Abstract
Given their increasingly frequent usage, understanding the chemical and structural properties which allow therapeutic nucleic acids to promote the death of cancer cells is critical for medical advancement. One molecule of interest is a 10-mer of FdUMP (5-fluoro-2′-deoxyuridine-5′-O-monophosphate) also called F10. To investigate causes of structural stability, we have computationally restored the 2′ oxygen on each ribose sugar of the phosphodiester backbone, creating FUMP[10]. Microsecond time-scale, all-atom, simulations of FUMP[10] in the presence of 150 mM MgCl2 predict that the strand has a 45% probability of folding into a stable hairpin-like secondary structure. Analysis of 16 μs of data reveals phosphate interactions as likely contributors to the stability of this folded state. Comparison with polydT and polyU simulations predicts that FUMP[10]’s lowest order structures last for one to 2 orders of magnitude longer than similar nucleic acid strands. Here we provide a brief structural and conformational analysis of the predicted structures of FUMP[10], and suggest insights into its stability via comparison to F10, polydT, and polyU.
Graphical Abstract

INTRODUCTION
The list of biological functions modified by oligonucleotide therapeutics grows ever larger,1–15 making them ideal candidates for treatments across a wide variety of diseases. A subset of these polymers has proven particularly useful in cytotoxic chemotherapy as both drug delivery systems and inhibitors of cancer growth.16–21 While some nucleobases are diffusible through cell membranes, e.g., 5-fluorouracil (5-FU),16,22 delivery systems for oligonucleotides, generally focusing on maintaining the stability of the polymer in vivo until reaching its target, are a major focus of current research.23–27
Given that F10, a 10-base polymer of FdUMP (5-fluoro-2′-deoxyuridine-5′-O-monophosphate), is cytotoxic,21,28–30 demonstrably more efficacious as a therapeutic, and better tolerated in vivo19,31–33 than the widely used 5-fluorouracil (5-FU),34,35 we are interested in methods for increasing both the structural and chemical stability of this polymer. We have previously investigated F10s response to magnesium36 and were curious about the polymer’s lack of structural stability in its presence. To gain further insight, we computationally restored the 2′ oxygen on each ribose sugar of the phosphodiester backbone, creating FUMP[10] (Figure 1), which all-atom MD simulations predicted to form stable hairpin conformations (Figure 2) in the presence of magnesium. This structural response is starkly different from that of F10, which is unstructured and unstable in the presence of magnesium chloride.36
Figure 1.

Nucleotides of the 10mer 5-fluorouridine-5′-monophosphate (FUMP[10]) is produced computationally by replacing a thymidine monophosphate’s methyl group with fluorine and adding a hydroxyl group at C2′. Throughout this article, we use the sugar numbering, base numbering and atom colors shown here.
Figure 2.
For each macrostate, (A) folded, (B) partially folded, and (C) extended, the structure with the smallest RMSD from the average structure is shown as solid with shows representing one standard deviation (by RMSD). These shadows serve as a conformation space uncertainty sphere, revealing the variety of conformations in each macrostate.59
Our particular interest in magnesium with respect to this polymer comes from previous experimental results and first principal calculations from FdU-substituted DNA in the presence of Mg2+ ions.37 Here we use molecular dynamics (MD) force field parameters informed by and supplemented with such experimental results regarding FdU37–39 and F10.18–20,28–31,33 Furthermore, ion-dependent behavior of nucleic acid strands has been of ongoing and longstanding interest in many contexts40–52 with particular interest in the role of magnesium.53–58
Here we propose a 3-state model of FUMP[10] folding and unfolding (Figure 2) along with comparisons to a 10-nucleotide strands of thymine (polydT), uracil (polyU), and F10 itself. From this comparison, we present insights into FUMP[10]’s relatively high level of stability. We have performed a similar analysis of F10 in the presence of magnesium elsewhere.36 However, given the current interest in F10 as an antileukemic therapeutic and a general need for greater understanding of the structural and chemical properties that allow it and other therapeutic nucleic acid polymers to promote the death of malignant cells we consider this analysis of this chemically perturbed polymer’s structure and kinetics deserving of its own presentation.
METHODS
Force Field Parameters
The CHARMM27 force field used here is based on the interaction energies of small model systems determined by both quantum mechanics computations and direct experiment.60–62 For MD simulations in general, the strength of the simulation depends on the reliability of the force field parameters. Obviously, the closer the model system used in fitting the force field parameters is to the system to be simulated, the more trustworthy the resulting structural and kinetic predictions will be. To this end, we use a modified version of the CHARMM27 force field with additional parameters for FdU based on experimental data and quantum mechanics computations done by Ghosh, 2011,37 and previously validated on simulations of DNA topoisomerase 1 (Top1) and FdU-substituted DNA.63
Solvation Conditions
These refined force field parameters informed our choice of neutralizing ions used in computationally solvating our systems, as the experimental work and quantum mechanics calculations referenced above compared interactions of FdU with zinc and magnesium, respectively.37 We solvated each nucleic acid strand in a cubic explicit TIP3P water box, 50 Å on a side, using the “Add Solvation Box” plugin in VMD.64 The addition of counterions to the solvent is particularly important for polyelectrolytes, such as oligonucleotides.
In the protonation state used in our work, each polymer FUMP[10], F10, polyU, and polydT carries a net −9 fundamental charges throughout the simulation, one from each of the phosphate groups in the respective backbones. This protonation state of backbone phosphates was selected for ease of comparison with previous computational work,36–38,63 including calculations validated in joint computational-experimental studies.37,38,63 As in most molecular dynamics simulations, we simulate the most probable protonation state, this is an approximation but should be a minor approximation for this system. In order to simulate a constant-pH, see for example Lee et al., 2004,65 we would have to sacrifice long-time sampling enabled in ACEMD, which is a more serious concern given the need for extensive sampling.
Additionally, in vivo, in vitro, and in silico interactions among residues are constrained by repulsive forces between these charges. The addition of positively charged magnesium ions in the form of magnesium chloride screens these negative charges from one another. Neutralizing the simulation’s total charge and setting the concentration to 0.150 M MgCl2 using VMD’s “Add Ions” tool resulted in an ionic strength of 0.2790 M. For a discussion of nucleic-acid ion interactions with commentary on the implications for MD simulations, see recent review by Lipfert, 2014.52 In the simulations conducted here, imino nitrogens on all nucleic acid strands were fully protonated. For a discussion of physiological protonation states and their effects on such polymers, see Melvin et al., 2016.36
Simulation Protocols
Simulations were run under the canonical ensemble (NVT), which is the thermodynamic ensemble recommended for the ACEMD66 simulation software used. As ACEMD’s is optimized for NVT66 on GPUs, selecting this ensemble allowed for longer simulation times given the computational resources available. Furthermore, solvated systems each contain more than 10 000 atoms, all statistical ensembles should converge to the same result. For a discussion of the viability of this ensemble for ACEMD production runs, see Harvey et al., 2009.66 Hydrogen mass repartitioning as implemented in ACEMD allowed us to use 4 fs time steps in our production runs. Before beginning these production runs, systems underwent 1000 steps of conjugate-gradient minimization. During simulation, systems were held at 300 K using a Langevin thermostat. For VdW and electrostatic forces, we applied a 9 Å cutoff and 7.5 Å switching distance, calculating long-range electrostatics with a smooth particle mesh Ewald (SPME) summation method.67,68 These simulations were run on Titan GPUs in metrocubo workstations produced by Acellera. The combination of this hardware and ACEMD, which is optimized for GPUs, allowed for traditional MD simulation rates of greater than 300 ns/day on the oligomers discusses here. Due to these relatively quick simulation times, we chose traditional MD for all simulations.
