Abstract
The proteome composition of Echis carinatus carinatus venom (ECV) from India was studied for the first time by tandem mass spectrometry analysis. A total of 90, 47, and 22 distinct enzymatic and non-enzymatic proteins belonging to 15, 10, and 6 snake venom protein families were identified in ECV by searching the ESI-LC-MS/MS data against non-redundant protein databases of Viperidae (taxid 8689), Echis (taxid 8699) and Echis carinatus (taxid 40353), respectively. However, analysis of MS/MS data against the Transcriptome Shotgun Assembly sequences (87 entries) of conger E. coloratus identified only 14 proteins in ECV. Snake venom metalloproteases and snaclecs, the most abundant enzymatic and non-enzymatic proteins, respectively in ECV account for defibrinogenation and the strong in vitro pro-coagulant activity. Further, glutaminyl cyclase, aspartic protease, aminopeptidase, phospholipase B, vascular endothelial growth factor, and nerve growth factor were reported for the first time in ECV. The proteome composition of ECV was well correlated with its biochemical and pharmacological properties and clinical manifestations observed in Echis envenomed patients. Neutralization of enzymes and pharmacological properties of ECV, and immuno-cross-reactivity studies unequivocally point to the poor recognition of <20 kDa ECV proteins, such as PLA2, subunits of snaclec, and disintegrin by commercial polyvalent antivenom.
Introduction
According to the World Health Organization, snakebite is a major health challenge in tropical and sub-tropical countries including India, and therefore, it is considered as a neglected tropical disease1. The “big four” venomous snakes – Daboia russelli, Naja naja, Echis carinatus, and Bungarus caeruleus account for the most snakebite mortality and morbidity in the Indian subcontinent2. Notably, envenomation by the saw scaled viper (E. carinatus; EC) (Fig. 1a) also requires immediate medical attention; and therefore, it is considered as a category I medically important snake in India. Because the clinical symptoms and pathophysiological manifestations following envenomation may vary depending on the geographical origin of the snake, unveiling the complex venom proteome of a snake from a particular locale is extremely important for correlating venom composition with pharmacological properties and the pathophysiology of envenomation. Echis carinatus is divided into two sub-species namely E. c. sochureki and E. c. carinatus 3 of which the latter is confined to peninsular India and is responsible for many fatalities in parts of western and southern India4. Although Casewell and co-workers reported venom proteome composition of E. c. sochureki (from United Arab Emirates) by combination of transcriptomics and proteomics approaches5; however, to the best of our knowledge, no work has yet explored the venom proteome profile of E. c. carinatus from peninsular India or compared the data with pharmacological properties as well as clinical manifestations in Echis envenomed patients.
Figure 1.
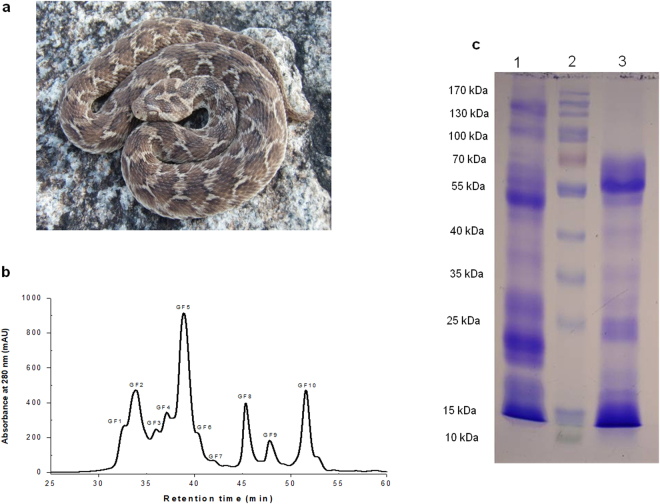
(a) Photograph of an Indian saw scaled viper (Echis carinatus). Photograph courtesy of Romulus Whitaker, Nov, 2010. (b) Fractionation of ECV (2.0 mg dry weight) on a Shodex KW- 803 gel filtration column coupled to Dionex Ultimate 3000 UHPLC system (Thermo Fisher Scientific, USA). The flow rate was maintained at 10 ml/h at room temperature (~23 °C) and fractions of 0.25 ml were collected. (c) SDS-PAGE analysis of crude ECV. Lane 1, 2 and 3 represents crude ECV (non-reduced), protein molecular markers and crude ECV (reduced) respectively.
Although traditional methods (chromatographic separation of venom proteins followed by biochemical assays) have long been used to investigate snake venom composition6–8, they have been inadequate to unravel the detail composition of complex venom proteomes. In contrast, the modern proteomic approach has eliminated most shortcomings of classical chromatographic techniques when followed by biochemical assays to identify the venom proteins9–13. Although tandem mass spectrometry-based protein identification is limited because of the absence of sufficient sequences in the protein database repository, it can contribute to the quantitative identification of venom proteomes14.
The administration of polyvalent antivenom (raised against the big four venomous snakes in India) is the only choice of therapy against snake envenomation15. For an antivenom therapy to be completely efficacious, all toxins of the venom would need to be recognized by the antivenom with neutralization of the toxicity and venom-induced lethality10–12,16. Because clinical manifestations of snake envenoming depend primarily on venom composition, the efficacy of commercial equine polyvalent antivenom (PAV) also depends on the geographical source of venom used for antivenom production. Therefore, the commercial antivenom may lack its potency to neutralize venom induced lethality in geographical locales situated distant from the source of immunizing venoms17. Consequently, the venom-antivenom cross-reactivity is important for the information it provides to improve the efficacy of commercial antivenom and the subsequent efficient treatment of snakebites11,12.
In this study, the previously unexplored venom proteome composition of E. c. carinatus venom (ECV) from southern India was unraveled by proteomic analysis and the venom toxin profile was correlated with biochemical and pharmacological properties of venom and the clinical manifestations that follow ECV envenomation in southern India. An attempt has also been made to determine the venom-antivenom cross-reactivity and potency of commercial PAVs in neutralizing the enzymatic activities and some pharmacological properties of ECV and its fractions.
Results and Discussion
Proteomic analysis reveals the occurrence enzymatic and non-enzymatic proteins in ECV
De-complexation of ECV by U-HPLC gel filtration chromatography on Shodex KW-803 resolved it into 10 peaks (Fig. 1b). SDS-PAGE analyses of crude venom under reduced and non-reduced conditions confirmed the dynamic range of proteins present in the venom under study (Fig. 1c). The analysis of the protein bands from crude ECV suggested the predominance of proteins in the mass ranges of 55–90 kDa and 10–20 kDa. Viperidae snake venom proteins in the mass range of 55–90 kDa are usually represented by snake venom metalloprotease (SVMP), L-amino acid oxidase (LAAO), and nucleotidase (NT) or adenosine monophosphatase (AMPase)18–20, whereas the venom proteins in the mass range of 10–20 kDa include phospholipase A2 (PLA2), disintegrins, and snaclec monomers21–23. The MALDI-TOF-MS analysis of gel filtration fractions of ECV at different mass ranges suggested the presence of wide range of distinct ions (307) in the m/z range of 5.0–150.2 kDa (Supplementary Table S1). However, several of these ECV proteins may not be the real gene products but at least some of these may have produced by autocatalysis of venom proteins. Noteworthy, MALDI-TOF-MS data representing a “fingerprint” of ECV from southern India would be useful for comparing venom proteins from congeneric snakes from different locales. ECV proteins in the mass range of 41–100 kDa represent 47.5% of venom proteins followed by very high molecule weight (>100 kDa) proteins (23.5%) whereas the low molecular mass proteins (5–20 kDa) are the least abundant ECV proteins (Supplementary Table S1).
Tandem mass spectrometry coupled to protein database search has evolved as a gold standard for snake venom protein identification11–13. Although, several isotope labeling methods such as SILAC, ICAT, and iTRAQ, are applied for mass spectrometry-based protein quantification24, albeit some limitations of these techniques may restrict their use for quantification of venom proteins24,25. On the contrary, the label free protein quantification techniques such as spectral count (MS2) and area-based (MS1) methods in addition to circumvent the above disadvantages are also comparable to other approaches of protein quantification including isotope labeling and mass spectral peak intensities26. Nevertheless, label free protein quantification methods are not devoid of technical difficulties like protein size, availability of trypsin cleavage sites along the protein, and lack of a comprehensive snake venom protein databases24,27. Moreover, the relative abundance of a protein in snake venom invariable depends on the number of peptides identified which in turn is influenced by the degree of sequence similarity between homologous sequences present in the database27. Therefore, in this study to avoid unambiguous protein identification as well as to improve the protein quantification, the semi-tryptic sequences were also considered, the spectral count as well as the area under peptides were normalized by the number of theoretical peptides13,24,27, and at least one overlapping distinct peptide was considered for ECV protein identification13. Further, to enhance the relevance of the venom proteomic analysis we have compared the ECV proteome with the published reports on venom proteome composition of congeneric snakes from other locales5.
In the present study, approximately 10% of the collected MS/MS spectra were matched to peptide sequences with high confidence. The collected MS/MS spectra usually show 10–30% high confidence matching to peptide sequences and several factors, for example existence of MS/MS spectra with non-canonical fragment ions that are excluded by database search engines, absence of the peptide sequences in the target database, and unanticipated post-translational modifications can be attributed to this effect28. The ESI-LC-MS/MS analyses of tryptic peptides generated from the gel filtration fractions of ECV showed sequence similarities with more than 1500 proteins deposited in Viperidae venom protein databases. Our stringent protein identification process; however, eliminated redundancies and led to a final recognition of 90 distinct enzymatic and non-enzymatic proteins belonging to 15 snake venom protein families of ECV (Table 1, Fig. 2a, Supplementary Table S2). Conversely, analysis of LC-MS/MS data against the genus (Echis) database entries in the NCBI returned with 665 protein entries; however, only 47 distinct proteins belonging to 10 snake venom families were identified in ECV proteome (Fig. 2b, Supplementary Table S3a,b). Further, only 22 distinct proteins distributed across 6 venom protein families were identified in the target Echis carinatus database (Fig. 2c, Supplementary Table S4a,b). Noteworthy, searching the LC-MS/MS data against the Transcriptome Shotgun Assembly (TSA) sequences of congeneric Echis coloratus venom resulted in identification of only 14 proteins belonging to 8 venom protein families (Fig. 2d, Supplementary Table S5a,b). Because mass-spectrometry-based proteomic analysis is database-dependent29; therefore, it is quite reasonable to understand that the difference in number of proteins identified in ECV proteome by searching the different protein databases was dependent on the database entry of genus or species-specific snake venom proteins. Consequently, several protein families such as vascular endothelial growth factor (VEGF), Kunitz-type protease inhibitor (KSPI), aminopeptidase (APase), NT, and glutaminyl cyclase (GC) of ECV identified by searching the Viperidae databases were not identified in Echis as well as E. carinatus venom protein databases due to limitations in genus and species-specific database entries.
Table 1.
