Abstract
Peroxiredoxins (Prxs) are vital regulators of intracellular reactive oxygen species levels in all living organisms. Their activity depends on one or two catalytically active cysteine residues, the peroxidatic Cys (CP) and, if present, the resolving Cys (CR). A detailed catalytic cycle has been derived for typical 2-Cys Prxs, however, little is known about the catalytic cycle of 1-Cys Prxs. We have characterized Prx6 from the cyanobacterium Anabaena sp. strain PCC7120 (AnPrx6) and found that in addition to the expected peroxidase activity, AnPrx6 can act as a molecular chaperone in its dimeric state, contrary to other Prxs. The AnPrx6 crystal structure at 2.3 Å resolution reveals different active site conformations in each monomer of the asymmetric obligate homo-dimer. Molecular dynamic simulations support the observed structural plasticity. A FSH motif, conserved in 1-Cys Prxs, precedes the active site PxxxTxxCp signature and might contribute to the 1-Cys Prx reaction cycle.
Introduction
Peroxiredoxins (Prxs) are an essential class of peroxidases (EC 1.11.1.15) ubiquitously found in all known species1,2. They efficiently reduce a variety of peroxides, are vital regulators of reactive oxygen species (ROS) levels and play important roles in cell signalling3–6. The strictly conserved active site peroxidatic cysteine residue (CP) is often located between residues 40 to 603. A common Prx classification is based on the presence and location of a second, the so called resolving cysteine residue (CR). The 1-Cys Prxs contain CP, but not CR, whereas the 2-Cys Prxs have both CP and CR. 2-Cys Prxs are further divided into the “atypical” and the “typical” 2-Cys Prxs, depending on whether CR is found in a central position or closer to the C-terminus, and also whether it forms an inter- or intra-molecular disulphide bond with CP during the reaction cycle3. Recently, a classification based on Deacon Active Site Profiling (DASP) was introduced by Nelson et al.7,8 that divides the Prx proteins into six subfamilies: (I) the Prx1/AhpC (“typical” 2-Cys Prxs) and (II) the Prx6 (mainly 1-Cys Prxs) subfamilies, form dimers via the “B” interface through β-strand interactions, whereas (III) the Prx5 (includes “atypical” 2-Cys Prxs) and (IV) Tpx (bacterial periplasmic thiol peroxidase, also includes “atypical” 2-Cys Prxs) subfamilies form dimers across the “A” interface (“A” for alternative or ancestral), (V) the members of the PrxQ subfamily (bacterioferritin co-migrating protein, BCP, and plant PrxQ; includes “atypical” 2-Cys Prxs) remain monomeric, and last (VI) a small subfamily called AhpE (for Mycobacterium tuberculosis AhpE). The DASP classification as implemented in the PREX database8,9 will be used throughout the text.
The first reference to a Prx was made in 1988 when Kim et al. described a thiol specific antioxidant (TSA) protein in Saccharomyces cerevisiae that acted as a “Protector Protein”10. They then identified a mammalian homologue in the brain of rats with a comparable protective function11. Since then, Prx have been associated with multiple metabolic pathways and were linked to a diverse range of processes such as scavenging of ROS and peroxides12–14, cell signalling5, inflammation, cancer and innate immunity15–17, apoptosis18, virulence19, and colonization20 in aerobic and anaerobic bacteria12,13,19,20. In addition, the oxidation state of Prx proteins is tightly linked to circadian rhythms in all domains of life21. Moreover, unlike other Prx proteins, members of the Prx6 family were shown to carry the additional Ca2+-independent phospholipase A2 (PLA2) activity22,23, reviewed by Fisher24.
Peroxiredoxins from the Prx1/AhpC and Prx6 subclasses assemble into antiparallel, obligate (α2) homo-dimers via the B-type dimer interface by fusing their central monomer β-sheet into one continuous β-sheet spanning the dimer25. The Prx1/AhpC and Prx6 members with two redox active cysteines, can further oligomerize into ring shaped pentamers of dimers (α2)5, when exposed to oxidative stress or increasing levels of heat. The ring formation is accompanied by a functional switch whereby the proteins lose peroxidase activity and gain molecular chaperone activity26–30. Decameric rings were observed, among others, in yeast27, Helicobacter pylori28, Schistosoma mansoni30 and Homo sapiens31,32.
Cyanobacteria are ancient and ubiquitous oxygenic prokaryotes33 and their Prxs have been studied to some extend (for reviews see6,7). Perez-Perez et al. suggested that the five different peroxiredoxins found in Synechocystis sp. PCC 6803 have additional functions beyond the peroxide detoxification activity34. Chaperone activity could be one such function. Dekker et al. support the hypothesis that the origin of the Ur-Chaperonin is derived from a thioredoxin/peroxiredoxin fold35. Until now however, there has been no report on the three-dimensional structure or functional analysis of a cyanobacterial Prx.
Using two-dimensional gel electrophoresis Mishra et al. found that under heat stress a particular protein accumulated in cyanobacteria, which was identified to be AhpC using mass spectrometry36. Increased expression of the Anabaena sp. PCC7120 AhpC protein (hereafter referred to as AnPrx6) in E. coli conferred enhanced resistance to a variety of abiotic stresses, suggesting that AnPrx6 plays an important role in the regulation of cellular processes in addition to the enzymes’ peroxidase activity37. These findings and the observation that AnPrx6 contains a sequence motif highly conserved in Prx6 proteins not found in other Prx families, raised the questions: (i) does AnPrx6 possess unique functional attributes, and (ii) can the high-resolution crystal structure provide a molecular level understanding of the function of AnPrx6 and perhaps of Prx6 proteins in general?
To address these questions, we have crystallized AnPrx638 and present here the first three-dimensional structure of a cyanobacterial Prx6 protein together with results obtained from bioinformatics analysis and from molecular dynamics simulations. Further, biochemical analyses provide evidence that AnPrx6 is unique among the Prx proteins, since it can act as a molecular chaperone in its dimeric state, contrary to other Prxs.
Results
Sequence analysis
Multiple sequence alignments (MSA) of Prx proteins and phylogenetic analysis with Clustal Omega39 established AnPrx6 as a member of the Prx6 subfamily. A BLASTp search40 with AnPrx6 as query against the Protein Data Bank (PDB)41 shows that the human Prx6 protein hORF6 (PDB code 1PRX42) shares 52% sequence identity with AnPrx6 and is the highest scoring orthologue with a BLASTp bits score of 544. hORF6 is the founding member of the Prx6 subfamily25 and carries a single cysteine residue, similar to AnPrx6. Additional high scoring orthologues include Prx6 from Arenicola marina (PDB code 2V2G43) and Prx6 protein from Plasmodium yoelii (PDB code 1XCC44) with 48% and 41% sequence identity, respectively. The sequence identity shared with two members of the Prx6 family that form (α2)5 decamer rings is slightly lower: 39% for the protein from the aerobic hyperthermophilic Crenarchaeon Aeropyrum pernix K1 (PDB code 2CV445) and 38% for the protein from the anaerobic hyperthermophilic archaeon Pyrococcus horikoshii (PDB code 3W6G46). Sequence similarities and differences are further discussed in the context of structural comparisons.
X-ray crystal structure determination and analysis
The crystal structure of AnPrx6 was determined from orthorhombic crystals in space group P21212138. Phase information and an initial atomic model were obtained by molecular replacement (MR) using the monomer A of human Prx6 (PDB code 1PRX42), as the search model. After multiple rounds of model building and refinement against data extending to a d-spacing of 2.3 Å the values for Rwork and Rfree converged at 20.4% and 24.5%, respectively. (For details see Supplementary Table S1). The crystallographic asymmetric unit contains four copies of the AnPrx6 protein, monomers A to D. In the case of monomers A and C, all native amino acids (Met1 to Lys212) could be built into the electron density map, whereas monomers B and D lack the N-terminal methionine. The four monomers associate into two dimers, AB and CD. Superimposition of the peptide backbone atoms of dimer AB onto dimer CD results in a low root mean square deviation (RMSD) of 0.22 Å which means that the two dimers are basically identical. We therefore refer to dimer AB in all further discussions, unless indicated otherwise.
