Abstract
Chronic inflammatory disorders such as rheumatoid arthritis are characterized by excessive pro-inflammatory or “M1” activation of macrophages, the primary cells of the innate immune system. Current treatments include delivery of glucocorticoids (e.g. dexamethasone – Dex), which reduce pro-inflammatory M1 behaviour in macrophages. However, these treatments have many off-target effects on cells other than macrophages, resulting in broad immunosuppression. To limit such side effects, drug-incorporated nano- and microparticles may be used to selectively target macrophages via phagocytosis, because of their roles as highly effective phagocytes in the body. In this study, surface-modified nanodiamond (ND) was explored as a platform for the delivery of dexamethasone to macrophages because of ND’s rich surface chemistry, which contributes to ND’s high potential as a versatile drug delivery platform. After finding that octadecylamine-functionalized nanodiamond (ND-ODA) enhanced adsorption of Dex compared to carboxylated ND, the effects of Dex, ND-ODA, and Dex-adsorbed ND-ODA on primary human macrophage gene expression were characterized. Surprisingly, even in the absence of Dex, ND-ODA had strong anti-inflammatory effects, as determined by multiplex gene expression via NanoString and by protein secretion analysis via ELISA. ND-ODA also inhibited expression of M2a markers yet increased the expression of M2c markers and phagocytic receptors. Interestingly, the adsorption of Dex to ND-ODA further increased some anti-inflammatory effects, but abrogated the effect on phagocytic receptors, compared to its individual components. Overall, the ability of ND-ODA to promote anti-inflammatory and pro-phagocytic behaviour in macrophages, even in the absence of loaded drugs, suggests its potential for use as an anti-inflammatory therapeutic to directly target macrophages through phagocytosis.
Introduction
Macrophages are innate immune cells that exhibit a broad range of behaviours, allowing them to act as key players in all aspects of the immune response, including tissue repair and disease. For example, in the normal healing response to an injury, pro-inflammatory M1 macrophages stimulate the inflammatory cascade, which signals the start of the healing process.1 2–3 days post-injury, M1 macrophages switch to the pro-healing/anti-inflammatory M2 phenotype.2 The M2 phenotype can be further divided into two major subgroups, each playing a distinct role in tissue repair: M2a macrophages, which are involved in tissue synthesis and maturation,3 and M2c macrophages, which are involved in the phagocytosis and clearance of apoptotic cells.4,5 The M2c phenotype is also believed to be involved in tissue remodelling and angiogenesis at early stages of wound healing.5 While it is now understood that macrophages exist in vivo on a spectrum of diverse phenotypes, including hybrid phenotypes, biomaterials that can modulate macrophage phenotype toward a particular set of behaviours would be highly beneficial for orchestrating tissue repair through the body’s natural healing mechanisms.
Chronic inflammatory conditions such as rheumatoid arthritis and chronic diabetic ulcers are associated with excessive M1 activation of macrophages.6–9 These chronic M1 macrophages overproduce inflammatory cytokines, leading to the destruction of surrounding tissues, and, consequently, chronic pain.8 Therefore, strategies that inhibit M1 polarization can be used to treat chronic inflammation.10,11 Moreover, biomaterials that promote the M2a or M2c phenotype may promote specific aspects of tissue repair through the actions of host macrophages. Conversely, an extended presence of M2a macrophages can also be detrimental. As part of the foreign body response to an implanted biomaterial, macrophages exhibiting characteristics of both M1 and M2a phenotypes fuse together to form multinucleated giant cells that promote fibrous capsule formation around the implant.12,13 As a result, the implant is isolated from the rest of the body, thus leading to a lack of biointegration and ultimately failure.14 Therefore, it is essential to strategically engineer biomaterials that promote or inhibit specific phenotypes of macrophages to restore a balance in their behaviour.
Delivery of glucocorticoids like dexamethasone (Dex) has been employed to reduce both M1-mediated inflammation and fibrous capsule formation15–17 as well as enhance phagocytosis of apoptotic cells18,19 and bacteria.20 However, glucocorticoid receptors are found in the cytoplasm of nearly all cells. Consequently, glucocorticoids react with many different cell types and produce off-target effects, including decreased drug accumulation at the affected site and broad suppression of the adaptive immune system by inducing lymphocyte apoptosis.21,22 Therefore, there is a need to develop a targeted drug delivery system that specifically delivers Dex to macrophages in order to effectively increase drug potency and reduce off-target effects. Because macrophages are highly phagocytic cells, delivering these drugs via nano- or microparticles is potentially an efficient way to target macrophages.23
Detonation nanodiamond (ND) is a commercially-available carbon nanomaterial24 that has attracted much attention from the biomedical field because of its many unique material properties, including its small primary particle size (~5 nm), rich surface chemistry, and cytocompatibility with various cell lines.25–27 As a result, ND has successfully demonstrated its ability to mechanically strengthen biopolymers28,29 and function as a platform for the delivery of therapeutics with diverse chemistries.26,30–33 While several studies have investigated the use of drug-loaded ND as an anti-inflammatory agent, the effects of ND on macrophage behaviour are poorly understood; some reports indicate no effects on inflammatory gene expression,34–37 but at least one report shows reduced expression of inflammatory cytokines by murine macrophages.38 The response of human macrophages, which can behave very differently from murine macrophages,39 to ND has not been investigated. Thus, the purpose of this study was to evaluate ND as a platform for the delivery of Dex to macrophages, including its direct effects on human macrophage behaviour.