The initial structure for all simulations was an extended state. Using default parameters in ACEMD, atoms are randomly assigned initial velocities. While the initial state certainly has an effect on simulations, the simulations presented here are of sufficient length to overcome such effects. This assertion is bolstered by Figure S1, showing that simulations have time to sample structures distinct from the starting state.
Processing and Analysis
Prior to structural and kinetic analysis, we concatenated data from four FUMP[10] simulations (one 1 μs and three 5 μs) totaling 16 μs in one trajectory. We then resampled the data at a rate of 1 frame every 5n s for a final trajectory of 3200 frames. This sampling rate was chosen to simplify visualization of clustering data and to expedite memory-intensive analysis techniques. For both polydT and polyU, we ran two 1 μs simulations, concatenating the resulting data into one trajectory for each respective oligonucleotide and resampling at a rate of 1 frame per 1 ns for a total of 2000 frames in each trajectory. In the process of concatenating these trajectories, we also removed the water molecules and metal ions used in the original solvation. For construction of Figure 7, showing the difference in ion interactions with radial distribution functions, we returned to the raw simulation outputs and resampled each system’s data at a rate of 1 ns per frame without removing any water or ion atoms. Due to memory limitations, only the first 11 μs of F10 data are included in this analysis. These trajectories including water and ions were also used in the analysis of each nucleic acid’s formation of hydrogen bonds with water.
Figure 7.
PolydT forms stable but not structured conformations (A) that have lifespans on the order of 80 ns. PolyU’s long-lived states (B) have lifespans on the order of 10 ns, compared to F10s macrostates (Figures 2 and 3), which last for microseconds at a time. (C) F10 shows both short medium range interactions with magnesium ions. (D) PolydT has the tightest binding with magnesium. (E) PolyU shows the lowest propensity to bind closely with magnesium.
For the purpose of clustering and Markov analysis, we used the quality threshold (QT)69 RMSD clustering method in VMD64 for partitioning the conformations of each system. We initially defined each frame in the F10 trajectory to be a microstate (i.e., 3200 microstates) and searched for 50 macrostates with RMSD QT clustering with a 51st macrostate used as a catchall for any structures VMD could not place in these 50 bins. We adjusted the RMSD cutoff parameter to minimize the number of singleton states outside state 51, resulting in final choice of 5 Å. For comparison of conformation stability, we applied this same cutoff and number of clusters to polydT and polyU. Based on visual inspection, RGYR and measurement of end-to-end distance of the 51 FUMP[10] states, we determined that there were only three structurally distinct macrostates for FUMP[10]. The lowest order macro-state of the 51 (Figure 2A) is clearly a folded state. The second macrostate of the 51 (Figure 2C) is an extended state. The remaining 49 states are all intermediate or partially folded states (Figure 2B). That is, they are neither compact and structured nor fully extended. Therefore, we collapsed the 49 highest order states into one macrostate. Note that the QT clustering algorithm produces the same three macrostates if the desired number of clusters is set at 2–1 folded, 1 extended and third catchall state for any microstates not fitting in the requested 2 bins. In-house Matlab scripts, which we have made available for free online via figshare,70 enabled the subsequent Markov analysis.
Admittedly, more specific structural and kinetic analysis would be available to us in a 51-state model rather than a 3-state model. However, our goal is to provide general insights into structural stability of FUMP[10]. Therefore, we present instead three distinct structural states predicted by the simulations and the statistics of transitions among them. For those interested, we provide analysis of the 51-state model in Figures S1 and 2 and Table S1.
When investigating the stability of each oligonucleotide, we used VMD’s hydrogen bond plugin with a donor–acceptor distance cutoff of 4 Å and a bond angle cutoff of 60 degrees. Phosphate interactions in the RNA and DNA structures were analyzed using DSSR, a new component of the 3DNA suite of software programs.71–74 By a phosphate interaction, we mean an electrostatic interaction in which phosphate group accepts a hydrogen bond from a nucleotide. This effect has been observed in several RNA crystal structures in the RCSB database,75 as shown in a meta-study across several such structures by Zirbel et al., 2009.76
We defined a native contact distance of 12 Å between backbone phosphorus atoms. This cutoff was chosen by finding the largest distance between backbone phosphorus atoms on two bases undergoing a phosphate interaction as detected by DSSR. Using the microstate with the largest number of phosphate interactions (Figure 3) as a reference structure and ignoring native contacts found between any residues i and j where j < i + 3 (for residue numbering, see Figure 3), we executed the native contacts search using the Python package MDAnalysis.77
Figure 3.

Most common base interactions across and within all macrostates are phosphate interactions wherein an oxygen atom in one phosphate group accepts a hydrogen bond from a nucleotide. The numbered residues are those referenced in Table 2.
RESULTS AND DISCUSSION
FUMP[10] Macrostates and Kinetics
From RMSD clustering on 16 μs of FUMP[10] simulation data, we identified three structurally distinct macrostates (Figure 2). The highest probability state (44.81% likely) is a hairpin structure (Figure 2A) All microstates, i.e., frames in this macrostate, maintain a hairpin-like conformation. The second most likely macrostate (41.63%) is the most varied. Conformations in this macrostate are neither compact nor extended but are intermediate states in the folding and unfolding processes FUMP[10] undergoes. The assembly of these macrostates does not provide a clear median structure for this intermediate state (see Methods Section). Therefore, for all figures, a representative for this most varied state was chosen at random from the constituent microstates. The extended or unfolded macrostate (13.56%) has as its median structure a nearly fully extended conformation. All structures within this macrostate exhibit a similarly extended shape.