The AnPrx6 monomers and the asymmetric dimer
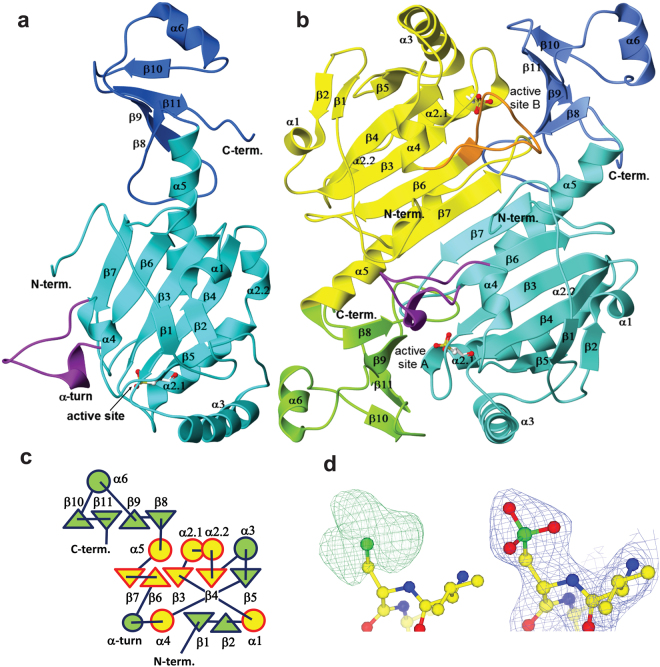
The overall structure of an AnPrx6 monomer (Fig. 1a) and dimer (Fig. 1b) is similar to known structures of 1-Cys and 2-Cys Prxs from the Prx1/AhpC and Prx6 subfamilies. Each monomer consists of two domains: an N-terminal peroxiredoxin domain (residues Met1 to Asn162) and a C-terminal dimerization domain (residues Tyr163 to Lys212).
Figure 1.
The Crystal structure of AnPrx6. (a) Ribbon diagram of monomer A. The catalytic N-terminal peroxiredoxin domain (M1-N162) is coloured light blue and the C-terminal dimerization domain (Y163-K212) dark blue. The termini and secondary structure elements are labelled. The active site CP is shown in ball-and-stick style. (b) Ribbon diagram of the AnPrx6 dimer. Monomer A is coloured as before. The N-terminal domain of monomer B is shown in yellow and the C-terminal domain in light green. The residues linking helix α4 to strand β6 fold into an α-helical turn in monomers A and C (purple) while they adopt an irregular loop in monomers B and D (orange). (c) Topology diagram of AnPrx6. The core secondary structure elements of the Trx fold are highlighted in yellow and red. Note that the α-helical turn is only present in AnPrx6 monomers A and C. (d) Left panel: A positive feature in the difference electron density map (m|Fo| − D|Fc|), calculated with a Cys at the active site and contoured at 3 RMSD, arises due to excess electrons (green mesh) covering the sulphur atom (green ball). This suggests that the active site Cys45 (CP) is triple oxidized to cysteine-sulfonic acid (Cys-O2-OH). Right panel: The quality of the (2 m|Fo| − D|Fc|) electron density map, contoured at 1 RMSD (blue mesh), after structure refinement with cysteine-sulfonic acid at position 45. All atoms and their electrons are accounted for and no extra difference electron density features are visible in the map. (|Fo| and |Fc| are the modulus of the observed and calculated structure factors, respectively, m is a figure of merit and D is a scale factor).
The N-terminal catalytic domain comprises seven β-strands and five α-helices. Five strands (β7, β6, β3, β4, and β5, from left to right in monomer A, Fig. 1a) assemble into the central β-sheet that is sandwiched between a β-hairpin, formed by strands β1 and β2, and helices α1 and α4 on one face and helices α2, α3 and α5 on the opposite side. The core of the N-terminal domain corresponds to the conserved thioredoxin fold1,25, consisting of beta strands β7, β6, β3 and β4 and α-helices α1, α4, α2 and α5 (highlighted in yellow and red in the topology diagram of Fig. 1c). Helix α2 contains a kink introduced at Pro57 that divides α2 into two sections denoted α2.1 and α2.2. The first turn of helix α2.1 contains the strictly conserved peroxidatic cysteine (CP). In this structure, the sulphur atom of CP (Cys45) is triple oxidized to cysteine-sulfonic acid (Cys45-O2-OH), which is evident from three additional features in the (m|Fo| - D|Fc|) difference electron density map (Fig. 1d).
The C-terminal dimerization domain folds into a four stranded anti-parallel β-sheet formed by strands β8, β9, β10 and β11 and a helix, α6, located between strands β9 and β10, that caps the sheet. The dimerization domain stabilizes the assembly of two monomers into an antiparallel obligate domain-swapped dimer as illustrated in Fig. 1b.
In all Prx6 structures determined so far, the monomers constituting the dimers are almost identical, with RMSD values below 0.5 Å and frequently below 0.3 Å. This is not the case for AnPrx6. Superimposing monomer A onto B and C onto D results in RMSD values of 0.87 Å and 0.94 Å, respectively, which is due to differences distributed throughout the two monomers. However, superimposing monomers A onto C and monomer B onto D results in almost perfect matches with RMSD values of about 0.2 Å in both cases.
In order to place the AnPrx6 protein into a greater structural context, its structure was simultaneously superimposed with thirteen representative Prx structures and a structure anchored multiple sequence alignment (saMSA) was obtained (Supplementary Fig. S1). The saMSA shows that residues Pro38, Thr42 and Cys45 of AnPrx6 map to the invariant Prx active site pattern P-X(3)-[TS]-X(2)-C, where Ser is observed in rare cases, X marks any amino acid and C is the peroxidatic Cys (CP). In addition, Arg122 matches the strictly conserved active site arginine found in all known peroxiredoxins.
Although the crystal structures of the fourteen proteins are mutually similar, with RMSD values ranging between 1.1 Å and 1.7 Å, they clearly differ in several key regions. One such region involves helix α3 and the loop connecting α3 with strand β5 that varies both in length and conformation among Prx6 family members and is distinctly different compared to the corresponding region of (α2)5 decameric ring forming Prxs, where helix α3 is approximately two turns shorter and the α3-β5 loop can form a 310 helical turn. (Supplementary Fig. S2).
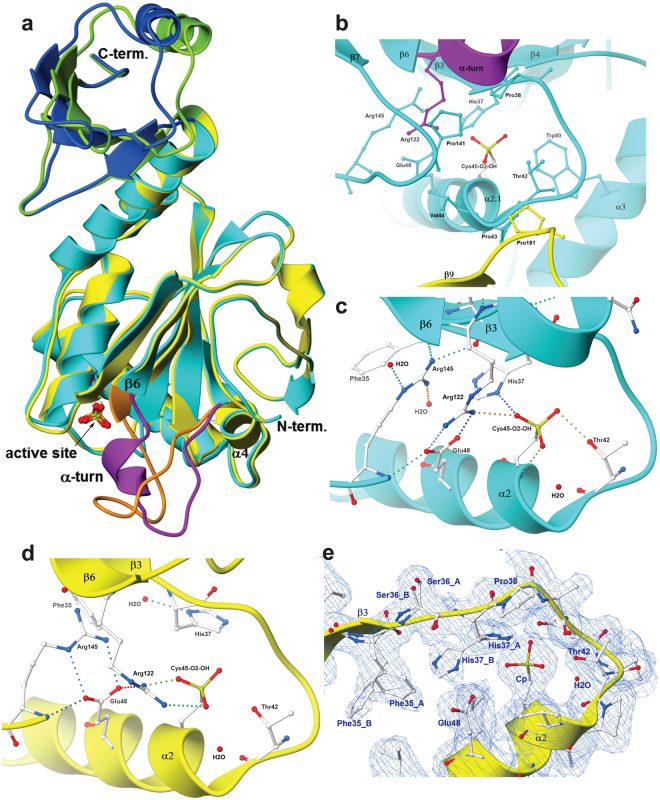
The second region of striking structural variability affects the amino acids between Asp109 and the conserved Arg122 that connect helix α4 with strand β6. This region is highlighted in purple and orange in monomer A and B, respectively in Fig. 1a,b and Supplementary Fig. S2. In some Prx proteins this region adopts an extended, irregular loop, e.g. in monomers B and D of AnPrx6; in hORF6 (PDB entry 1PRX42) and in Aeropyrum pernix Prx6 (PDB entry 2CV445), whereas it folds into a short two stranded β-sheet in others, e.g. Arenicola marina Prx6 (PDB entry 2V2G43) and Trypanosoma cruzi, Prx1 (PDB entry 4LLR47). In contrast, residues Ala117 to Thr120 of AnPrx6 monomers A and C fold into an α-helical turn (marked purple in Fig. 1a,b), while the same region of monomers B and D forms an extended loop (orange in Figs 1b, 2a and Supplementary Fig. S2). The α-helical turn is a novel feature which was not previously observed in any Prx structure. Additional minor structural variations propagate throughout the monomers A and B and become visible as colour differences between the light blue and yellow ribbons (Fig. 2a). The differences are emphasized in loop regions and are clearly visible in the displacement of the C-terminal domains.