Carboxylated ND (ND-COOH) has been previously used as a drug delivery vehicle for Dex.32,37 Huang et al.37 assessed the effects of Dex-adsorbed ND-COOH-embedded polymer film on M1-like RAW 264.7 murine macrophages. Their results showed that Dex’s bioactivity was retained, in that the expression of several pro-inflammatory genes (Tnf, Il6, and Nos2) was significantly downregulated, indicating that Dex’s bioactivity was retained. The effects of the recently described octadecylamine-functionalized ND (ND-ODA),40 with or without Dex loading, are yet to be explored. Compared to ND-COOH, which is negatively charged and hydrophilic, ND-ODA is slightly positively charged and hydrophobic,40 and thus would be expected to have major effects on both Dex adsorption as well as macrophage behaviour.41,42 Indeed, uptake of highly negatively charged microparticles has been shown to induce macrophage apoptosis.43 Thus, the goal of this study was to determine how ND-ODA and Dex-loaded ND-ODA affect human macrophages, including those that are first polarized to phenotypes associated with tissue healing and disease (M1 and M2a). We investigated the adsorption of Dex onto both ND-COOH and ND-ODA, and then characterized the response of primary human M0, M1, and M2a macrophages to Dex, ND-ODA, and Dex-loaded ND-ODA using gene expression of a panel of macrophage phenotype markers as well as angiogenesis-related genes. Finally, a subset of our findings was confirmed with protein secretion analysis.
Methods
Experimental design
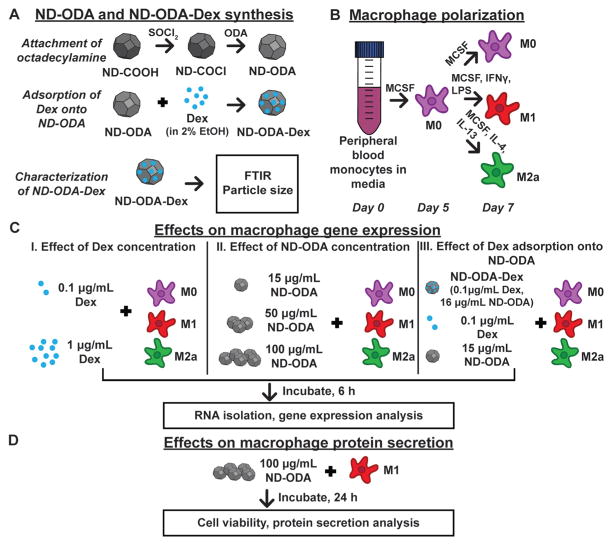
Fig. 1 illustrates the overall experimental design of this study. First, two ND platforms, ND-COOH and ND-ODA, were synthesized and characterized for their Dex adsorption capacities (Fig. 1A). After confirming ND-ODA’s adsorption properties, particle size and surface chemistry were analysed for the ND-ODA-Dex complexes.
Fig. 1.
Overview of the experimental design. (A) ND-ODA was synthesized by first chlorinating carboxylated ND, and then replacing the active chlorine group with ODA. Then, Dex was adsorbed onto ND-ODA. The ND-ODA-Dex complexes were then further characterized by FTIR and particle size analysis. (B) Peripheral blood monocytes were differentiated into M0 macrophages. At day 5, the cells were either kept as M0 or further polarized into M1 and M2a phenotypes in ultra-low attachment well-plates. (C) After polarization was complete at day 7, either Dex (at low and high concentrations), ND-ODA (at low, medium, or high concentrations), or Dex-loaded (ND-ODA-Dex; at low Dex and low ND concentrations) was added to cells and incubated for 6 h prior to RNA isolation and gene expression analysis. (D) To confirm the gene expression data on the protein level, the high dose of ND-ODA was incubated with M1 macrophages for 24 h. Then, the cells were counted and the conditioned media was analysed for protein secretion.
Then, human monocytes derived from peripheral blood were differentiated into unactivated macrophages (M0) and either kept as M0 or polarized into M1 or M2a (Fig. 1B). Three studies were simultaneously performed to determine the effects of Dex and/or ND-ODA on macrophage gene expression (Fig. 1C). In the first study, the effect of Dex concentration was explored by separately adding low and high doses of Dex to M0, M1, and M2a macrophages. In the second study, the effect of ND-ODA dose was similarly studied, using low, medium, and high doses of ND-ODA. Finally, in the third study, Dex-adsorbed ND-ODA was evaluated using the low doses of both Dex and ND-ODA from the first two studies. The results of the gene expression analysis represent the full-scale investigation using a single donor of human macrophages after pilot studies showed similar trends of both Dex and ND-ODA on macrophage behaviour using multiple donors of human macrophages.
