Abstract
Together, the three human rhinovirus (RV) species are the most frequent cause of the common cold. Because of their high similarity with other viral species of the genus Enterovirus, within the large family Picornaviridae, studies on RV infectious activities often offer a less pathogenic model for more aggressive enteroviruses, e.g. poliovirus or EV71. Picornaviruses enter via receptor mediated endocytosis and replicate in the cytosol. Most of them depend on functional F-actin, Rab proteins, and probably motor proteins. To assess the latter, we evaluated the role of myosin light chain kinase (MLCK) and two myosin V isoforms (Va and Vb) in RV-B14 infection. We report that ML-9, a very specific MLCK inhibitor, dramatically reduced RV-B14 entry. We also demonstrate that RV-B14 infection in cells expressing dominant-negative forms of myosin Va and Vb was impaired after virus entry. Using immunofluorescent localization and immunoprecipitation, we show that myosin Va co-localized with RV-B14 exclusively after viral entry (15 min post infection) and that myosin Vb was present in the clusters of newly synthesized RNA in infected cells. These clusters, observed at 180 min post infection, are reminiscent of replication sites. Taken together, these results identify myosin light chain kinase, myosin Va and myosin Vb as new players in RV-B14 infection that participate directly or indirectly in different stages of the viral cycle.
Introduction
The common cold is normally limited to the upper respiratory tract1. The economic costs associated with non-influenza respiratory infections have been estimated to approach 40 billion dollars per year in the United States alone2. While various viruses can cause symptoms of a common cold, about 50% of all the cases are caused by human rhinoviruses (RVs), members of the family Picornaviridae in the genus Enterovirus 3. When associated with other pathogens or conditions such as asthma, chronic obstructive pulmonary disease or cystic fibrosis, RV infections can lead to severe and life threatening airway diseases4. Due to the high similarity presented by the thirteen species of the genus Enterovirus (including rhinovirus genera A, B, and C) with respect to their genome sequences, the 3D-structure of their protein shells and based on observations with poliovirus, it is believed that the release of the RNA genome from a virion only occurs after internalization or at least after the virus becomes sheltered from the extracellular medium5.
Following entry by receptor-mediated endocytosis along diverse pathways6–9, the virus first converts into the subviral A-particle that is expanded by 4% with respect to the native virion. The conformation adopted is more permeable for the exit of the RNA genome, probably through one of the pores on the 2-fold axes or at the pseudo 3-fold axes10–13. Cryo-electron microscopy image reconstructions of heated poliovirus suggest that the highly structured RNA unfolds and starts exiting as a single-strand though one of the holes along the 2-fold axes14 leaving an empty capsid (subviral B-particle), which might then be shuttled to lysosomes for degradation15. Snug attachment of the viral capsid to the endosomal membrane might establish a tight seal16 allowing the genome to cross the lipid bilayer without exposure to the degradative milieu of the (late) endosome17.
The viral RNA appears in the cytosol as soon as 10 min after the initiation of infection18. Recently, it was demonstrated that the genomic RNA of poliovirus remains RNAse-protected during exit from the capsid19 suggesting a specific mechanism of viral genome delivery that not only directs, but also protects the RNA such that it reaches the cytoplasm intact and functional. Despite intense studies with several drugs targeting host proteins/mechanisms, this early step in infection is not fully understood. Compounds that increase endosomal pH have proven very effective at interfering with a majority of RVs20–22, different intracellular routes are likely to be used23. Internalization of RV-A2 and RV-B14 were reduced in cells expressing dominant-negative dynamin II (K44A)24–27 probably due to disturbance of proper clathrin-coated pit maturation28. Regardless of actin dependence of RVs9,29, RV-A2, RV-A16, RV-B3, and RV-B14 infections were not impaired in nocodazole treated cells8,18,21,30. Nonetheless, RV-B89 infection seems to be different, as ciliobrevin and nocodazole can reduce infection8,9.
The current paper reports that RV-B14 infection requires myosin light chain kinase (MLCK) for cell entry while myosin Va and Vb are required for intracellular events prior to replication. MLCK possesses a critical role in regulating the cell membrane tension and protrusion, thereby stabilizing the membrane skeleton through F-actin-binding31. The molecular motors myosin Va and Vb are described as complementary intracellular cargo transporters32. Myosin Va can transport endogenous mRNA33 and various intracellular vesicles34. Myosin Vb, on the other hand, is involved with endocytic membranes recycling35. Therefore, we propose that RV-B14 uses a “phagocytic-like” mechanism for entry due to its MLCK dependency, intracellular degradation evasion in dominant-negative expressing cells (Va and Vb) and the presence of RV-B14 (protein and RNA) in structures similar to a phagocytic cup in the initial steps of infection. Further, myosin Va appears to have an early role in replication and myosin Vb a late role.
Results
1-(5-Chloronaphthalenesulfonyl)homopiperazine-HCl (ML-9) impacts RV-B14 entry
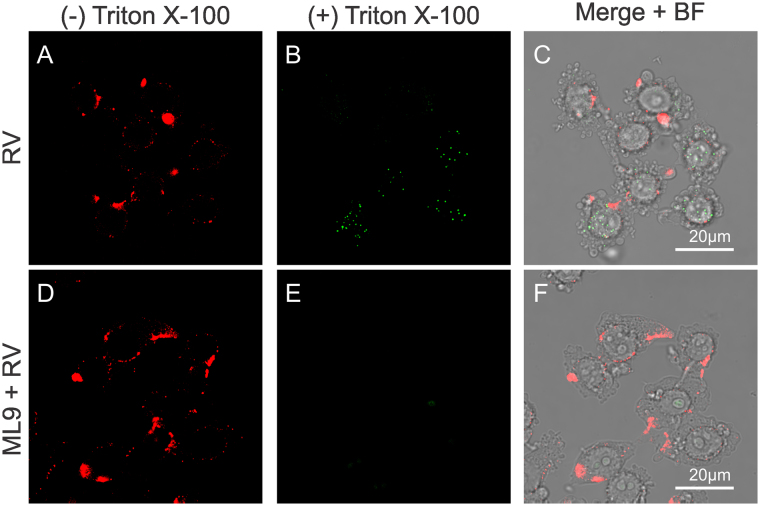
Wortmannin blocks phosphatidylinositol 3-kinases (PI3K) and delays infection of RV-A2 after entry by retarding the transfer of virions from early to late endosomes36. However, wortmannin can also impair MLCK function by directly binding at or near to the catalytic site of the enzyme in an irreversible manner37. Furthermore, MLCK is a calmodulin activated kinase responsible for myosin light chain phosphorylation, thus modulating myosin II binding to F-actin, and is described to be essential in endocytic routes in non-muscular cells38. To study whether MLCK might also be involved in RV-B14 entry, cells were pretreated with 10 µM ML9 30 min before infection. ML9 is a very specific MLCK inhibitor, structurally different from wortmannin, that binds at or near the ATP-binding site causing a competitive inhibition of the kinase39. Sixty minutes post infection (PI), cells were fixed and labeled with anti-RV-B14 followed by IRDye® 680LT conjugated anti-Rabbit IgG (Red). Next, cells were permeabilized and again labeled with anti-RV-B14, but followed by fluorescein isothiocyanate (FITC) conjugated anti-Rabbit (Green) to distinguish extracellular (plasma membrane-bound) and intracellular virus40. ML-9 pretreatment significantly reduced RV-B14 entry (Fig. 1). This effect differed from that observed for wortmannin in RV-A2 infection and could be explained by different sites of action of wortmannin36 and ML-9.
Figure 1.
ML9 impacts on RV-B14 entry. Cells were incubated without (A–C; control) or with 10 µM ML9 (D–F) for 30 min prior to infection with RV-B14 (MOI = 10) for 1 h at 34 °C. Cells were processed for confocal microscopy before and after permeabilization with Triton X-100 as indicated. Extracellular (A and D) and intracellular (B and E) viruses were visualized with anti-RV-B14 plus IRDye® 680LT anti-Rabbit IgG (red) and anti-RV-B14 plus anti-Rabbit IgG H&L (FITC, green), respectively. Juxtaposition of images after permeabilization with Triton X-100 (B vs E) reveals that ML9 treatment prevents RV-B14 entry.
Expression of myosin Va or Vb tail domains prevents RV-B14 propagation
Initial evidence for the importance of myosin V in viral infections came from the mild inhibition of respiratory syncytial virus in MDCK cells constitutively expressing the myosin Vb tail from a doxycycline controlled cassette41. Indeed, myosin V is involved in intracellular cargo trafficking42,43. To evaluate the potential roles of myosin Va and Vb function in RV-B14 infections, dominant-negative mutants of each fused to eGFP (DN-Va and DN-Vb) were obtained. These GFP-tail fusion proteins possess the cargo-binding motifs without the motor domain, and can compete for cargo binding with the respective endogenous full-length myosin V, disrupting its transport44. In addition, the DN-Va construct contained the alternatively spliced DDK exon, which can be bound by dynein light chain, and allow DN-Va to sequester this adaptor protein45. HeLa-H1 cells were transfected with individual constructs (DN-Va or DN-Vb) and infected with RV-B14 (wild type without labeling) at 34 °C for 20 h. Control experiments demonstrated that neither mock transfection (Fig. 2A–C) nor transfection with a plasmid encoding eGFP alone (Fig. 2D,E) interfered with RV-B14 infection (Fig. 2, Anti-RV-B14 column). In contrast, cells expressing either DN-Va (Fig. 2G–I) or DN-Vb (Fig. 2J–L), which are fused to eGFP, were virtually unaffected by RV-B14 infection. Nevertheless, endocytosed virus particles were observed in the cells transfected with DN-Va or DN-Vb (Fig. 2I and L; arrows). Further, we noted that infections by RV-B14 could occur in cells with the lowest observable expression of DN-Vb (Fig. 2L; arrow head). The number of cells doubly positive for eGFP and viral protein were analyzed in 10 different fields (Fig. 2M), which revealed that the presence of eGFP had no effect on viral replication; nearly 100% of the cells expressing eGFP were also positive for RV-B14 proteins. In contrast, expression of either DN-Va or DN-Vb (Fig. 2M) had a significant impact on virus infection (*p value < 0.0001).