Figure 2.
The asymmetric monomers. (a) Superimposition of monomer A (light and dark blue) and B (yellow and green) highlights their overall conformational differences especially the loop connecting α4 and β6 (Asp109 to the conserved Arg122). Residues Ala117 to Thr120 of monomer A fold into an α-helical turn, while they form an irregular loop in monomer B, thus affecting the geometry of the active site. (b) View of the residues lining the active site environment in monomer A. The α-helical turn is oriented such that the carbonyl oxygen atoms and the helical axis point towards CP. Part of strand β9 and Pro181 from monomer B are marked in yellow. (c) Hydrogen bonding network of monomer A and monomer C. The Cys45-O2-OH forms a H-bond (dotted line) through its Oδ2 atom with the oxygen (Oγ1) of Thr42, and through its Oδ1 atom an H-bond with the Nδ1 atom of His37. While Arg122 mainly engages in a salt-bridge with Glu48, it also forms a H-bond through its Nη1 to the Oδ1 of Cys-O2-OH. Downstream in the sequence but close in space lies Arg145 whose main chain nitrogen atom H-bonds to the Oε1 atom of Glu48 and its Nη2 atom forms a bifurcated H-bond with the carbonyl oxygen atoms of Ser36 and Arg122. In addition, the Nε and Nη1 of Arg145 bind one water molecule each. A third water molecule is held in place by the main-chain nitrogen atom of Val44 and does not directly H-bond with Cys45-O2-OH. A corresponding water molecule is found in many Prx structures. (d) The active site of monomer D. Here, the side chain of His37 flips away from CP and forms an H-bond with a water molecule. Arg122 forms a salt bridge with the oxygen atoms Oδ1 and Oδ3 of Cys45-O2-OH, and maintains a H-bond to Glu48. The side chain of Arg145 rotates into a position where its Nε and Nη2 atoms can form a new salt bridge with the Oε1 and Oε2 atoms of Glu48. The rearrangement leads to a shift of the CP-loop, which disrupts the H-bond between CP and Thr42. (e) The electron density (blue mesh) covering the active site residues of monomer B clearly suggests alternate conformations for Phe35, Ser36 and His37, which reflects the dynamics of these residues.
Conformational asymmetry of the active site environment
The helix α2 contains the peroxidatic cysteine (CP) and is part of the β3-α2-β4 motif, common to all Prx proteins. The strictly conserved residues Pro38 and Thr42 of the catalytic PxxxTxxCP motif are part of the loop connecting β3 with α2. The so-called region I and the CP-loop26 overlap with this loop and continue into the first two turns of helix α2. The CP-loop adopts a “fully folded” (FF) conformation4,25 in all four AnPrx6 monomers, thereby positioning CP at the bottom of the active site pocket bounded by residues Phe35, His37, Pro38, Thr42, Pro43, Val44, Glu48, Trp80, Arg122, Pro141 and Arg145 of the N-terminal domain and Pro181 from the C-terminal domain of the adjacent monomer (Fig. 2b). The histidine residue directly preceding the β3-α2 loop (His37 in AnPrx6) is conserved in all Prx6 family members, while 2-Cys Prx proteins contain a Tyr or, in rare cases, a Trp at the corresponding position.
Despite apparently identical CP oxidation states in all four monomers, the positions and hydrogen bonding patterns of key active site residues, including Phe35, His37, Cys45-O2-OH (CP), Arg122 and Arg145, differ in the two monomers within the dimer. In monomer A and similarly in monomer C, the oxygen atom Oδ1 of Cys45-O2-OH forms hydrogen bonds with the Nδ1 atom of His37 and the Nη1 atom of the strictly conserved Arg122 (Fig. 2c). Moreover, the guanidinium group of Arg122 forms a salt-bridge with the side chain carboxylic acid of Glu48. The guanidinium group of the conserved Arg145 participates in a hydrogen bond network that includes the carboxyl oxygen atoms of Ser36 and Arg122 as well as two water molecules that are bound to its Nη1 and Nε atoms. In monomer A, Arg145 interacts through the main chain N atom with Glu48, which is positioned one turn away from CP on helix α2.1. An active site water molecule, which is coordinated by the main chain N atom of Val44, is present in all four AnPrx6 monomers and aligns almost linearly with Sγ of CP and Nδ1 of His37. It was proposed for the human hORF6 protein (PDB code 1PRX42) that an equivalent water molecule participates in the Prx6 reaction mechanism42. The arrangement of the active site residues, the water molecules and hydrogen bonds observed in monomers A and C of AnPrx6 are nearly identical to the side chain orientations and H-bond patterns found in the human hORF6 protein (PDB code 1PRX42) and in the active site arrangement of the hypervalent sulphur intermediate of Aeropyrum pernix K1 (PDB code 2ZCT48) where only Phe40 deviates from the orientation of Phe35 in monomer A of AnPrx6.
Close inspection of the active site residues of monomer D (Fig. 2d), shows variations compared to monomer B, as discussed in the next paragraph, and clear differences in orientation compared to monomer A. Conformational changes affect the peptide backbone, the side chain positions and, consequently, the H-bond pattern. The most striking difference is the placement of His37, whose side chain dihedral angle changed by about −130° (clockwise rotation), compared to its orientation in monomer A. His37 adopts a position away from CP, where its Nδ1 atom forms a hydrogen bond with a water molecule. The new His37 orientation becomes possible due to a movement of strand β4 and the following loop, which allows a counter clock wise rotation of the Oγ of Ser70 thus providing a space for His37. Due to the formation of an α-helical turn in the loop connecting α4 to β6 of monomer A, Arg122 rotates away from Glu48 and engages in a salt-bridge interaction with the Oδ1 and Oδ3 of the triple oxidized Cys45. Being released from the interaction with Arg122, Glu48 forms a new salt-bridge with the Nε and Nη2 atoms of Arg145. It is noteworthy that Phe35 and Ser36 follow the move of His37. Especially the side chain dihedral angle of Phe35 changes by about −65° (clockwise rotation) and the Oγ of Ser36 rotates about + 125° (counter clockwise) compared to monomer A, respectively. The three residues form the FSH motif that is strictly conserved in all known Prx6 proteins. It is striking that the active site arrangement of amino acids in monomer D is almost identical to the active site of the reduced (SH) and the sulfinic acid (SO2H) form of human Prx6 (PDB codes 5B6M49 and 5B6N49) with an overall RMSD of 0.42 Å to both structures. Likewise, the active site of monomer D is reminiscent of the active sites of Plasmodium yoelii Prx6 (PDB code 1XCC44), Arenicola marina Prx6 (PDB code 2V2G43) as well as the fully folded active sites of the decameric 2-Cys peroxiredoxins from Human Erythrocytes (PDB code 1QMV50) and Schistosoma mansoni (PDB code 3ZTL30). In addition, the orientation of active site residues in monomer D match even the corresponding amino acid arrangement found in 2-Cys Prxs with locally unfolded CP-loops, except for the position of CP itself (1E2Y51, 3TUE52, 4FH853).
Unexpectedly, the electron density surrounding the active site of monomer B (Fig. 2e) suggests two clearly discernible alternative conformations for Phe35, and His37, while the two conformations of Ser36 are both comparable to the one observed in monomer D. The orientations of Phe35 and His37 assigned to the stronger electron density were refined to an occupancy of about 0.6, and the occupancy for the second orientation, corresponding to the weaker electron density, amounted to 0.4. The positions of Phe35 and His37 with the higher occupancy match those observed in monomer D, where His37 is pointing away from CP. The positions with the lower occupancy are similar but not identical to those of Phe35 and His37 observed in monomer A and C. The crystallographic data alone do not provide a conclusive answer as to whether the alternate positions were due to dynamic motions or due to distinct static positions. In case of dynamic motion, it is conceivable that the movements of the FSH motif residues, Phe35, Ser36 and His37, are correlated. It is also possible that the plasticity of the peptide main chain, including the formation of the helical turn are necessary for the movements of Phe35, Ser36, His37, Arg122 and Arg145 and hence for the catalytic activity of AnPrx6.