The expression levels of 14 genes indicative of macrophage phenotype and that encode proteins with roles in angiogenesis and tissue healing were analysed to determine changes in macrophage behaviour. Because of their roles in propagating inflammation, TNF, IL1B, and CCR7 were selected as M1 markers.44 TIMP3, MRC1, and CCL22 were selected as M2a markers because of their involvement in tissue remodelling and inflammation resolution.45,46 Additionally, 5 M2c markers were used. CD163 and MERTK were selected because of their roles in phagocytosis.4,17 MMP7 and MMP8 were selected because of their roles in extracellular matrix remodelling.47,48 VCAN, which contributes to cell adhesion properties, was also selected as an M2c marker.5 Because of their roles in angiogenesis and tissue healing, VEGFA, PDGFB, and MMP9 were also investigated. Additionally, VEGFA can be classified as an M1 marker; PDGFB can be classified as an M2a marker.45
In order to confirm a subset of the gene expression results on the protein level, the effects of ND-ODA on macrophage protein secretion was investigated using macrophages derived from an additional donor (Fig. 1D). After adding the high dose of ND-ODA to M1 macrophages, culture media was analysed after 24 h for secretion of pro-inflammatory factors tumor necrosis factor-α (TNF-α) and interleukin-1β (IL-1β). Finally, secretion of the anti-inflammatory factors IL-10 and transforming growth factor-β1 (TGF-β1) was analysed as part of a preliminary investigation into the mechanisms behind observed changes in macrophage behaviour.
Synthesis of ND-COOH and ND-ODA
As-received UD90 grade ND (donated by sp3 Diamond Technologies) was first air oxidized at 425 °C for 5 h and then acid purified by refluxing with an nitric acid/hydrochloric acid mixture at 90 °C for 24 h in order to simultaneously decrease the sp2-bonded carbon content and attach carboxyl (COOH) surface groups (ND-COOH).49 To produce octadecylamine-functionalized ND (ND-ODA), 1.5 g ND-COOH was reacted with thionyl chloride in presence of anhydrous N,N-dimethylformamide (DMF), which is as a catalyst for this reaction, at 70 °C for 24 h to produce ND-COCl. The highly reactive –Cl groups were then replaced with –NH2 groups, connected to a long aliphatic chain, through reacting with 1 g octadecylamine at 90 °C for 96 h, and then rinsing with methanol to remove excess reactant.40 All chemical reagents were purchased from Sigma Aldrich.
Adsorption activity of Dex onto ND-COOH and ND-ODA
To characterize adsorption activity, 5 mL samples of increasing concentrations of dexamethasone (Sigma Aldrich, D4902) in 2% ethanol were adsorbed to ND. Because of Dex’s low solubility even with the addition of ethanol, the range of initial dexamethasone concentrations was between 5 and 120 μg mL−1. ND-COOH or ND-ODA (2 ± 0.1 mg) was added to solutions of varied Dex concentrations, and bath sonicated for ~1 min in order to break up large aggregates. Then, the samples were placed on an orbital shaker in the dark at room temperature and were left to shake overnight at 200 rpm (ThermoFisher MaxQ 4450). The samples were then centrifuged at 4000 rpm (3220g, Eppendorf 5810R) for 2 h. The supernatant, which consisted of unbound Dex, was collected for UV-visible spectrophotometry (UV-VIS) analysis (Thermoscientific Nanodrop 1000, 243 nm), and the concentration was calculated from the measured absorbance using a calibration curve. The calculated concentrations of non-adsorbed drug in the supernatant, also known as the equilibrium concentrations, were subtracted from the initial concentrations to determine the mass of the drug adsorbed. Then, the adsorption activities for each sample were calculated by dividing the individual calculated masses of the drug (mg) adsorbed by the known masses of ND (g) that were used to adsorb the drug in each sample. To construct the adsorption isotherms, the experimental adsorption activities for each concentration of Dex were plotted for each corresponding equilibrium concentration. These data points were subjected to fitting using two common adsorption isotherm models, Langmuir and Freundlich, in order to determine the adsorption mechanism (mono- or multilayer, hetero- or homogeneous adsorption, etc.).50,51 The models differ with respect to their key assumptions and their mathematical representations. The Langmuir isotherm assumes monolayer adsorption with a distinct number of available adsorption sites, which are all equivalent and independent. It also assumes that the adsorbate (drug) is immobilized upon contact. Mathematically, Langmuir adsorption is represented as:
where A is the calculated adsorption activity for each equilibrium concentration (Ceq), while Amax is the predicted maximum possible adsorption capacity for a single monolayer, and KL is a predicted value corresponding to the bond strength between the adsorbent and adsorbate.50 On the other hand, the empirical Freundlich isotherm assumes that adsorption can be either multi- or monolayer, and that the adsorption sites are heterogeneous. Mathematically, Freundlich adsorption is represented as:
where K and n are arbitrary constants that do not provide any information about the adsorption capacity of a monolayer or its bond strength.51 The experimental adsorption isotherm data were fit to both Langmuir and Freundlich models by means of non-linear least-squares fitting using Microsoft Excel Data Solver. Using Excel, Pearson’s correlation coefficient (R) was also calculated and used to determine goodness of fit.