Figure 2.
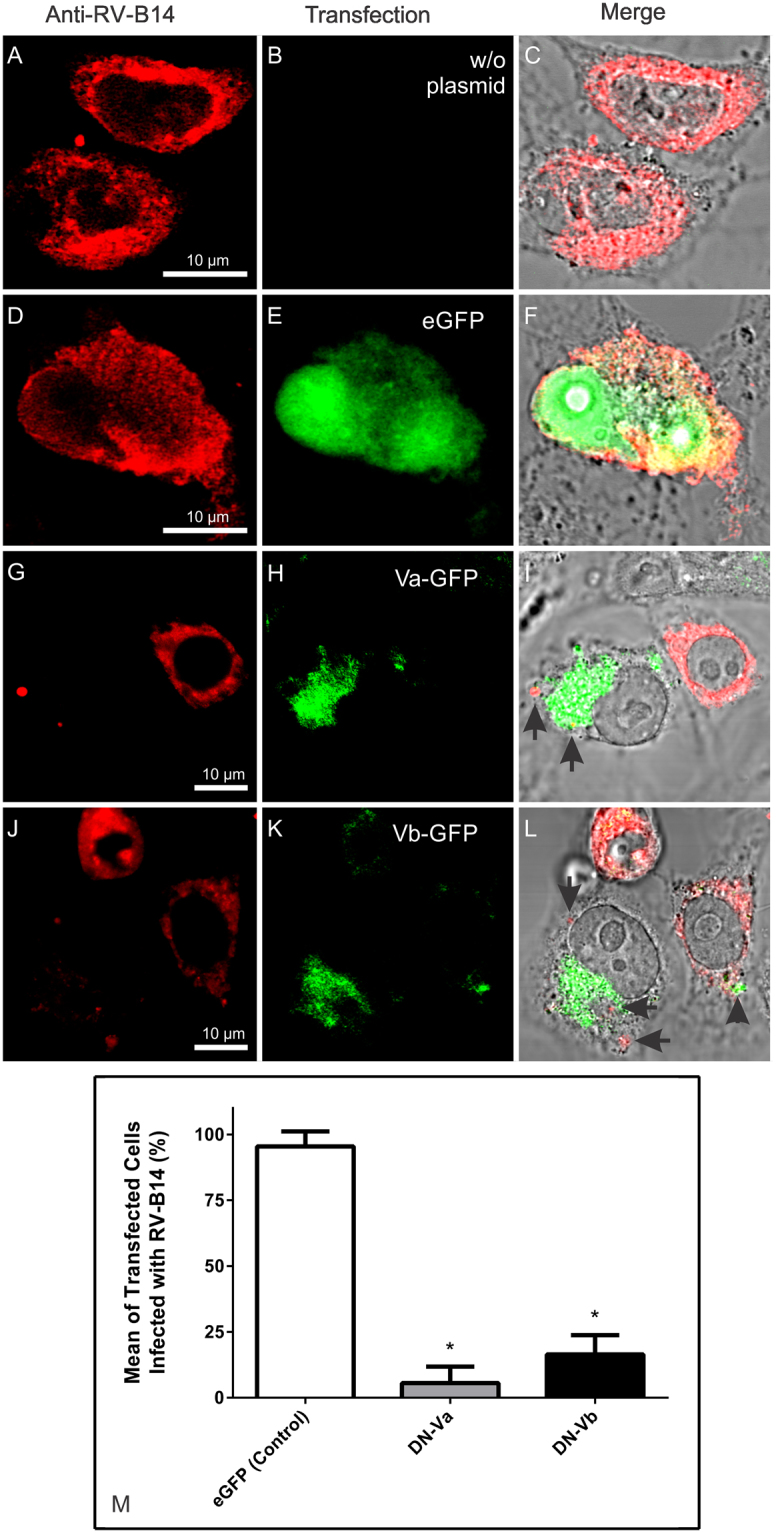
Overexpression of myosin Va or Vb tail domains prevents virus infection. HeLa-H1 cells transfected with plasmids encoding dominant-negative forms of myosin Va (DN-Va) or myosin Vb (DN-Vb), fused to eGFP, were infected with RV-B14 (MOI = 1) for 20 h at 34 °C. Mock transfected cells (w/o plasmid) and cells transfected with an eGFP plasmid alone (eGFP) were similarly infected. In the upper panel a representative field for each transfection is shown (eGFP expression in green): mock (A–C); eGFP (D–F); DN-Va (G–I), and DN-Vb (J–L). After infection, cells were fixed and labeled with anti-RV-B14 plus anti-Rabbit IgG H&L (TRITC, red). Fluorescence images were merged with bright field images for better distinction of cells boundaries (C,F,I and L). RV-B14 particles persisted at the periphery of cells expressing DN-Va or -Vb, (arrows in I and L, respectively). Note that some cells present low expression of DN-Vb (arrowhead in L). (M) Quantification of transfected cells infected with RV-B14 (% of transfected cells in 10 different fields). *p value < 0.0001 (vs. control).
Myosin Va colocalizes with viral protein and BrU-RNA in phagocytic cup-like structures
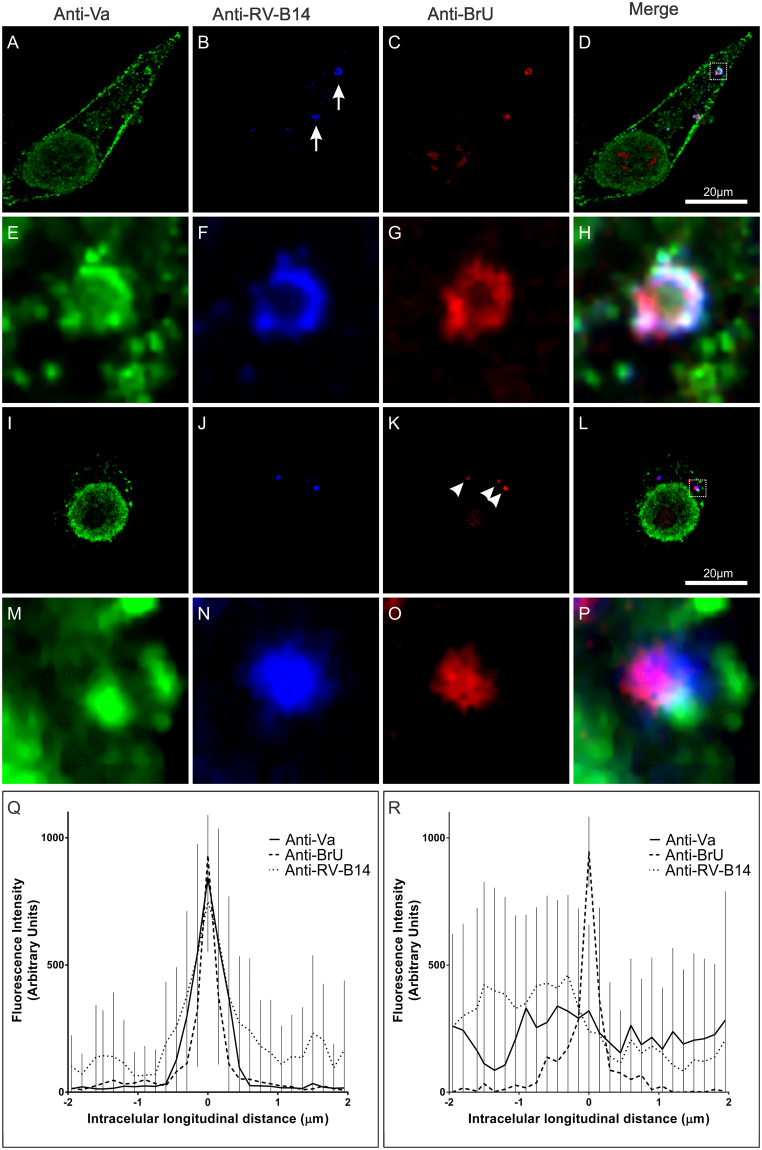
To study the temporal aspects of myosin Va function potentially contributing to RV-B14 infections, the position of viral RNA and viral protein in relation to endogenous myosin Va was determined at 15 min and 60 min PI. To specifically detect virus genomes, viral RNA was metabolically labeled with bromouridine (BrU)5,46,47 during virus stock production. The BrU labeling showed no impact on the infectivity of purified virus. HeLa-H1 cells were infected with the BrU-labeled RB-14 (BrU-RV-B14) for either 15 min (Fig. 3A–H) or 60 min (Fig. 3I–P) before fixation to separately visualize viral protein, viral RNA, and myosin Va with suitable antibody combinations. At 15 min PI, RV-B14 protein accumulated in small dots and clusters (Fig. 3B; arrows), most of which coinciding with viral RNA (Fig. 3C). Colocalization of the myosin Va signal (Fig. 3A and D) with both RV-B14 protein and BrU-RNA was immediately apparent, and could be even better distinguished at higher magnification (Fig. 3E–H; inset of D dashed square). The subcellular distribution of myosin Va coincided spatially with both the RV-B14 RNA genome and protein (Fig. 3D and H). The morphological shape of the labeled structures was reminiscent of phagocytic cups, a protrusion of the cell surface that extends along and finally closes around the particle or nutrient to be engulfed48. At 60 min PI, multiple viral RNA foci (Fig. 3K; arrowheads) were detected in the perinuclear region (Fig. 3L). However, the colocalization of myosin Va, RV-B14 protein and BrU-RNA signals was less clear, even at higher magnification (Figs. 3M–P; inset of L, dashed square). A signal intensity histogram was measured for the anti-myosin Va (anti-Va), anti-RV-B14, and the anti-BrdU antibody (anti-BrU) localizations along a 4 µm line that was 0.1 µm wide and centered on the region with the maximum anti-BrU intensity (examples in Supplementary Fig. S1). Fluorescent signals from 10 different experiments were grouped and plotted for the experiments performed at 15 (Fig. 3Q) and 60 min PI (Fig. 3R). The intensities of the peaks were well matched at 15 min PI, but not at 60 min. From this analysis, we conclude that, at 15 min PI, vesicles contain virus capsids and partially released RNA bound to myosin Va, and that RNA is fully accessible to translation and replication at 60 min PI.
