that Myc mutant cells are arrested in metaphase/anaphase, which may secondarily lead to cell death. This possibility deserves further examination.
Finally, it is well-established that Wnt/β-catenin signaling is involved in a number of normal and pathological processes in embryonic and adult metazoans. The role of Wnt/β-catenin in these varied processes and tissues involves activation of unique sets of target genes. This would suggest that other co-factors interact with β-catenin to determine target gene activation. This is further supported by the observation that although several ‘universal’ β-catenin target genes have been identified, none of these genes is expressed in all Wnt/β-catenin responsive tissues. For example, Axin2, Nkd1 and Nkd2 are commonly referred to as universal β-catenin targets; however, they each show unique expression patterns in the developing kidney suggesting co-regulation by cell type-specific factors. Thus, although β-catenin is necessary for their expression, it is not sufficient. Therefore, it is somewhat surprising that Lef/Tcf sites are sufficient to drive expression of reporter genes in some cells and tissues. Indeed, in flies Groucho (the ortholog of Tcf)-binding elements are not sufficient. Although Lef/Tcf reporters do seem to be reporting a subset of β-catenin activity in mice, they are clearly not reporting all activity. This raises the question of whether a truly ‘universal’ β-catenin target gene exists in mammals and suggests that caution should be taken when interpreting β-catenin activity based on reporter expression (Barolo, 2006).
MATERIALS AND METHODS
Plasmid constructs and transgenic mouse lines
The Fam19a5-GRE, Fam19a5-GRE-1-2160 and Fam19a5-GRE-2161-2934 constructs with the Fam19a5 enhancer elements were obtained by polymerase chain reaction (PCR) technology. The fragments were cloned into the optimized transgene vector 8 (pOTV8) containing a β-galactosidase (β-gal) gene (lacZ) and a minimal promoter from the human elastase 1 gene (ELA1) (Rose and MacDonald, 1997). The enhancer constructs were linearized with SalI and NotI to release the ampicillin gene. The backbone with enhancer elements were purified using Elutip D columns (Whatman) and microinjected into the pronuclei of one-cell embryos. The surviving embryos were implanted into B6SJLF1 pseudopregnant foster mothers.
Characterization of the Fam19a5-GRE
Conserved transcription factor-binding sites within Fam19a5 were identified using ECR browser (https://ecrbrowser.dcode.org/). Wnt9b microarray data (Karner et al., 2011) were analyzed using the over-enrichment analysis method/network database/x transcription factor target method at Webgestalt.org.
For functional analysis of the Fam19a5-GRE, E12.5 G0 embryos were dissected from B6SJLF1 foster mothers. DNA from embryo tails was collected and analyzed by PCR. PCR of genomic DNA determined that 18 out of 88 E12.5 G0 embryos were positive for the transgene. All 18 embryos were further analyzed by whole-mount β-galactosidase staining. Five out of 18 showed β-galactosidase activity within the developing metanephric kidney, the brain, dorsal root ganglia, somites and lung (Fig. 1B, Fig. 3B; data not shown). The remaining 13 transgenic embryos showed no staining or sporadic staining in random tissues (not shown).
Based on the results above, an additional round of pronuclear injection was performed. Founder pups were genotyped. Fam19a5-GRE-positive mice were saved and bred with Swiss Websters (SW) to generate individual lines. P1 embryonic kidneys were collected from offspring from each line and X-Gal staining was performed. Eight lacZ-positive lines were detected and two lines were saved and crossed with SW mice to maintain the line. All lines showed staining in the NPCs. All subsequent characterization was performed with line M550.
Mouse genotyping
The PCR primers for the Fam19a5 enhancer constructs were: Fam19a5-GRE forward 5'-AAGAGAGGCTTGAGGCTTA-3′, reverse 5'-GAAAGAGGAGGTCCTTATG-3′; Fam19a5-GRE-1-2160 forward 5'-AAGAGAGGCTTGAGGCTTA-3′, reverse 5'-GGAAGCCATTCTGTCTTGATAC-3′; Fam19a5-GRE-2161-2934 forward 5'-AAGGAGGAGGCTTACATCCCC-3′, reverse 5'-GAAAGAGGAGGTCCTTATGGACTTC-3′. The genotyping primers were: Fam19a5-GRE and Fam19a5-GRE-2161-2934 forward 5'-ATGACCTTCGACACACCACTC-3′, reverse 5'-TGGGCAATATCGCGGCTCAG-3′; Fam19a5-GRE-1-2160 forward 5'-AAGAGAGGCTTGAGGCTTA-3′, reverse 5'- TGGGCAATATCGCGGCTCAG-3′.