Characterization of ND-ODA and ND-ODA-Dex
Because of its superior adsorption properties, further analysis was carried out on ND-ODA and maximally loaded (~25 mg Dex per g ND-ODA) Dex-adsorbed ND-ODA complexes (ND-ODA-Dex). FTIR (PerkinElmer Spectrum One) analysis of ND-COOH, ND-ODA, and ND-ODA-Dex was performed in ambient environment. All samples were freeze-dried, finely ground with potassium bromide (KBr) powder, and pressed into a pellet prior to FTIR analysis. Dynamic light scattering (DLS) was used to measure particle size (Malvern Zetasizer Nano ZS). FTIR was also used to qualitatively monitor batch-to-batch variations in functional group modification and drug adsorption, and the small variations were found to insignificantly impact drug adsorption capacity. Prior to DLS analysis, ND-ODA and maximally-loaded ND-ODA-Dex were dispersed in PBS, bath sonicated for 1 min, and then filtered using a 10 μm cell strainer (pluriSelect) in order to remove aggregates that are too large for phagocytosis.52 The average particle size distribution was calculated from 5 repeated experiments.
Differentiation and polarization of primary human macrophages
Primary human monocytes derived from peripheral blood were purchased from the Human Immunology Core at the University of Pennsylvania (Philadelphia, PA). Cells from one donor were used for gene expression analysis, and follow up studies for confirmation on the protein level were conducted using cells from a different donor. Monocytes were differentiated into macrophages (M0, M1, and M2a) as previously described.45 By Briefly, monocytes were cultured in ultra-low attachment flasks with RPMI 1640 media supplemented with 10% heat-inactivated human serum (from human male AB plasma, Sigma Aldrich), 1% penicillin/streptomycin, and 20 ng mL−1 recombinant human macrophage colony stimulating factor (MCSF). Cells were incubated at 37 °C at 5% CO2. On day 3, the media was refreshed. On day 5, the M0 macrophages were gently scraped, counted, and plated with fresh MCSF-supplemented media into 24 well plates at a concentration of 106 cells per mL. Polarization was then performed by adding 100 ng mL−1 recombinant human interferon-gamma (IFNγ) and 100 ng mL−1 lipopolysaccharide (LPS) for M1 or 40 ng mL−1 recombinant human interleukin-4 (IL-4) and 20 ng mL−1 recombinant human interleukin-13 (IL-13) for M2a.45 After 2 days of polarization, the media was replaced with cytokine-free media containing ND samples as described below. All cytokines were purchased from PeproTech.
Treatment of macrophages with Dex, ND-ODA, and ND-ODA-Dex
To determine the effect of Dex on macrophages, a stock solution of 20 μg mL−1 in cytokine-free media with 2% ethanol was prepared. Then, the concentrations were diluted in media to either 1 μg mL−1 (high Dex) or 0.1 μg mL−1 (low Dex), and added to the macrophages for 6 h (n = 4). To determine the effect of bare ND-ODA on macrophages, 330 μg mL−1 ND-ODA was dispersed in cytokine-free media, bath sonicated for 1 min, and filtered using a 10 μm cell strainer. In order to calculate the average mass loss after filtering ND-ODA and ND-ODA-Dex, the mass of the cell strainer was weighed before and after filtering, and the difference in mass was compared to the original mass of ND-ODA or ND-ODA-Dex. ND-ODA concentration was assumed to be ~100 μg mL−1 after filtering, due to the average mass loss calculated as ~70%. This stock solution was either used directly (100 μg ml−1, high ND-ODA) or diluted in cytokine-free media to produce concentrations of 50 μg mL−1 (medium ND-ODA) and 15 μg mL−1 (low ND-ODA). These high, medium, and low doses of ND-ODA were added to the macrophages for 6 h prior to RNA extraction for gene expression analysis (n = 3–4).