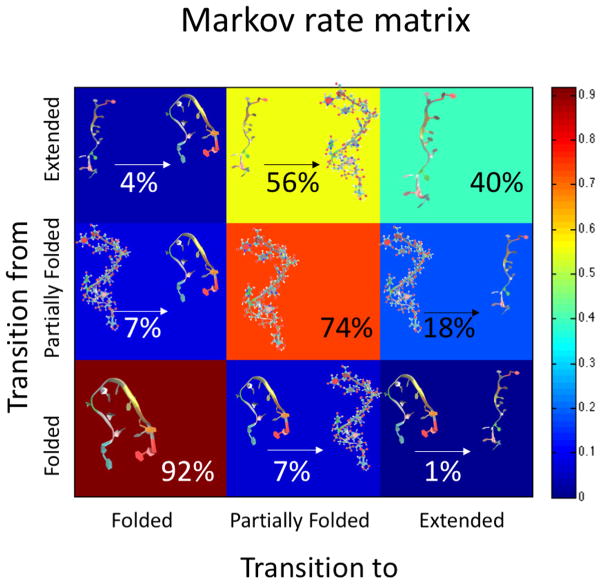
Using this three-state model, we recover the kinetics of the simulation by constructing a first-order (memoryless) Markov rate matrix (Figure 4). From calculations of these transition probabilities, we see that the folded and partially folded states serve as kinetic traps, i.e., once entering that state, FUMP[10] is most likely to stay there (91.91% for folded and 74.40% for partially folded). We also see that when in an extended conformation, FUMP[10] is most likely (55.99%) to transition into the partially folded state. However, the transition probability from partially folded to folded is small (7.43%), meaning that while the folded state is preferred there is a large barrier to folding. Starting from any initial state, these transition probabilities converge within 40 steps (200 ns) to steady state populations that are within 0.1% of the equilibrium probability distribution of the trajectory. That is, based on the Markov model built from our simulations, in the ensemble of all possible simulations, we expect to see populations similar to those we observed after 200 ns on average. Based on the transition probabilities shown here, the expected transition times are 13 ns for extended to partially folded, 75 ns for partially folded to folded, and 78 ns for extended to folded. On the other hand, the unfolding times for the fully and partially folded states are 101 and 49 ns, respectively.
Figure 4.
Starting from any initial state (i.e., folded, partially folded, or extended), these transition probabilities converge within 40 steps (200 ns) to steady state populations of macrostates that is within 0.1% of the populations derived from RMSD clustering on the MD trajectories. This recovery of kinetics via Markov analysis reveals that the transitions folded< – > partially folded and unfolded< – > partially folded are more likely than direct transitions between folded and unfolded. Based on the transition probabilities shown here, the expected transition times are 13 ns for extended to partially folded, 75 ns for partially folded to folded, and 78 ns for extended to folded. On the other hand, the unfolding times for the fully and partially folded states are 101 and 49 ns, respectively.
Using these population and transition statistics, we calculated the free energy profiles for the most likely folding and unfolding pathway indicated by the Markov rate matrix folded< – > partially folded and unfolded< – > partially folded (Figure 5). While already seen from transition probabilities, these calculations show from an energetics perspective that while folded and partially folded are roughly equally likely there is a large energy barrier to transitioning between the two states. We also see that moving from partially folded to extended is not favorable but moving from extended to partially folded is favorable.
Figure 5.

A combination of RMSD clustering, RGYR-based histograms, and visual inspection indicates 3 macrostates: folded, partially folded, and extended. Free energy calculations show that while folded and partially folded are equally likely there is a large energy barrier to transitioning between the two states. Furthermore, while moving from partially folded to extended is not favorable, moving from extended to partially folded is favorable. This apparent asymmetry comes from our simplified presentation of the pathway as unidimensional.
Folded Macrostates
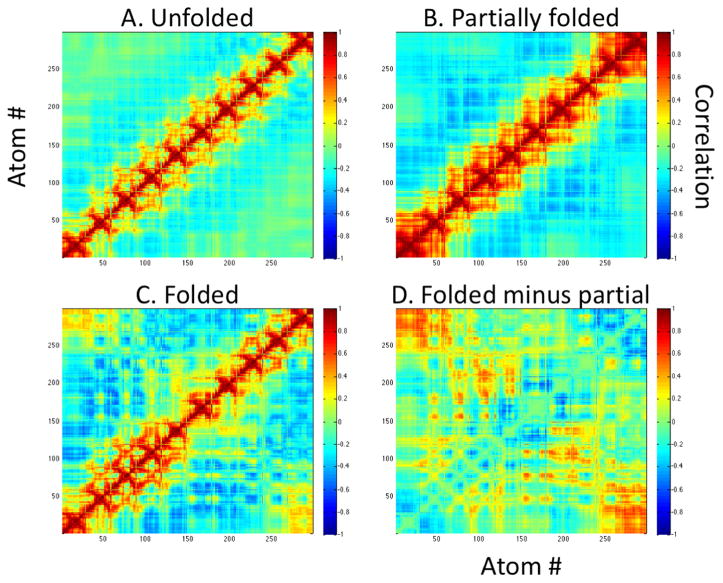
To understand the behavioral differences in the three structural macrostates of FUMP[10], we examined the correlated motion of atoms in each macrostate separately (Figure 6B). From this analysis, we observe little correlation other than within residues for atoms in the unfolded macrostate (Figure 6A). Some medium range, strong anticorrelation appears between some atoms in the partially folded state (Figure 6B). Long-range moderate correlation and medium range strong anticorrelation appear between many atoms in the folded state (Figure 6C). The difference in the folded and partially folded states is seen more clearly in Figure 6D where we have displayed the quantitative difference between the two correlation matrices.
Figure 6.
(A) Little correlated motion occurs between atoms other self-residue interactions. (B) In the partially folded macrostate, some interactions occur among neighboring residues. (C) Neighboring residue interactions and interactions across the hairpin conformation seen in Figure 2A appear in the folded macrostate. The difference in partially folded and folded correlations is shown in (D) revealing that the greatest differences occur in residue interactions across the hairpin structure. That is, the greatest differences occur along and near the antidiagonal elements of the folded and partially folded correlation matrices.
Via analysis of the hydrogen bonds occurring in each macrostate with both VMD64 and DSSR71–73 (the latter of which categorizes the types of hydrogen bond interations), we determined that the most common interactions among residues involved an oxygen atom from one nucleotide’s phosphate group accepting a bond from another nucleotide (Figure 3). The statistics for the phosphate interactions in each macrostate are summarized in Table 1.
Table 1.
Phosphate Interactions Occur when an Oxygen Atom from One Nucleotide’s Phosphate Group Accepts a Hydrogen Bond from Another Nucleotidea
| folded | partially folded | extended | |
|---|---|---|---|
| mean phosphate interactions | 4.9686 | 3.3829 | 2.5991 |
| standard deviation | 1.1777 | 1.5604 | 1.3847 |
| standard error | 3.108 × 10−2 | 4.275 × 10−2 | 6.647 × 10−2 |
Based on these averages, the shape of FUMP[10]’s folded macrostates owes its structure to phosphate interactions. Two-sample t-test without assuming equal variance reveals that each mean number of phosphate interactions is significantly different from the other two with p-values less than 10−16.