The fact that the His37 directly precedes the CP-loop and is strictly conserved within the Prx6 subfamily, suggests an important role in the catalytic cycle of Prx6 proteins. It has been suggested that the conserved histidine together with a strictly conserved arginine residue (His37 and Arg122 in AnPrx6) facilitate the deprotonation of CP by lowering its pKa and stabilizing the thiolate form, which is essential for catalysis42,54,55. Despite the accumulated knowledge, the exact catalytic cycle for AnPrx6 is not yet known and to which extend Phe35, Ser36 and His37 are involved will be the focus of further studies. To date, no single electron donor has been conclusively identified for the reduction of the oxidized CP in members of the 1-Cys Prx6 subfamily. Various suggestions were put forward on how the oxidized CP is returned to the reduced state. The protected location of CP within the environment of the active site pocket seemingly prevents its reduction by a protein agent such as thioredoxin. Instead, various small reducing agents, such as glutathione (GSH)24,56 and ascorbate (vitamin C)57, have been suggested.
Molecular dynamics simulations
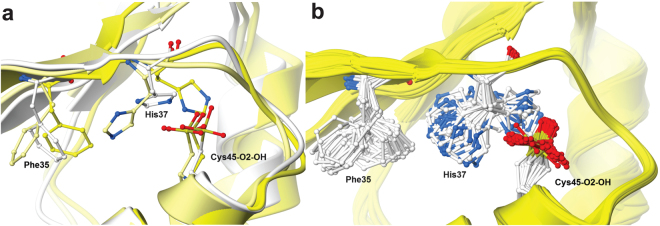
To test the hypothesis of dynamic, correlated movement of Phe35 and His37 and to examine whether the asymmetry of the AnPrx6 dimer observed in the crystal is a feature maintained in solution, molecular dynamics (MD) simulations were performed. The isolated AnPrx6 dimer (monomers A and B) was surrounded by a periodic boundary box of water molecules and simulated for 150 ns. To confirm the simulation results, the simulations were performed in triplicate, starting from different velocity distributions. The MD trajectory describes a dynamic dimer and indicates that the structural features of each monomer (i.e., the α-helical turn between α4 and β6 in monomer A and the extended loop in the same region of monomer B) can fluctuate around the positions observed in the X-ray structure throughout the simulation (Supplementary Fig. S3). The results imply that the asymmetry is a feature of the dimer which is maintained in solution and is not due to crystal packing interactions. The MD simulations also showed the correlated interconversion of the side chain orientations of Phe35 and His37 in monomer B (Fig. 3a, Supplementary Fig. S3 and the Supplementary simulations Sim1 and Sim3, corresponding to Simulation [1] and Simulation [3] in Supplementary Fig. S3e), whereas the side chains of the two residues in monomer A remained close to their positions observed in the X-ray structure (Simulation [2] in Supplementary Fig. S3e). The variations of the His37-Cys45 and Arg122-Cys45 distances are different for monomer A and B (Supplementary Fig. S4). Monomer A shows larger distance fluctuations and longer Arg122-Cys45 H-bond distances compared to those of monomer B, whereas in monomer B the His37-Cys45 distance fluctuations are more pronounced and the H-bond distances are longer compared to monomer A. The opposite trend between the two H-bond interactions might occur because the Arg122-Cys45 interaction is strong (or more stable) in monomer B, and allows His37 to be released from Cys45 and move more freely. In contrast, in monomer A, as Arg122 interacts weakly with Cys45, Cys45 interacts more strongly with His37, thus limiting its motion. This difference suggests a correlation between the two CP interactions and the increased mobility of His37 (via concerted motion with Phe35).
Figure 3.
Visualisation of the dynamic movement of Phe35 and His37. (a) The active site of monomer B at three different time points along the MD trajectory (t = 1 ns, white, 20 ns, light yellow and 75 ns, yellow) visualizes the coordinated motion of Phe35 and His37. The His37 side chain dihedral angle of about −180° corresponds to the conformation found in monomer A and the value of −57° to that found in monomer D. The active site CP (Cys45-O2-OH) does not move significantly. (b) Active site of monomer B after Ensemble Refinement. The final Ensemble contained 45 models of the AnPrx6 structure. While Phe35 and His37 display multiple alternative orientations, the orientation of the oxidized CP does not vary significantly. Although the MD simulations and the Ensemble refinement are independent methods, they both suggest large conformational oscillations for His37 and Phe35 in monomer B.
Taken together the MD simulations strengthen the hypothesis that the α-helical turn between α4 and β6 in monomer A (and C) leads to a more restricted and hence more ordered active site, as depicted in Fig. 2c, where His37 is hydrogen bonded to the oxidized CP. The simulations further indicate that the unstructured loop connecting α4 and β6 in monomer B facilitates the correlated movement of Phe35 and His37 and, while His37 is pointing away from CP, Arg122 forms a salt bridge with the active site cysteine-sulfonic acid.
Synergistic effect upon dimer formation
Two AnPrx6 monomers associate into an asymmetric, antiparallel obligate homo-dimer. To highlight the conformational asymmetry between the two monomers, we refer to the dimer as an (αα*) type homo-dimer, in order to distinguish it from commonly observed symmetric (α2) type homo-dimers. The AnPrx6 monomers interact through hydrogen bonds across an extended interface. The central contacts are formed through four main-chain hydrogen bonds across the B-type dimer interface, involving Thr136 and Thr138 of strands β7 from each monomer. The dimer is further stabilized through a hydrogen bond network created by main- and side chain hydrogen bonds of residues from the loop between β7 and α5 (Tyr139, Ser142, Thr143, Gly144, Arg145) as well as residues from α5 (Phe147, Glu149, R152). Additional interfacial H-bond contacts are provided by residues from the “domain swapped” C-terminal domains (amino acids Tyr163 to Lys212). Specifically, residues from the loop connecting β8 and β9 (Thr167, Ala169, Asp170) of one monomer form hydrogen bonds with residues of the loop linking β7 and α5 (Tyr139, Ser142, Gly144, Asn146) from the second monomer and with residues T47 on helix α2.2. Even Lys201 and Tyr203 of the C-terminal loop connecting β10 and β11 interact with Asp83 and Glu86, respectively, from α3 of the second monomer. Finally, Arg4 of the N-terminal tail of one monomer interacts with the backbone oxygen of Asp109 from the other monomer. Taken together, these hydrogen bonds are part of the extended dimerization interface that buries about 2185 Å2 of the surface area from monomer A and 2199 Å2 from monomer B, as determined by PDBePISA58, which amounts for approximately 21% of the total surface area of each monomer.
AnPrx6 does not form higher molecular weight oligomers
Members of the Prx1/AhpC and some members of the Prx6 family are able to assemble into (α2)5 decameric rings in response to increasing oxidative stresses25,26. To experimentally verify the oligomeric state of AnPrx6 in solution, analytical size exclusion chromatography (SEC) analysis was carried out at three different protein concentrations (0.1 mg ml−1, 1 mg ml−1 and 10 mg ml−1) in the absence or presence of 20 mM H2O2. Each chromatogram contained a single retention peak. In the absence of H2O2 the peak positions were observed at retention volumes of 15.38 ml, 15.34 ml and 15.40 ml. In the presence of 20 mM H2O2 the retention volumes were 15.53 ml, 15.37 ml and 15.18 ml (Supplementary Fig. S5). Using a standard curve (BioRad Gel Filtration Standard) to convert the retention volumes to molecular weights, one finds that the average molecular weight in the absence of H2O2 is 44,2 kDa and 44,7 kDa in the presence of 20 ml H2O2. These values are in good agreement with the calculated value for dimeric AnPrx6 (48.4 kDa).
No peaks were detected at lower retention volumes. In particular there was no peak at a retention volume of 12 ml, which would correspond to the molecular weight of a (α2)5 decamer (242 kDa). The results from the SEC measurements confirm that AnPrx6 is dimeric in the native, reduced state as well as under oxidative conditions and, unlike the Prx1/AhpC proteins from yeast27 and Helicobacter pylori28, does not undergo oxidative stress-dependent oligomerization.
In the crystallographic asymmetric unit (a.s.u.) of AnPrx6 crystals in space group P212121, two AnPrx6 dimers pack against each other. They associate through helix α6 of the C-terminal domain, through residues from the N-terminal loop and the loop between helix α4 and strand β6, thereby burying an interface of 690 Å2 (PDBePISA58) (Supplementary Fig. S6). Considering the small size of the contact area and that the two dimers are related by a non-crystallographic twofold symmetry axis, one can conclude that the dimer-dimer interaction is not biologically relevant. Further contacts between neighbouring asymmetric units are propagated through strands β10 of the C-terminal extensions (Supplementary Fig. S6) and neither of the packing contacts is mediated through the A-type interface and will not lead to decameric rings.