Mouse lines
The Wnt9b, Six2-eGFPCre BAC transgenic, catnb flox, c-Myc and N-Myc floxed mice have all been previously described (Carroll et al., 2005; Kobayashi et al., 2008; Trumpp et al., 2001; Knoepfler et al., 2002). All animals were housed, maintained and used according to protocols approved by the Institutional Animal Care and Use Committees at University of Texas Southwestern Medical center and following the guidelines from NCI-Frederick Animal Care and Use Committee.
Ex vivo organ culture
Organ culture was as previously described (Pan et al., 2015). Briefly, embryonic kidneys were dissected out at E11.5 and grown on transwell filters (Whatman) at the air/media interface. The small chemical inhibitors 10058-F4 (c-Myc inhibitor), IWP2 (Wnt inhibitor; Chen et al., 2009), fludarabine (STAT1 inhibitor, Sigma) and S3I-201 (STAT3 inhibitor, Sigma) were added to the culturing medium at the concentration indicated for 48 h. The tissues were fixed and embedded in OCT or analyzed by X-Gal staining.
X-Gal staining
Whole embryos or kidneys were fixed with 1% formaldehyde and 0.2% gluteraldedyde for 30 min. After washing with 0.2% NP40 in PBS, the samples were incubated with 5-bromo-4-chloro-3-indolyl-beta-D-galactoside (X-Gal; Molecular Probes) staining solution for 1 h. Whole-mount images were taken on a Zeiss NeoLumar stereoscope using an Olympus DP71 camera. The stained kidneys were embedded in OCT and sectioned. The sections were mounted and photographed. All experiments were repeated at least three times with a minimum of six individual kidneys from six distinct embryos assayed per replicate.
H&E histology
Kidneys from P1 pups were fixed with 4% paraformaldehyde (PFA) and embedded in paraffin. The kidneys were then sectioned and stained with H&E using standard techniques.
In situ hybridization
Whole-mount and section in situ hybridization was performed as previously described (Carroll et al., 2005). Tissues were hybridized with antisense probes for the following genes: Six2, Cited1, Fam19a5, Pla2g7, C1qdc2, Pax8, c-Myc and N-Myc.
Luciferase reporter assay
Fam19a5 enhancer elements were cloned into the pGL4 minP vector (9PIE841, Promega). HEK 293 cells (originally from ATCC) were seeded in a 24-well plate 24 h prior to transfection. Wild-type and/or mutant Fam19a5 reporters with or without c-Myc cDNA plasmid were transfected in HEK 293 cells using Fugene HD (E2311, Promega) following the manufacturer's protocol. After 24 h, media was replaced and cells were treated with or without LiCl for another 18 h. CHIR99021 (CHIR; Stemgent) was re-suspended in DMSO to make a 10 mM stock. Cells were harvested and luciferase assays were performed using a Dual-Luciferase Reporter Assay System (E1910, Promega). All experiments were repeated at least three times and the average fold changes are presented. Significance was determined using a Student's t-test.
Immunostaining
Kidneys were fixed overnight in 4% PFA at 4°C with gentle agitation. After cryoprotection in 30% sucrose, the specimens were embedded in OCT and cryosectioned at 10 µm thickness. Sections were washed three times in 0.1% Triton X-100/PBS for 5 min each wash and blocked for a minimum of 1 h at room temperature in 5% fetal bovine serum, 0.1% Triton X-100 in PBS. Sections were incubated in primary antibody at 4° overnight followed by three washes in 0.1% Triton-X in PBS. Sections were then incubated in secondary antibody for 2 h at room temperature, washed three times in PBS, mounted with Vectashield, and examined by scanning laser confocal microscopy (Zeiss LSM-510). The primary antibodies used were chicken anti-β-gal (1:2000, ab9361, Abcam), mouse anti-cytokeratin (1:500, C2562, Sigma), rabbit anti-Six2 (1:500, 11562-1-AP, ProteinTech), biotinylated anti-DBA (1:500, B1035, Vector Laboratories), biotinylated anti-LTL (1:500, B1325, Vector Laboratories), rabbit anti-Cited1 (1:500, RB-9219, NeoMarkers; note that this antibody is no longer available), rabbit anti-pHH3 (1:500, H0412, Sigma), rabbit anti-active caspase-3 (1:500, G748A, Promega). Nuclei were counterstained using either TO-PRO-3 (1:1000, Invitrogen) or DAPI (Sigma).