A major commonality of the conformations in the folded macrostate is their apparent structure. That is, the polymer appears structured in each of the constituent frames. To quantify this seeming orderliness, we calculated native contacts between residues. The native contacts that occur in more than 50% of the folded conformations are listed in Table 2. The unfolded and partially folded macrostates have no such contacts occurring in more than 50% of their frames.
Table 2.
Using the Microstate with the Largest Number of Phosphate Interactions (Figure 3) as a Reference Structure and Ignoring Native Contacts Found between Any Residues i and j where j < i + 3, We Calculate the Percent of Native Contacts Occurring in the Folded Macrostatea
| residue i | residue j | native contact ij (%) |
|---|---|---|
| 2 | 9 | 88.97 |
| 3 | 8 | 89.88 |
| 3 | 9 | 52.90 |
| 4 | 8 | 57.43 |
Residues listed here are labeled in Figure 3. Only native contacts occurring in more than 50% of the folded frames are shown here.
Furthermore, in the folded macrostate, there are 3 specific phosphate interactions that occur in more than 50% of the frames included in the folded macrostate. Using the base numbering in Figure 3, these frequent phosphate interactions occur between (in the form donor and acceptor) bases 5 and 6, 8 and 4, and 9 and 3, found using VMD’s hydrogen bond tool. The prevalence of these particular phosphate interactions implicate these residues in particular in FUMP[10]’s relative structural stability. We have included a table of all such phosphate interactions in the folded macrostate along with specific atoms involved and frequency of occurrence in Table S2.
Comparison to polyU and polydT
Whereas FUMP[10] has stable structures existing for microseconds at a time, the longest lived structures for polydT (Figure 7A) have lifetimes on the order of 80 ns (Figure S3). PolyU’s structures have even shorter lifetimes, which the most stable (Figure 7B) lasting on the order of 10 ns (Figure S4). The correlation matrix for polyU (Figure S5) closely resembles that of FUMP[10]’s extended state, indicating little to no correlation in atomic motions. The correlation matrix for polydT (Figure S6) shows medium range correlations similar to those in FUMP[10]’s partially folded state. Both polyU and polydT form fewer phosphate interactions than FUMP[10], with polydT forming the fewest phosphate interactions. The statistics for phosphate interactions in all three systems are summarized in Table 3.
Table 3.
Lack of Phosphate Interactions in the polydT Simulation Is Surprising Given the Stability of polydT’s Conformations Relative to Those of polyUa
| F10 | polyU | polydT | |
|---|---|---|---|
| mean phosphate interactions | 3.987 | 2.954 | 0.4920 |
| standard deviation | 1.655 | 1.480 | 0.6612 |
| standard error | 2.926 × 10−2 | 3.308 × 10−2 | 1.478 × 10−2 |
A two-sample t-test without assuming equal variance reveals that each mean number of phosphate interactions is significantly different from the other two with p-values less than 10−16.
To better understand these differences in stability, we examined the interactions of each nucleic acid strand with the local ion environment (Figure 7C–E). FUMP[10] has both short-range and medium-range interactions with the magnesium ion solution, indicating the formation of an ion cloud or “atmosphere”43,47,54 in addition to the binding events indicated by the early spike in radial distribution. PolyU shows some binding interactions with magnesium, as indicated by the spike in the radial distribution function at 1.15 Å (Figure 7B). PolydT forms even tighter bonds with the magnesium, with a slightly larger spike occurring at 0.05 Å (Figure 7C).
Given fluorine’s capacity as a hydrogen bond acceptor, we compared the three oligonucleotides’ propensity to form hydrogen bonds with water. The statistics for hydrogen bond formation with water are shown in Table 4.
Table 4.
Similarity in FUMP[10] and polyU’s Propensity To Form Hydrogen Bonds with Water Is on the One Hand Surprising Due To Its Lack of the Additional Acceptor Compared To FUMP[10] but Also Sensible as FUMP[10] Is a polyU Derivativea
| F10 | polyU | polydT | |
|---|---|---|---|
| mean hydrogen bonds with water | 226.2 | 197.2 | 228.9 |
| standard deviation | 25.87 | 19.46 | 22.24 |
| standard error | 0.2446 | 0.4350 | 0.4972 |
A two-sample t-test without assuming equal variance reveals that each mean number of hydrogen bonds is significantly different from the other two with p-values less than 10−16.
Comparison of correlation matrices for the three systems (Figure 6 and Figures S5 and S6) show similarities between FUMP[10]’s extended state and polyU along with similarities between FUMP[10]’s partially folded state and polydT. By naive induction, these relationships might suggest that polyU prefers an extended state and polydT a partially folded but mostly unstructured state. Indeed, the lowest order state of polyU (5% likely) is a mostly extended conformation (Figure 7A) and the lowest order state of polydT (4% likely) is a partially folded, unstructured conformation (Figure 7B).
All three oligonucleotides enter stable conformations. However, the time scales over which these states survive are widely varied. FUMP[10]’s stable states survive for up to microseconds at a time, whereas neither polyU nor polydT have structures achieving even 100 ns in lifespan. Judging by the statistics in Table 3, F10s long-lived stable states, e.g., Figure 2A, and polyU’s short-lived stable states, e.g., Figure 7B, are likely due to weak electrostatic interactions in the form of phosphate interactions. However, this explanation clearly does not hold for polydT.
One might suggest that polydT’s higher level of stability relative to polyU is due to hydrogen bond interactions with water. Quantitative analysis of the hydrogen bonds with water in each system confirms that FUMP[10] and polyU both have a higher capacity for forming such bonds than does polydT. For a similar reason that polyU might be thought likely to form hydrogen bonds with surrounding water molecules, FUMP[10]’s additional hydrogen bond acceptor in the form of fluorine suggests a higher capacity for the formation of such bonds. However, an analysis of the MD simulations indicates this is not the case. FUMP[10], a derivative of polyU, and polyU have similar propensities for forming hydrogen bonds with water with polydT forming the fewest hydrogen bonds with water of the three, Table 4.
Therefore, for understanding the stability of polydT, we turned to interactions with magnesium ions. Judging from the tight magnesium binding emerging in polydT simulations (Figure 7E) we conclude it is short-range metal binding interactions that stabilize the conformations of the thymine polymer for up to 80 ns at at time. Both FUMP[10] and polyU have fewer of such interactions with polyU having the fewest. Furthermore, FUMP[10] uniquely shows some medium-range interactions, see the 5–10 Å range of Figure 7C, indicating that the formation of an ion cloud near FUMP[10] provides additional stability for its conformations.