Figure 3.
Myosin Va colocalizes with viral BrU-RNA containing vesicle in an early phase of infection. HeLa-H1 cells infected with BrU-RV-B14 (MOI = 10) for 15 min (A–H) or 60 min (I–P) at 34 °C. After the infection, cells were labeled for anti-myosin Va (A,E,I, and M; anti-Va - green), anti-RV-B14 (B,F,J, and N; anti-RV-B14 - blue), and anti-BrU-RNA (C,G,K, and O; anti-BrU - red). (D,H,L, and P) Merged images demonstrating the intracellular disposition of myosin Va, RV-B14 protein, and BrU-RNA. (E–H and M–P) Enlarged regions correspond to the dashed boxes in panels D and L, respectively. (B; arrows) Internalized viral particles present in the periphery of the cell. (K; arrowheads) Perinuclear BrU-RNA foci. (Q and R) Intensity histogram of signals (10 different experiments) collected along a line (4 µm long and 0.1 µm wide centered on the anti-BrU maximum intensity) for 15 and 60 min, respectively.
BrU-labeled viral genomes co-precipitate with myosin Va in a time-dependent manner
To assess whether myosin Va directly or indirectly interacts with the RV-B14 genome, HeLa-H1 cells were challenged with BrU-RV-B14 (similar to Fig. 3), and the infection was halted at different times by crosslinking with formaldehyde49. Cells were subsequently lysed with RIPA buffer and myosin Va was immunoprecipitated with antibodies bound to agarose beads. The presence of BrU-RNA was then ascertained on a dot-blot with anti-BrdU antibody and the quantity of BrU-RNA, relative to the anti-Va signal, was plotted against time (Fig. 4). The BrU-RNA signal was highest at 15 min, decreased at 30 min, and returned to near control levels at later times. These findings corroborate the colocalization data (Fig. 3), as RV-B14 RNA release appears to be highly coincident with the presence of myosin Va. This suggests that myosin Va could contribute to the transport and proper localization of the viral RNA genome from the cell periphery to the replication sites, similar to its previously described role in mRNA transport and proper localization in dendritic spines33,50.
Figure 4.
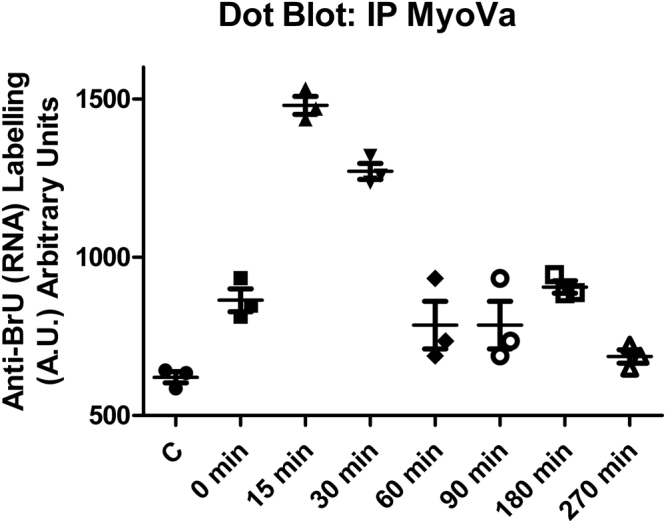
BrU labeled viral genomes co-precipitate with myosin Va in a time-dependent manner. Hela-H1 cells were infected with BrU-RV-B14 (MOI = 10) at 0, 15, 30, 60, 90, 180, and 240 minutes. Cells were lightly crosslinked (1% formaldehyde for 2 min) and lysed with RIPA buffer. Extracts were centrifuged and 4 h incubated with (mouse) anti-myosin Va antibody followed by overnight incubation with anti-Mouse antibody linked to agarose beads. Clarified complexes were immobilized on PVDF membrane, probed with (rabbit) anti-BrdU antibody (plus IRDye® 800LT anti-Rabbit IgG), striped, and probed with (mouse) anti-myosin Va antibody (plus IRDye® 680LT anti-Mouse IgG). Quantification from three independent experiments (different symbols in the graph) showing the signal intensity of anti-BrU (normalized by anti-myosin Va signal) at different times PI.
Myosin Va is absent from newly synthesized RNA clusters in late events during RV-B14 infection
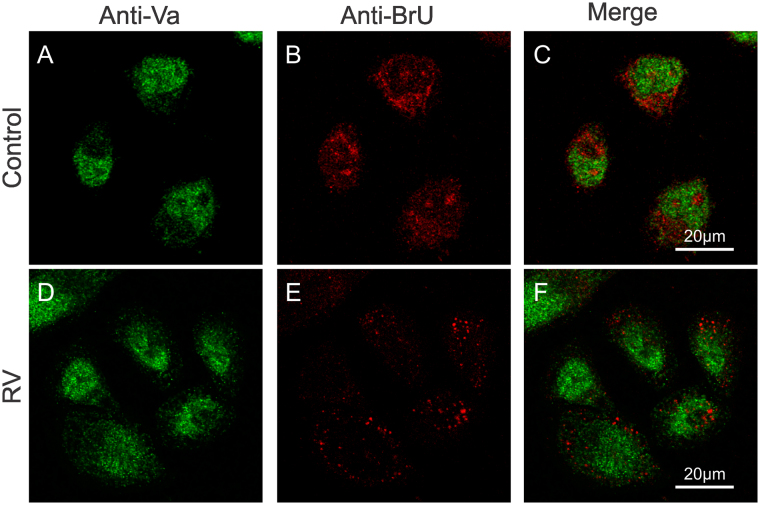
To study the onset of RV-B14 replication, we examined the distribution of newly synthesized RNA at 180 min PI. In previous reports, the de novo synthesis of RV RNA genomes was detected in HeLa cells through fluorescent in situ hybridization and 3H-uridine incorporation within 180–210 min PI18,51. Here, mock-infected controls (Fig. 5A–C) and RV-B14 (wild type without labeling) infected cells (Fig. 5D–F) were pulse-labeled with BrU at 30 min PI until 180 min PI. Subsequently, the cells were fixed, labeled with anti-myosin Va (Fig. 5A,C,D, and F; green) and anti-BrU (Fig. B,C,E, and F; red). At 180 min PI, the distribution of myosin Va was similar between the control (Fig. 5A) and infected cells (Fig. 5D) with no evidence of its colocalization with the BrU signal (Fig. 5C and F), restricting our proposed role for myosin Va to the initial steps of RV-B14 infection.
Figure 5.
Identification of putative RV-B14 replication sites. HeLa-H1 cells were mock infected (A–C) or infected (D–E) with RV-B14 (MOI = 50) for 180 min at 34 °C. At 30 min PI, cells were pulse labeled with 2.5 mM BrU to incorporate BrU into nascent RNA. The cells were then processed for confocal microscopy with (rabbit) anti-myosin Va antibody plus anti-Rabbit IgG H&L (FITC) (A and D; green) and Alexa Fluor 555® anti-BrdU (B and E; red). (B) Nascent BrU-RNA. (E) BrU-RNA clusters presumably representing viral RNA. (C and F) Merged images from (A,B and D–E) respectively.
The observations of the intracellular BrU localization revealed that in mock infected cells, the BrU-labled RNAs were evenly distributed through the cytosol (Fig. 5B). On the other hand, in RV-B14 infected cells, the BrU-RNAs were concentrated in clusters unevenly distributed through the cytosol (Fig. 5E). The observed pattern of the de novo synthesized RNA in infected cells was compatible with previously reported structures for rhinovirus that were defined as replication sites (see Fig. 1 in Jurgeit, et al.52). Since specific markers for rhinovirus replication sites were not employed, we refer to this pattern of RNA clusters, which was both time and infection-state dependent, as putative RV-B14 replication sites.