For cell proliferation and apoptosis, E15.5 sections of Six2Cre;c-Mycflox/flox;N-Mycflox/flox (mutant) or c-Mycflox/flox;N-Mycflox/flox (control) kidneys were stained with antibodies to Six2, cytokeratin, phospho-histone H3 or cleaved caspase 3 and DAPI. The ratio of proliferation was calculated as the number of Six2/pHH3 or Six2/Casp3 double-positive nuclei divided by the total number of Six2-positive nuclei. The total number of cells counted for active caspase 3 was 203 in wild type and 78 in mutants. The total number of cells for pHH3 was 652 in wild type and 241 in mutants. All images were captured on a Zeiss LSM 510 or 710 confocal microscope.
Tissue analysis
All data presented in figures are representative examples from one of at least three different experiments on at least three different embryos/organs. No significant variability was noted in tissues of the same genotype.
Supplementary Material
Acknowledgements
We thank members of the Carroll lab for comments on this manuscript and the Cleaver and Marciano labs for helpful discussions. The c- and N-Myc floxed mice were generously provided by Dr Allan Balmain (University of California, San Francisco, USA). The pOTV8 plasmid was provided by Dr Ray MacDonald (UT Southwestern Medical Center, TX, USA).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Methodology: C.M.K.; Data curation: X.P., C.M.K.; Writing - original draft: X.P., T.J.C.; Writing - review & editing: T.J.C.; Supervision: T.J.C.; Funding acquisition: T.J.C.
Funding
Work in this study was supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (DK080004, DK095057, DK106743 to T.J.C.) and from the National Institutes of Health to the UTSW George O'Brien Kidney Research Core Center (P30DK079328). Deposited in PMC for release after 12 months.
Data availability
Microarray data have been deposited in Gene Expression Omnibus under accession numbers GSE106155 and GSE106195.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.153700.supplemental
References
- Barolo S. (2006). Transgenic Wnt/TCF pathway reporters: all you need is Lef? Oncogene 25, 7505-7511. 10.1038/sj.onc.1210057 [DOI] [PubMed] [Google Scholar]
- Bates C. M., Kharzai S., Erwin T., Rossant J. and Parada L. F. (2000). Role of N-myc in the developing mouse kidney. Dev. Biol. 222, 317.- . [DOI] [PubMed] [Google Scholar]
- Benzing T., Simons M. and Walz G. (2007). Wnt signaling in polycystic kidney disease. J. Am. Soc. Nephrol. 18, 1389-1398. 10.1681/ASN.2006121355 [DOI] [PubMed] [Google Scholar]
- Boyle S., Misfeldt A., Chandler K. J., Deal K. K., Southard-Smith E. M., Mortlock D. P., Baldwin H. S. and deCaestecker M. (2008). Fate mapping using Cited1-CreERT2 mice demonstrates that the cap mesenchyme contains self-renewing progenitor cells and gives rise exclusively to nephronic epithelia. Dev. Biol. 313, 234-245. 10.1016/j.ydbio.2007.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. C., Muthukrishnan S. D., Fetting J. L. and Oxburgh L. (2015). A synthetic niche for nephron progenitor cells. Dev. Cell 34, 229-241. 10.1016/j.devcel.2015.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll T. J., Park J.-S., Hayashi S., Majumdar A. and McMahon A. P. (2005). Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev. Cell 9, 283-292. 10.1016/j.devcel.2005.05.016 [DOI] [PubMed] [Google Scholar]
- Chen B., Dodge M. E., Tang W., Lu J., Ma Z., Fan C. W., Wei S., Hao W., Kilgore J., Willians N. S. et al. (2009). Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and caner. Mat. Chem. Biol. 5, 100-107. 10.1038/nchembio.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillard M. and Trudel M. (2009). C-Myc as a modulator of renal stem/progenitor cell population. Dev. Dyn. 238, 405-414. 10.1002/dvdy.21841 [DOI] [PubMed] [Google Scholar]
- Cowling W. H. and Cole M. D. (2007). Turning the tables: Myc activates Wnt in breast cancer. Cell Cycle 6, 2625-2627. 10.4161/cc.6.21.4880 [DOI] [PubMed] [Google Scholar]
- Cowling V. H., D'Cruz C. M., Chodosh L. A. and Cole M. D. (2007). c-Myc transforms human mammary epithelial cells through repression of the Wnt inhibitors DKK1 and SFRP1. Mol. Cell. Biol. 27, 5135-5146. 10.1128/MCB.02282-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currier N., Chea K., Hlavacova M., Sussman D. J., Seldin D. C. and Dominguez I. (2010). Dynamic expression of a LEF-EGFP Wnt reporter in mouse development and cancer. Genesis 48, 183-194. 10.1002/dvg.20604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta R. and Fuchs E. (1999). Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development 126, 4557-4568. [DOI] [PubMed] [Google Scholar]