Small angle X-ray scattering (SAXS) measurements were carried out at two protein concentrations (1.6 mg ml−1 and 4.8 mg ml−1) to verify whether AnPrx6 exists exclusively in its dimeric state in solution. The resulting SAXS scattering curves coincided and show variation only in the level of noise (Supplementary Fig. S7). From Guinier plot analysis the radius of gyration was determined to be Rg = 2.98 ± 0.03 nm for both concentrations. This value corresponds to a hydrodynamic diameter 2r ≈ 7.7 nm, which is in good agreement with the X-ray structure, where the largest distance within the AnPrx6 dimer is approximately 8,1 nm. The molecular weight of the scattering particles in solution was estimated to be 44 kDa, using Hen Egg White Lysozyme as the molecular weight standard. Similarly, using the Porod volume, the molecular weight was estimated to be 48 kDa. Considering that the calculated molecular weight of the AnPrx6 monomer is 24.2 kDa, one can conclude from the SAXS data that AnPrx6 is a dimer in solution.
AnPrx6 acts both as a peroxidase and as molecular chaperone
Peroxidase activity
Enzymatic peroxidase activity of AnPrx6 was determined according to the FOX assay as described in Jiang et al.59 with hydrogen peroxide (H2O2) as the substrate. The enzyme was incubated in 600 μM H2O2 and the reaction was terminated after one minute by addition of FOX reagent. Based on the observed absorption at 560 nm, the H2O2 consumption can be determined from a standard curve and the peroxidase activity of AnPrx6 was calculated to be 230 ± 12 nmol H2O2 min−1 (mg of AnPrx6)−1. These results suggest that the overexpressed and purified AnPrx6 protein has a moderate peroxidase activity as compared to PrxQ that showed 30 times higher activity measured under the same conditions. In order to assess if the low peroxidase activity could be due to over-oxidation of the enzyme, the activity was measured at increasing concentrations of H2O2, starting from 0 to 600 μM H2O2. The peroxidase activity increased almost linearly up to 100 μM H2O2, where it levelled out at 230 ± 12 nmol H2O2 min−1 (mg of AnPrx6)−1 (Supplementary Fig. S8). This suggests that the peroxidase activity is saturated at approximately 100 µM H2O2 and that the enzyme is able to function also at higher H2O2 concentrations under incubation times less than 1 minute. Furthermore, a linear dependency of the rate of H2O2 consumption was observed when increasing the AnPrx6 concentrations between 0 μM to 60 μM and using a fixed, saturating amount H2O2 (Supplementary Fig. S9).
Chaperone activity
Chaperone activity for 1-Cys Prx6 proteins has not yet been reported. In order to enquire potential chaperone activity of AnPrx6, a chaperone assay that uses Hen Egg White Lysozyme (HEWL) as substrate was employed60. Lysozyme is a monomeric protein of 14.6 kDa with four disulphide bonds stabilizing its tertiary structure. Reduction of a lysozyme solution with dithiothreitol (DTT) and heating at 43 °C causes it to partially unfold, which leads to aggregation and precipitation and results in increased light scattering. The thermal aggregation of HEWL at 43 °C in the presence of DTT and AnPrx6 or BSA was monitored by measuring the light scattering of the reaction mixture at a wavelength of 360 nm. In control experiments without AnPrx6, the scattering intensity increased exponentially over time, indicating that the rate of aggregation and precipitation increased as the assay proceeded. The addition of AnPrx6 at a molar ratio of 1:32 (AnPrx6 to Lysozyme) inhibited the thermal aggregation of HEWL (Fig. 4). Replacing AnPrx6 with BSA had a slight inhibitory effect on the aggregation of HEWL (Fig. 4). This was expected since BSA is known to possess moderate chaperone activity61,62. Additional experiments, where the molar ratio of AnPrx6 to lysozyme was adjusted to 1:22 and 1:16 revealed a chaperone activity independent of the AnPrx6 concentration within the tested range of molar ratios, whereas BSA showed little or no chaperone activity (Supplementary Fig. S10).
Figure 4.
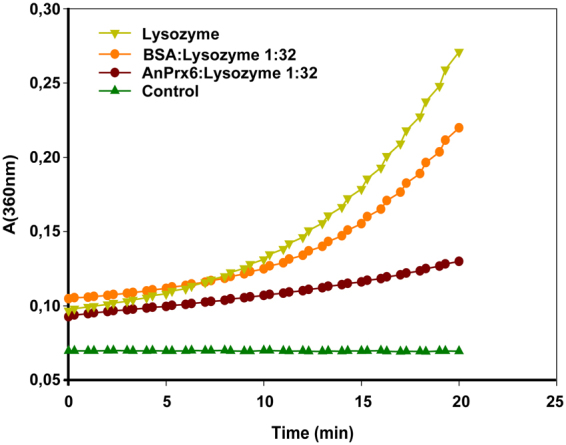
Chaperone activity assay. Thermal aggregation of HEWL at 43 °C was monitored at a wavelength of 360 nm by measuring the degree of light scattering at different time points. The reference level of scattering was determined with a reaction mixture containing only buffer, HEWL and DTT (light green triangles). Aggregation is strongly reduced in the presence of dilute AnPrx6 (brown dots), whereas the same dilution of BSA (positive control) has almost no effect on aggregation (orange dots). Buffer and DTT (dark green triangles) were used as negative control (blank).
Discussion
The crystal structure of AnPrx6 provides a first glimpse of a cyanobacterial 1-Cys Prx protein and is an unique example of an asymmetric Prx6 homo-dimer. The structural asymmetry within the so called αα* type homo-dimer was confirmed through molecular dynamics simulations, which showed that the motions of Phe35 and His37, of the FSH motif, are correlated. The asymmetry has an impact on the positions and interactions of key active site residues, in particular the placement of His37 and Arg122, which will affect the pKa value of the active site cysteine (CP) and hence its reactivity. Remarkably, the two different active site conformations observed within the AnPrx6 dimer, each resemble an active site conformation that had been previously reported for symmetric (α2) Prx dimers from different species and with different oxidation states. The asymmetry together with backbone fluctuations around the active site could be a common feature of 1-Cys Prx6 proteins and crucial for their activity.
We propose that, in the particular case of AnPrx6 and perhaps the Prx6 subclass, the asymmetric structural fluctuations propagate through the dimer and accompany or possibly induce the movement of the conserved FSH motif and particularly the conserved His37 residue that precedes the CP-loop. These fluctuations, together with the movements of Arg122, are likely to contribute to a shift of the pKa value of the peroxidatic CP, such that one active site is primed for catalysis while the other can be reduced. Yuan et al.63 and Salsbury et al.64 predicted similar asymmetric Prx dimers from MD simulations. However, the AnPrx6 structure is so far the only example of an experimentally observed asymmetry within a Prx6 dimer.
Furthermore, our functional analyses provide evidence that the AnPrx6 homo-dimer harbours dual functions. It acts as a peroxidase and, in its dimeric state as a molecular chaperone. The absence of high molecular weight oligomers was confirmed by SAXS measurements and size exclusion chromatography, the latter carried out under native and H2O2 stress conditions.
We speculate that AnPrx6 is a unique and ancient member of the Prx family that has been retained in cyanobacteria and might represent an early state of Prxs evolution, capable of both peroxidase and chaperone functions, without the need to change its oligomeric state.
Organisms that perform oxygenic photosynthesis face unique challenges in terms of ROS management and protein folding because photosystem II is both a powerful generator of ROS and requires assembly and frequent regeneration due to the very rapid turnover of the D1 subunit, even under normal physiological conditions65. Although the immediate effect of AnPrx6 and other peroxiredoxins on photosystem II is not yet understood66 it is possible that the dual functions of AnPrx6 and its unique oligomerization behaviour evolved to cope with these challenges, especially since cyanobacteria frequently inhabit extreme environments and emerged at a time when the earth’s atmosphere contained only a small fraction of oxygen. Indeed, the cyanobacterial photosynthesis was the key driver in changing the planet’s atmosphere from reducing to oxidizing. It is intriguing to speculate that the first adaptations to life under highly oxidizing conditions may have occurred inside the cells of oxygen-producing cyanobacteria and that this may be the origin of the unique characteristics of AnPrx6, a cyanobacterial 1-Cys Prx6.