Myosin Vb is present in putative RV-B14 replication sites
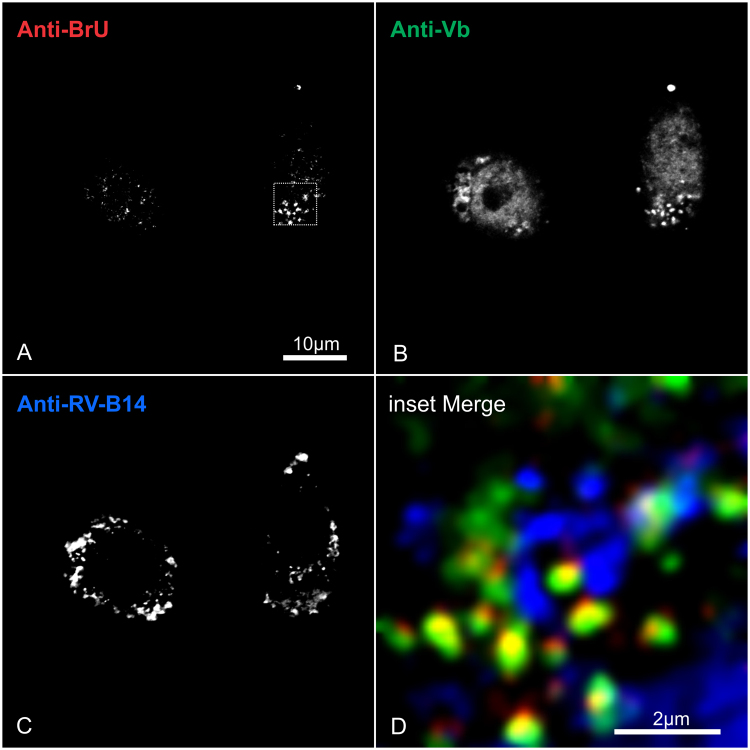
Previous data on the localization of myosin Vb to nuclear transcription centers53 led us to hypothesize that myosin Vb could be sequestered during RV-B14 infection to compose the cytosolic replication complex. To test this hypothesis, HeLa-H1 cells were infected with RV-B14 (wild type without labeling) and pulse-labeled with BrU at 30 min PI (similar to Fig. 5). Cells were fixed at 180 min PI and examined for the distribution of the de novo synthesized BrU-containing RNA (Fig. 6A), myosin Vb (Fig. 6B), and of de novo synthesized RV-B14 capsid proteins (Fig. 6C). The signal distributions were better distinguished at a higher magnification (Fig. 6D inset of 6 A, dashed square). The RNA clusters induced by infection could be promptly identified (Fig. 6D – red, similar to Fig. 5E), and showed a strong colocalization with myosin Vb (Fig. 6D – green). Remarkably, we observed a well-defined crown of RV-B14 proteins surrounding the RNA clusters (Fig. 6D – blue), which supports our proposition that the structures labeled by BrU in infected cells represent RV-B14 replication sites (Fig. 5E).
Figure 6.
Putative RV-B14 replication sites recruit myosin Vb protein. HeLa-H1 cells were infected with RV-B14 (MOI = 50) for 180 min at 34 °C. At 30 min PI, cells were pulse labeled with 2.5 mM BrU to incorporate it into nascent RNA. Cells were subsequently processed for confocal microscopy with Alexa Fluor 555® anti-BrdU (A; red), (mouse) anti-myosin Va antibody plus Alexa Fluor 488® goat anti-Mouse (B; green) and (rabbit) anti-RV-B14 plus IRDye® 680LT anti-Rabbit IgG (C; blue). (D) Enlargement of dashed box in A.
Three-dimensional imaging of a representative cell infected with RV-B14 (wild type without labeling) and pulse labeled with BrU 30 min PI (similar as Figs 5 and 6) revealed colocalization of myosin Vb with the putative RV-B14 replication sites in all three dimensions (Fig. 7, white arrow). To quantify the fluorescent signals of anti-BrU and anti-myosin Vb for comparison in 3D, we measured the fluorescence intensities along a line (4 µm long and 0.1 µm wide centered on the anti-BrU maximum intensity) from 10 different experiments, which were plotted in each dimension (Fig. 8). In all dimensions, the maximum intensity of myosin Vb correlated with the maximum BrU signal. This 3D colocalization suggested that myosin Vb was definitively present in the putative RV-B14 replication sites.
Figure 7.
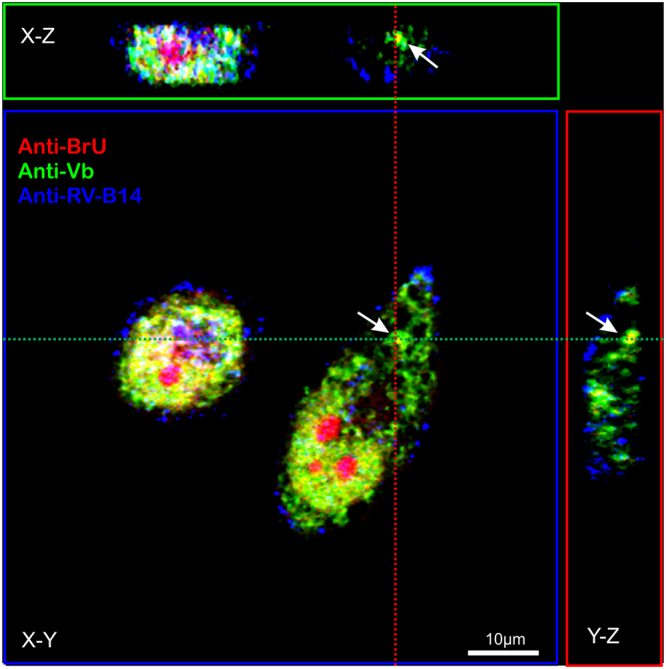
The three dimensional distribution of myosin Vb coincides with putative RV-B14 replication sites. HeLa-H1 cells were infected with RV-B14 (MOI = 50) for 180 min at 34 °C. At 30 min PI, cells were pulse labeled with 2.5 mM BrU to incorporate BrU into nascent RNA. At the end of infection, cells were then processed for 3D confocal microscopy with Alexa Fluor 555® anti-BrdU (anti-BrU; red), (mouse) anti-myosin Vb antibody plus Alexa Fluor 488® goat anti-Mouse (anti-Vb; green), and (rabbit) anti-RV-B14 plus IRDye® 680LT anti-Rabbit IgG (anti-RV-B14; blue). A representative cell was imaged with z-series (Zen Software – LSM510) to capture several image planes from the top to the bottom of the cell. The X-Z axis (green dashed line) was projected to the upper box (green) and Y-Z axis (red dashed line) was projected to the right-side box (red). Nucleolus was nonspecifically labeled with Alexa Fluor 555® anti-BrdU.
Figure 8.
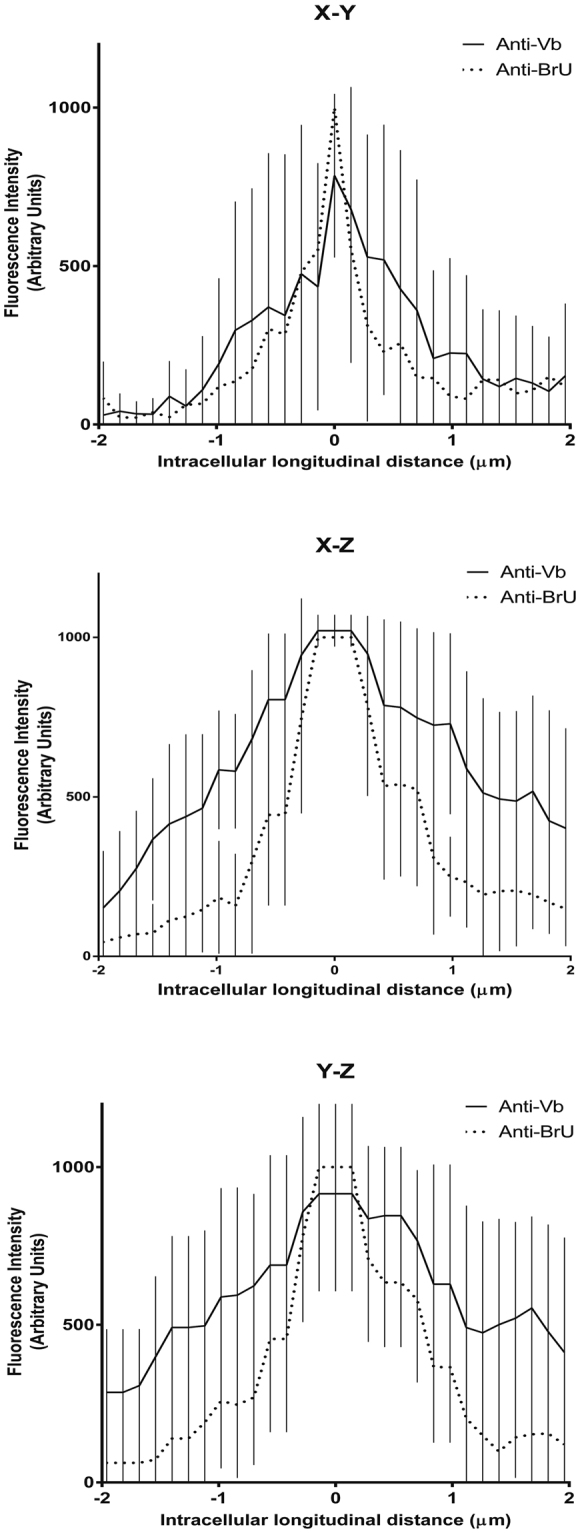
Fluorescence intensity 3D quantification confirms the presence of myosin Vb within putative RV-B14 replication sites. The intensity histogram of signals from BrU-RNA and myosin Vb (10 different experiments similar to Fig. 7) were measured along a line (4 µm long and 0.1 µm wide centered on the anti-BrU maximum intensity). Both individual fluorescent signal (anti-BrU and anti-Vb) were grouped and plotted for each axis. The plateau observed along the X-Z and Y-Z axis is due to lower microscope resolution in Z as compared to the resolution along the X-Y axis.
Discussion
The MLCK inhibitor, ML9, affected RV-B14 entry (Fig. 1) to a greater degree than that observed previously with wortmannin during an RV-A2 infection36. Although not mentioned by Brabec et al., the RV-A2 inhibitory effect of wortmannin could also be explained by a MLCK inhibition37, in addition to the PI3K inhibition described in the report36. MLCK is, together with Rho-kinase, fundamental for stress fiber dynamics54, which can be exploited by viruses for their intracellular transport to the site of replication55. Furthermore, PI3K and MLCK inhibitors have been demonstrated to impair non-opsonic phagocytosis56 and phagocytosis in Kupffer cells57, respectively. Our observed inhibition of RV-B14 entry in the presence of ML9 strongly suggests a role for MLCK in virus internalization, most likely involving actin dynamics and phagocytosis-like mechanisms.