Summary of different proteins identified in Indian Saw Scaled Viper (E. c. carinatus) venom by tandem mass spectrometry analysis of gel filtration peaks against Viperidae family of proteins.
| S. No. | Accession No. | Description | Source organism | −10lgP | Coverage (%) | Distinct Peptides | Avg. Mass (Da) | GF peak(s) |
|---|---|---|---|---|---|---|---|---|
| Enzymatic proteins | ||||||||
| P- I Metalloprotease (1 protein) | ||||||||
| 1 | gi|727360735 | Snake venom metalloproteinase K, partial | Echis coloratus | 102.69 | 8 | 1 | 48851 | 1,3–7 |
| P- II Metalloprotease (5 proteins) | ||||||||
| 1 | gi|297594086 | Metalloproteinase | Echis carinatus sochureki | 114.32 | 34 | 2 | 12578 | 3–8,10 |
| 2 | gi|297593820 | Metalloproteinase, partial | Echis carinatus sochureki | 113.74 | 15 | 3 | 28319 | 1,3–7 |
| 3 | gi|297594078 | Metalloproteinase | Echis carinatus sochureki | 100.95 | 9 | 2 | 53640 | 3,7–10 |
| 4 | gi|297594068 | Metalloproteinase | Echis coloratus | 98.47 | 31 | 2 | 13935 | 3,4–8 |
| 5 | gi|297594070 | Metalloproteinase, partial | Echis carinatus sochureki | 77.02 | 8 | 1 | 27679 | 5,7–10 |
| P- III Metalloprotease (18 proteins) | ||||||||
| 1 | gi|297593950 | Metalloproteinase | Echis coloratus | 165.72 | 13 | 3 | 68942 | 1–3,7 |
| 2 | gi|300079900 | Factor X activator heavy chain | Daboia russellii russellii | 153.11 | 11 | 2 | 69521 | 1–7,9,10 |
| 3 | gi|297593790 | Metalloproteinase, partial | Echis carinatus sochureki | 149.47 | 17 | 3 | 63999 | 1–8,9 |
| 4 | gi|297593830 | Metalloproteinase, partial | Echis carinatus sochureki | 147.78 | 22 | 3 | 35963 | 1,3–8 |
| 5 | gi|297593798 | Metalloproteinase, partial | Echis carinatus sochureki | 145.07 | 25 | 5 | 39004 | 3,7.8,10 |
| 6 | gi|297593862 | Metalloproteinase, partial | Echis coloratus | 130.71 | 14 | 1 | 57183 | 1,3,7 |
| 7 | gi|162329887 | Chain A, Crystal Structure Of Russell’s Viper Venom Metalloproteinase | Daboia russellii russellii | 124.48 | 13 | 1 | 47646 | 1–3,4–8 |
| 8 | gi|83523634 | Group III snake venom metalloproteinase | Echis ocellatus | 119.25 | 15 | 2 | 69598 | 1–3,7–8 |
| 9 | gi|297593842 | Metalloproteinase, partial | Echis carinatus sochureki | 116.46 | 26 | 2 | 27821 | 1,3,7–8 |
| 10 | gi|297593794 | Metalloproteinase | Echis carinatus sochureki | 115.08 | 7 | 1 | 68205 | 1,3–7 |
| 11 | gi|297593836 | Metalloproteinase, partial | Echis carinatus sochureki | 107.73 | 9 | 3 | 41105 | 1,7–8 |
| 12 | gi|297593852 | Metalloproteinase, partial | Echis coloratus | 103.55 | 10 | 1 | 56407 | 3,7,8,10 |
| 13 | gi|320579375 | Group III snake venom metalloproteinase, partial | Echis ocellatus | 103.03 | 9 | 1 | 63254 | 3,8 |
| 14 | gi|297593822 | Metalloproteinase, partial | Echis carinatus sochureki | 99.09 | 11 | 2 | 35935 | 1,2 |
| 15 | gi|297593854 | Metalloproteinase | Echis coloratus | 91.01 | 5 | 1 | 69286 | 1,3,7,8,10 |
| 16 | gi|297593858 | Metalloproteinase | Echis coloratus | 72.99 | 5 | 1 | 68727 | 3 |
| 17 | gi|52000738 | Snake venom metalloproteinase HT-1 | Crotalus ruber ruber | 62.04 | 7 | 1 | 23601 | 3,7 |
| 18 | gi|297593984 | Metalloproteinase, partial | Echis pyramidum leakeyi | 53.13 | 5 | 1 | 49052 | 3,7 |
| Phospholipase A 2 (12 proteins) | ||||||||
| 1 | gi|298351762 | Basic phospholipase A2 3 | Daboia russellii russellii | 198.21 | 69 | 2 | 13687 | 7,8,10 |
| 2 | gi|81174981 | Basic phospholipase A2 VRV-PL-V | Daboia russellii russellii | 183.48 | 46 | 1 | 13587 | 8,10 |
| 3 | gi|71912229 | Phospholipase A2 | Daboia russellii limitis | 183.45 | 75 | 4 | 15353 | 7,8,9,10 |
| 4 | gi|82096307 | Acidic phospholipase A2 EC-I | Echis carinatus | 178.05 | 59 | 3 | 15523 | 6,7 |
| 5 | gi|149243451 | Chain A, Crystal Structure Of A New Isoform Of Phospholipase A2 From Russells Viper | Daboia russellii russellii | 121.82 | 60 | 2 | 13452 | 3,10 |
| 6 | gi|87130858 | Phospholipase A2-III | Daboia russellii russellii | 83.27 | 27 | 2 | 13441 | 7 |
| 7 | gi|163311140 | Ecarpholin S | Echis carinatus | 69.3 | 25 | 2 | 13819 | 10 |
| 8 | gi|59727030 | D1E6b Phospholipase A2 | Cerrophidion godmani | 68.37 | 10 | 1 | 15970 | 7 |
| 9 | gi|1041577114 | Phospholipase A2 1 h | Agkistrodon contortrix contortrix | 63.46 | 10 | 1 | 15961 | 7,10 |
| 10 | gi|1041577423 | Phospholipase A2 1 h | Agkistrodon piscivorus | 60.88 | 6 | 1 | 15748 | 7 |
| 11 | gi|71912223 | Basic phospholipase A2 | Daboia russelii | 43.99 | 13 | 1 | 15461 | 10 |
| 12 | gi|157834128 | Chain A, Anticoagulant Class Ii Phospholipase A2 | Vipera russelli russelli | 33.79 | 25 | 1 | 13626 | 10 |
| Serine protease (11 proteins) | ||||||||
| 1 | gi|134129 | Factor V activator RVV-V alpha | Daboia siamensis | 115.68 | 34 | 6 | 26182 | 3–7,9,10 |
| 2 | gi|311223824 | Serine beta-fibrinogenase-like protein precursor | Daboia siamensis | 91.84 | 11 | 2 | 28035 | 2–7 |
| 3 | gi|297593766 | Serine protease, partial | Echis coloratus | 76.47 | 8 | 2 | 28863 | 2–3,6–8 |
| 4 | gi|293491172 | Serine protease, partial | Echis ocellatus | 73.07 | 14 | 1 | 28236 | 2,3,6,7 |
| 5 | gi|306756038 | Serine protease VLSP-3 precursor | Macrovipera lebetina | 66.96 | 14 | 1 | 28352 | 3 |
| 6 | gi|297593764 | Serine protease, partial | Echis carinatus sochureki | 63.79 | 10 | 2 | 26218 | 7,8,10 |
| 7 | gi|13959655 | Venom serine proteinase-like protein 2 | Macrovipera lebetina | 57.74 | 6 | 1 | 28894 | 3,7 |
| 8 | gi|297593786 | Serine protease, partial | Echis coloratus | 56.82 | 5 | 1 | 28184 | 3,7,8 |
| 9 | gi|306756034 | Serine protease VLSP-1 precursor | Macrovipera lebetina | 54.18 | 6 | 1 | 28702 | 3 |
| 10 | gi|298351881 | Snake venom serine protease rhinocerase | Bitis rhinoceros | 53.95 | 28 | 2 | 9978 | 3,7 |
| 11 | gi|297593736 | Serine protease, partial | Echis coloratus | 51.71 | 8 | 2 | 26001 | 3 |
| L-amino acid oxidase (3 proteins) | ||||||||
| 1 | gi|194400545 | Secreted L-amino acid oxidase precursor | Daboia russelii | 212.00 | 29 | 10 | 56888 | 1–10 |
| 2 | gi|727360693 | L-amino acid oxidase B variant 2 | Echis coloratus | 115.20 | 15 | 1 | 35024 | 1 |
| 3 | gi|1002590400 | PREDICTED: L-amino-acid oxidase isoform × 1 | Protobothrops mucrosquamatus | 104.80 | 8 | 1 | 57159 | 1,3,7,10 |
| Phosphodiesterase (1 protein) | ||||||||
| 1 | gi|586829527 | Phosphodiesterase | Macrovipera lebetina | 124.14 | 8 | 1 | 96181 | 1,3 |
| Nucleotidase (1 protein) | ||||||||
| 1 | gi|211926756 | Ecto-5’-nucleotidase | Gloydius brevicaudus | 102.06 | 9 | 1 | 64434 | 1,3,6 |
| Glutaminyl cyclase (1 protein) | ||||||||
| 1 | gi|380846517 | Glutaminyl-peptide cyclotransferases | Daboia russelii | 84.26 | 6 | 2 | 42116 | 6,9 |
| Aspartic proteases (1 protein) | ||||||||
| 1 | gi|109287596 | Renin-like aspartic protease | Echis ocellatus | 66.41 | 5 | 2 | 43872 | 2,3 |
| Aminopeptidase (1 protein) | ||||||||
| 1 | gi|743538216 | Endoplasmic reticulum aminopeptidase 1-like protein | Crotalus horridus | 86.52 | 5 | 4 | 107397 | 1–2,4,6 |
| Phospholipase B (1 protein) | ||||||||
| 1 | gi|727360709 | Phospholipase B | Echis coloratus | 80.51 | 6 | 3 | 64541 | 1,2,3 |
| Non enzymatic proteins | ||||||||
| Snaclec (20 proteins) | ||||||||
| 1 | gi|55670410 | Chain B, Structure Of Ems16-Alpha2-I Domain Complex | Echis multisquamatus | 123.97 | 45 | 6 | 15099 | 1,2–9 |
| 2 | gi|300490474 | P68 alpha subunit | Daboia russellii limitis | 122.92 | 23 | 1 | 18108 | 1 |
| 3 | gi|727360769 | C-type lectin J | Echis coloratus | 116.83 | 23 | 2 | 18279 | 1–8,10 |
| 4 | gi|758377456 | C-type lectin-like protein 3A | Macrovipera lebetina | 115.87 | 37 | 2 | 18079 | 1–3,7–8 |
| 5 | gi|38493055 | Chain A, Crystal Structure Of Ems16 | Echis multisquamatus | 114.95 | 49 | 4 | 15874 | 1,2,3,7,8 |
| 6 | gi|300490472 | P68 beta subunit | Daboia siamensis | 110.56 | 32 | 1 | 16910 | 1–3,5–8 |
| 7 | gi|802148 | Echicetin beta subunit | Echis carinatus | 104.7 | 31 | 1 | 14869 | 3–7,9 |
| 8 | gi|32452854 | Echicetin A-chain | Echis carinatus | 103.17 | 31 | 5 | 15363 | 1,2–9 |
| 9 | gi|40889261 | Chain B, Echicetin | Echis carinatus | 101.88 | 29 | 1 | 14922 | 3,4,6–9 |
| 10 | gi|758377450 | C-type lectin-like protein 3B | Macrovipera lebetina | 94.95 | 26 | 1 | 17043 | 1,5–8 |
| 11 | gi|998226344 | C-type lectin 2 | Bitis arietans | 93.67 | 32 | 1 | 17891 | 1,3,4,6,7 |
| 12 | gi|300490462 | Dabocetin alpha subunit | Daboia russellii russellii | 86.53 | 27 | 2 | 17493 | 3,4,7 |
| 13 | gi|218526484 | Snaclec A13 | Macrovipera lebetina | 77.52 | 15 | 2 | 15308 | 3,5–8 |
| 14 | gi|82175557 | Snaclec salmorin subunit A | Gloydius brevicaudus | 53.46 | 6 | 1 | 17293 | 1,3,7,8 |
| 15 | gi|300490478 | P31 alpha subunit | Daboia russellii limitis | 52.92 | 14 | 1 | 18179 | 3,4 |
| 16 | gi|578004418 | Lebecin B, partial | Macrovipera lebetina | 52.69 | 18 | 1 | 17553 | 4–6,8 |
| 17 | gi|538259843 | C-type lectin factor IX/X binding protein B subunit, partial | Protobothrops flavoviridis | 43.47 | 8 | 1 | 12795 | 3,4–7 |
| 18 | gi|205275155 | C-type lectin | Echis ocellatus | 36.75 | 9 | 1 | 16882 | 3 |
| 19 | gi|2829697 | Snaclec echicetin subunit alpha | Echis carinatus sochureki | 36.05 | 14 | 1 | 15803 | 3,7 |
| 20 | gi|82174836 | Snaclec coagulation factor IX/factor X-binding protein subunit B | Echis carinatus | 34.43 | 7 | 1 | 14372 | 1,7,9 |
| Disintegrin (6 proteins) | ||||||||
| 1 | gi|82194569 | Disintegrin | Echis carinatus | 151.29 | 89 | 1 | 7127 | 3–8,10 |
| 2 | gi|544584743 | Disintegrin EC6 subunit alpha | Echis carinatus sochureki | 114.32 | 34 | 2 | 12578 | 3–8,10 |
| 3 | gi|182705265 | Disintegrin schistatin-like subunit B | Echis carinatus | 104.43 | 52 | 2 | 6961 | 1,3,6–8,10 |
| 4 | gi|82203514 | Disintegrin gabonin-1 | Bitis gabonica | 94.79 | 18 | 2 | 13792 | 4–5,7 |
| 5 | gi|182705262 | Disintegrin VLO4 | Macrovipera lebetina obtusa | 91.67 | 46 | 1 | 7108 | 7 |
| 6 | gi|110346540 | RTS-containing short disintegrin, partial | Echis ocellatus | 46.91 | 23 | 1 | 4588 | 10 |
| Vascular endothelial growth factor (1 protein) | ||||||||
| 1 | gi|48429241 | Snake venom vascular endothelial growth factor toxin ICPP | Macrovipera lebetina | 57.74 | 10 | 1 | 12574 | 3,5–6 |
| Cystine rich secretory proteins (3 proteins) | ||||||||
| 1 | gi|190195321 | Cysteine-rich seceretory protein Dr-CRPK | Daboia russelii | 125.59 | 22 | 4 | 26688 | 7 |
| 2 | gi|803374854 | Cysteine-rich venom protein | Echis coloratus | 116.84 | 19 | 1 | 24699 | 7 |
| 3 | gi|1041577503 | Cysteine-rich secretory protein 1 | Agkistrodon piscivorus | 84.05 | 12 | 1 | 26681 | 7 |
| Kunitz type serine protease inhibitor (3 proteins) | ||||||||
| 1 | gi|159883522 | Trypsin inhibitor-3 precursor | Daboia siamensis | 118.27 | 51 | 2 | 9443 | 10 |
| 2 | gi|239977245 | Kunitz-type serine protease inhibitor B1 | Daboia siamensis | 69.75 | 31 | 1 | 9318 | 10 |
| 3 | gi|123913154 | Kunitz-type serine protease inhibitor 4 | Daboia russellii russellii | 67.17 | 14 | 1 | 9145 | 7–8,10 |
| Nerve growth factor (1 protein) | ||||||||
| 1 | gi|400499 | Venom nerve growth factor | Daboia russellii russellii | 61.53 | 21 | 2 | 13283 | 3 |
Figure 2.