Comparison to F10
In a previous MD study, we observed F10 to be unstable and exhibit occasional structure when solvated in 150 mM MgCl2.36 In those calculations, the 5 highest population RMSD-based QT clusters comprised only 4.5% of simulated frames across 4 trajectories totaling 16 μs of data. None of these states existed for more than a few nanoseconds at a time. Even though F10 exhibited no long-term stable conformation in the presence of magnesium, 26.7% of MD frames contained at least 1 base–base interaction.36 It should be noted that in this previous study36 all F10 phosphate groups carried a net −1 charge, informing the choice of protonation state in the current study.
FUMP[10] forms base–base interactions in 6.2% of frames. However, interactions among nucleotides involving a phosphate group forms in 98.12% of frames. We see that the prevalence of phosphate interactions is responsible for the higher level of structural stability observed for FUMP[10] relative to F10 with the same phosphate-group protonation states.
FUMP[10] Structure and Stability
The nearly equal free energy (and, therefore, probabilities) for the folded and partially folded states of FUMP[10] along with the favorable change in free energy from extended to partially folded demonstrate that FUMP[10] prefers a compact structure over an extended structure. Markov analysis confirms this bias toward compact states, showing that when in an extended conformation, FUMP[10] is most likely (55.99%) to transition out of the extended state into a partially folded state. This preference is not surprising given the hydrophobic nature of nucleic basis and the hydrophilic nature of the backbone, resulting from the net negative charge on each phosphate group.
Less expected, though, is the large free energy barrier between the partially folded and folded macrostate. This high barrier likely exists due to the structured nature of the folded macrostate. There is a clear hairpin shape to all of the microstates within this macrostate. This secondary structure is stabilized primarily by phosphate interactions. Alignment of the phosphate groups and nucleic bases into the ordered structure seen in Figure 2 carries a high entropic cost. Once in this state, though, the stabilizing weak electrostatic interactions make FUMP[10] only 8% likely to leave the macrostate.
Native contact analysis, Table 2, indicates which residue interactions are most responsible for the folded state’s stability. Phosphate interactions by hydrogen bonds across the hairpin hold the hairpin shape together. Beyond this stability, FUMP[10]’s folded state shows more long-range communication between atoms in the form of correlated motions than the other two macrostates. The difference in the folded and partially folded macrostates’ correlation matrices show a key distinction in these two states. The interactions stabilizing the macrostate also lead to long-range communication between atom pairs. While the partially folded state exhibits some correlated motion, it is clearly not to the extent seen in the folded state.
Comparison with Experimental Literature
Experimental work on single-stranded oligonucleotides indicates that nucleic acid hairpins are in general stable structures.78–83 Work by Ma, 2006,78 on 8mer hairpins containing UUUU and UUCG tretraloops shows the free energy barrier of moving from a folded to unfolded state to be 2 and 3 kcal/mol, which falls in the range of an earlier review of experimental work on RNA hairpins in general by Varani, 1995,80 who reports the free energy of hairpin formation to be between −5.7 and −1.7 kcal/ mol. The hairpins discussed in these works owe their stability to multiple intramolecular interactions including Watson–Crick base pairs, base pair stacking, and phosphate interactions.78,80
However, FUMP[10]’s intermolecular interactions appear limited to base-phosophate interactions. This difference along with FUMP[10]’s composition of only fluorouridine make direct comparison to FUMP[10] somewhat spurious. However, the range of free energies of (un)folding, does locate FUMP[10]’s stability (1.5 kcal/mol barrier to unfolding) on the lower end of oligonucleotide hairpin structures.
CONCLUSIONS
FUMP[10] prefers a compact, structured state in the presence of magnesium. However, the free energy barrier for transitioning from a partially folded, unstructured state into the lowest order hairpin-like state is large. Conversely, FUMP[10] prefers to not be in a mostly extended state, having a favorable free energy of transition from the extended macrostate to the partially folded macrostate. Phosphate interactions forming across the hairpin structure are responsible for the stability of this state, seen in its persistence for up to microseconds at a time. Furthermore, long-range communication in the form of correlated motion occurs between atoms in the folded, hairpin macrostate more so than in the partially folded and extended states.
Unlike polyU, which MD calculations predicted to prefer an extended state, and polydT, predicted to prefer a partially folded state, FUMP[10] prefers a compact, structured conformation. The stability of short-lived states of polyU is likely due to the formation of hydrogen bonds with water, while polydT owes its stability to short-range bond formation with magnesium. FUMP[10]’s stability comes from a combination of both hydrogen bonds with water and short to medium range interactions with magnesium. Additionally, FUMP[10] shows the highest propensity for intramolecular phosphate interactions, providing a mechanism for the one to 2 orders of magnitude longer lifespan of its lowest order state relative to those of polyU and polydT.
Supplementary Material
Acknowledgments
Crystallography & Computational Biosciences services were supported by the Comprehensive Cancer Center of Wake Forest University NCI CCSG P30CA012197 grant. This work was partially supported by National Institutes of Health grant T32-GM095440, supporting R.L.M. This work was partially supported by National Institutes of Health grant R01CA129373 to F.R.S. Some computations were performed on the Wake Forest University DEAC Cluster, a centrally managed resource with support provided in part by the University. F.R.S. also acknowledges a Reynolds Research leave from Wake Forest University.
Footnotes
Notes
The authors declare the following competing financial interest(s): Dr. Gmeiner is an owner of Salzburg Therapeutics, the company holding licenses for FdUMP-[N].
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpcb.7b04724.