Rhinovirus infection strongly depends on cytoskeletal proteins9. Overexpression of the Rab11 mutants Rab11-GTP (constitutively active) and Rab11-GDP (a dominant-negative) revealed that uncoating of RV-A89 depends on Rab118. Other Rab11 dependent viruses include type I parainfluenza virus58, respiratory syncytial virus41, and human immunodeficiency virus 1, which are also dependent on myosin Vb during infection59. Conzemius et al. demonstrated that RV-B14 infection progressed in cells expressing a dominant-negative Rab118. This demonstrated that RV-B14 does not enter the endosome recycling pathway. However, RV-B14 infection could require other factors, including Rab5 and Rab7 to transit from early endosomes to late endosomes (reviewed by Hutagalung and Novick60), and a demand for myosin Vb was not excluded.
The myosin Vb requirement for type I parainfluenza virus and respiratory syncytial virus infection prompted us to study whether myosin Vs were also playing a role in RV-B14 infection. We began by determining whether myosin Va, which can interact differently with various Rab proteins61,62, has a role in RV-B14 entry. From the data shown in Fig. 2, it is clear that interfering with the function of myosin Va and/or Vb dramatically reduced RV-B14 replication, as deduced from the virtual absence of viral proteins in cells expressing the DN tail domains and challenged with RV-B14. Additionally, the internalized RV-B14 (Fig. 2I and L – arrows) appeared to remain intact, inferred by the shape of the fluorescent signal, and presumably infectious. Unfortunately, we were not able to test whether it was indeed infectious, because it was not possible for us to exclusively extract these particles. Interestingly, the intracellular preservation of RV-B14 in the presence of DN-Va resembled the Mycobacterium tuberculosis that persists within the host cells in the immature phagosomal compartment due to several evasion strategies63.
Next, we determined the distribution of RV-B14 structural proteins, its RNA genome, and endogenous myosin Va at different times PI by immunofluorescence microscopy. A major difficulty to overcome in these experiments was to specifically identify the viral genome with high sensitivity during the early stages of infection. Previously, fluorescence in situ hybridization probes were used to demonstrate that the RV genome became accessible in the cytoplasm 10–12 min after the virus was presented to the host cell18. Molecular beacons have also been used to track the RNA genome of another enterovirus, coxsackievirus B6, inside host cells64. However, both approaches require unique reagents and highly involved protocols. For molecular beacons, the background fluorescence from non-functional beacons can negatively impact the sensitivity of detection. Other probes have been attempted, but were generally ineffective for localizing the viral RNA after it reaches the cytosol16, most likely due to a dilution of the probe, intracellular competition with endogenous RNA, or degradation18.
Here, we used BrU to directly label the viral RNA genome metabolically during production of viral stocks. It did not interfere with replication, since viral stocks were readily generated (BrU-RV-B14). This approach allowed the incorporation of multiple BrU labels into each genome that appeared to provide high sensitivity in the identification of the RNA genome, specifically from the internalized virus after release from the capsid and arrival in the cytoplasm. This advancement was important to successfully observe the colocalization of myosin Va with the RV-B14 genome and the capsid proteins that occurred within 15 min PI (Fig. 3A–F). Similar kinetics were described in the recruitment of ceramide and glycosphingolipid to the cell membrane during RV-B14 infection, which showed an accumulation that peaked at 15 min PI65. The same ceramide and glycosphingolipids were also accumulated during Pseudomonas aeruginosa, Staphylococcus aureus, and Neisseriae gonorrhoeae infection66–69. Additionally, the shape of this BrU-RNA and RV-B14-protein containing structures resemble myosin Va phagocytic cups70.
By 60 min, myosin Va was no longer localized to the same subcellular regions as the RV-B14 BrU labeled genome and its capsid proteins; neither did the capsid proteins colocalize with the viral genome, indicating that (complete) uncoating had occurred (Fig. 3G–L), and capsid proteins could enter the degradation route (as seen for RV-A2 in Fig. 1 from Neubauer, et al.22). Quantification of the intensity histograms (Fig. 3M and N) corroborated the convergence of all three fluorescent signals at 15 min and their separation at 60 min PI. To further explore the interaction of all these components, we immunoprecipitated myosin Va from cells infected with BrU-RV-B14 (similar as in Fig. 3) and probed the material with antiserum against BrU (Fig. 4). The BrU-RNA peak at 15 min and the return to baseline at 60 min was consistent with the colocalization results (Fig. 3). The immunoprecipitation assay included a crosslink to stabilize protein-RNA complexes, and to prevent myosin Va from dissociating from the BrU-RV-B14 genome during the procedure71. Our observations of uncoated BrU-RNA (Figs 3 and 4) did not exclude the possibility that we actually detected A-particles with partially released RNA72. Nevertheless, our data strongly argue that myosin Va was playing an essential role in the release of the RV-B14 genome. The possible interaction between virus RNA and myosin Va, according to the immunoprecipitation assay (Fig. 4), could be direct or indirect. The indirect interaction could use dynein light chain 1 (DLC1) as an adaptor, similar to what was observed in drosophila, where DLC1 binding to gurken mRNA is essential for its proper localization in oocytes73,74. Another possibility is that the myosin plays a role in mechanical destabilization of the capsid75,76. None of these possibilities can be refuted without further investigation.
BrU incorporation into de novo synthesized RNA between 30 and 180 min PI allowed the identification of RNA clusters in infected cells (Figs 5–7). Approximately 1 hour PI, RVs express 2Apro, a protease that degrades eIF4G and polyA binding protein, to dismantle cellular cap-dependent translation that predominates for host mRNA. Viral translation continues through internal ribosome entry sites, which is a viral RNA-specific sequence that allows the recruitment of ribosomes in a cap-independent manner77. Furthermore, 2Apro cleaves Phe/Gly-containing nucleoporin proteins within nuclear pore complexes, which impairs nuclear export78. Consequently, these groups of events could help to explain the differences observed in the patterns of de novo synthesized RNA (Fig. 5) between mock and infected cells, which we propose to represent RV replication sites in infected cells.
Different groups showed that replicated RV RNA genomes could be detected in HeLa cells through fluorescent in situ hybridization and 3H-uridine incorporation within 180–210 min PI18,51. We observed in our work that at 180 min PI, infected HeLa cells were presenting well-defined RNA clusters. These RNA clusters possess a size and intracellular distribution similar to those observed for replication sites from diverse enteroviruses79,80. Since our images do not include labeling for the viral replication machinery, we can only describe these structures, defined by the BrU incorporation (Fig. 5E), as putative RV-B14 replication sites.
Triple labeling (BrU-RNA/Myosin Vb/RV-B14 capsid proteins) demonstrated the involvement of myosin Vb in the putative RV-B14 replication sites (Figs 6 and 7). According to the literature, overexpression of the myosin Vb tail domain led to the accumulation of early endosomal antigen 1 positive endosomes near myosin Vb clusters located in the vicinity of the nucleus81. Myosin Vb is required for transit out of plasma membrane recycling systems32, and the hijacking of its functions by RV-B14 infection could reroute the endosomal membranes to replication sites in an autophagosome-like manner82. If what we observed were replication sites (Figs. 5–7), the presence of myosin Vb could help to explain the diverse origins of the membranes found in replication sites of different enteroviruses, and the specific conjunct of proteins peculiar for each type of virus83.
Lastly, more tests are needed to identify the adaptor proteins that, together with MLCK, myosin Va, and Vb, support viral entry, genome release and interaction with the RNA clusters observed here that resemble viral replication sites.
Materials and Methods
Reagents
1 - (5 - Chloronaphthalenesulfonyl) homopiperazine - HCl (ML-9), Bromouridine (BrU), DMEM,, protease inhibitor cocktail, RNAse Inhibitor ProtectRNA™, and deoxynuclease I were obtained from Sigma (St. Louis, MO, USA). Fetal bovine serum (FBS), Opti-MEM, rabbit anti-BrdU, Alexa Fluor 555® mouse anti-BrdU, rabbit anti-myosin Va, mouse anti-myosin Va, mouse anti-myosin Vb FITC, and Alexa Fluor 488® goat anti-Mouse were from Life Technologies (Carlsbad, CA, USA). Pluronic F68 was from Merck (Rahway, NJ, USA). FuGENE HD was from Promega (Madison, WI, USA). IRDye® 680LT Donkey anti-Rabbit IgG, IRDye® 800LT Donkey anti-Rabbit IgG, and IRDye® 680LT Donkey anti-Mouse IgG were from LI-COR Biosciences (Lincoln, NE, USA). Goat Anti-Rabbit IgG H&L (FITC), Goat Anti-Rabbit IgG H&L (Tetramethylrhodamine (TRITC)), and Goat Anti-Mouse IgG H&L (Agarose) were from ABCAM (Cambridge, UK).
Cell Culture
Human cervix carcinoma cells expressing ICAM-1 (HeLa-H1, CRL 1958; ATCC, Manassas, VA) were maintained in suspension culture or as attached cells. For suspension cultures, cells were maintained in complete DMEM (8% FBS, 1 mM EDTA, 1% pluronic F68, and 1% penicillin/streptomycin) within Erlenmeyer flasks shaken at 100 rpm at 37 °C in a 5% CO2 atmosphere. Cultures were seeded at a concentration of 5 × 105 cells/mL and subdivided every 3–4 days. Monolayer cell cultures were maintained in DMEM without phenol red supplemented with 10% FBS, 30 mM MgCl2, and 1% penicillin/streptomycin. Cells were subdivided before confluence.