Materials and Methods
Cloning, expression, purification and crystallization of AnPrx6
The native AnPrx6 protein harbours 212 amino acids (accession code: WP_010998539.1). The ahpC gene (accession code: alr4404) that codes for the AnPrx6 protein from Anabaena sp. PCC 7120, also known as Nostoc sp. PCC7120, was extracted from genomic DNA by PCR. The purified PCR fragment was digested with EcoRI and NotI and ligated into a pGEX-5X-2 GST expression vector (GE Healthcare) digested with the same enzymes37. Escherichia coli (E. coli) strains DH5α and BL21(DE3) were used as hosts for vector amplification and protein expression, respectively. The recombinant AnPrx6 protein was purified and crystallized as described by Mishra et al. (2011)38. Further details on expression, purification, crystallization of AnPrx6 are provided in the Supporting Information (SI) Material and Methods.
X-ray data collection and processing
Crystals of AnPrx6 were vitrified in a nitrogen gas stream maintained at 100 K (Oxford CryoSystems Ltd, UK) and tested for diffraction at the in-house X-ray source (X8Proteum system, Bruker AXS). The best diffracting crystals were preserved in liquid nitrogen for later use. X-ray diffraction data sets were collected from cryo-cooled AnPrx6 crystals at a wavelength λ = 0.9793 Å onto an ADSC Q315R detector at beam-line ID23-167 of the European Synchrotron Radiation Facility, Grenoble, France. The diffracted intensities were indexed and integrated with XDS68, prepared for scaling with pointless69. Aimless70 was used to scale and merge the intensities which were converted to structure factors with cTruncate71. About 5% of the data was marked for Rfree calculation. (For crystallographic details and statistics see Supplementary Table 1).
Structure determination and refinement
Phases were obtained by molecular replacement (MR) using the on-line version of Auto-Rickshaw72 (http://www.embl-hamburg.de/Auto-Rickshaw/) using monomer A of human Prx6 (hORF6, PDB code 1PRX42) as the search model. The CCP4 program suite71 was used for data analysis and manipulation. Iterative rounds of manual model building with Coot73 (version 0.8.2-pre rev. 5817) and refinement with phenix.refine74,75 of the Phenix program package (version 1.10.1–2155) were carried out until the R-free factor and R-factor converged. The data processing and refinement statistics are listed in the Supplementary Table 1. The atomic coordinates and structure factors for AnPrx6 have been deposited with the Worldwide Protein Data Bank (wwPDB) under code 5M6T. Additional ensemble refinement was carried out using the Ensemble refinement [alpha] program76 within the Phenix program package (version 1.10.1-2155).
Sequence and structure analysis
Sequence searches against the non-redundant protein sequence database (nr) at NCBI77 and the Protein Data Bank (PDB)41 were performed with BLASTp40. Multiple sequence alignments of Prx proteins and subsequent phylogenetic tree analysis were carried with Clustal Omega39 at the European Bioinformatics Institute, EBI, (http://www.ebi.ac.uk/Tools/msa/clustalo/). For Prx subfamily classification the PREX database9 was consulted (http://csb.wfu.edu/prex.test/).
Structural analysis was carried out with Coot73,78 and ICM-Pro (MolSoft LLC)79,80. ICM-Pro was further used for multiple global structure superimpositions and to create the structure anchored multiple sequence alignment (saMSA), for which only structures with at least 30% sequence identity to AnPrx6 were considered. The PDB codes and further details about the superimposed structures are provided in the SI Material and Methods section.
All molecular graphic rendering was carried out with ICM-Pro (MolSoft LLC) or CCP4MG78.
Molecular dynamics simulations
The simulation system was prepared based on the determined X-ray structure of AnPrx6 (PDB ID: 5M6T), in which the first dimer (i.e., chains A and B) was used. As suggested by the electron density map the peroxidatic Cys (Cys45) was modelled as the triply oxidized cysteine-sulfonic acid in both monomers. Protonation states of all ionisable residues were assigned based on their pKa values or hydrogen bond interactions deduced from the X-ray structure. The positions of hydrogen atoms were determined using the HBUILD facility of the CHARMM program81 (version c38a2). All crystal waters were included and the resulting system was further solvated with a 99.0 Å rhombic dodecahedron (RHDO) box of TIP3P waters82 which gives at least 9 Å of solvation between the edge of the protein to the simulation box boundary along the longest protein axis. Then, 53 Na+ and 41 Cl− ions were added at random positions to neutralize and to make the total ionic concentration of the final system to 0.15 M, resulting in a total of 67,003 atoms.
The resulting system was first energy minimized to avoid starting the molecular dynamics (MD) simulations with bad contacts between atoms in the system. A total of 5,000 steps of energy minimizations were performed with varying constraints and restraints: (1) 500 steps of minimization of all hydrogen atoms, (2) 1000 minimization steps with all protein heavy atoms fixed at the position of the X-ray structure, (3) 2000 steps of minimization with the harmonic position restraints applied to all protein heavy atoms, and (4) finally, 1500 steps of minimization without any restraints. The system was then heated slowly from 0 K to 300 K over 24 ps, followed by 150 ns production MD simulations. In all calculations, the all-atom CHARMM22 force fields83 with the CMAP correction84 applied to the protein backbone dihedrals were used to represent the protein and ions. The water molecules were represented by the TIP3P water model82. The force fields parameters for the oxidized Cys45 were developed based on the CHARMM General force fields85. All simulations were carried out using the RHDO periodic boundary conditions. Electrostatic interactions were evaluated using the smooth particle mesh Ewald (PME) summation method86 with 100 × 100 × 100 fast Fourier transform grid and real space interactions evaluated using the 11.0 Å cut-off. The same cut-off distance was used in the evaluation of van der Waals interactions, in which the interactions were switched off smoothly to zero between 9.0 Å and 11.0 Å. The MD simulations were performed with 2 fs integration time step, during which the temperature was maintained at 300 K using the Langevin thermostat (i.e., the NVT condition) with the 1 ps−1 friction coefficient, and the SHAKE-like algorithm87 applied to constrain bonds involving hydrogens. All MD simulations were performed using the NAMD program88 and the system preparation, energy minimizations, and trajectory analysis using the CHARMM program, respectively.
Chaperone activity assay
The assay is based on the thermal aggregation of lysozyme and follows the protocol established by Ferreira et al.60. We used 360 μl of an aqueous solution containing 50 mM NaPi buffer pH 7.1 and 30 mM DTT and placed it for 5 minutes in a water bath maintained at 43 °C. Thereafter, a mixture of AnPrx6 and Hen Egg White Lysozyme were added to the solution at a final AnPrx6:Lysozyme molar ratio adjusted to 1:32 and a final reaction volume of 400 μl. After the addition of AnPrx6 and Lysozyme, the reaction temperature was maintained at 43 °C and the level of light scattered off the reaction mixture was measured every 30 seconds over a period of 20 minutes at a wavelength of 360 nm using a micro-plate spectrophotometer (SPECTRA max, 190 USA). Negative control experiments, determining the baseline for scattering, were performed using 400 μl of the buffer solution including DTT but excluding AnPrx6 or Lysozyme. Positive control experiments were performed by replacing AnPrx6 with BSA.
Data Availability
The atomic coordinates and structure factors for AnPrx6 from Anabaena sp. PCC 7120 have been deposited with the Worldwide Protein Data Bank (wwPDB) under code 5M6T.
Electronic supplementary material
Acknowledgements
This work was supported by a Post-doctoral grant from the Lawski foundation (to Y.M.); a grant from the Swedish Research Council (Vetenskapsrådet, to S.J. and K.N.); a grant from the Kempestiftelserna (to S.J.); a grant from the Carl Tryggers foundation (CTS 08: 343, to U.H.S); a 50% faculty funded PhD position (to U.H.S.); a start-up grant from the University of Texas in Arlington (to K.N.); and L. C. Rai is thankful to Dept. of Science and Technology (DST) for financial support in the form of a project and JC Bose national fellowship. K.N. gratefully acknowledges the Swedish National Infrastructure for Computing (SNIC) at High Performance Computing Center at North (HPC2N) and the National Energy Research Scientific Computing Center (NERSC) for providing computational resources. The Umeå Protein Expertise Platform, PEP, is acknowledged for protein production and purification. The authors are grateful for the MX beamtime granted at the ESRF in Grenoble, France and at MAX IV Laboratory in Lund, Sweden, where the SAXS experiments were conducted with the help of beamline scientist Sylvio Haas. The authors thank Prof. B. Buchanan, University of California Berkeley, USA, for critical reading and comments on the manuscript.