Cluster time series for 51-state models of FUMP10, PolyU, and PolydT; Markov rate matrix for 51-state model of FUMP10; correlation matrices for PolyU and PolydT; occupancy and survival rates for FUMP10; detailed phosphate interactions (PDF)
References
- 1.Skutella T, Stöhr T, Probst JC, Ramalho-Ortigao FJ, Holsboer F, Jirikowski GF. Antisense oligodeoxynucleotides for in vivo targeting of corticotropin-releasing hormone mRNA: comparison of phosphorothioate and 3′-inverted probe performance. Horm Metab Res. 1994;26:460–464. doi: 10.1055/s-2007-1001733. [DOI] [PubMed] [Google Scholar]
- 2.Souza P, Sedlackova L, Kuliszewski M, Wang J, Liu J, Tseu I, Liu M, Tanswell AK, Post M. Antisense oligodeoxynucleotides targeting PDGF-B mRNA inhibit cell proliferation during embryonic rat lung development. Development. 1994;120:2163–2173. doi: 10.1242/dev.120.8.2163. [DOI] [PubMed] [Google Scholar]
- 3.Biro S, Fu YM, Yu ZX, Epstein SE. Inhibitory effects of antisense oligodeoxynucleotides targeting c-myc mRNA on smooth muscle cell proliferation and migration. Proc Natl Acad Sci U S A. 1993;90:654–658. doi: 10.1073/pnas.90.2.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young SL, Krawczyk SH, Matteucci MD, Toole JJ. Triple helix formation inhibits transcription elongation in vitro. Proc Natl Acad Sci U S A. 1991;88:10023–10026. doi: 10.1073/pnas.88.22.10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konopka K, Rossi JJ, Swiderski P, Slepushkin Va, Düzgüneş N. Delivery of an anti-HIV-1 ribozyme into HIV-infected cells via cationic liposomes. Biochim Biophys Acta, Biomembr. 1998;1372:55–68. doi: 10.1016/s0005-2736(98)00046-7. [DOI] [PubMed] [Google Scholar]
- 6.Karikó K, Megyeri K, Xiao Q, Barnathan ES. Lipofectin-aided cell delivery of ribozyme targeted to human urokinase receptor mRNA. FEBS Lett. 1994;352:41–44. doi: 10.1016/0014-5793(94)00914-7. [DOI] [PubMed] [Google Scholar]
- 7.Dai W, Wang C, Wang F, Wang Y, Shen M, Chen K, Cheng P, Zhang Y, Yang J, Zhu R, et al. Anti-miR-197 inhibits migration in HCC cells by targeting KAI 1/CD82. Biochem Biophys Res Commun. 2014;446:541–548. doi: 10.1016/j.bbrc.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Meng W, Jiang L, Lu L, Hu H, Yu H, Ding D, Xiao K, Zheng W, Guo H, Ma W. Anti-miR-155 oligonucleotide enhances chemosensitivity of U251 cell to taxol by inducing apoptosis. Cell Biol Int. 2012;36:653–659. doi: 10.1042/CBI20100918. [DOI] [PubMed] [Google Scholar]
- 9.Pu M, Garrahan JP, Hirst JD. Comparison of implicit solvent models and force fields in molecular dynamics simulations of the PB1 domain. Chem Phys Lett. 2011;515:283–289. [Google Scholar]
- 10.Kim TG, Kim CH, Won EH, Bae SM, Ahn WS, Park JB, Sin JI. CpG-ODN-stimulated dendritic cells act as a potent adjuvant for E7 protein delivery to induce antigen-specific antitumour immunity in a HPV 16 E7-associated animal tumour model. Immunology. 2004;112:117–125. doi: 10.1111/j.1365-2567.2004.01851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prince Ga, Mond JJ, Porter DD, Yim KC, Lan SJ, Klinman DM. Immunoprotective activity and safety of a respiratory syncytial virus vaccine: mucosal delivery of fusion glycoprotein with a CpG oligodeoxynucleotide adjuvant. Journal of virology. 2003;77:13156–13160. doi: 10.1128/JVI.77.24.13156-13160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discovery. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mercatante DR, Mohler JL, Kole R. Cellular response to an antisense-mediated shift of Bcl-x pre-mRNA splicing and antineo-plastic agents. J Biol Chem. 2002;277:49374–49382. doi: 10.1074/jbc.M209236200. [DOI] [PubMed] [Google Scholar]
- 14.Sazani P, Kole R. Modulation of alternative splicing by antisense oligonucleotides. Prog Mol Subcell Biol. 2003;31:217–239. doi: 10.1007/978-3-662-09728-1_8. [DOI] [PubMed] [Google Scholar]
- 15.Wang C, Vernon R, Lange O, Tyka M, Baker D. Prediction of structures of zinc-binding proteins through explicit modeling of metal coordination geometry. Protein Sci. 2010;19:494–506. doi: 10.1002/pro.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostertag D, Amundson KK, Espinoza FL, Martin B, Buckley T, Da Silva APG, Lin AH, Valenta DT, Perez OD, Ibañez CE, et al. Brain tumor eradication and prolonged survival from intratumoral conversion of 5-fluorocytosine to 5-fluorouracil using a nonlytic retroviral replicating vector. Neuro-Oncology. 2012;14:145–159. doi: 10.1093/neuonc/nor199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller CR, Williams CR, Buchsbaum DJ, Gillespie GY. Intratumoral 5-fluorouracil produced by cytosine deaminase/5-fluorocytosine gene therapy is effective for experimental human glioblastomas. Cancer Res. 2002;62:773–80. [PubMed] [Google Scholar]
- 18.Gmeiner WH, Lema-Tome C, Gibo D, Jennings-Gee J, Milligan C, Debinski W. Selective anti-tumor activity of the novel fluoropyrimidine polymer F10 towards G48a orthotopic GBM tumors. J Neuro-Oncol. 2014;116:447–54. doi: 10.1007/s11060-013-1321-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pardee TS, Gomes E, Jennings-Gee J, Caudell D, Gmeiner WH. Unique dual targeting of thymidylate synthase and topoisomerase1 by FdUMP[10] results in high efficacy against AML and low toxicity. Blood. 2012;119:3561–70. doi: 10.1182/blood-2011-06-362442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pardee TS, Stadelman K, Jennings-Gee J, Caudell DL, Gmeiner WH. The poison oligonucleotide F10 is highly effective against acute lymphoblastic leukemia while sparing normal hematopoietic cells. Oncotarget. 2014;5:4170–9. doi: 10.18632/oncotarget.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gmeiner WH, Reinhold WC, Pommier Y. Genome-wide mRNA and microRNA profiling of the NCI 60 cell-line screen and comparison of FdUMP[10] with fluorouracil, floxuridine, and topoisomerase 1 poisons. Mol Cancer Ther. 2010;9:3105–14. doi: 10.1158/1535-7163.MCT-10-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Tahtawy A, Wolf W. In vivo measurements of intratumoral metabolism, modulation, and pharmacokinetics of 5-fluorouracil, using 19F nuclear magnetic resonance spectroscopy. Cancer Res. 1991;51:5806–5812. [PubMed] [Google Scholar]