Virus production, purification, and metabolic labeling with BrU
Suspension cell cultures of HeLa-H1 were transferred to DMEM without phenol red supplemented with 1% FBS, 30 mM MgCl2, and 1% penicillin/streptomycin at 34 °C. RV-B14 was added at an multiplicity of infection (MOI) of 0.1 and cells remained in culture for 10 h. Cells (2 L) were separated from the media by centrifugation (1,000 × g for 5 min at 25 °C), resuspended in 5–10 mL DMEM without phenol red, transferred to 50 mL round bottom tubes, and subjected to five freeze-thaw cycles using liquid nitrogen and 20 °C water bath. Cell debris was removed by centrifugation (10,000 × g for 30 min) and the supernatants were combined. Sarkosyl (to 1%) and 10 mM EDTA were added prior centrifugation (200,000 × g for 2 h @ 4 °C). The pellet, containing virus, was suspended in TE buffer (10 mM Tris-HCl and 1 mM EDTA; pH 7.5), layered on top of a 5–45% discontinuous sucrose gradient made with TE buffer, and centrifuged (250,000 × g for 90 min @ 4 °C). Banded virus was detected in collected fractions by western blot analysis using anti-RV-B14 sera (see details below). Positive fractions were pooled and diluted with two volumes of TE buffer for storage at −70 °C. All virus fractions were tested for purity (electron microscopy and dynamic light scattering assay) and infectivity. Metabolic labeling of the viral genome with BrU (BrU-RV-B14) was accomplished by adding 10 mM BrU to the growth media at 30 min PI. Beside BrU pulse, the production of BrU-RV-B14 was the same for RV-B14 described above.
Analysis of RV-B14 infectivity
The infectivity of RV-B14 virus samples was measured according to the 50% tissue culture infective dose (TCID50) using HeLa-H1 cells. Adherent cells were infected with serial dilutions of virus stocks ranging from 10−1 to 10−8, with the infection medium described previously. After 48 h incubation at 34 °C, the viability of the cells was measured via crystal violet staining. The TCID50 was calculated according to the method of Reed and Muench84.
Experimental Infections of adherent HeLa-H1
Synchronized infections with RV-B14 were achieved by a 15 min pre-incubation of cells at 4 °C, addition of virus at designated MOIs, and an additional 15 min incubation at 4 °C. Next, cells were washed with ice-cold PBS, to remove nonattached virus particles, and internalization was initiated by the addition of pre-warmed DMEM without phenol red (35 °C). Infections were allowed to proceed as described in the text, and halted by transfer into a 3.8% formaldehyde solution.
Plasmid expression vectors and cell transfections
The carboxy-terminal coding sequence (aa 955 - aa 1852) from the brain isoform of mouse myosin Va85 was amplified for fusion to the carboxy-terminus of eGFP in the mammalian expression vector pEGFP-C2 (Takara Bio, Mountain View, CA). The myosin Vb tail fused to eGFP was a gift from Jim Goldenring32. The GFP expression control used the mammalian expression vector pEGFP-C2 alone (Takara Bio, Mountain View, CA). Cells were grown on 20 mm glass coverslips until ~80% confluence and transfected (1 µg of DNA and 2 µL of FuGENE HD diluted in Opti-MEM). After 1 hour, complete growth medium was added to the plates. Experiments were performed 18 hours after transfection.
Anti-RV-B14 antibody generation and immunolabeling
A high titer polyclonal sera was generated against RV-B14 according to the method described previously86 in partnership with the APCBiotecnologia (IBfCCF – UFRJ). Briefly, two rabbits were injected three times over the course of 45 days with 1 µg of high purity RV-B14. Two weeks after the last injection, total serum was obtained and filtered (0.22 µm, low-binding). Antibody titer was determined against RV-B14 by dot blot assay and antigenicity specificity was tested by western blot.
For immunofluorescence assays, cells were fixed with 3.8% formaldehyde in PBS, washed three times with PBS for 5 min each, permeabilized in PBS with 0.1% Triton X-100, washed three times in PBS, and blocked (5% BSA and 0.05% Tween 20 in PBS) for 1 h before incubation with antibodies. To detect myosin Va, myosin Vb, RV-B14 proteins, and BrU (according to each assay), a sequential labeling protocol was used. Initially, mouse anti-myosin Va or mouse anti-myosin Vb (1:500 dilution) were co-incubated with anti-RV-B14 rabbit sera (1:1000 dilution) for 1 h in PBS with 5% BSA and 0.05% Tween 20. Next, the coverslips were washed three times in PBS for 5 min followed by incubation with the appropriate fluorescent secondary antibody. After 1 h, the coverslips were washed three times with PBS and then incubated with the directly labeled antibody against BrU (1:250 dilution). Cells were prepared according Rietdorf40 to label virus outside the cell (Fig. 1A–C) and inside the cell (Fig. 1D–F). Finally, the coverslips were washed with PBS three times and mounted for microscopy.
Confocal fluorescence microscopy and image analysis
Images of the fluorescent signals were acquired at room temperature with a LSM510 META Microscope (Carl Zeiss, Oberkochen, Germany) equipped with a 100X oil immersion objective (N.A. 1.42). Images were captured with the iris set for one airy unit and 1024 × 1024 pixel resolution. Following acquisition, images were quantified for various parameters as described in the text. In all instances, the pixel intensities were obtained from the raw images.
Myosin Va immunoprecipitation and DOT blot assay
Cells were incubated in DMEM without phenol red supplemented with 1% FBS, 30 mM MgCl2, and 1% penicillin/streptomycin at 4 °C with BrU-RV-B14 (MOI = 10) for 15 min, to allow viral attachment. To start the infection, the medium was exchanged for pre-warmed medium (35 °C). Infections were continued for 0 (infection was stopped after virus attachment), 15, 30, 60, 90, 180 or 270 min. Samples were cross-linked with 1% formaldehyde for 2 min to halt the infection and to stabilize intracellular complexes. RIPA buffer was then added to the cells and protein-RNA complex extraction proceeded according to the RIP protocol87. Briefly, cells were treated with 200 mM glycine-PBS then washed with ice-cold PBS. Cells were kept on ice and scrapped into lysis buffer (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA pH 8, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, protease Inhibitor cocktail, RNAse Inhibitor ProtectRNA™, DNase I, and 1% BSA). Homogenate were collected and centrifuged (5,000 × g for 30 min at 4 °C). The supernatant was then incubated for 4 h with mouse anti-myosin Va antibody at 4 °C with rocking. Agarose beads coupled with anti-Mouse antibody were added for an overnight incubation with rocking at 4 °C. The next day, beads were collected by centrifugation and washed six times with lysis buffer. Precipitated complexes were released with elution buffer (6 M urea, 20 mM Tris-HCl pH 7.5, 100 mM NaCl) and samples (10 µg total protein) were subsequently applied by vacuum to a low-fluorescence PVDF membrane (LI-COR Biosciences, Lincoln, NE) with the aid of a Dot Blot Microfiltration Apparatus (BioRad, Hercules, CA, US). Membranes were washed and blocked with Odyssey Blocking Solution (LI-COR Biosciences) for 4 h. Next, the membrane was incubated with a solution containing rabbit anti-BrU antibody (1:1000 dilution) diluted in Blocking Buffer for 2 h, washed 3 times, and incubated with anti-Rabbit IRDye 800 nm antibody (1:5000 dilution) for 2 h. Membranes were washed and scanned with Odyssey (LI-COR Biosciences). Membranes were stripped using 10X EK-Away Stripping Buffer (Life Technologies, Carlsbad, CA), according to the manufacturer’s protocol, followed by incubation with loading control (mouse anti-myosin Va and anti-Mouse IRDye 680), washing, and scanning. Results are shown as bars of the anti-BrU (anti-Rabbit IRDye 800) signal normalized to the anti-myosin Va (anti-Mouse IRDye 680) signal.
Ethics statement
Antibody generation by partnership with APC Biotecnologia (Instituto de Biofísica Carlos Chagas Filho - UFRJ) involving animals were carried out in accordance with the recommendations of the National Council for the Control of Animal Experimentation (CONCEA), and were approved by the institutional Ethics Committee on Animal Use (CEUA).
Statistical analysis
Analyzes were performed using GraphPad Prisma 5.01 statistical software. The variables of the analyzed groups were compared using a non-parametric Kruskal Wallis ANOVA test. Multiple comparisons were made using Dunn’s multiple test. A probability value of less than 0.05 was regarded as significant.
Data availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Electronic supplementary material
Acknowledgements
The authors would like to thank Dr. Dieter Blaas (Max F. Perutz Laboratories - Medical University of Vienna) for his insightful critics and helpful discussions. We thank Dr. James R. Goldenring (Epithelial Biology Center - Vanderbilt University School of Medicine) for kindly providing the myosin Vb plasmids. The myosin Va tail construct (DN-Va) was created in the lab of Dr. John A. Mercer (Institute for Stem Cell Biology and Regenerative Medicine - McLaughlin Research Institute). We are grateful to Dr. Jerson Lima Silva (Instituto de Bioquímica Médica Leopoldo de Meis – Universidade Federal do Rio de Janeiro) for all the support provided during the development of this work. We thank Emerson R. Gonçalves and Vivian Neuza dos Santos for competent technical assistance.