- Ferrer-Vaquer A., Piliszek A., Tian G., Aho R. J., Dufort D. and Hadjantonakis A. K. (2010). A sensitive and bright single-cell resolution live imaging reporter of Wnt/β-catenin signaling in the mouse. BMC Dev. Biol. 10, 121 10.1186/1471-213X-10-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karner C. M., Das A., Ma Z., Self M., Chen C., Lum L., Oliver G. and Carroll T. J. (2011). Canonical Wnt9b signaling balances progenitor cell expansion and differentiation during kidney development. Development 138, 1247-1257. 10.1242/dev.057646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A., Valerius M. T., Mugford J. W., Carroll T. J., Self M., Oliver M. and McMahon A. P. (2008). Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem. Cell 3, 169-181. 10.1016/j.stem.2008.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler P. S., Cheng P. F. and Eisenman R. N. (2002). N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 16, 2699-2712. 10.1101/gad.1021202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little M. H. and McMahon A. P. (2012). Mammalian kidney development: principles, progress, and projections. Cold Spring Harb. Perspect. Biol. 4, a008300 10.1101/cshperspect.a008300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretto S., Cordenonsi M., Dupont S., Braghetta P., Broccoli V., Hassan A. B., Volpin D., Bressan G. M. and Piccolo S. (2003). Mapping Wnt/b-catenin signaling during mouse development and in colorectal tumors. Proc. Natl. Acad. Sci. USA 100, 3299-3304. 10.1073/pnas.0434590100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marose T. D., Merkel C. E., McMahon A. P. and Carroll T. J. (2008). Beta-catenin is necessary to keep cells of ureteric bud/Wolffian duct epithelium in a precursor state. Dev. Biol. 314, 112-126. 10.1016/j.ydbio.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Schnell U., Karner C. M., Small E. V. and Carroll T. J. (2015). A Cre-inducible fluorescent reporter for observing apical membrane dynamics. Genesis 53, 285-293. 10.1002/dvg.22848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. S., Ma W., O'Brien L. L., Chung E., Guo J. J., Cheng J. G., Valerius M. T., McMahon J. A., Wong W. H. and McMahon A. P. (2012). Six2 and Wnt regulate self-renewal and commitment of nephron progenitors through shared gene regulatory networks. Dev. Cell 23, 637-651. 10.1016/j.devcel.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulkkinen K., Murugan S. and Vainio S. (2008). Wnt signaling in kidney development and disease. Organogenesis 4, 55-59. 10.4161/org.4.2.5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose S. D. and MacDonald R. J. (1997). Evolutionary silencing of the human Elastase I Gene (ELA1). Hum. Mol. Genet. 6, 897-903. 10.1093/hmg/6.6.897 [DOI] [PubMed] [Google Scholar]
- Stark K., Vainio S., Vassileva G. and McMahon A. P. (1994). Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature 372, 679-683. 10.1038/372679a0 [DOI] [PubMed] [Google Scholar]
- Trowe M.-O., Airik R., Weiss A.-C., Farin H. F., Foik A. B., Bettenhausen E., Schuster-Gossler K., Taketo M. M. and Kispert A. (2012). Canonical Wnt signaling regulates smooth muscle precursor development in the mouse ureter. Development 139, 3099-3108. 10.1242/dev.077388 [DOI] [PubMed] [Google Scholar]
- Trumpp A., Refaeli Y., Oskarsson T., Gasser S., Murphy M., Martin G. R. and Bishop J. M. (2001). c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 414, 768-773. 10.1038/414768a [DOI] [PubMed] [Google Scholar]
- Tycko B., Li C.-M. and Buttyan R. (2007). The Wnt/β-catenin pathway in Wilms tumors and prostate cancers. Curr. Mol. Med. 7, 479-489. 10.2174/156652407781387118 [DOI] [PubMed] [Google Scholar]
- Xu J., Wong E. Y., Cheng C., Li J., Sharkar M. T., Xu C. Y., Chen B., Sun J., Jing D. and Xu P. X. (2014). Eya1 interacts with Six2 and Myc to regulate expansion of the nephron progenitor pool during nephrogenesis. Dev. Cell 31, 434-447. 10.1016/j.devcel.2014.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Cunningham M., Arnold H., Chasse D., Monteith T., Ivaldi G., Hahn W. C., Stukenberg P. T., Shenolikar S., Uchida T. et al. (2004). A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 6, 308-318. 10.1038/ncb1110 [DOI] [PubMed] [Google Scholar]
- Yu J., Carroll T. J., Rajagopal J., Kobayashi A., Ren Q. and McMahon A. P. (2009). A Wnt7b-dependent pathway regulates the orientation of epithelial cell division and establishes the cortico-medullary axis of the mammalian kidney. Development 136, 161-171. 10.1242/dev.022087 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.