- 23.Wang Y, Miao L, Satterlee A, Huang L. Delivery of oligonucleotides with lipid nanoparticles. Adv Drug Delivery Rev. 2015;87:68–80. doi: 10.1016/j.addr.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen J, Szoka FC. Nucleic acid delivery: The missing pieces of the puzzle? Acc Chem Res. 2012;45:1153–1162. doi: 10.1021/ar3000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones CH, Chen CK, Ravikrishnan A, Rane S, Pfeifer Ba. Overcoming nonviral gene delivery barriers: Perspective and future. Mol Pharmaceutics. 2013;10:4082–4098. doi: 10.1021/mp400467x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konishi M, Kawamoto K, Izumikawa M, Kuriyama H, Yamashita T. Gene transfer into guinea pig cochlea using adeno-associated virus vectors. J Gene Med. 2008;10:610–618. doi: 10.1002/jgm.1189. [DOI] [PubMed] [Google Scholar]
- 27.Nishikawa M, Huang L. Nonviral vectors in the new millennium: delivery barriers in gene transfer. Hum Gene Ther. 2001;12:861–870. doi: 10.1089/104303401750195836. [DOI] [PubMed] [Google Scholar]
- 28.Liao ZY, Sordet O, Zhang HL, Kohlhagen G, Antony S, Gmeiner WH, Pommier Y. A novel polypyrimidine antitumor agent FdUMP[10] induces thymineless death with topoisomerase I-DNA complexes. Cancer Res. 2005;65:4844–51. doi: 10.1158/0008-5472.CAN-04-1302. [DOI] [PubMed] [Google Scholar]
- 29.Bijnsdorp IV, Comijn EM, Padron JM, Gmeiner WH, Peters GJ. Mechanisms of action of FdUMP[10]: metabolite activation and thymidylate synthase inhibition. Oncol Rep. 2007;18:287–91. doi: 10.3892/or.18.1.287. [DOI] [PubMed] [Google Scholar]
- 30.Gmeiner WH, Skradis A, Pon RT, Liu J. Cytotoxicity and in-vivo tolerance of FdUMP[10]: a novel pro-drug of the TS inhibitory nucleotide FdUMP. Nucleosides Nucleotides. 1999;18:1729–30. doi: 10.1080/07328319908044836. [DOI] [PubMed] [Google Scholar]
- 31.LIU J, KOLATH J, ANDERSON J, KOLAR C, LAWSON TA, TALMADGE J, GMEINER WH. Positive interaction between 5-FU and FdUMP[10] in the inhibition of human colorectal tumor cell proliferation. Antisense Nucleic Acid Drug Dev. 1999;9:481–486. doi: 10.1089/oli.1.1999.9.481. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Skradis A, Kolar C, Kolath J, Anderson J, Lawson T, Talmadge J, Gmeiner WH. Increased Cytotoxicity and Decreased In Vivo Toxicity of FdUMP[10] Relative to 5-FU. Nucleosides Nucleotides. 1999;18:1789–1802. doi: 10.1080/07328319908044843. [DOI] [PubMed] [Google Scholar]
- 33.Liu C, Willingham M, Liu J, Gmeiner WH. Efficacy and safety of FdUMP[10] in treatment of HT-29 human colon cancer xenografts. Int J Oncol. 2002;21:303–8. [PubMed] [Google Scholar]
- 34.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–8. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 35.HEIDELBERGER C, CHAUDHURI NK, DANNEBERG P, MOOREN D, GRIESBACH L, DUSCHINSKY R, SCHNITZER RJ, PLEVEN E, SCHEINER J. Fluorinated pyrimidines, a new class of tumour-inhibitory compounds. Nature. 1957;179:663–6. doi: 10.1038/179663a0. [DOI] [PubMed] [Google Scholar]
- 36.Melvin RL, Gmeiner WH, Salsbury FR., Jr All-atom molecular dynamics reveals mechanism of zinc complexation with therapeutic F10. J Phys Chem B. 2016;120:10269–10279. doi: 10.1021/acs.jpcb.6b07753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh S, Salsbury FR, Jr, Horita Da, Gmeiner WH. Zn2+ selectively stabilizes FdU-substituted DNA through a unique major groove binding motif. Nucleic Acids Res. 2011;39:4490–4498. doi: 10.1093/nar/gkr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosh S, Salsbury FR, Jr, Horita Da, Gmeiner WH. Cooperative stabilization of Zn 2+:DNA complexes through netropsin binding in the minor groove of FdU-substituted DNA. J Biomol Struct Dyn. 2013;31:1301–1310. doi: 10.1080/07391102.2012.732343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stuart CH, Horita Da, Thomas MJ, Salsbury FR, Lively MO, Gmeiner WH. Site-specific DNA-doxorubicin conjugates display enhanced cytotoxicity to breast cancer cells. Bioconjugate Chem. 2014;25:406–13. doi: 10.1021/bc4005427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stein A, Crothers DM. Conformational changes of transfer RNA. The role of magnesium(II) Biochemistry. 1976;15:160–8. doi: 10.1021/bi00646a025. [DOI] [PubMed] [Google Scholar]
- 41.Chen H, Meisburger SP, Pabit Sa, Sutton JL, Webb WW, Pollack L. Ionic strength-dependent persistence lengths of single-stranded RNA and DNA. Proc Natl Acad Sci U S A. 2012;109:799–804. doi: 10.1073/pnas.1119057109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen H, Meisburger SP, Pabit Sa, Sutton JL, Webb WW, Pollack L. Ionic strength-dependent persistence lengths of single-stranded RNA and DNA. Proc Natl Acad Sci U S A. 2012;109:799–804. doi: 10.1073/pnas.1119057109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bukhman YV, Draper DE. Affinities and selectivities of divalent cation binding sites within an RNA tertiary structure. J Mol Biol. 1997;273:1020–1031. doi: 10.1006/jmbi.1997.1383. [DOI] [PubMed] [Google Scholar]
- 44.Mills JB, Vacano E, Hagerman PJ. Flexibility of single-stranded DNA: use of gapped duplex helices to determine the persistence lengths of poly(dT) and poly(dA) J Mol Biol. 1999;285:245–57. doi: 10.1006/jmbi.1998.2287. [DOI] [PubMed] [Google Scholar]
- 45.Draper DE. RNA folding: thermodynamic and molecular descriptions of the roles of ions. Biophys J. 2008;95:5489–95. doi: 10.1529/biophysj.108.131813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson RC, Doudna Ja. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–39. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Draper DE. A guide to ions and RNA structure A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Šponer J, Banáš P, Jurečka P, Zgarbová M, Kührová P, Havrila M, Krepl M, Stadlbauer P, Otyepka M. Molecular dynamics simulations of nucleic acids. from tetranucleotides to the ribosome. J Phys Chem Lett. 2014;5:1771–1782. doi: 10.1021/jz500557y. [DOI] [PubMed] [Google Scholar]
- 49.Schuetz JD, Wallace HJ, Diasio RB. 5-Fluorouracil incorporation into DNA of CF-1 mouse bone marrow cells as a possible mechanism of toxicity. Cancer Res. 1984;44:1358–63. [PubMed] [Google Scholar]
- 50.McFail-Isom L, Sines CC, Williams LD. DNA structure: cations in charge? Curr Opin Struct Biol. 1999;9:298–304. doi: 10.1016/S0959-440X(99)80040-2. [DOI] [PubMed] [Google Scholar]
- 51.Chin K, Sharp Ka, Honig B, Pyle aM. Calculating the electrostatic properties of RNA provides new insights into molecular interactions and function. Nat Struct Biol. 1999;6:1055–1061. doi: 10.1038/14940. [DOI] [PubMed] [Google Scholar]