Author Contributions
A.R.H., D.W.P.J., A.C.O., V.P.S., and A.M.O.G., designed the study. A.R.H., R.B.G., and C.B.D. conducted experiments. A.R.H., D.W.P.J., R.B.G., C.B.D., and A.M.O.G. analyzed and interpreted the data. A.R.H., D.W.P.J., and A.M.O.G. wrote the manuscript. A.C.O. and V.P.S. approved the manuscript for submission.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-17501-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jacobs SE, Lamson DM, St George K, Walsh TJ. Human rhinoviruses. Clinical microbiology reviews. 2013;26:135–162. doi: 10.1128/CMR.00077-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fendrick AM, Monto AS, Nightengale B, Sarnes M. The economic burden of non-influenza-related viral respiratory tract infection in the United States. Archives of internal medicine. 2003;163:487–494. doi: 10.1001/archinte.163.4.487. [DOI] [PubMed] [Google Scholar]
- 3.Makela MJ, et al. Viruses and bacteria in the etiology of the common cold. Journal of clinical microbiology. 1998;36:539–542. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gern JE. The ABCs of rhinoviruses, wheezing, and asthma. Journal of virology. 2010;84:7418–7426. doi: 10.1128/JVI.02290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandenburg B, et al. Imaging poliovirus entry in live cells. PLoS biology. 2007;5:e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergelson JM, Coyne CB. Picornavirus entry. Advances in experimental medicine and biology. 2013;790:24–41. doi: 10.1007/978-1-4614-7651-1_2. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs R, Blaas D. Productive entry pathways of human rhinoviruses. Advances in virology. 2012;2012:826301. doi: 10.1155/2012/826301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conzemius R, Ganjian H, Blaas D, Fuchs R. ICAM-1 Binding Rhinoviruses A89 and B14 Uncoat in Different Endosomal Compartments. Journal of virology. 2016;90:7934–7942. doi: 10.1128/JVI.00712-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganjian, H., Zietz, C., Mechtcheriakova, D., Blaas, D. & Fuchs, R. ICAM-1 Binding Rhinoviruses Enter HeLa Cells via Multiple Pathways and Travel to Distinct Intracellular Compartments for Uncoating. Viruses9, 10.3390/v9040068 (2017). [DOI] [PMC free article] [PubMed]
- 10.Shingler KL, et al. The enterovirus 71 A-particle forms a gateway to allow genome release: a cryoEM study of picornavirus uncoating. PLoS pathogens. 2013;9:e1003240. doi: 10.1371/journal.ppat.1003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butan C, Filman DJ, Hogle JM. Cryo-electron microscopy reconstruction shows poliovirus 135S particles poised for membrane interaction and RNA release. Journal of virology. 2014;88:1758–1770. doi: 10.1128/JVI.01949-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pickl-Herk A, et al. Uncoating of common cold virus is preceded by RNA switching as determined by X-ray and cryo-EM analyses of the subviral A-particle. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20063–20068. doi: 10.1073/pnas.1312128110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, et al. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nature structural & molecular biology. 2012;19:424–429. doi: 10.1038/nsmb.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bostina M, Levy H, Filman DJ, Hogle JM. Poliovirus RNA is released from the capsid near a twofold symmetry axis. Journal of virology. 2011;85:776–783. doi: 10.1128/JVI.00531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schober D, Kronenberger P, Prchla E, Blaas D, Fuchs R. Major and minor receptor group human rhinoviruses penetrate from endosomes by different mechanisms. Journal of virology. 1998;72:1354–1364. doi: 10.1128/jvi.72.2.1354-1364.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harutyunyan S, et al. Viral uncoating is directional: exit of the genomic RNA in a common cold virus starts with the poly-(A) tail at the 3’-end. PLoS pathogens. 2013;9:e1003270. doi: 10.1371/journal.ppat.1003270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bilek G, et al. Liposomal nanocontainers as models for viral infection: monitoring viral genomic RNA transfer through lipid membranes. Journal of virology. 2011;85:8368–8375. doi: 10.1128/JVI.00329-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brabec-Zaruba M, Pfanzagl B, Blaas D, Fuchs R. Site of human rhinovirus RNA uncoating revealed by fluorescent in situ hybridization. Journal of virology. 2009;83:3770–3777. doi: 10.1128/JVI.00265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groppelli E, et al. Picornavirus RNA is protected from cleavage by ribonuclease during virion uncoating and transfer across cellular and model membranes. PLoS pathogens. 2017;13:e1006197. doi: 10.1371/journal.ppat.1006197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jurgeit A, et al. Niclosamide is a proton carrier and targets acidic endosomes with broad antiviral effects. PLoS pathogens. 2012;8:e1002976. doi: 10.1371/journal.ppat.1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nurani G, Lindqvist B, Casasnovas JM. Receptor priming of major group human rhinoviruses for uncoating and entry at mild low-pH environments. J Virol. 2003;77:11985–11991. doi: 10.1128/JVI.77.22.11985-11991.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neubauer C, Frasel L, Kuechler E, Blaas D. Mechanism of entry of human rhinovirus 2 into HeLa cells. Virology. 1987;158:255–258. doi: 10.1016/0042-6822(87)90264-9. [DOI] [PubMed] [Google Scholar]
- 23.Blaas D. Viral entry pathways: the example of common cold viruses. Wiener medizinische Wochenschrift. 2016;166:211–226. doi: 10.1007/s10354-016-0461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snyers L, Zwickl H, Blaas D. Human rhinovirus type 2 is internalized by clathrin-mediated endocytosis. Journal of virology. 2003;77:5360–5369. doi: 10.1128/JVI.77.9.5360-5369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huber M, Brabec M, Bayer N, Blaas D, Fuchs R. Elevated endosomal pH in HeLa cells overexpressing mutant dynamin can affect infection by pH-sensitive viruses. Traffic. 2001;2:727–736. doi: 10.1034/j.1600-0854.2001.21007.x. [DOI] [PubMed] [Google Scholar]
- 26.Bayer N, Schober D, Huttinger M, Blaas D, Fuchs R. Inhibition of clathrin-dependent endocytosis has multiple effects on human rhinovirus serotype 2 cell entry. The Journal of biological chemistry. 2001;276:3952–3962. doi: 10.1074/jbc.M004722200. [DOI] [PubMed] [Google Scholar]
- 27.DeTulleo L, Kirchhausen T. The clathrin endocytic pathway in viral infection. The EMBO journal. 1998;17:4585–4593. doi: 10.1093/emboj/17.16.4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mettlen M, Pucadyil T, Ramachandran R, Schmid SL. Dissecting dynamin’s role in clathrin-mediated endocytosis. Biochemical Society transactions. 2009;37:1022–1026. doi: 10.1042/BST0371022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saba TG, et al. Rhinovirus-induced macrophage cytokine expression does not require endocytosis or replication. American journal of respiratory cell and molecular biology. 2014;50:974–984. doi: 10.1165/rcmb.2013-0354OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bayer N, et al. Effect of bafilomycin A1 and nocodazole on endocytic transport in HeLa cells: implications for viral uncoating and infection. Journal of virology. 1998;72:9645–9655. doi: 10.1128/jvi.72.12.9645-9655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C, et al. Myosin light chain kinase (MLCK) regulates cell migration in a myosin regulatory light chain phosphorylation-independent mechanism. The Journal of biological chemistry. 2014;289:28478–28488. doi: 10.1074/jbc.M114.567446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lapierre LA, et al. Myosin vb is associated with plasma membrane recycling systems. Molecular biology of the cell. 2001;12:1843–1857. doi: 10.1091/mbc.12.6.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salerno VP, et al. Myosin-Va mediates RNA distribution in primary fibroblasts from multiple organs. Cell motility and the cytoskeleton. 2008;65:422–433. doi: 10.1002/cm.20272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lombardo AT, et al. Myosin Va molecular motors manoeuvre liposome cargo through suspended actin filament intersections in vitro. Nature communications. 2017;8:15692. doi: 10.1038/ncomms15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, et al. Myosin Vb mobilizes recycling endosomes and AMPA receptors for postsynaptic plasticity. Cell. 2008;135:535–548. doi: 10.1016/j.cell.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brabec M, Blaas D, Fuchs R. Wortmannin delays transfer of human rhinovirus serotype 2 to late endocytic compartments. Biochemical and biophysical research communications. 2006;348:741–749. doi: 10.1016/j.bbrc.2006.07.125. [DOI] [PubMed] [Google Scholar]
- 37.Nakanishi S, et al. Wortmannin, a microbial product inhibitor of myosin light chain kinase. The Journal of biological chemistry. 1992;267:2157–2163. [PubMed] [Google Scholar]
- 38.Li L, Wu X, Yue HY, Zhu YC, Xu J. Myosin light chain kinase facilitates endocytosis of synaptic vesicles at hippocampal boutons. Journal of neurochemistry. 2016;138:60–73. doi: 10.1111/jnc.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saitoh M, Ishikawa T, Matsushima S, Naka M, Hidaka H. Selective inhibition of catalytic activity of smooth muscle myosin light chain kinase. The Journal of biological chemistry. 1987;262:7796–7801. [PubMed] [Google Scholar]
- 40.Rietdorf J, et al. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nature cell biology. 2001;3:992–1000. doi: 10.1038/ncb1101-992. [DOI] [PubMed] [Google Scholar]