Author Contributions
Y.M. and U.H.S. conceived the project. Y.M., M.H., R.L., P.S., and U.H.S. designed and performed the experiments. S.J., W.P.S., N.C., L.C.R and U.H.S. contributed reagents. K.N. performed molecular dynamics simulations. Y.M., M.H., K.N., R.L., C.A.G.S. and U.H.S analysed the data. Y.M., M.H., K.N., C.A.G.S., S.J., W.P.S. and U.H.S. wrote the article.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
A correction to this article is available online at https://doi.org/10.1038/s41598-018-26715-8.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-17044-3.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/30/2018
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has been fixed in the paper.
References
- 1.Wood ZA, Schroder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 2003;28:32–40. doi: 10.1016/S0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 2.Knoops B, Loumaye E, Van Der Eecken V. Evolution of the peroxiredoxins. Subcell. Biochem. 2007;44:27–40. doi: 10.1007/978-1-4020-6051-9_2. [DOI] [PubMed] [Google Scholar]
- 3.Hofmann B, Hecht H-J, Flohe L. Peroxiredoxins. Biol. Chem. 2002;383:347–364. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- 4.Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 5.Neumann CA, Cao J, Manevich Y. Peroxiredoxin 1 and its role in cell signaling. Cell Cycle. 2009;8:4072–4078. doi: 10.4161/cc.8.24.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dietz KJ. Peroxiredoxins in plants and cyanobacteria. Antioxid. Redox Signal. 2011;15:1129–1159. doi: 10.1089/ars.2010.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson KJ, et al. Analysis of the peroxiredoxin family: using active-site structure and sequence information for global classification and residue analysis. Proteins. 2011;79:947–964. doi: 10.1002/prot.22936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poole, L. B. & Nelson, K. J. Distribution and Features of the Six Classes of Peroxiredoxins. Mol. Cells (2016). [DOI] [PMC free article] [PubMed]
- 9.Soito L, et al. PREX: PeroxiRedoxin classification indEX, a database of subfamily assignments across the diverse peroxiredoxin family. Nucleic Acids Res. 2011;39:D332–337. doi: 10.1093/nar/gkq1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim K, Kim IH, Lee KY, Rhee SG, Stadtman ER. The isolation and purification of a specific “protector” protein which inhibits enzyme inactivation by a thiol/Fe(III)/O2 mixed-function oxidation system. J. Biol. Chem. 1988;263:4704–4711. [PubMed] [Google Scholar]
- 11.Chae HZ, et al. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc. Natl. Acad. Sci. USA. 1994;91:7017–7021. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seaver LC, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 2001;183:7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosgrove K, et al. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J. Bacteriol. 2007;189:1025–1035. doi: 10.1128/JB.01524-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lien YC, Feinstein SI, Dodia C, Fisher AB. The roles of peroxidase and phospholipase A2 activities of peroxiredoxin 6 in protecting pulmonary microvascular endothelial cells against peroxidative stress. Antioxid. Redox Signal. 2012;16:440–451. doi: 10.1089/ars.2011.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shichita T, et al. Peroxiredoxin family proteins are key initiators of post-ischemic inflammation in the brain. Nat. Med. 2012;18:911–917. doi: 10.1038/nm.2749. [DOI] [PubMed] [Google Scholar]
- 16.Nystrom T, Yang J, Molin M. Peroxiredoxins, gerontogenes linking aging to genome instability and cancer. Genes Dev. 2012;26:2001–2008. doi: 10.1101/gad.200006.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishii T, Warabi E, Yanagawa T. Novel roles of peroxiredoxins in inflammation, cancer and innate immunity. J. Clin. Biochem. Nutr. 2012;50:91–105. doi: 10.3164/jcbn.11-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hampton, M. B. & O’Connor, K. M. Peroxiredoxins and the Regulation of Cell Death. Mol. Cells (2016). [DOI] [PMC free article] [PubMed]
- 19.Heym B, et al. Effects of overexpression of the alkyl hydroperoxide reductase AhpC on the virulence and isoniazid resistance of Mycobacterium tuberculosis. Infect. Immun. 1997;65:1395–1401. doi: 10.1128/iai.65.4.1395-1401.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynolds CM, Meyer J, Poole LB. An NADH-dependent bacterial thioredoxin reductase-like protein in conjunction with a glutaredoxin homologue form a unique peroxiredoxin (AhpC) reducing system in Clostridium pasteurianum. Biochemistry. 2002;41:1990–2001. doi: 10.1021/bi011802p. [DOI] [PubMed] [Google Scholar]
- 21.Edgar RS, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim TS, et al. Identification of a human cDNA clone for lysosomal type Ca2 + -independent phospholipase A2 and properties of the expressed protein. J. Biol. Chem. 1997;272:2542–2550. doi: 10.1074/jbc.272.4.2542. [DOI] [PubMed] [Google Scholar]
- 23.Kang SW, Baines IC, Rhee SG. Characterization of a mammalian peroxiredoxin that contains one conserved cysteine. J. Biol. Chem. 1998;273:6303–6311. doi: 10.1074/jbc.273.11.6303. [DOI] [PubMed] [Google Scholar]
- 24.Fisher AB. Peroxiredoxin 6: a bifunctional enzyme with glutathione peroxidase and phospholipase A(2) activities. Antioxid. Redox Signal. 2011;15:831–844. doi: 10.1089/ars.2010.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall A, Nelson K, Poole LB, Karplus PA. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox Signal. 2011;15:795–815. doi: 10.1089/ars.2010.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood ZA, Poole LB, Hantgan RR, Karplus PA. Dimers to doughnuts: redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry. 2002;41:5493–5504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- 27.Jang HH, et al. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Chuang MH, et al. The antioxidant protein alkylhydroperoxide reductase of Helicobacter pylori switches from a peroxide reductase to a molecular chaperone function. Proc. Natl. Acad. Sci. USA. 2006;103:2552–2557. doi: 10.1073/pnas.0510770103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim JC, et al. Irreversible oxidation of the active-site cysteine of peroxiredoxin to cysteine sulfonic acid for enhanced molecular chaperone activity. J. Biol. Chem. 2008;283:28873–28880. doi: 10.1074/jbc.M804087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saccoccia F, et al. Moonlighting by different stressors: crystal structure of the chaperone species of a 2-Cys peroxiredoxin. Structure. 2012;20:429–439. doi: 10.1016/j.str.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon JC, et al. Oxidative stress-dependent structural and functional switching of a human 2-Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2-induced cell death. J. Biol. Chem. 2005;280:28775–28784. doi: 10.1074/jbc.M505362200. [DOI] [PubMed] [Google Scholar]
- 32.Cao Z, Tavender TJ, Roszak AW, Cogdell RJ, Bulleid NJ. Crystal structure of reduced and of oxidized peroxiredoxin IV enzyme reveals a stable oxidized decamer and a non-disulfide-bonded intermediate in the catalytic cycle. J. Biol. Chem. 2011;286:42257–42266. doi: 10.1074/jbc.M111.298810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schopf JW. The paleobiological record of photosynthesis. Photosynth. Res. 2011;107:87–101. doi: 10.1007/s11120-010-9577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez-Perez ME, Mata-Cabana A, Sanchez-Riego AM, Lindahl M, Florencio FJ. A comprehensive analysis of the peroxiredoxin reduction system in the Cyanobacterium Synechocystis sp. strain PCC 6803 reveals that all five peroxiredoxins are thioredoxin dependent. J. Bacteriol. 2009;191:7477–7489. doi: 10.1128/JB.00831-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dekker C, Willison KR, Taylor WR. On the evolutionary origin of the chaperonins. Proteins. 2011;79:1172–1192. doi: 10.1002/prot.22952. [DOI] [PubMed] [Google Scholar]
- 36.Mishra Y, Bhargava P, Chaurasia N, Rai LC. Proteomic evaluation of the non-survival of Anabaena doliolum (Cyanophyta) at elevated temperatures. Europ J Phycol. 2009;44:551–565. doi: 10.1080/09670260902947001. [DOI] [Google Scholar]
- 37.Mishra Y, Chaurasia N, Rai LC. AhpC (alkyl hydroperoxide reductase) from Anabaena sp. PCC 7120 protects Escherichia coli from multiple abiotic stresses. Biochem. Biophys. Res. Commun. 2009;381:606–611. doi: 10.1016/j.bbrc.2009.02.100. [DOI] [PubMed] [Google Scholar]