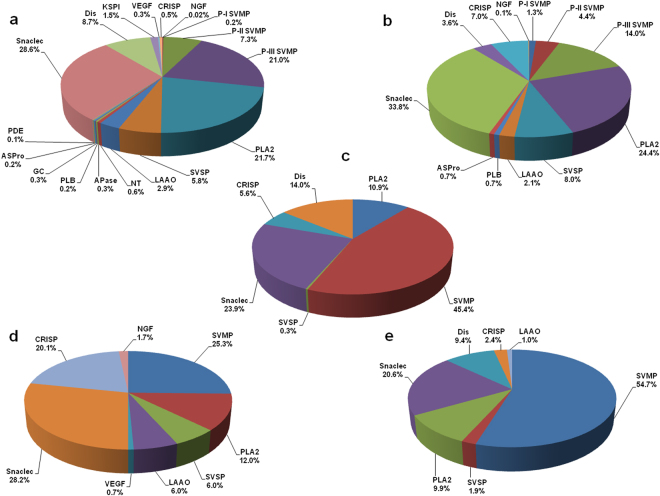
Protein family composition of ECV proteome. The pie chart represents the relative occurrence of different enzymatic and non-enzymatic protein families of ECV when the data was searched against NCBI protein entries with taxonomy set to (a) Viperidae (taxid 8689), (b) Echis (taxid 8699), (c) E. carinatus (taxid 40353), and (d) Transcriptome Shotgun Assembly (TSA) sequences of E. coloratus (BioProject: PRJEB2884). (e) Composition of E. c. sochurecki (UAE) venom as predicted from venom gland transcriptomic analysis5.
The proteomic composition of E. c. carinatus venom, when searched against species-specific protein databases (Fig. 2c) corroborates well with the published transcriptomic data of E. c. sochureki venom gland (Fig. 2e)5; nevertheless, the minor variation in the relative abundances of venom proteins in these two congeneric snakes reflects the sub-species-specific difference and/or geographical variation in snake venom composition. Notably, the LAAO identified in E. c. sochureki venom by transcriptomic approach5 was not identified by species-specific database analysis of ECV, although this enzyme was identified in the venom under study by biochemical analysis as well as by genus-specific database search (Fig. 2b, Supplementary Table S3a,b). This result suggests the absolute necessity of species-specific transcriptomic database in proteomic study29,30. Further, identification of few homologous protein isoforms in ECV by searching the TSA sequences of E. coloratus may presumably be explained on the basis of venom variability between the two species of Echis.
Due to lack of species-specific (E. c. carinatus) transcriptomic database we have applied both MS1 and MS2-based quantitative approaches for determining the relative protein abundance in ECV and compared their results with each-other. The relative abundance of each protein class calculated by both MS1 (area based) and MS2 (spectral count)-based methods did not show significant deviation (Supplementary Fig. S1a–h); therefore, the relative abundance of ECV proteins is presented as an average data determined from MS1 and MS2-based methods (Fig. 2a–d). The alignments of MS/MS-derived peptide sequences with the homologous proteins from Viperidae, Echis, E. carinatus and translated E. coloratus TSA sequence databases are shown in Supplementary Fig. S2a–d. Some of the MS/MS spectra corresponding to mass (Da) 1250.5966 (2+), 118.5608 (3+), 733.3646 (2+), 798.3912 (2+), 929.4719 (2+), 1741.8192 (2+), 2942.252 (2+), 907.4837 (2+) and 1103.5645 (2+) are shown in Supplementary Fig. S3.
It is noteworthy to mention that among the LC-MS/MS-identified protein families, GC, aspartic protease (ASPro), APase, phospholipase B (PLB), VEGF, and nerve growth factor (NGF) were shown to be present in ECV for the first time. By proteomic analysis, the enzymatic proteins identified in ECV include SVMP, PLA2, snake venom serine proteases (SVSP), LAAO, NT, APase, PLB, GC, ASPro, and phosphodiesterase (PDE) (Fig. 2c).
Proteases of snake venoms are broadly classified as SVMP and SVSP31. The proteomic analysis suggested the presence of enzymes from both groups of proteases in ECV; nevertheless, the species-specific database search demonstrated predominance of SVMP group in ECV (Fig. 2c) which corroborates well with the venom proteins composition of E. c. sochureki determined by transcriptomic analysis (Fig. 2e). Remarkably, the relative abundance of SVMP in ECV (Fig. 2c) was found to be lower as compared to SVMP content determined by proteomic analysis in the venoms of its congeneric species such as E. coloratus, E. ocellatus, E. pyramidum leakeyi and E. c. sochureki 5. Interestingly, the relative abundance of SVMP in ECV varies significantly depending on the target databases (Fig. 2a–d) owing to different number of entries of venom proteins in these databases. The biochemical analyses suggested that GF 1, followed by GF 2, exhibited the highest metalloprotease activity (Table 2), which was supported by LC-MS/MS analysis of the gel filtration peaks (Table 1). Further, SVMPs are grouped to PI, PII and PIII classes based on their size and domain structure32. Analysis of LC-MS/MS data against Viperidae venoms showed that ECV was predominated by PIII-SVMPs (17.9% of the ECV proteome) (Table 1; Fig. 2c). Because SVMP is the most abundant component in Echis venom, therefore the significant variation in SVMP content among Echis spp. may lead to differences in severity of clinical manifestations and pharmacological activity post Echis spp. bite.
Table 2.
In vitro pharmacological properties and associated enzymatic activities of crude ECV and its GF fractions. Values are mean ± SD of triplicate determinations. Average PT and APTT of control PPP was determined at 18 ± 0.0 s and 35.0 ± 0.9 s, respectively. Platelet count of control (untreated) sample was 2.5 ± 0.1 × 106 cells/ml. Significance of difference with respect to control (ap < 0.05). The ( + ) sign indicates presence whereas (−) sign indicates absence of fibrinogen clotting activity.
| Venom fraction | Protein content (% yield) | Pharmacological Properties | Enzyme activity | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasma clotting activity (Ca2+-clotting time) U/mg (x104) | Coagulant (C) or Anticoagulant (AC) | PT (s) | APTT (s) | Percent decrease in platelet count (%) | Fibrinogen clotting activity | PLA2 (U/mg × 103) | SVMP (U/mg) | TAME (U/mg) | BAEE (U/mg × 103) | ||
| ECV | 100.0 ± 0.4 | 4.3 ± 0.0004 | C | 15.5 ± 0.7a | 15.5 ± 2.12a | 62.1 ± 3.1a | + | 7.2 ± 0.6 | 0.6 ± 0.01 | 5.0 ± 0.2 | 0.8 ± 0.04 |
| GF-1 | 6.9 ± 0.4 | 32.3 ± 0.0006 | C | 17.0 ± 0.0a | 17.0 ± 1.41a | 56.7 ± 2.5a | − | 0.4 ± 0.02 | 2.2 ± 0.06 | − | 1.6 ± 0.006 |
| GF-2 | 9.0 ± 0.2 | 26.7 ± 0.0007 | C | 17.5 ± 0.4 | 24.5 ± 2.12a | 66.3 ± 2.9a | + + | 0.3 ± 0.01 | 1.4 ± 0.04 | 103.3 ± 5.2 | 4.1 ± 0.3 |
| GF-3 | 4.7 ± 0.3 | 26.3 ± 0.0010 | C | 16.0 ± 0.0a | 21.5 ± 0.7a | 31.8 ± 1.1a | + | 0.5 ± 0.03 | 0.9 ± 0.04 | 140.0 ± 7.0 | 3.5 ± 0.1 |
| GF-4 | 6.9 ± 0.3 | 20.3 ± 0.0010 | C | 16.5 ± 0.1a | 24.5 ± 0.7a | 35.7 ± 1.6a | + | 0.1 ± 0.05 | 0.7 ± 0.01 | 61.7 ± 3.1 | 2.6 ± 0.09 |
| GF-5 | 15.2 ± 0.5 | 18.3 ± 0.0005 | C | 18.5 ± 0.7 | 30.5 ± 0.7 | 25.7 ± 0.8a | + | 0.3 ± 0.2 | 0.3 ± 0.02 | 80 ± 3.6 | 1.9 ± 0.07 |
| GF-6 | 5.1 ± 0.3 | 14.3 ± 0.0008 | C | 15.5 ± 0.7a | 26.0 ± 1.4a | 26.4 ± 0.7a | − | 9.9 ± 0.4 | 0.5 ± 0.09 | − | 1.2 ± 0.04 |
| GF-7 | 11.3 ± 0.8 | 15.3 ± 0.0008 | AC | 18.0 ± 0.1 | 32.0 ± 0.2 | 34.5 ± 1.2a | − | 12.1 ± 0.7 | 0.4 ± 0.02 | − | 0.8 ± 0.01 |
| GF-8 | 15.1 ± 0.3 | 134.3 ± 0.0014 | AC | 18.0 ± 0.2 | 30.5 ± 0.7 | 51.1 ± 1.9a | − | 18.1 ± 0.9 | 0.5 ± 0.03 | − | 0.5 ± 0.03 |
| GF-9 | 7.0 ± 0.2 | 128.0 ± 0.0014 | AC | 18.0 ± 0.2 | 38.5 ± 0.7 | 12.7 ± 0.3 | − | 12.1 ± 0.7 | 0.2 ± 0.02 | − | 0.2 ± 0.01 |
| GF-10 | 4.4 ± 0.1 | 11.7 ± 0.0007 | AC | 18.0 ± 0.1 | 42.5 ± 2.12 | 50.3 ± 2.1a | − | 5.9 ± 0.2 | 0.5 ± 0.02 | − | 0.04 ± 0.01 |
Significant esterolytic activity exhibited by proteins was eluted in GF 2 to GF 5 (Table 2), which may be correlated with the presence of quantitatively higher amounts of pro-coagulant serine proteases, in these fractions (Table 1). Depending on target database search the relative abundance of SVSPs was found to vary in ECV proteome (Fig. 2a–d, Table 1). The relative abundance of SVSP (determined by searching the species-specific protein databases) in ECV was found to be comparable with the relative abundance of this class of protease determined by proteomic analysis in the venom of other species of Echis 5.