To determine the effect of ND-ODA-Dex on macrophages, ND-ODA-Dex samples were prepared to concentrations of low ND (15 μg mL−1) and low Dex (0.1 μg mL−1). Briefly, 4 mg ND-ODA was added to 10 μg mL−1 Dex in 2% ethanol, bath sonicated, and mixed overnight. Using UV-VIS as described above, the ND-ODA-Dex were determined to have an adsorption activity of ~6 mg Dex per g ND-ODA. Therefore, 1 mg of ND-ODA has approximately 6 μg Dex adsorbed to its surface. After separating the ND-ODA-Dex complexes by centrifugation and freeze-drying, 2 mg ND-ODA-Dex was rinsed once in PBS to ensure all unbound Dex had been removed. To make a dispersion with 0.1 μg mL−1 Dex, the 2 mg ND-ODA-Dex were dispersed in 37.5 mL, bath sonicated for 1 min, and filtered using a 10 μm cell strainer. With 70% mass loss, the final concentration of DEX was 0.1 μg mL−1 and the final concentration of ND-ODA was 16 μg mL−1. This dispersion was then added to the macrophages (n = 3–4). Cytokine-free media without ND-ODA was added to the controls (n = 3). Following 6 h incubation, all macrophage samples were scraped, collected, and centrifuged. The supernatant was then discarded, and the cells were placed in 1 mL TRIzol (Invitrogen, USA) and stored at −80 °C for later RNA extraction. RNA was isolated using a TRIzol:chloroform method53 in combination with the RNeasy Micro Kit (Qiagen) according to the manufacturer’s instructions.
NanoString analysis
Expression of a custom-designed set of 20 genes (14 markers of the M1, M2a, and M2c phenotypes identified and validated in ref. 5 and 45 as well as angiogenic genes plus 6 housekeeping genes, ESI Table 1†) was analyzed using a NanoString nCounter Analysis System (NanoString Technologies). Prior to analysis, Agilent RNA 6000 Nano Kits were used in combination with an Agilent 2100 Bioanalyzer to determine RNA concentration. A standardized mass of RNA (100 ng) RNA from each sample was mixed with reporter/capture probe pairs for each gene, which included 8 negative and 6 positive controls, according to the manufacturer’s instructions. The raw gene counts were normalized across the entire data set according to NanoString’s nCounter Expression Data Analysis Guide. Briefly, the average geometric mean of all of the positive control counts was divided by the individual geometric means of the positive controls for each sample to determine sample-specific scaling factors. The data were then corrected by multiplying sample-specific scaling factors by the individual counts for each gene in each sample. The data were then log-transformed to achieve normal distribution, and the average log-transformed negative control values for each lane were subtracted from each sample. Negative values were regarded as zero. Statistical analysis was performed in GraphPad Prism using a one-way ANOVA and Tukey’s multiple comparisons test. In order to apply one-way ANOVA to discern significant effects within phenotypes, the log-transformed data were first checked for normality by visualising the sample distribution. Then, the Brown–Forsythe’s test was used to confirm that the variances were homogenous. Finally, Tukey’s multiple comparisons test was applied to discern significant differences between individual sample sets at a significance (alpha) level of p < 0.01. Data are represented as mean ± standard deviation (SD).
Cell viability and protein secretion analysis
To validate a subset of the gene expression results on the protein level, primary human M1 macrophages were cultured with 100 μg mL−1 ND-ODA (the high dose) as described above and compared to M0 and M1 controls (n = 4 per group). Cells (obtained from a different donor than the gene expression experiments) were incubated at 37 °C at 5% CO2 in 1 mL cytokine-free media with or without ND-ODA as indicated for 24 h. Then, cells were scraped from the plate and collected. A small aliquot was used to count viable cells using the trypan blue exclusion assay.54 To collect the conditioned media for protein secretion analysis, the remainder of the cell suspension was centrifuged for 7 min at 400g, and supernatant was collected. Protein content in the conditioned media from each sample group (M0, M1, M1 + ND-ODA) was analysed using enzyme-linked immunosorbent assays (ELISAs) according to the manufacturer’s instructions for human interleukin-10 (IL-10), interleukin-1β (IL-1β), tumor necrosis factor-α (TNF-α) (PeproTech, Rocky Hill, NJ, USA), and transforming growth factor-β1 (TGF-β1) (Affymatrix eBiosciences, Wein, Austria). Statistical analysis was performed using an unmatched one-way ANOVA with a Tukey’s post-hoc multiple comparisons analysis. p < 0.05 was considered significant.
Results
Adsorption activity of Dex onto ND-ODA
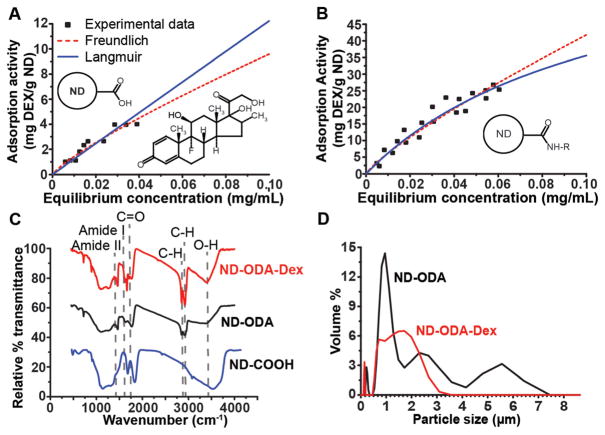
The experimental results for ND-COOH (Fig. 2A) and ND-ODA (Fig. 2B) were in agreement with both the Langmuir and Freundlich isotherm models, as determined by the Pearson’s correlation coefficients. While we cannot draw any definitive conclusions about the adsorption mechanisms, we can still use this information to compare the predicted adsorption properties between ND-COOH and ND-ODA. Since empirical Freundlich’s fit parameters do not reveal any information about the adsorption mechanism, the Langmuir fit parameters for adsorption and bond strength (Amax and KL respectively) were used to compare the adsorption properties of ND-COOH and ND-ODA. In comparing the predicted strength of binding (KL) for the Langmuir model (Table 1), Dex showed markedly stronger adsorption to ND-ODA compared to ND-COOH. Conversely, ND-COOH showed a much higher theoretical monolayer adsorption capacity than ND-ODA. However, it is important to note that this theoretical monolayer adsorption capacity cannot be realized in this system because of the limited solubility of Dex.