- 52.Lipfert J, Doniach S, Das R, Herschlag D. Understanding nucleic acid-ion interactions. Annu Rev Biochem. 2014;83:813–41. doi: 10.1146/annurev-biochem-060409-092720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Misra V, Draper D. On the role of magnesium ions in RNA stability. Biopolymers. 1998;48:113–135. doi: 10.1002/(SICI)1097-0282(1998)48:2<113::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 54.Misra VK, Draper DE. The interpretation of Mg(2+) binding isotherms for nucleic acids using Poisson-Boltzmann theory. J Mol Biol. 1999;294:1135–1147. doi: 10.1006/jmbi.1999.3334. [DOI] [PubMed] [Google Scholar]
- 55.Misra VK, Draper DE. Mg(2+) binding to tRNA revisited: the nonlinear Poisson-Boltzmann model. J Mol Biol. 2000;299:813–25. doi: 10.1006/jmbi.2000.3769. [DOI] [PubMed] [Google Scholar]
- 56.Misra VK, Draper DE. The linkage between magnesium binding and RNA folding. J Mol Biol. 2002;317:507–521. doi: 10.1006/jmbi.2002.5422. [DOI] [PubMed] [Google Scholar]
- 57.Grilley D, Misra V, Caliskan G, Draper DE. Importance of partially unfolded conformations for Mg(2+)-induced folding of RNA tertiary structure: structural models and free energies of Mg2+ interactions. Biochemistry. 2007;46:10266–78. doi: 10.1021/bi062284r. [DOI] [PubMed] [Google Scholar]
- 58.Zheng H, Shabalin IG, Handing KB, Bujnicki JM, Minor W. Magnesium-binding architectures in RNA crystal structures: validation, binding preferences, classification and motif detection. Nucleic Acids Res. 2015;43:3789–3801. doi: 10.1093/nar/gkv225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Melvin RL, Salsbury FR., Jr Visualizing ensembles in structural biology. J Mol Graphics Modell. 2016;67:44–53. doi: 10.1016/j.jmgm.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Foloppe N, MacKerell AD., Jr All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J Comput Chem. 2000;21:86–104. [Google Scholar]
- 61.MacKerell AD, Banavali NK. All-atom empirical force field for nucleic acids: II. Application to molecular dynamics simulations of DNA and RNA in solution. J Comput Chem. 2000;21:105–120. [Google Scholar]
- 62.MacKerell AD, Banavali N, Foloppe N. Development and current status of the CHARMM force field for nucleic acids. Biopolymers. 2000;56:257–265. doi: 10.1002/1097-0282(2000)56:4<257::AID-BIP10029>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 63.Gmeiner WH, Salsbury FR, Jr, Olsen CM, Marky La. The stability of a model substrate for topoisomerase 1-mediated DNA religation depends on the presence of mismatched base pairs. J Nucleic Acids. 2011;2011:631372. doi: 10.4061/2011/631372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graphics. 1996;14:33–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 65.Lee MS, Salsbury FR, Jr, Brooks CL. Constant-pH molecular dynamics using continuous titration coordinates. Proteins: Struct, Funct Genet. 2004;56:738–52. doi: 10.1002/prot.20128. [DOI] [PubMed] [Google Scholar]
- 66.Harvey MJ, Giupponi G, Fabritiis GD. ACEMD: accelerating biomolecular dynamics in the microsecond time scale. J Chem Theory Comput. 2009;5:1632–1639. doi: 10.1021/ct9000685. [DOI] [PubMed] [Google Scholar]
- 67.Darden T, York D, Pedersen L. Particle mesh Ewald: An N log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089. [Google Scholar]
- 68.Harvey MJ, De Fabritiis G. An implementation of the smooth particle mesh Ewald method on GPU hardware. J Chem Theory Comput. 2009;5:2371–2377. doi: 10.1021/ct900275y. [DOI] [PubMed] [Google Scholar]
- 69.Heyer LJ, Kruglyak S, Yooseph S. Exploring expression data: identification and analysis of coexpressed genes. Genome Res. 1999;9:1106–1115. doi: 10.1101/gr.9.11.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Melvin R, Godwin R, Xiao J, Salsbury FR., Jr Markov cluster analysis in Matlab. figshare. 2015 doi: 10.6084/m9.fig-share.1566809. [DOI] [Google Scholar]
- 71.Lu XJ, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc. 2008;3:1213–27. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu XJ, Olson WK, Bussemaker HJ. The RNA backbone plays a crucial role in mediating the intrinsic stability of the GpU dinucleotide platform and the GpUpA/GpA miniduplex. Nucleic Acids Res. 2010;38:4868–76. doi: 10.1093/nar/gkq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic acids research. 2003;31:5108–21. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu HC, Chung SS, Fornili A, Fraternali F. Anatomy of protein disorder, flexibility and disease-related mutations. Frontiers in Molecular Biosciences. 2015;2:1–8. doi: 10.3389/fmolb.2015.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berman HM. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zirbel CL, Šponer JE, Šponer J, Stombaugh J, Leontis NB. Classification and energetics of the base-phosphate interactions in RNA. Nucleic Acids Res. 2009;37:4898–4918. doi: 10.1093/nar/gkp468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Michaud-Agrawal N, Denning EJ, Woolf TB, Beckstein O. MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. J Comput Chem. 2011;32:2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma H, Proctor DJ, Kierzek E, Kierzek R, Bevilacqua PC, Gruebele M. Exploring the energy landscape of a small RNA hairpin. J Am Chem Soc. 2006;128:1523–1530. doi: 10.1021/ja0553856. [DOI] [PubMed] [Google Scholar]
- 79.Varani G, Cheong C, Tinoco I. Structure of an unusually stable RNA hairpin. Biochemistry. 1991;30:3280–3289. doi: 10.1021/bi00227a016. [DOI] [PubMed] [Google Scholar]
- 80.Varani G. Exceptionally stable nucleic acid hairpins. Annu Rev Biophys Biomol Struct. 1995;24:379–404. doi: 10.1146/annurev.bb.24.060195.002115. [DOI] [PubMed] [Google Scholar]
- 81.Shu Z, Bevilacqua PC. Isolation and characterization of thermodynamically stable and unstable RNA hairpins from a triloop combinatorial library. Biochemistry. 1999;38:15369–15379. doi: 10.1021/bi991774z. [DOI] [PubMed] [Google Scholar]
- 82.Nakano M, Moody EM, Liang J, Bevilacqua PC. Selection for thermodynamically stable DNA tetraloops using temperature gradient gel electrophoresis reveals four motifs: d(cGNNAg), d(cGNABg),d(cCNNGg), and d(gCNNGc) Biochemistry. 2002;41:14281–92. doi: 10.1021/bi026479k. [DOI] [PubMed] [Google Scholar]
- 83.Proctor DJ, Schaak JE, Bevilacqua JM, Falzone CJ, Bevilacqua PC. Isolation and characterization of stable tetraloops with the motif YNMG that participates in tertiary interactions. Biochemistry. 2002;41:12062–12075. doi: 10.1021/bi026201s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.