The molecular mass of PLA2 from snake venoms is reported to be in the range of 10–15 kDa33. In accordance with its molecular weight, it was mostly eluted in the GF 7–9 fractions (Table 1). In the case of Russell’s viper venom (RVV), PLA2s have been shown to interact with high molecular weight components of venom leading to their elution in the initial gel filtration fractions of RVV11,12. Interestingly, the present study also found the elution of trace quantities of PLA2 in GF 3 (Table 1). The relative abundance of PLA2 determined by analyzing data against Viperidae and Echis venoms (2a-b) was found to be two-fold then its relative abundance determined by species-specific database search (2c) which may presumably due to less entry of this important class of venom protein in latter database. The relative abundance of PLA2 in ECV (10.9%) determined by species-specific database search (Fig. 2c) was comparable to the relative abundance of the same enzyme in venoms of E. coloratus, E. ocellatus, and E. c. sochureki albeit PLA2 content of E. pyramidum leakeyi venom was found to be higher5 which may have an evolutionary significance34.
ATPase, ADPase, AMPase, and PDE are poorly characterized, high molecular mass (>50 kDa) proteins of snake venom20,35. Our biochemical assays showed that ECV contains ATPase (6.3 ± 0.006 × 104 U/mg), ADPase (3.6 ± 0.300 × 104 U/mg), AMPase/nucleotidase (8.2 ± 0.4 × 103 U/mg), and PDE (18.6 ± 0.08 U/mg) enzyme activities. Nevertheless, ATPase and ADPase from ECV could not be identified by proteomic analysis (in the present study) owing to a limited protein database deposition11,12 or by transcriptomic analysis of venom glands of Echis spp 5,30. Occurrence of PDE in ECV could not be identified when the LC-MS/MS data were searched against the genus and species-specific databases (Fig. 2b,c) or by the transcriptomic analysis of E. c. sochureki venom gland for the reason stated above; however, searching the MS/MS data against Viperidae venom proteins resulted in identification of PDE as a minor component of the ECV (Table 1, Fig. 2a)11,12,36.
LAAOs are high molecular weight (60–150 kDa) snake venom enzymes19,37. ECV demonstrated an LAAO specific activity of 0.3 ± 0.01 U/mg. Depending upon the venom protein / transcriptomic database search the relative abundance of LAAO isoenzymes in ECV proteome demonstrated significant variation (2.1–6%) (Fig. 2a,b,d, Table 1, Supplementary Tables 2–5). However, due to limited database entry LAAO could not be identified in ECV proteome by species-specific database search (Fig. 2c, Supplementary Table S4a,b). The LAAO was also reported to be a minor component in venom of other species of Echis 5,30.
Hyaluronidase activity, at the tested dose of 3 µg/ml, was not found by the biochemical assay or by the LC-MS/MS analysis of ECV or by proteomic and transcriptomic analyses of venoms of Echis spp 5. Nevertheless, Urs et al.38 and Girish et al.39 demonstrated the presence of hyaluronidase in ECV, albeit they used a much higher concentration of venom (200 µg/ml) in their enzyme assay.
Proteomic analysis identified non-enzymatic proteins in ECV, including snaclec, disintegrin, Kunitz-type protease inhibitor (KSPI), VEGF, cysteine-rich secretory protein (CRISP), and NGF (Fig. 2a–d).
C-type lectin (snaclec), the Ca2+-dependent non-enzymatic proteins9,40 of snake venom, are found to be the most abundant non-enzymatic group of proteins in the ECV proteome; however, depending on non-redundant protein / TSA sequence database search their relative occurrence in ECV was demonstrated to vary between 24–34% (Fig. 2a–d, Table 1, Supplementary Tables 2–5). A comparison shows that the snaclec content of ECV proteome (Fig. 2c) was 3–4 folds higher as compared to the snaclec content in other species of Echis 5. Because snaclec are one of the major components of Echis venom responsible for adverse pharmacological effects in bite victims (see below) therefore, this species-specific variation in snaclec content in different species of Echis may have a profound clinical significance.
The disintegrins are cysteine rich non-enzymatic proteins characterized with a conserved arginine-glycine-aspartic acid (RGD) motif41. They were the second most abundant non-enzymatic proteins in ECV (Fig. 2c) and among the identified disintegrins, 5 isoforms (gi|82194569, gi|544584743, gi|182705265, gi|82203514, and gi|182705262) belong to dimeric disintegrin sub-family. The disintegrin content of ECV (14%), determined by species-specific protein database search (Fig. 2c), was found to be slightly higher than the disintegrin content determined by proteomic as well as transcriptomic analysis of E. c. sochureki venom5.
Apart from the above mentioned proteins, single isoforms of actin and myosin were also identified by proteomic analysis against Viperidae family of venom proteins (data not shown). Nevertheless, they are not venom components and may be contamination in the ECV from the venom extraction. Considering their likely origin and low abundance (0.3%), they were not considered in calculating the relative abundance of proteins in ECV.
The proteome composition of ECV is well correlated with its in vitro pharmacological properties (coagulopathy) and clinical manifestations after envenomation
Venoms of the Viperidae family of snakes are reported to influence the hemostatic system of the victim, a leading cause of death among viper bite patients7,11,42. Nevertheless, blood clotting effects are more pronounced in cases of Echis bite, compared to Russell’s viper bite43. Local symptoms of envenomation including persistent local swelling, edema, bleeding/haemorrhage, and pain at the bite site and systemic envenoming symptoms such as hemotoxicity and coagulopathy have been observed as severe clinical symptoms in E. carinatus bite patients in southern India and other parts of the Indian subcontinent43–45 (personal communication from Dr. A. Zachariah, Christian Medical College, Vellore, and R. Whitaker, herpetologist, Madras Snake Park, Chennai).
The yield of venom per milking from an adult saw-scaled viper (0.8–1.0 feet in length) is approximately 12 mg43. Therefore, the concentration of ECV in the blood of an adult human after a full bite would be expected to be in the range of 2.5–3.0 µg/ml. The assays of blood coagulation activity suggest that crude ECV (3.0 µg/ml) and GF 1–6 fractions were strongly pro-coagulant, whereas the proteins from GF 8 and 9 demonstrated anticoagulant activity (Table 2). The pro-coagulant activity of ECV is also more pronounced than the activity displayed by crude RVV12,43. SVMPs and SVSPs have been shown to influence the hemostatic system by activation of blood coagulation factor Xa, factor V, prothrombin, and by virtue of thrombin-like activity11,46. The abundance of SVMPs and SVSPs in the ECV proteome (Fig. 2c), compared to the RVV proteome12, makes this venom more pro-coagulant. The SVMPs and SVSPs are usually mid- and high-molecular mass proteins (>33 kDa) of Viperidae venoms18,47,48, and they were separated in the GF 1–6 fractions (Table 2), which is supported by previous reports11,12. Subsequently, the low molecular mass proteins (<30–10 kDa) that are eluted in GF 7–10 gel filtration fractions, exhibited anticoagulant activity (Table 2). These fractions are predominated by PLA2s, KSPIs, and disintegrins (Table 1), and their anticoagulant properties have been well characterized23,49,50.
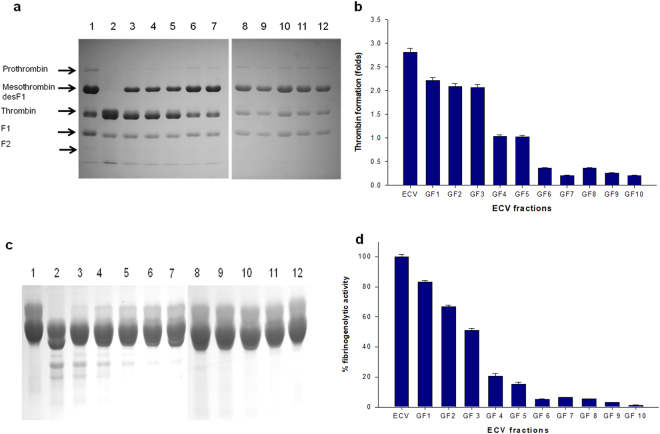
SVMPs and SVSPs primarily interfere with the blood coagulation cascade of victim/prey42,46 that ultimately leads to consumption coagulopathy and the prolongation of whole blood clotting time of E. carinatus bite patients in southern India45 (personal communication from Dr. A. Zachariah, CMC, Vellore). Interestingly, SVMPs were detected in all gel filtration fractions by proteomic analysis (Table 1) and by biochemical assay (Table 2). Carinactivase18 and Ecarin50 possessing prothrombin activation property have been well characterized as high molecular weight (>50 kDa) SVMPs from ECV. Although our stringent protein identification process did not identify the above proteases in ECV, the remarkable prothrombin activation exhibited by ECV and its gel filtration fractions, GF 1–3 (Fig. 3a,b), may have been due to the presence of several pro-coagulant SVMPs and RVV-X activator-like proteins in ECV (Tables 1 and 2).
Figure 3.
Prothrombin activation (thrombin formation) and fibrinogenolytic activity by ECV and GF fractions. (a) 12.5% SDS-PAGE analysis of prothrombin activation by ECV (3 µg/ml) and gel filtration fractions (0.5 µg/ml) under non-reducing conditions. Lane 1, prothrombin incubated with 1X PBS for 3 h at 37 °C (control); lanes 2–12, products of prothrombin hydrolysis by crude ECV (3 µg/ml) and GF 1–10 (0.5 µg/ml), respectively. Two 12.5% SDS-PAGE gels run under identical conditions were cropped and fused together. The full length unedited gels images are shown in Supplementary Figure S4. (b) The trace quantity of thrombin formed by auto degradation of control prothrombin was considered as 1 and other values (fold increase in thrombin formation determined by densitometry scanning of gel using Image Quant TL software 8.1) were compared to that. Values are mean ± SD of triplicate determinations. (c) 12.5% SDS-PAGE analysis of fibrinogen degradation by ECV and its gel filtration fractions. Lane 1 contains fibrinogen incubated with 1X PBS for 3 h at 37 °C (control), lanes 2–12 contain fibrinogen incubated with ECV (3 µg/ml) and GF 1–10 (0.5 µg/ml), respectively. Two 12.5% SDS-PAGE gels run under identical conditions were cropped and fused together. The full length unedited gels images are shown in Supplementary Figure S5. (d) Percentage of fibrinogenolytic activity was calculated by measuring the degradation of the Aα band of fibrinogen. The disappearance of Aα band of fibrinogen by crude ECV was considered as 100% activity and the band intensities of the treated samples were compared to that. Values are mean ± SD of triplicate determinations.
Fibrinogenolytic activity was prominent in ECV, and especially, the GF 1–5 fractions due to elution of proteases (SVMPs and SVSPs) in these fractions (Fig. 3c,d). The occurrence of these enzymes was also evident from biochemical and proteomic analyses (Tables 1 and 2). Furthermore, the above fractions are rich in esterase activities, predominantly BAEE-esterase activity (Table 2), which is often exhibited by SVSPs that possess fibrinogenolytic activity47,48. The crude venom (or GF 1–5) proteins degraded the Aα chain of fibrinogen leaving β and γ chains intact (Fig. 3c), which suggests that ECV is predominated by α-fibrinogenase. Further, it has been well documented that prothrombin-activating SVMPs and procoagulant SVSPs together play a predominant role in inducing coagulation and in in vivo defibrinogenating activities42,47,48. Interestingly, tryptic peptides VIGGDECNIN and TSTYIAPLSLPSSPPR of Factor V activator pro-coagulant serine protease isoenzymes47 and a thrombin-like serine protease (Russelobin)48, detected in this ECV proteome, have also been shown to cause in vivo defibrinogenation in mice47,48. Further, crude ECV and its gel filtration fractions (GF 2–5) exhibited remarkable fibrinogen clotting activity leading to the formation of tenuous fibrin clots (Table 2). This pharmacological property can be correlated to the presence of Russelobin-like serine proteases (gi|311223824)48 in these fractions (Table 1). Taken together, defibrinogenation (consumption of fibrinogen) and prothrombin as well as factor V (FV) activation, induced by the abundant SVMPs and SVSPs in ECV, may be responsible for the prolongation of blood coagulation in bite victims, which is a major clinical symptom of E. c. carinatus envenomation46.