Fig. 2.
(A, B) Adsorption isotherms of dexamethasone (Dex) onto ND-COOH (A) and ND-ODA (B), fit to Langmuir and Freundlich isotherm models. The chemical structure of Dex is shown as an inset in A. (C) FTIR spectra showing the successful carboxylation of ND (ND-COOH), ODA attachment to ND (ND-ODA), and adsorption of Dex onto ND-ODA (ND-ODA-Dex). (D) Particle size distribution of ND-ODA and ND-ODA-Dex after being bath sonicated for 1 min and filtered using a 10 μm cell strainer.
Table 1.
Langmuir and Freundlich isotherm fit parameters. Calculated fit parameters and corresponding Pearson correlation coefficients for Langmuir and Freundlich adsorption isotherms fit to Dex adsorption onto ND-COOH and ND-ODA
| ND type | Langmuir isotherm | Freundlich isotherm | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Amax (mg g−1) | KL (mL g−1) | R | Kf | n | R | |
| ND-COOH | 1232.6 | 0.1 | 0.95 | 64.5 | 0.83 | 0.96 |
| ND-ODA | 75.1 | 9.0 | 0.96 | 282.4 | 0.83 | 0.96 |
FTIR of ND and ND-Dex
In comparing the FTIR spectra for ND-COOH and ND-ODA, the reduction of O–H bonds (~3400 cm−1) and the disappearance of C═O bonds (~1700 cm−1) in addition to the rise of amide I (1650 cm−1), amide II (1550 cm−1), and C–H (~2800–3000 cm−1) bonds on ND-ODA indicate that octadecylamine (-ODA) successfully replaced the –COOH groups on ND-COOH (Fig. 2C). FTIR analysis of ND-ODA-Dex showed that Dex was successfully adsorbed onto ND-ODA because of the appearance of C═O unconjugated ketone bonds (~1700 cm−1), strengthening of C–H bonds (~2800–3000 cm−1), and the narrowing of the–OH stretch (~3400 cm−1) (Fig. 2C).
Particle size of ND-ODA and ND-ODA-Dex
Although the combined use of bath sonication and the 10 μm cell strainer removed large aggregates, particle size analysis of ND-ODA post-treatment revealed a multi-modal distribution of aggregates up to 7 μm in diameter, with the largest population centered around 1 μm (Fig. 2D). However, Dex adsorption seemed to break apart the larger (>3 μm) aggregates, instead forming a prominent, broad population centered around 1–2 μm. Additionally, both materials showed the presence of a smaller population centered around 150 nm.
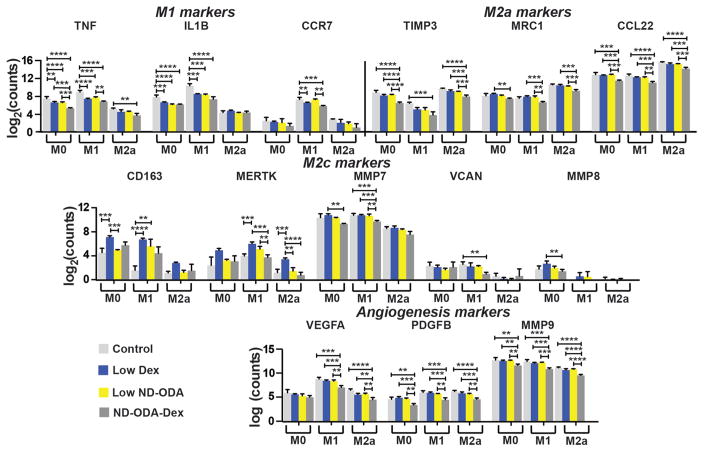
Effects of Dex on macrophage gene expression
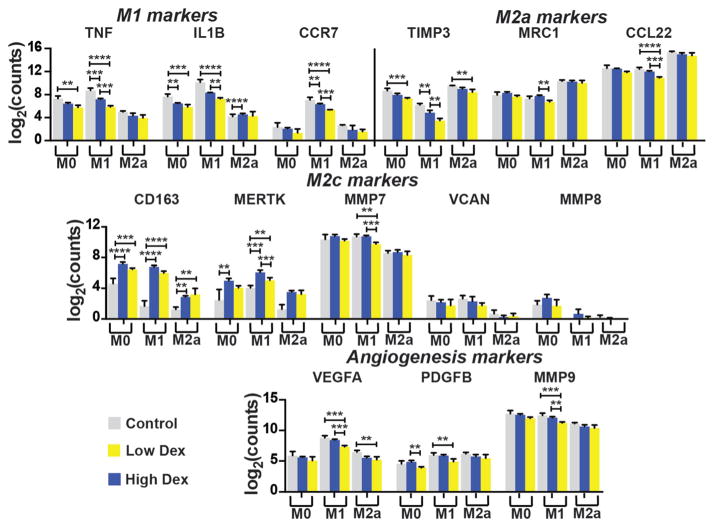
In keeping with its known anti-inflammatory effects, Dex treatment resulted in the dose-dependent reduction of pro-inflammatory M1-associated genes, including TNF, IL1B, and CCR7, in M0 and M1 macrophages (Fig. 3). Dex also decreased expression of M2a markers TIMP3, CCL22, and MRC1, in M1 macrophages. Dex treatment also caused the upregulation of M2c-associated CD163 and MERTK, which are both receptors involved in phagocytosis, in all three phenotypes, although these effects were not dose-dependent. Dex treatment did not significantly affect expression of non-phagocytosis-related M2c markers MMP7, MMP8, and VCAN, with the exception of small yet significant decreases in MMP7 expression by M1 macrophages. Finally, with respect to the potent angiogenic factors VEGFA, PDGFBB, and MMP9, the high dose of Dex decreased expression, especially in M1 macrophages.