- 38.Mishra Y, et al. Expression, purification, crystallization and preliminary X-ray crystallographic studies of alkyl hydroperoxide reductase (AhpC) from the cyanobacterium Anabaena sp. PCC 7120. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011;67:1203–1206. doi: 10.1107/S1744309111025747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 41.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi HJ, Kang SW, Yang CH, Rhee SG, Ryu SE. Crystal structure of a novel human peroxidase enzyme at 2.0 A resolution. Nat. Struct. Biol. 1998;5:400–406. doi: 10.1038/nsb0598-400. [DOI] [PubMed] [Google Scholar]
- 43.Smeets A, et al. The crystal structure of the C45S mutant of annelid Arenicola marina peroxiredoxin 6 supports its assignment to the mechanistically typical 2-Cys subfamily without any formation of toroid-shaped decamers. Protein Sci. 2008;17:700–710. doi: 10.1110/ps.073399308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vedadi M, et al. Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms. Mol. Biochem. Parasitol. 2007;151:100–110. doi: 10.1016/j.molbiopara.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 45.Mizohata E, et al. Crystal structure of an archaeal peroxiredoxin from the aerobic hyperthermophilic crenarchaeon Aeropyrum pernix K1. J. Mol. Biol. 2005;354:317–329. doi: 10.1016/j.jmb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura T, et al. Structure of peroxiredoxin from the anaerobic hyperthermophilic archaeon Pyrococcus horikoshii. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013;69:719–722. doi: 10.1107/S1744309113014036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pineyro MD, et al. Crystal structure of the tryparedoxin peroxidase from the human parasite Trypanosoma cruzi. J. Struct. Biol. 2005;150:11–22. doi: 10.1016/j.jsb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 48.Nakamura T, et al. Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate. Proc. Natl. Acad. Sci. USA. 2008;105:6238–6242. doi: 10.1073/pnas.0709822105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim KH, Lee W, Kim EE. Crystal structures of human peroxiredoxin 6 in different oxidation states. Biochem. Biophys. Res. Commun. 2016;477:717–722. doi: 10.1016/j.bbrc.2016.06.125. [DOI] [PubMed] [Google Scholar]
- 50.Schroder E, et al. Crystal structure of decameric 2-Cys peroxiredoxin from human erythrocytes at 1.7 A resolution. Structure. 2000;8:605–615. doi: 10.1016/S0969-2126(00)00147-7. [DOI] [PubMed] [Google Scholar]
- 51.Alphey MS, Bond CS, Tetaud E, Fairlamb AH, Hunter WN. The structure of reduced tryparedoxin peroxidase reveals a decamer and insight into reactivity of 2Cys-peroxiredoxins. J. Mol. Biol. 2000;300:903–916. doi: 10.1006/jmbi.2000.3881. [DOI] [PubMed] [Google Scholar]
- 52.Fiorillo A, Colotti G, Boffi A, Baiocco P, Ilari A. The crystal structures of the tryparedoxin-tryparedoxin peroxidase couple unveil the structural determinants of Leishmania detoxification pathway. PLoS Negl. Trop. Dis. 2012;6:e1781. doi: 10.1371/journal.pntd.0001781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen JB, et al. Peroxiredoxin-1 from the human hookworm Ancylostoma ceylanicum forms a stable oxidized decamer and is covalently inhibited by conoidin A. Chem. Biol. 2013;20:991–1001. doi: 10.1016/j.chembiol.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nelson KJ, Parsonage D, Hall A, Karplus PA, Poole LB. Cysteine pK(a) values for the bacterial peroxiredoxin AhpC. Biochemistry. 2008;47:12860–12868. doi: 10.1021/bi801718d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Portillo-Ledesma S, et al. Deconstructing the catalytic efficiency of peroxiredoxin-5 peroxidatic cysteine. Biochemistry. 2014;53:6113–6125. doi: 10.1021/bi500389m. [DOI] [PubMed] [Google Scholar]
- 56.Manevich Y, Feinstein SI, Fisher AB. Activation of the antioxidant enzyme 1-CYS peroxiredoxin requires glutathionylation mediated by heterodimerization with pi GST. Proc. Natl. Acad. Sci. USA. 2004;101:3780–3785. doi: 10.1073/pnas.0400181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Monteiro G, Horta BB, Pimenta DC, Augusto O, Netto LE. Reduction of 1-Cys peroxiredoxins by ascorbate changes the thiol-specific antioxidant paradigm, revealing another function of vitamin C. Proc. Natl. Acad. Sci. USA. 2007;104:4886–4891. doi: 10.1073/pnas.0700481104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 59.Jiang ZY, Woollard AC, Wolff SP. Hydrogen peroxide production during experimental protein glycation. FEBS Lett. 1990;268:69–71. doi: 10.1016/0014-5793(90)80974-N. [DOI] [PubMed] [Google Scholar]
- 60.Ferreira RM, et al. Purification and characterization of the chaperone-like Hsp26 from Saccharomyces cerevisiae. Protein Expr. Purif. 2006;47:384–392. doi: 10.1016/j.pep.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 61.Marini I, Moschini R, Del Corso A, Mura U. Chaperone-like features of bovine serum albumin: a comparison with alpha-crystallin. Cell. Mol. Life Sci. 2005;62:3092–3099. doi: 10.1007/s00018-005-5397-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finn TE, Nunez AC, Sunde M, Easterbrook-Smith SB. Serum albumin prevents protein aggregation and amyloid formation and retains chaperone-like activity in the presence of physiological ligands. J. Biol. Chem. 2012;287:21530–21540. doi: 10.1074/jbc.M112.372961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuan Y, Knaggs M, Poole L, Fetrow J, Salsbury F., Jr. Conformational and oligomeric effects on the cysteine pK(a) of tryparedoxin peroxidase. J. Biomol. Struct. Dyn. 2010;28:51–70. doi: 10.1080/07391102.2010.10507343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salsbury FR, Jr, Yuan Y, Knaggs MH, Poole LB, Fetrow JS. Structural and electrostatic asymmetry at the active site in typical and atypical peroxiredoxin dimers. J. Phys. Chem. B. 2012;116:6832–6843. doi: 10.1021/jp212606k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sundby C, McCaffery S, Anderson JM. Turnover of the photosystem II D1 protein in higher plants under photoinhibitory and nonphotoinhibitory irradiance. J. Biol. Chem. 1993;268:25476–25482. [PubMed] [Google Scholar]
- 66.Muthuramalingam M, et al. Multiple redox and non-redox interactions define 2-Cys peroxiredoxin as a regulatory hub in the chloroplast. Mol Plant. 2009;2:1273–1288. doi: 10.1093/mp/ssp089. [DOI] [PubMed] [Google Scholar]
- 67.Nurizzo D, et al. The ID23-1 structural biology beamline at the ESRF. J Synchrotron Radiat. 2006;13:227–238. doi: 10.1107/S0909049506004341. [DOI] [PubMed] [Google Scholar]
- 68.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Panjikar S, Parthasarathy V, Lamzin VS, Weiss MS, Tucker PA. Auto-rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr. D Biol. Crystallogr. 2005;61:449–457. doi: 10.1107/S0907444905001307. [DOI] [PubMed] [Google Scholar]
- 73.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Afonine PV, et al. Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2010;66:1153–1163. doi: 10.1107/S0907444910026582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Burnley BT, Afonine PV, Adams PD, Gros P. Modelling dynamics in protein crystal structures by ensemble refinement. Elife. 2012;1:e00311. doi: 10.7554/eLife.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McNicholas S, Potterton E, Wilson KS, Noble ME. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. D Biol. Crystallogr. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abagyan RT, Totrov M, Kuznetsov M. ICM - A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J. Comp. Chem. 1994;15:488–506. doi: 10.1002/jcc.540150503. [DOI] [Google Scholar]
- 80.Abagyan RA, Batalov S. Do aligned sequences share the same fold? J. Mol. Biol. 1997;273:355–368. doi: 10.1006/jmbi.1997.1287. [DOI] [PubMed] [Google Scholar]
- 81.Brooks BR, et al. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 83.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 84.Mackerell AD, Jr, Feig M, Brooks CL., III. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 85.Vanommeslaeghe K, et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Essmann U, Perera L, Berkowitz ML. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 87.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:219–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 88.Phillips JC, et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors for AnPrx6 from Anabaena sp. PCC 7120 have been deposited with the Worldwide Protein Data Bank (wwPDB) under code 5M6T.