SVMPs are often associated with additional cysteine-rich, and disintegrin domains that are cleaved off by proteolysis or autolysis during venom secretion32. Further, in some cases, C-type lectin domains (snaclec) are found to be linked with PIII-SVMPs by disulfide linkage32. Among them, the CRISP and snaclec domains are reported to trigger inflammation by recruiting inflammatory cells at the bite site51. Further, Ecarpholin (accession no. gi|163311140), a basic PLA2 isolated and characterized from ECV was predicted to be myotoxic in nature21. It was also detected by LC-MS/MS analysis in the current study (Table 1). Since this basic myotoxic PLA2 can induce edema in the footpad of experimental Swiss albino mice52, the persistent local swelling and edema observed in Echis bite patients43,44 can be correlated with the presence of significant amounts of CRISPs, snaclecs and PLA2s in the ECV proteome (Fig. 2a–c).
Further, other symptoms of envenomation, such as hemoptysis, haematemesis, and haematuria, observed in a few patients bitten by E. carinatus43,44 (personal communication from Dr. A. Zachariah, CMC, Vellore) may be correlated to the existence of PLA2s, SVMPs, and SVSPs in ECV (Table 1, Fig. 2c). Hemorrhage and local tissue damage is yet another important clinical symptom observed in Echis bite patients43,44,53. Among the SVMP classes, PII and PIII SVMPs are associated with non-metalloproteinase domains (disintegrin and cysteine-rich) that are reported to target extracellular matrix (ECM) and cause hemorrhage54. Therefore, this clinical feature can be correlated with the abundance of both the sub-classes of SVMPs in southern India ECV proteome. Hypotension and shock, the two other prominent clinical symptoms of Echis envenomation43 are probably attributed to venom ATPase and VEGF (Fig. 2c)55 as well as coagulopathy and bleeding.
A decrease in the circulatory platelet count (thrombocytopenia) after Echis bite is a very common clinical finding;43,44 and ECV and its gel filtration fractions induced loss of platelet integrity in vitro, albeit this effect was more pronounced by GF 1 and 2 proteins (Table 2) containing abundant SVMPs and snaclecs (Tables 1 and 2). Because the above two proteins collectively account for snake venom-induced thrombocytopenia12,56,57, it could be concluded that a high proportion of the SVMPs and snaclecs in ECV (Fig. 2c) are liable for the observed thrombocytopenia induced by E. carinatus venom in human (Table 2). Although ECV did not exhibit significant aggregation or deaggregation of washed platelets (data not shown), it rapidly (<60 s) induced agglutination of platelet rich plasma (PRP). A heterodimeric snaclec Echicetin, purified from venom of E. carinatus from unknown regions of India and identified in southern India ECV (accession nos. gi|802148, gi|32452854, gi|40889261, gi|2829697) (Table 1) binds specifically to platelet glycoprotein (GP)Ib and causes platelet agglutination22.
Assessment of immuno-cross-reactivity of ECV with commercial polyvalent antivenom indicated poor recognition of low molecular weight venom proteins
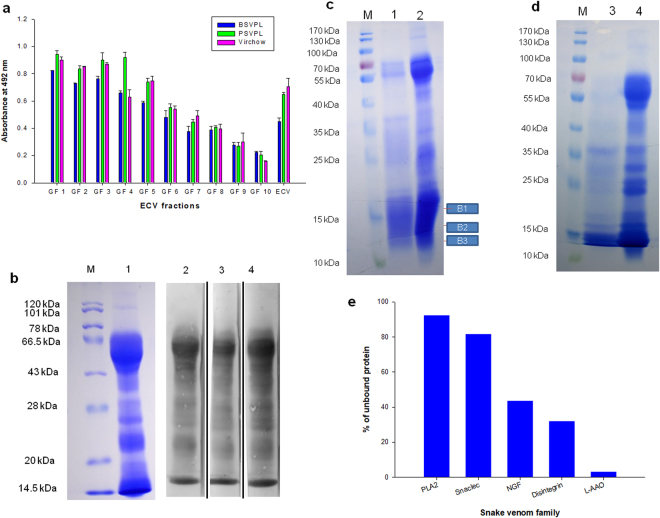
To date, antivenom remains the only choice for the treatment of snakebite. Nevertheless, the safety and efficacy of antivenoms are major concerns for efficient antivenom treatment. Therefore, the immuno-recognition of different commercial polyvalent antivenoms towards ECV and its gel filtration fractions was studied by ELISA, Western blot, and immuno-chromatographic (second generation antivenomics) analyses. Results from the ELISA experiment indicated that immuno-recognition of high (>45 kDa) and mid molecular weight (20–45 kDa) proteins was significantly higher (p < 0.05) than the recognition of low molecular mass proteins (<20 kDa) of ECV for all tested PAVs (Fig. 4a). This finding essentially reflects the poor immunogenicity of low molecular weight ECV proteins, or alternatively, the geographical source of ECV used for raising antivenom was different than the locale of the ECV used in this study. The corroboration of results from ELISA and Western blotting analysis also showed that ECV proteins ranging in mass from 10–20 kDa were less recognized by the tested antivenoms (Fig. 4b). The immuno-chromatographic approach also showed that mostly the low molecular weight (<20 kDa) venom proteins were not efficiently captured by PAV (Fig. 4c,d). Subsequently, by LC-MS/MS analysis (antivenomics study) these low molecular mass ECV proteins (Fig. 4c) were identified as PLA2s, α- and β-subunits of snaclecs, disintegrins, and NGFs (Table 3), and the percent of these proteins did not bind to PAV-immuno-affinity column is shown in Fig. 4e. Because PLA2s as well as snaclecs play a significant role in ECV-induced toxicity and adverse pharmacological effects in bite victim;44,58,59 therefore, least neutralization and/or recognition of these components of ECV by commercial PAV is a serious concern for effective antivenom therapy.
Figure 4.
(a) Immunological cross-reactivity of ECV and GF fractions with commercial PAVs by ELISA. Values are mean ± SD of triplicate determinations. (b) Immunoblot analysis of ECV against commercial PAVs. Lanes M and 1 contain protein molecular markers and ECV (100 µg, reduced), respectively. Lanes 2, 3, and 4 represent blots immuno-detected by PAVs – BSVPL, PSVPL and Virchow, respectively. (c) 12.5% SDS-PAGE analysis of affinity column unbound fraction. Lanes 1 and 2 represent unbound proteins and crude ECV (500 µg, reduced), respectively. Lane M contains protein molecular markers. (d) 12.5% SDS-PAGE analysis of immunoaffinity column bound ECV proteins. Lanes 3 and 4 represent immunoaffinity column bound ECV proteins and crude ECV, respectively. Lane M contains protein molecular markers. (e) Percentage of unbound protein less recognized by PAV (PSVPL) coupled with immunoaffinity column.
Table 3.
List of proteins identified by in-gel trypsin digestion and subsequent ESI-LC-MS/MS analysis of the ECV proteins poorly recognized by commercial PAVs. To enhance the ECV protein coverage, the data was searched against Viperidae family of proteins.
| Sl no. | Accession | Description | Protein family | Source organism | Score | Coverage | Unique peptides | MW [kDa] | SDS-PAGE band |
|---|---|---|---|---|---|---|---|---|---|
| 1 | gi|149243451 | Basic phospholipase A2 | PLA2 | Daboia russelii | 1136.5 | 79.3 | 7 | 13.6 | 3 |
| 2 | gi|82096307 | Acidic phospholipase A2 | PLA2 | Echis carinatus | 1050.4 | 61.7 | 10 | 15.5 | 1–3 |
| 3 | gi|38493055 | Snaclec EMS16 subunit alpha | Snaclec | Echis multisquamatus | 428.8 | 49.6 | 5 | 18.2 | 1 |
| 4 | gi|298351762 | Basic phospholipase A2 | PLA2 | Daboia russelii | 307.3 | 52.8 | 5 | 13.7 | 1–3 |
| 5 | gi|802148 | Snaclec echicetin subunit beta | Snaclec | Echis carinatus | 305.9 | 24.3 | 2 | 14.9 | 3 |
| 6 | gi|32452854 | Snaclec echicetin subunit alpha (fragment) | Snaclec | Echis carinatus | 292.8 | 20.0 | 2 | 15.7 | 1–3 |
| 7 | gi|300490462 | Snaclec dabocetin subunit alpha | Snaclec | Daboia russellii russellii | 253.9 | 46.1 | 3 | 17.5 | 3 |
| 8 | gi|400499 | Venom nerve growth factor | NGF | Daboia russelii | 236.4 | 21.3 | 2 | 13.3 | 1 |
| 9 | gi|55670410 | Snaclec EMS16 subunit beta | Snaclec | Echis multisquamatus | 184.4 | 15.5 | 3 | 17.7 | 1 |
| 10 | gi|82194569 | Disintegrin (Fragment) | Disintegrin | Echis carinatus | 91.7 | 43.7 | 2 | 7.1 | 1,3 |
| 11 | gi|194400545 | L-amino-acid oxidase (fragments) | LAAO | Daboia russelii | 88.3 | 6.6 | 2 | 46.3 | 1–2 |
| 12 | gi|163311140 | Basic phospholipase A2 homolog ecarpholin | PLA2 | Echis carinatus | 67.6 | 25.4 | 2 | 13.8 | 3 |
The major challenge in managing Echis bite patients is the efficient treatment of local swelling, which does not subside even after several vials of antivenom treatment, thus leaving the victims partially crippled (personal communication from R. Whitaker, Chennai). PLA2s are responsible for edema induction or local swelling44, whereas snaclecs can induce thrombocytopenia12,56. Therefore, the poor immunogenicity of these ECV components may be one of the reasons that explain the persistent local swelling in bite victims even after several vials of antivenom therapy. Henceforth, strategies must be designed either by inclusion of antibodies specifically raised against these low molecular weight toxic venom components or improving their antigenicity so as to mitigate the deleterious effect of the proteins for better hospital management of Echis bite patients. Further, presence of hemorrhagic SVMPs in ECV contributes to the formation of stable neutrophil extracellular trap (NET) at the bite site which forms a barrier for the free flow of blood and prevents antivenom from reaching the damaged site. Therefore, although high molecular weight SVMPs are well recognized and neutralized, antivenom treatment fails in efficient reversal of ECV induce local toxicity and swelling60.
Neutralization of enzymatic activity and pharmacological properties of ECV by commercial polyvalent antivenom highlights the requirement of a well-designed immunological protocol for antivenom production
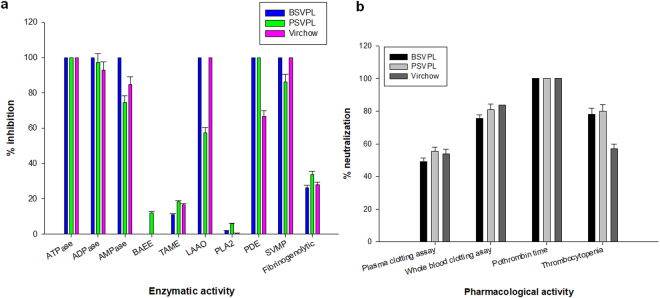
Our previous studies have demonstrated a good correlation between enzyme function and the pathophysiology of snake envenomation11–13. The efficient treatment of ECV bite will depend on effective neutralization of enzymatic activities and pharmacological properties of this venom. Commercial PAVs showed variable inhibitions of the enzymatic and pharmacological properties of ECV, which again, may be correlated to the source of ECV used for antivenom production. The enzymatic activities exhibited by high molecular mass proteins (>50 kDa), such as LAAO, PDE, SVMP, ATPase, ADPase, AMPase were efficiently neutralized by the antivenom (Fig. 5a); however, the enzymatic activities of mid and low molecular weight proteins such as PLA2, SVSP, Nα-p-Tosyl-L-arginine methyl ester hydrochloride (TAME), and Nα-Benzoyl-L-arginine ethyl ester hydrochloride (BAEE) exhibited mostly by SVSPs were least neutralized by antivenom (Fig. 5a). Interestingly, these proteins (except PLA2) were well recognized by PAV (Fig. 4b,c,d), suggesting that the catalytic sites of the enzymes may be poor immunogens. Further, the negligible neutralization of PLA2 activity suggests that the low molecular mass (~14–15 kDa) pharmacologically active proteins of ECV also serve as poor antigens (Fig. 5a).