Fig. 3.
Effect of Dex on macrophage gene expression. Log-transformed data are presented as mean ± SD. n = 3–4 from a single donor. **p < 0.01, ***p < 0.001, ****p < 0.0001. Significant differences in marker expression between M0, M1, and M2a groups are not noted for the sake of clarity.
Effects of ND-ODA on macrophage gene expression
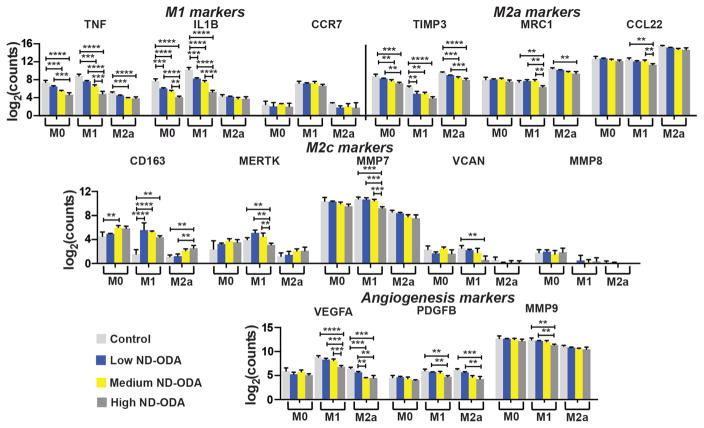
Interestingly, treatment with ND-ODA caused significant dose-dependent downregulation of the genes encoding pro-inflammatory cytokines TNF and IL1B in both M0 and M1 macrophages (Fig. 4). ND-ODA also significantly reduced expression of M2a-associated TIMP3 in all three macrophage phenotypes. The highest dose of ND-ODA reduced expression of the other M2a markers MRC1 and CCL22, especially in M1 macrophages. In contrast, ND-ODA treatment resulted in increased M2c-associated CD163 expression, with the effect most pronounced in M1 macrophages. Furthermore, high ND-ODA caused a significant downregulation of M2c markers MMP7 and VCAN in M1 macrophages. The high dose of ND-ODA also caused a significant reduction in angiogenic factors VEGFA in M1 macrophages, as well as PDGFB and MMP9 in both M1 and M2a macrophages.
Fig. 4.
Effect of ND-ODA on macrophage gene expression. Gene expression of markers associated with M1, M2a, and M2c macrophages and angiogenesis were analysed. Log-transformed data are presented as mean ± SD. n = 3–4 from a single donor. **p < 0.01, ***p < 0.001, ****p < 0.0001. Significant differences in marker expression between M0, M1, and M2a groups are not noted for the sake of clarity.
Effect of ND-ODA-Dex on macrophage gene expression
The effect of ND-ODA-Dex on macrophage gene expression was compared to the sum of its parts: the low dose of Dex and the low dose of ND-ODA (Fig. 5). Compared to both low Dex and low ND-ODA, ND-ODA-Dex further reduced TNF expression in M0 macrophages. The addition of DEX to ND-ODA also increased the effect of ND-ODA on downregulation of TNF expression in M1 macrophages. The combination of Dex and ND-ODA did not significantly affect the ability of either individual component to further reduce IL1B expression, but ND-ODA-Dex did significantly reduce CCR7 expression compared to low ND-ODA in M1 macrophages. Compared to both low Dex and low ND-ODA, ND-ODA-Dex also significantly reduced expression of the M2a markers TIMP3 and CCL22. While downregulation of TIMP3 was only significant in M0 and M2a macrophages, downregulation of CCL22 was significant in all three phenotypes. Surprisingly, unlike its components, ND-ODA-Dex did not increase expression of the M2c-associated phagocytic markers CD163 or MERTK in any macrophage phenotype compared to untreated controls. However, ND-ODA-Dex did significantly decrease expression of the M2c marker MMP7 in M1 macrophages and of the angiogenic factors VEGFA, PDGFB, and MMP9 in both M0 and M1 macrophages compared to both low Dex and low ND-ODA.