- 41.Brock SC, Goldenring JR, Crowe JE., Jr. Apical recycling systems regulate directional budding of respiratory syncytial virus from polarized epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15143–15148. doi: 10.1073/pnas.2434327100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donovan KW, Bretscher A. Head-to-tail regulation is critical for the in vivo function of myosin V. The Journal of cell biology. 2015;209:359–365. doi: 10.1083/jcb.201411010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hehnly H, Stamnes M. Regulating cytoskeleton-based vesicle motility. FEBS letters. 2007;581:2112–2118. doi: 10.1016/j.febslet.2007.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshimura A, et al. Myosin-Va facilitates the accumulation of mRNA/protein complex in dendritic spines. Current biology: CB. 2006;16:2345–2351. doi: 10.1016/j.cub.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 45.Izidoro-Toledo TC, et al. A myosin-Va tail fragment sequesters dynein light chains leading to apoptosis in melanoma cells. Cell death & disease. 2013;4:e547. doi: 10.1038/cddis.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munyon W, Salzman NP. The incorporation of 5-fluoro-uracil into poliovirus. Virology. 1962;18:95–101. doi: 10.1016/0042-6822(62)90181-2. [DOI] [PubMed] [Google Scholar]
- 47.Salzman NP, Lockart RZ, Jr., Sebring ED. Alterations in HeLa cell metabolism resulting from poliovirus infection. Virology. 1959;9:244–259. doi: 10.1016/0042-6822(59)90118-7. [DOI] [PubMed] [Google Scholar]
- 48.Gerisch G, et al. Self-organizing actin waves as planar phagocytic cup structures. Cell adhesion & migration. 2009;3:373–382. doi: 10.4161/cam.3.4.9708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Au PC, Helliwell C, Wang MB. Characterizing RNA-protein interaction using cross-linking and metabolite supplemented nuclear RNA-immunoprecipitation. Molecular biology reports. 2014;41:2971–2977. doi: 10.1007/s11033-014-3154-1. [DOI] [PubMed] [Google Scholar]
- 50.McCaffrey MW, Lindsay AJ. Roles for myosin Va in RNA transport and turnover. Biochemical Society transactions. 2012;40:1416–1420. doi: 10.1042/BST20120172. [DOI] [PubMed] [Google Scholar]
- 51.Yin FH, Knight E., Jr. In vivo and in vitro synthesis of human rhinovirus type 2 ribonucleic acid. Journal of virology. 1972;10:93–98. doi: 10.1128/jvi.10.1.93-98.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jurgeit A, et al. An RNA replication-center assay for high content image-based quantifications of human rhinovirus and coxsackievirus infections. Virology journal. 2010;7:264. doi: 10.1186/1743-422X-7-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindsay AJ, McCaffrey MW. Myosin Vb localises to nucleoli and associates with the RNA polymerase I transcription complex. Cell motility and the cytoskeleton. 2009;66:1057–1072. doi: 10.1002/cm.20408. [DOI] [PubMed] [Google Scholar]
- 54.Katoh K, Kano Y, Amano M, Kaibuchi K, Fujiwara K. Stress fiber organization regulated by MLCK and Rho-kinase in cultured human fibroblasts. American journal of physiology. Cell physiology. 2001;280:C1669–1679. doi: 10.1152/ajpcell.2001.280.6.C1669. [DOI] [PubMed] [Google Scholar]
- 55.Taylor MP, Koyuncu OO, Enquist LW. Subversion of the actin cytoskeleton during viral infection. Nature reviews. Microbiology. 2011;9:427–439. doi: 10.1038/nrmicro2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tachado SD, Samrakandi MM, Cirillo JD. Non-opsonic phagocytosis of Legionella pneumophila by macrophages is mediated by phosphatidylinositol 3-kinase. PloS one. 2008;3:e3324. doi: 10.1371/journal.pone.0003324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watanabe S, Hirose M, Ueno T, Kominami E, Namihisa T. Integrity of the cytoskeletal system is important for phagocytosis by Kupffer cells. Liver. 1990;10:249–254. doi: 10.1111/j.1600-0676.1990.tb00466.x. [DOI] [PubMed] [Google Scholar]
- 58.Stone R, Hayashi T, Bajimaya S, Hodges E, Takimoto T. Critical role of Rab11a-mediated recycling endosomes in the assembly of type I parainfluenza viruses. Virology. 2016;487:11–18. doi: 10.1016/j.virol.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Varthakavi V, et al. The pericentriolar recycling endosome plays a key role in Vpu-mediated enhancement of HIV-1 particle release. Traffic. 2006;7:298–307. doi: 10.1111/j.1600-0854.2005.00380.x. [DOI] [PubMed] [Google Scholar]
- 60.Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lindsay AJ, et al. Identification and characterization of multiple novel Rab-myosin Va interactions. Molecular biology of the cell. 2013;24:3420–3434. doi: 10.1091/mbc.E13-05-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pylypenko O, et al. Structural basis of myosin V Rab GTPase-dependent cargo recognition. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20443–20448. doi: 10.1073/pnas.1314329110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jamwal SV, et al. Mycobacterial escape from macrophage phagosomes to the cytoplasm represents an alternate adaptation mechanism. Scientific reports. 2016;6:23089. doi: 10.1038/srep23089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yeh HY, Yates MV, Mulchandani A, Chen W. Visualizing the dynamics of viral replication in living cells via Tat peptide delivery of nuclease-resistant molecular beacons. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17522–17525. doi: 10.1073/pnas.0807066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dreschers S, et al. Infections with human rhinovirus induce the formation of distinct functional membrane domains. Cell Physiol Biochem. 2007;20:241–254. doi: 10.1159/000104170. [DOI] [PubMed] [Google Scholar]
- 66.Grassme H, Riehle A, Wilker B, Gulbins E. Rhinoviruses infect human epithelial cells via ceramide-enriched membrane platforms. The Journal of biological chemistry. 2005;280:26256–26262. doi: 10.1074/jbc.M500835200. [DOI] [PubMed] [Google Scholar]
- 67.Grassme H, et al. Host defense against Pseudomonas aeruginosa requires ceramide-rich membrane rafts. Nature medicine. 2003;9:322–330. doi: 10.1038/nm823. [DOI] [PubMed] [Google Scholar]
- 68.Abraham SN, Duncan MJ, Li G, Zaas D. Bacterial penetration of the mucosal barrier by targeting lipid rafts. J Investig Med. 2005;53:318–321. doi: 10.2310/6650.2005.53609. [DOI] [PubMed] [Google Scholar]
- 69.Grassme H, et al. Acidic sphingomyelinase mediates entry of N. gonorrhoeae into nonphagocytic cells. Cell. 1997;91:605–615. doi: 10.1016/S0092-8674(00)80448-1. [DOI] [PubMed] [Google Scholar]
- 70.Diakonova M, Bokoch G, Swanson JA. Dynamics of cytoskeletal proteins during Fcgamma receptor-mediated phagocytosis in macrophages. Molecular biology of the cell. 2002;13:402–411. doi: 10.1091/mbc.01-05-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gerstberger S, Hafner M, Ascano M, Tuschl T. Evolutionary conservation and expression of human RNA-binding proteins and their role in human genetic disease. Advances in experimental medicine and biology. 2014;825:1–55. doi: 10.1007/978-1-4939-1221-6_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harutyunyan S, Kowalski H, Blaas D. The Rhinovirus subviral a-particle exposes 3’-terminal sequences of its genomic RNA. Journal of virology. 2014;88:6307–6317. doi: 10.1128/JVI.00539-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mhlanga MM, et al. In vivo colocalisation of oskar mRNA and trans-acting proteins revealed by quantitative imaging of the Drosophila oocyte. PloS one. 2009;4:e6241. doi: 10.1371/journal.pone.0006241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rom I, Faicevici A, Almog O, Neuman-Silberberg FS. Drosophila Dynein light chain (DDLC1) binds to gurken mRNA and is required for its localization. Biochimica et biophysica acta. 2007;1773:1526–1533. doi: 10.1016/j.bbamcr.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 75.Greber UFV. and Host Mechanics Support Membrane Penetration and Cell Entry. Journal of virology. 2016;90:3802–3805. doi: 10.1128/JVI.02568-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ortega-Esteban A, et al. Fluorescence Tracking of Genome Release during Mechanical Unpacking of Single Viruses. ACS nano. 2015;9:10571–10579. doi: 10.1021/acsnano.5b03020. [DOI] [PubMed] [Google Scholar]
- 77.Aumayr M, Schrempf A, Uzulmez O, Olek KM, Skern T. Interaction of 2A proteinase of human rhinovirus genetic group A with eIF4E is required for eIF4G cleavage during infection. Virology. 2017;511:123–134. doi: 10.1016/j.virol.2017.08.020. [DOI] [PubMed] [Google Scholar]
- 78.Watters, K. et al. Differential Disruption of Nucleocytoplasmic Trafficking Pathways by Rhinovirus 2A Proteases. Journal of virology91, 10.1128/JVI.02472-16 (2017). [DOI] [PMC free article] [PubMed]
- 79.Schlegel A, Giddings TH, Jr., Ladinsky MS, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. Journal of virology. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Limpens, R. W. et al. The transformation of enterovirus replication structures: a three-dimensional study of single- and double-membrane compartments. mBio2, 10.1128/mBio.00166-11 (2011). [DOI] [PMC free article] [PubMed]
- 81.Provance DW, Jr., et al. Myosin-Vb functions as a dynamic tether for peripheral endocytic compartments during transferrin trafficking. BMC cell biology. 2008;9:44. doi: 10.1186/1471-2121-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Longatti A, Tooze SA. Recycling endosomes contribute to autophagosome formation. Autophagy. 2012;8:1682–1683. doi: 10.4161/auto.21486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Netherton C, Moffat K, Brooks E, Wileman T. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Advances in virus research. 2007;70:101–182. doi: 10.1016/S0065-3527(07)70004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pourianfar HR, Javadi A, Grollo L. A Colorimetric-Based Accurate Method for the Determination of Enterovirus 71 Titer. Indian journal of virology: an official organ of Indian Virological Society. 2012;23:303–310. doi: 10.1007/s13337-012-0105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Seperack PK, Mercer JA, Strobel MC, Copeland NG, Jenkins NA. Retroviral sequences located within an intron of the dilute gene alter dilute expression in a tissue-specific manner. The EMBO journal. 1995;14:2326–2332. doi: 10.1002/j.1460-2075.1995.tb07227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cooney MK, Kenny GE. Immunogenicity of rhinoviruses. Proceedings of the Society for Experimental Biology and Medicine. Society for Experimental Biology and Medicine. 1970;133:645–650. doi: 10.3181/00379727-133-34536. [DOI] [PubMed] [Google Scholar]
- 87.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).