Figure 5.
Neutralization of (a) enzyme activities and (b) pharmacological properties of ECV by PAVs at 1:10 (venom: antivenom) ratio. Values are mean ± SD of triplicate determinations.
Interference with the hemostatic system is the major pharmacological effect of ECV (Table 2); therefore, the potency of commercial antivenoms to neutralize this property of crude ECV was also investigated. All of the PAVs at 1:10 (venom:antivenom) efficiently neutralized the pro-coagulant activity and loss of platelet integrity induced by ECV (Fig. 5b). This may be correlated to the efficient neutralization of pro-coagulant SVMPs (Fig. 5b), indicating a good correlation between metalloprotease activity of Viperidae venom and the above pharmacological properties12.
Materials and Methods
E. c. carinatus (saw-scaled viper) venom (ECV) from Irula Snake Catchers Industrial Cooperative Society, Chennai, South India, was a gift from Premium Serum and Vaccines Pvt. Ltd, India. Lyophilized powders of polyvalent snake antivenom raised in horses against venom cocktail of Big four Indian snakes (Naja naja, Daboia russelli, Bungarus caeruleus and Echis carinatus), vials were purchased from Bharat serum and Vaccines Ltd. (BSVPL) (Batch no. A05315029, expiry date: January, 2019), Premium Serum and Vaccines Pvt. Ltd. (PSVPL) (Batch no. 012015, expiry date: December, 2018) and Virchow Biotech Pvt. Ltd. (Virchow) (Batch no. 012005, expiry date: May, 2018). Prothrombin time (PT) and activated partial thromboplastin time (APTT) were determined using Liquiplastin and Liquicelin-E kits purchased from Tulip Diagonstics, Mumbai, India. All other chemicals used were analytical grade and procured from Sigma Aldrich, USA.
Fractionation of ECV through gel filtration chromatography and SDS-PAGE analysis of crude ECV
2 mg of ECV (dry weight) was dissolved in 20 mM Tris-Cl, pH-7.4 buffer containing 150 mM NaCl (buffer A) and centrifuged at 10,000 rpm for 10 min. The supernatant was filtered through 0.2 µ membrane syringe filter and protein content was estimated by the Lowry method61. Then 0.1 ml filtrate containing 1.5 mg of protein was fractionated on a Shodex KW-803 gel filtration column (8 × 300 mm, 5 µm) equipped with Dionex Ultimate 3000 UHPLC system (Thermo Fisher Scientific, USA). The column was pre-equilibrated with the above buffer and the flow rate was adjusted to 15 ml/h at room temperature (~23 °C). The elution of protein was monitored at 280 nm and fractions of 0.25 ml were collected. The peaks were pooled for the enzymatic activity assay, pharmacological characterization, and LC-MS/MS analysis. Protein content of crude ECV and fractions was determined61 and from a standard curve of BSA, the concentration of unknown protein was determined at 660 nm.
The crude ECV was analyzed on 12.5% SDS-PAGE in both reduced and non-reduced conditions. Protein bands were visualized by PhastGel Blue R stain (GE Healthcare, Sweden). The molecular masses of the ECV proteins were also determined by MALDI-TOF-MS analysis (4800 MALDI TOF/TOF™ Analyzer, Applied Biosystems) as described earlier12,48. The masses were determined in the m/z ranges of 5–20, 21–40, 41–100, and >100 kDa.
Proteomic identification of ECV proteome by LC-MS/MS analysis of GF fractions
To identify the venom proteome, 80 µg of each gel filtration fraction was subjected to ESI-LC-MS/MS analysis11–13. The venom proteins were digested with sequencing grade trypsin (13 ng/μL in 10 mM ammonium bicarbonate containing 10% acetonitrile) at an enzyme substrate ratio of 1:30 at 37 °C for 18 h. The tryptic peptides were desalted, concentrated using ZipTip C18 (Merck, USA) and reconstituted in 0.1% formic acid. They were separated on a Zorbax 300SB-C18 analytical column (75 μm × 150 mm, 3.5 μm, Agilent) at a flow rate of 300 nL/min applying the following mobile phase gradient: from 11% B for 5 min, 11 to 25% B in 20 min, 25 to 53% B in 16 min, 53 to 100% B in 5 min, 100% B for 4 min, and then 11% B for 4 min. Solvent A and B were 0.1% formic acid and 80% acetonitrile containing 0.1% formic acid, respectively. The eluted peptides were then analysed on an LTQ Orbitrap Discovery hybrid mass spectrometer (ThermoFisher Scientific, Bremen, Germany) interfaced to an Agilent 1200 HPLC via a Nanomate Triversa (Advion BioSciences, Ithaca, NY). The ionization voltage was set at 1.7 kV.
The raw data was acquired and processed by Xcalibur software (ThermoFisher Scientific, Bremen, Germany) in a data-dependent acquisition (DDA) mode with 1 MS survey scan followed by 5 MS/MS scans. The full-scan MS spectra were acquired in the FT mode in the scan range of m/z 300−2000 (lock mass was set to 445.12 corresponding to polysiloxane) with a resolution of 30, 000 (full width at half-maximum). MS/MS fragmentation was collision-induced dissociation (CID) in linear mode with the following triggering conditions: minimum signal intensity, 10,000; charge state, +2, +3; maximum injection time for MS/MS, 500 ms; and isolation width, 2 amu.
The LC-MS/MS data were searched independently against the entries in non-redundant NCBI database with taxonomy set to (a) Viperidae (taxid: 8689 with 56,902 protein entries), (b) Echis (taxid: 8699 with 788 protein entries) and (c) Echis carinatus (taxid: 40353 with 162 protein entries). The data were analyzed by PEAKS 8.5 software (Bioinformatics Solutions Inc., Ontario, Canada). Carbamidomethylation of cysteine, oxidation of methionine, deamidation of asparagine and glutamine and pyro-glu of N-terminal glutamine residues were set as variable modifications. Precursor and fragment mass tolerances were set to 10 ppm and 0.8 Da, respectively, up to two missed cleavages were allowed and non-specific cleavage at one end (semi-tryptic) was considered. The false discovery rate (FDR) was kept very stringent (0.1%). To exclude the contaminating proteins from identification, the contaminant database with 115 protein entries (ftp://ftp.thegpm.org/fasta/cRAP/crap.fasta) was included in the database search process. All the redundant peptides were removed from the data set and thereafter each protein entry was manually verified. The following criteria were set for the purposes of protein identification: (a) only matching proteins and peptides showing a −10 log P value ≥ 30 and 20, respectively, and (b) occurrence of at least one overlapping distinct peptide.
The relative abundance of the venom proteins was determined by MS1 (area under peptide) as well as MS2 (spectral count) based label-free methods. For both the methods, the sum of areas or the spectral count was normalized by number of theoretical peptides and the normalized values were calculated using equation 1. The number of theoretical peptides for each identified protein was determined using MassSorter v3.1 software.
| 1 |
Thereafter, the relative abundance of a protein in a particular GF fraction was calculated using equation 2:
| 2 |
The average of the relative protein abundance of ECV determined by two different methods (MS1 areas-based and MS2 spectral count) was considered to represent the relative abundance of ECV venom proteome.
In addition, the MS/MS data were searched against Transcriptome Shotgun Assembly (TSA) sequences of Echis coloratus venom, the only translated Echis protein sequence database in NCBI (BioProject: PRJEB2884, 87 translated protein entries) and analyzed using PEAKS 8.5 software. The search parameters were the same as stated under LC-MS/MS data analysis. After removing the redundant peptides the translated protein entries were manually verified. The matching proteins and peptides showing a −10 log P value ≥ 30 and 20, respectively and occurrence of at least one overlapping distinct peptide were the pre-requisite for protein identification12,13. The relative abundances of ECV proteins were determined by MS2 mean spectral count and MS1 area-based methods (equations 1 and 2). The data was presented as an average of the relative protein abundance of ECV determined by two different methods.
Biochemical characterization
Esterolytic activity of crude ECV and its GF fractions were checked by the spectrophotometric method using Nα-Benzoyl-L-arginine ethyl ester hydrochloride (BAEE) and Nα-p-Tosyl-L-arginine methyl ester hydrochloride (TAME) as substrate48. One unit of TAME and BAEE-esterase activity is defined as an increase in absorbance of 0.01 at 254 nm and 244 nm, respectively, during the first 5 min of the reaction at 37 °C. PDE activity was assayed by the spectrophotometric method using bis-p-nitrophenyl phosphate as a substrate62. One unit of PDE activity is defined as micromoles of p-nitrophenol released per min. The protease activity of crude ECV and its GF fractions was determined by incubating 3.0 µg/ml of crude ECV or 0.5 µg/ml of GF fraction or 1X PBS (control) with fibrinogen (2.5 mg/ml) for 3 hours at 37 °C. The fibrinogen degradation products were separated by 12.5% SDS-PAGE (reduced) and percent fibrinogenolytic activity was calculated by measuring the degradation of the Aα chain of fibrinogen. The band intensity of Aα chain of fibrinogen molecule after crude ECV treatment was considered as 100% fibrinogenolytic activity and other values were compared to that. The protein band intensities were measured by Image Quant TL 8.1 software (GE Healthcare, Sweden).
PLA2 activity of crude ECV (3 µg/ml) and GF fractions (0.5 µg/ml) was assayed by the turbidometric method63. One unit of PLA2 activity is defined as a decrease in 0.01 absorbance at 740 nm after 10 min of incubation. L-kynurenine was used as a substrate for screening of L-amino acid oxidase (LAAO) activity of crude ECV (3 µg/ml) and GF peaks (0.5 µg/ml). The unit of LAAO activity was defined as nmol kynurenic acid produced/min under the assay conditions and specific LAAO activity was expressed as unit/mg protein11,12.
ATPase and ADPase activity of crude ECV (3 µg/ml) and GF fractions (0.5 µg/ml) was assayed by the method of Williams and Esnouf64 with slight modifications as described by Mukherjee et al.11 The 5′-nucleotidase (AMPase) activity was determined according to the protocol of Sinsheimer and Koerner65 with slight modifications described in our previous publication11. One unit of ATPase/ADPase/AMPase activity was defined as μM of Pi released per min at 37 °C.
Hyaluronidase activity of crude ECV (3 µg/ml) was assayed as described previously12. One unit of enzyme activity was defined as a decrease in turbidity by 1% as compared to the control and activity was expressed as U/mg protein66.
SVMP activity was assayed by using azocasein as a substrate. Crude ECV (3 µg/ml) or GF peaks (0.5 µg/ml) were added to reaction mixture and incubated at 37 °C for 10 min. The activity was checked by the spectrophotometric method and specific activity was expressed as ΔA450nm/min/mg protein11.
Pharmacological characterization
Goat blood obtained from slaughter house was collected in 3.8% tri sodium citrate and the blood was centrifuged at 4300 rpm for 10 min at 4 °C11,63. The pellet was discarded and the yellowish supernatant was termed as platelet poor plasma (PPP) and it was used within 4 hours after its collection. The anticoagulant activity of crude venom or its fraction on PPP was determined by Ca-clotting time11,63. For the control, 1X PBS instead of venom was added to PPP. One unit of coagulant or anticoagulant activity was defined as a decrease or an increase of 1 second of clotting time of PPP incubated with crude ECV or GF fractions, compared to control PPP63. Prothrombin time (PT) and activated partial thromboplastin time (APTT) of crude ECV (3 µg/ml) and GF peaks (0.5 µg/ml) were determined using commercial diagnostic kits following the instructions of the manufacturer63.
The prothrombin activation (FXa-like activity) property of crude ECV and GF peaks, if any, was analyzed by 12.5% SDS-PAGE of human prothrombin incubated with ECV or fractions or FXa (control)42. Formation of thrombin from prothrombin was quantified by measuring the density/intensity of thrombin (36 kDa) band in Image Quant TL software 8.1 (GE Healthcare, Sweden)49. Presence of thrombin-like enzyme activity was ascertained by measuring the fibrinogen clotting activity of crude ECV (3 μg/ml) and the GF peaks (0.5 μg/ml)48. Loss of platelet integrity induced by crude ECV or GF peaks was determined by incubation of crude ECV (3 µg/ml) or GF peaks (0.5 µg/ml) with washed platelet or Tyrode’s solution (control) at 37 °C for 6 hour in a CO2 incubator. The platelets were stained with trypan blue and counted in the hemocytometer using Motic Images plus 3.0 ML software12.
Immunological cross-reactivity, antivenomics, neutralization of enzymatic activities, and tested pharmacological properties of crude ECV by commercial polyvalent antivenom
The immunological cross-reactivity of commercial PAVs towards ECV and its GF fractions was studied by ELISA and western blot analysis, as described previously11,12. Briefly, for ELISA experiment, 100 ng protein of ECV or GF fractions was coated in 96 well microtiter ELISA plate (in triplicate) for overnight at 4 °C and washed three times with 1X PBS buffer containing 0.05% tween-20 (wash buffer). Two hundred ng of PAV was used as primary antibody and incubated for 2 h at room temperature followed by washing with wash buffer. Anti-horse IgG HRP conjugated secondary antibody (1:2000 dilution) was incubated for 2 h at room temperature to detect the primary antibody. Color was developed by adding substrate (1X TMB/H2O2) to the well for 30 minutes in dark condition and reaction was stopped by adding 50 µl of 2 M H2SO4. The absorbance was taken at 492 nm against blanks in Multiskan GO (Thermoscientific, USA) microplate reader.
Western blotting experiment was performed by running 100 µg protein of ECV (reduced) in 12.5% SDS-PAGE and transferred to PVDF membranes12. Non-specific bindings were blocked by 5% fat free skimmed milk for overnight at 4 °C. After that the membranes were washed with 1X TBS with 0.05% tween-20 (washing buffer). Primary antibodies (15 mg/ml PAVs) at a dilution of 1:1000 ratios were incubated for 1 h at room temperature. Thereafter the membranes were washed and ALP conjugated secondary antibody (1:15000 dilution) was incubated for 2 h at room temperature. Blots were developed using BCIP/NBT substrate kit (Sigma Aldrich, USA) and densitometry scanning (Epson America, Inc) was done.
Immunological cross-reactivity of crude ECV against PAV (PSVPL) was assessed by an immunoaffinity chromatographic approach67. Briefly, after coupling the NHS-activated Sepharose 4 Fast Flow column (GE Healthcare) with 15 mg PAV (PSVPL), the excess antivenom was washed with 1X PBS pH 7.4 buffer67. Then, 500 µg of crude ECV was loaded onto the column and incubated at 37 °C for 2 h in a shaker orbiter. Unbound venom proteins were eluted with 5 column volumes of 1X PBS, pH 7.4, desalted in PD 10 column (GE Healthcare, Sweden) and vacuum dried (Labconco, Model: 7670061, USA). Bound venom proteins were eluted by washing the column with 5 column volumes of 0.1 M glycine, pH 2.0. The pH of the eluted proteins was immediately neutralized by 1 M Tris-Cl, pH 9.0. The bound and unbound fractions, as well as 500 µg of crude ECV were separated by 12.5% SDS-PAGE. The poorly immunogenic, low molecular mass unbound proteins were excised from the gel and subsequently identified by LC-MS/MS analysis against Viperidae protein databases as described above. Percentage of unbound proteins was quantified by spectral count method; the total mean spectral count of PAV unbound proteins was compared with the total mean spectral count of corresponding ECV proteins (100%).
The neutralization potency of polyvalent antivenom (PAV) towards the pro-coagulant activity of crude ECV was determined by pre-incubating PAVs with crude ECV at 1:10 (protein: protein) for 30 min at 37 °C prior to the assay of Ca2+ clotting time of PPP, and enzymatic activities of venom11,12,68. The percent inhibition was calculated by comparing the pharmacological/enzymatic activity of crude ECV in the absence of PAVs (100% activity).
All the experiments were performed according to Tezpur University ethical committee and bio-safety committee guidelines.
Data accessibility
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE69 partner repository with Project Name “Echis carinatus carinatus (India) venom proteomics” and the dataset identifier PXD007980.
Statistical Analysis
Student’s t test using Sigma Plot 11.0 for Windows (version 10.0) was used to test the significance of difference in enzymatic activities and pharmacological properties of ECV and its GF fractions. A value of p ≤ 0.05 was considered significant.
Electronic supplementary material
Acknowledgements
The authors thank C-CAMP, NCBS, Bangalore for LC-MS/MS analysis. The authors also thank Prof. S. S. Ghosh and Ms. N. Arora, Indian Institute of Technology, Guwahati for MALDI-TOF-MS analysis. Authors also thank Dr. A. Zachariah, Christian Medical College, Vellore and Mr. R. Whitaker, Herpitologist, Madras Snake Park, Chennai for sharing information on clinical symptomps of Echis carinatus bite in Southern India. BK and AP received Junior Research Fellowships from DBT U-Excel project grant. AC is a recipient of a Junior Research fellowship award from a DBT Indo-Russia project grant. This study received grant from DBT, New Delhi sponsored Unit of Excellence in Biotechnology in NER of India (BT/412/NE/U-Excel/2013) and DBT National Bioscience Award grant (BT/HRD/ NBA/34/01/2012–13/(ix)) to AKM.
Author Contributions
A.K.M. conceived the idea, designed the study, and edited the final manuscript; A.P. and A.C. performed the experiments; A.K.M., A.P. and B.K. analyzed the data and wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-17227-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Organization, W. H. Rabies and envenomings: a neglected public health issue: report of a consultative meeting, World Health Organization, Geneva, 10 January 2007 (2007).
- 2.McNamee D. Tackling venomous snake bites worldwide. Lancet. 2001;357:1680. doi: 10.1016/S0140-6736(00)04868-6. [DOI] [PubMed] [Google Scholar]
- 3.Cherlin, V. A. & Hughes, B. New facts on the taxonomy of snakes of the genus Echis. (Division of Amphibians and Reptiles, National Museum of Natural History, Smithsonian Institution, 1984).
- 4.Alirol E, Sharma SK, Bawaskar HS, Kuch U, Chappuis F. Snake bite in SouthAsia: a review. PLoS neglected tropical diseases. 2010;4:e603. doi: 10.1371/journal.pntd.0000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casewell NR, et al. Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:9205–9210. doi: 10.1073/pnas.1405484111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mukherjee AK, Maity CR. The composition of Naja naja venom samples from three districts of West Bengal, India. Comparative biochemistry and physiology. Part A, Molecular & integrative physiology. 1998;119:621–627. doi: 10.1016/S1095-6433(97)00475-3. [DOI] [PubMed] [Google Scholar]
- 7.Mukherjee A, Ghosal S, Maity C. Some biochemical properties of Russell’s viper (Daboia russelli) venom from Eastern India: correlation with clinico-pathological manifestation in Russell’s viper bite. Toxicon. 2000;38:163–175. doi: 10.1016/S0041-0101(99)00125-7. [DOI] [PubMed] [Google Scholar]
- 8.Mukherjee AK, Maity CR. Biochemical composition, lethality and pathophysiology of venom from two cobras–Naja naja and N. kaouthia. Comparative biochemistry and physiology. Part B, Biochemistry & molecular biology. 2002;131:125–132. doi: 10.1016/S1096-4959(01)00473-0. [DOI] [PubMed] [Google Scholar]
- 9.Calvete JJ, Juarez P, Sanz L. Snake venomics. Strategy and applications. Journal of mass spectrometry: JMS. 2007;42:1405–1414. doi: 10.1002/jms.1242. [DOI] [PubMed] [Google Scholar]
- 10.Tan KY, Tan CH, Fung SY, Tan NH. Venomics, lethality and neutralization of Naja kaouthia (monocled cobra) venoms from three different geographical regions of Southeast Asia. Journal of proteomics. 2015;120:105–125. doi: 10.1016/j.jprot.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Mukherjee AK, Kalita B, Mackessy SP. A proteomic analysis of Pakistan Daboia russelii russelii venom and assessment of potency of Indian polyvalent and monovalent antivenom. Journal of proteomics. 2016;144:73–86. doi: 10.1016/j.jprot.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Kalita B, Patra A, Mukherjee AK. Unraveling the Proteome Composition and Immuno-profiling of Western India Russell’s Viper Venom for In-Depth Understanding of Its Pharmacological Properties, Clinical Manifestations, and Effective Antivenom Treatment. Journal of proteome research. 2017;16:583–598. doi: 10.1021/acs.jproteome.6b00693. [DOI] [PubMed] [Google Scholar]
- 13.Dutta S, et al. Proteomic analysis to unravel the complex venom proteome of eastern India Naja naja: Correlation of venom composition with its biochemical and pharmacological properties. Journal of proteomics. 2017;156:29–39. doi: 10.1016/j.jprot.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 14.Eichberg S, Sanz L, Calvete JJ, Pla D. Constructing comprehensive venom proteome reference maps for integrative venomics. Expert review of proteomics. 2015;12:557–573. doi: 10.1586/14789450.2015.1073590. [DOI] [PubMed] [Google Scholar]
- 15.Gutierrez JM, Leon G, Burnouf T. Antivenoms for the treatment of snakebite envenomings: the road ahead. Biologicals: journal of the International Association of Biological Standardization. 2011;39:129–142. doi: 10.1016/j.biologicals.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Pla D, et al. Proteomics and antivenomics of Papuan black snake (Pseudechis papuanus) venom with analysis of its toxicological profile and the preclinical efficacy of Australian antivenoms. Journal of proteomics. 2017;150:201–215. doi: 10.1016/j.jprot.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Warrell DA, Gutierrez JM, Calvete JJ, Williams D. New approaches & technologies of venomics to meet the challenge of human envenoming by snakebites in India. The Indian journal of medical research. 2013;138:38–59. [PMC free article] [PubMed] [Google Scholar]
- 18.Yamada D, Sekiya F, Morita T. Isolation and characterization of carinactivase, a novel prothrombin activator in Echis carinatus venom with a unique catalytic mechanism. The Journal of biological chemistry. 1996;271:5200–5207. doi: 10.1074/jbc.271.38.23418. [DOI] [PubMed] [Google Scholar]
- 19.Mukherjee AK, Saviola AJ, Burns PD, Mackessy SP. Apoptosis induction in human breast cancer (MCF-7) cells by a novel venom L-amino acid oxidase (Rusvinoxidase) is independent of its enzymatic activity and is accompanied by caspase-7 activation and reactive oxygen species production. Apoptosis. 2015;20:1358–1372. doi: 10.1007/s10495-015-1157-6. [DOI] [PubMed] [Google Scholar]
- 20.Trummal K, et al. 5′-Nucleotidase from Vipera lebetina venom. Toxicon. 2015;93:155–163. doi: 10.1016/j.toxicon.2014.11.234. [DOI] [PubMed] [Google Scholar]
- 21.Polgár J, Magnenat EM, Peitsch MC, Wells TN, Clemetson KJ. Asp-49 is not an absolute prerequisite for the enzymic activity of low-Mr phospholipases A2: purification, characterization and computer modelling of an enzymically active Ser-49 phospholipase A2, ecarpholin S, from the venom of Echis carinatus sochureki (saw-scaled viper) Biochemical Journal. 1996;319:961–968. doi: 10.1042/bj3190961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navdaev A, Dörmann D, Clemetson JM, Clemetson KJ. Echicetin, a GPIb-binding snake C-type lectin from Echis carinatus, also contains a binding site for IgMκ responsible for platelet agglutination in plasma and inducing signal transduction. Blood. 2001;97:2333–2341. doi: 10.1182/blood.V97.8.2333. [DOI] [PubMed] [Google Scholar]
- 23.Mukherjee AK. A major phospholipase A 2 from Daboia russelii russelii venom shows potent anticoagulant action via thrombin inhibition and binding with plasma phospholipids. Biochimie. 2014;99:153–161. doi: 10.1016/j.biochi.2013.11.026. [DOI] [PubMed] [Google Scholar]
- 24.Choi H, Fermin D, Nesvizhskii AI. Significance analysis of spectral count data in label-free shotgun proteomics. Molecular & cellular proteomics: MCP. 2008;7:2373–2385. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia Q, Wang T, Park Y, Lamont RJ, Hackett M. Differential quantitative proteomics of Porphyromonas gingivalis by linear ion trap mass spectrometry: non-label methods comparison, q-values and LOWESS curve fitting. International journal of mass spectrometry. 2007;259:105–116. doi: 10.1016/j.ijms.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu X, et al. Spectral index for assessment of differential protein expression in shotgun proteomics. Journal of proteome research. 2008;7:845–854. doi: 10.1021/pr070271+. [DOI] [PubMed] [Google Scholar]
- 27.Ziganshin RH, et al. Quantitative proteomic analysis of Vietnamese krait venoms: Neurotoxins are the major components in Bungarus multicinctus and phospholipases A2 in Bungarus fasciatus. Toxicon. 2015;107:197–209. doi: 10.1016/j.toxicon.2015.08.026. [DOI] [PubMed] [Google Scholar]
- 28.Houel S, et al. Quantifying the impact of chimera MS/MS spectra on peptide identification in large-scale proteomics studies. Journal of proteome research. 2010;9:4152–4160. doi: 10.1021/pr1003856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reeks T, et al. Deep venomics of the Pseudonaja genus reveals inter- and intra-specific variation. Journal of proteomics. 2016;133:20–32. doi: 10.1016/j.jprot.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 30.Wagstaff SC, Sanz L, Juarez P, Harrison RA, Calvete JJ. Combined snake venomics and venom gland transcriptomic analysis of the ocellated carpet viper, Echis ocellatus. Journal of proteomics. 2009;71:609–623. doi: 10.1016/j.jprot.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Matsui T, Fujimura Y, Titani K. Snake venom proteases affecting hemostasis and thrombosis. Biochimica et biophysica acta. 2000;1477:146–156. doi: 10.1016/S0167-4838(99)00268-X. [DOI] [PubMed] [Google Scholar]
- 32.Fox JW, Serrano SM. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon. 2005;45:969–985. doi: 10.1016/j.toxicon.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 33.Burke JE, Dennis EA. Phospholipase A2 structure/function, mechanism, and signaling. Journal of lipid research. 2009;50(Suppl):S237–242. doi: 10.1194/jlr.R800033-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogawa T, et al. Accelerated evolution of snake venom phospholipase A2 isozymes for acquisition of diverse physiological functions. Toxicon. 1996;34:1229–1236. doi: 10.1016/S0041-0101(96)00112-2. [DOI] [PubMed] [Google Scholar]
- 35.Mitra J, Bhattacharyya D. Phosphodiesterase from Daboia russelli russelli venom: purification, partial characterization and inhibition of platelet aggregation. Toxicon. 2014;88:1–10. doi: 10.1016/j.toxicon.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Santoro ML, Vaquero TS, Paes Leme AF, Serrano SM. NPP-BJ, a nucleotide pyrophosphatase/phosphodiesterase from Bothrops jararaca snake venom, inhibits platelet aggregation. Toxicon. 2009;54:499–512. doi: 10.1016/j.toxicon.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 37.Naumann GB, et al. Cytotoxicity and inhibition of platelet aggregation caused by an l-amino acid oxidase from Bothrops leucurus venom. Biochimica et biophysica acta. 2011;1810:683–694. doi: 10.1016/j.bbagen.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 38.Nanjaraj Urs AN, et al. Local and systemic toxicity of Echis carinatus venom: neutralization by Cassia auriculata L. leaf methanol extract. Journal of natural medicines. 2015;69:111–122. doi: 10.1007/s11418-014-0875-3. [DOI] [PubMed] [Google Scholar]
- 39.Girish KS, Mohanakumari HP, Nagaraju S, Vishwanath BS, Kemparaju K. Hyaluronidase and protease activities from Indian snake venoms: neutralization by Mimosa pudica root extract. Fitoterapia. 2004;75:378–380. doi: 10.1016/j.fitote.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 40.Mukherjee AK, Dutta S, Mackessy SP. A new C-type lectin (RVsnaclec) purified from venom of Daboia russelii russelii shows anticoagulant activity via inhibition of FXa and concentration-dependent differential response to platelets in a Ca 2+ -independent manner. Thrombosis research. 2014;134:1150–1156. doi: 10.1016/j.thromres.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 41.McLane MA, Vijay-Kumar S, Marcinkiewicz C, Calvete JJ, Niewiarowski S. Importance of the structure of the RGD-containing loop in the disintegrins echistatin and eristostatin for recognition of αIIbβ3 and αvβ3 integrins. FEBS letters. 1996;391:139–143. doi: 10.1016/0014-5793(96)00716-8. [DOI] [PubMed] [Google Scholar]
- 42.Thakur R, Chattopadhyay P, Ghosh SS, Mukherjee AK. Elucidation of procoagulant mechanism and pathophysiological significance of a new prothrombin activating metalloprotease purified from Daboia russelii russelii venom. Toxicon. 2015;100:1–12. doi: 10.1016/j.toxicon.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 43.Dass, B., Bhatia, R. & Singh, H. Venomous Snakes of India and Management of Snake Bites. Snakes in India: A Source Book, 257 (1998).
- 44.Gnanathasan A, Rodrigo C, Peranantharajah T, Coonghe A. Saw-scaled viper bites in Sri Lanka: is it a different subspecies? Clinical evidence from an authenticated case series. The American journal of tropical medicine and hygiene. 2012;86:254–257. doi: 10.4269/ajtmh.2012.11-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warrell DA. The clinical management of snake bites in the Southeast Asian region. Southeast Asian J Trop Med Public Health. 1999;30:1–67. [PubMed] [Google Scholar]
- 46.Thakur, R. & Mukherjee, A. K. Pathophysiological significance and therapeutic applications of snake venom protease inhibitors. Toxicon (2017). [DOI] [PubMed]
- 47.Mukherjee AK. The pro-coagulant fibrinogenolytic serine protease isoenzymes purified from Daboia russelii russelii venom coagulate the blood through Factor V activation: role of glycosylation on enzymatic activity. PloS one. 2014;9:e86823. doi: 10.1371/journal.pone.0086823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mukherjee AK, Mackessy SP. Biochemical and pharmacological properties of a new thrombin-like serine protease (Russelobin) from the venom of Russell’s Viper (Daboia russelii russelii) and assessment of its therapeutic potential. Biochimica et biophysica acta. 2013;1830:3476–3488. doi: 10.1016/j.bbagen.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 49.Mukherjee AK, Mackessy SP. Pharmacological properties and pathophysiological significance of a Kunitz-type protease inhibitor (Rusvikunin-II) and its protein complex (Rusvikunin complex) purified from Daboia russelii russelii venom. Toxicon. 2014;89:55–66. doi: 10.1016/j.toxicon.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 50.Kornalik F, Blombäck B. Prothrombin activation induced by Ecarin-a prothrombin converting enzyme from Echis carinatus venom. Thrombosis research. 1975;6:53–63. doi: 10.1016/0049-3848(75)90150-4. [DOI] [PubMed] [Google Scholar]
- 51.Escalante, T., Rucavado, A. & Gutiérrez, J. Snake venom metalloproteinases. Biological roles and participation in the pathophysiology of envenomation. Handbook of venoms and toxins of reptiles, 115–138 (2009).
- 52.Zhou X, et al. Structural characterization of myotoxic ecarpholin S from Echis carinatus venom. Biophysical journal. 2008;95:3366–3380. doi: 10.1529/biophysj.107.117747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nanjaraj Urs AN, et al. Progressive Hemorrhage and Myotoxicity Induced by Echis carinatus Venom in Murine Model: Neutralization by Inhibitor Cocktail of N,N,N’,N’-Tetrakis (2-Pyridylmethyl) Ethane-1,2-Diamine and Silymarin. PloS one. 2015;10:e0135843. doi: 10.1371/journal.pone.0135843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herrera C, et al. Tissue localization and extracellular matrix degradation by PI, PII and PIII snake venom metalloproteinases: clues on the mechanisms of venom-induced hemorrhage. PLoS neglected tropical diseases. 2015;9:e0003731. doi: 10.1371/journal.pntd.0003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warrell DA. Snake bite. The Lancet. 2010;375:77–88. doi: 10.1016/S0140-6736(09)61754-2. [DOI] [PubMed] [Google Scholar]
- 56.Rucavado A, et al. Characterization of aspercetin, a platelet aggregating component from the venom of the snake Bothrops asper which induces thrombocytopenia and potentiates metalloproteinase-induced hemorrhage. Thrombosis and haemostasis. 2001;85:710–715. [PubMed] [Google Scholar]
- 57.Gutierrez JM, Escalante T, Rucavado A. Experimental pathophysiology of systemic alterations induced by Bothrops asper snake venom. Toxicon. 2009;54:976–987. doi: 10.1016/j.toxicon.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 58.Saikia D, Thakur R, Mukherjee AK. An acidic phospholipase A(2) (RVVA-PLA(2)-I) purified from Daboia russelli venom exerts its anticoagulant activity by enzymatic hydrolysis of plasma phospholipids and by non-enzymatic inhibition of factor Xa in a phospholipids/Ca(2+) independent manner. Toxicon. 2011;57:841–850. doi: 10.1016/j.toxicon.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 59.Saikia, D. & Mukherjee, A. K. Anticoagulant and membrane damaging properties of snake venom phospholipase A 2 enzymes. Snake Venoms, 87–104 (2017).
- 60.Katkar GD, et al. NETosis and lack of DNase activity are key factors in Echis carinatus venom-induced tissue destruction. Nature communications. 2016;7:11361. doi: 10.1038/ncomms11361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. The Journal of biological chemistry. 1951;193:265–275. [PubMed] [Google Scholar]
- 62.Sulkowski E, Laskowski M. Inactivation of 5′-nucleotidase in commercial preparations of venom exonuclease (phosphodiesterase) Biochimica et Biophysica Acta (BBA)-Nucleic Acids and Protein Synthesis. 1971;240:443–447. doi: 10.1016/0005-2787(71)90538-7. [DOI] [Google Scholar]
- 63.Dutta S, Gogoi D, Mukherjee AK. Anticoagulant mechanism and platelet deaggregation property of a non-cytotoxic, acidic phospholipase A2 purified from Indian cobra (Naja naja) venom: inhibition of anticoagulant activity by low molecular weight heparin. Biochimie. 2015;110:93–106. doi: 10.1016/j.biochi.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 64.Williams WJ, Esnouf MP. The fractionation of Russell’s-viper (Vipera russellii) venom with special reference to the coagulant protein. The Biochemical journal. 1962;84:52–62. doi: 10.1042/bj0840052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sinsheimer RL, Koerner JF. Di-desoxyribonucleotides1. Journal of the American Chemical Society. 1952;74:283–283. doi: 10.1021/ja01121a531. [DOI] [Google Scholar]
- 66.Pukrittayakamee S, et al. The hyaluronidase activities of some Southeast Asian snake venoms. Toxicon. 1988;26:629–637. doi: 10.1016/0041-0101(88)90245-0. [DOI] [PubMed] [Google Scholar]
- 67.Pla D, Gutierrez JM, Calvete JJ. Second generation snake antivenomics: comparing immunoaffinity and immunodepletion protocols. Toxicon. 2012;60:688–699. doi: 10.1016/j.toxicon.2012.04.342. [DOI] [PubMed] [Google Scholar]
- 68.Mukherjee AK, Mackessy SP, Dutta S. Characterization of a Kunitz-type protease inhibitor peptide (Rusvikunin) purified from Daboia russelii russelii venom. International journal of biological macromolecules. 2014;67:154–162. doi: 10.1016/j.ijbiomac.2014.02.058. [DOI] [PubMed] [Google Scholar]
- 69.Vizcaino JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic acids research. 2016;44:D447–456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE69 partner repository with Project Name “Echis carinatus carinatus (India) venom proteomics” and the dataset identifier PXD007980.