Fig. 5.
Effect of ND-ODA-Dex on macrophage gene expression. Gene expression of markers associated with M1, M2a, and M2c macrophages and angiogenesis were analysed. Log-transformed data are presented as mean ± SD. n = 3–4 from a single donor. **p < 0.01, ***p < 0.001, ****p < 0.0001. Significant differences in marker expression between M0, M1, and M2a groups are not noted for the sake of clarity.
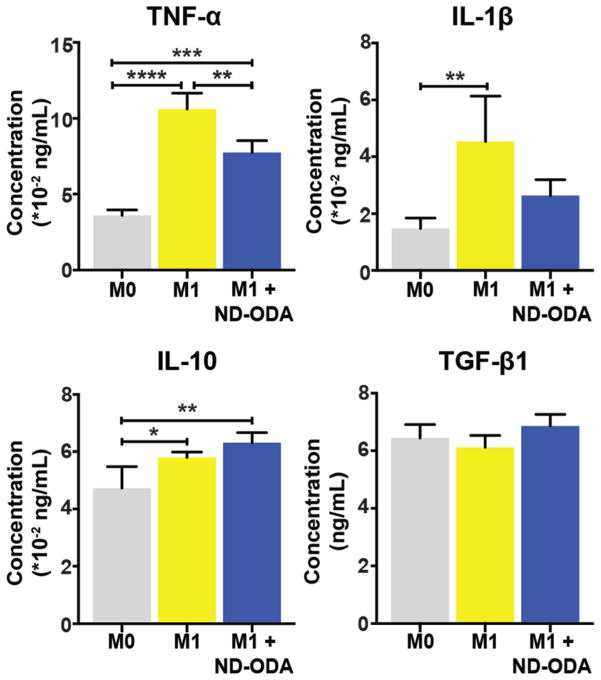
Effect of ND-ODA on protein secretion by M1 macrophages
As expected, M1 macrophages increased secretion of pro-inflammatory cytokines TNF-α and IL-1β compared to M0 (Fig. 6). The addition of 100 μg mL−1 ND-ODA caused a significant reduction in secretion of TNF-α by M1 macrophages, in keeping with gene expression results. IL-1β secretion was also reduced but this result was not significant. Secretion of the anti-inflammatory factors IL-10 and TGF-β1 was slightly increased due to the addition of ND-ODA, though these results were not significant. Cell viability tests confirmed that the numbers of cells in each condition were not different from one another, suggesting that the observed effects were due to changes in inflammatory behaviour, not cell number (ESI Fig. 1†). Cell viability of all groups was >95%, confirming that ND-ODA is not cytotoxic.
Fig. 6.
Protein content after 24 h of culture in conditioned media. Data are represented as mean ± SD of all experimental replicates (n = 4 per group). *p < 0.01, **p < 0.001, ***p < 0.0001, ****p < 0.00001.
Discussion
Overall, we present three major findings from this study: (1) the attachment of octadecylamine to the ND surface enhanced its adsorption activity for Dex; (2) even in the absence of Dex, ND-ODA showed anti-inflammatory effects on primary human macrophages in terms of gene expression and protein secretion and increased the gene expression of phagocytic receptors (Table 2); and (3) the combination of Dex and ND-ODA resulted in enhanced anti-inflammatory effects on gene expression, but abrogated effects on genes associated with phagocytosis. This study represents the first report of interactions between macrophages and ND-ODA. We are especially intrigued by ND-ODA’s anti-inflammatory properties. These results suggest that ND-ODA holds potential to be utilized as an anti-inflammatory therapeutic to directly target macrophages through phagocytosis, thus avoiding off-target effects presented by Dex or other anti-inflammatory drugs. Furthermore, patients have also sometimes exhibited resistance to Dex.55 Therefore, using ND-ODA as a macrophage-targeted drug-free platform has additional potential to avoid drug resistance. In addition, the potential anti-angiogenic effects of ND-ODA suggest that it would be particularly useful for inflammatory conditions characterized by excessive pathological angiogenesis, like rheumatoid arthritis.56 ND-ODA also inhibited expression of M2a markers, suggesting potential as a treatment for fibrosis.
Table 2.
Effects of Dex, ND-ODA, and ND-ODA-Dex. Summary of the effects of Dex, ND-ODA, and Dex on macrophage gene expression
| Dex | ND-ODA | ND-ODA-Dex | |
|---|---|---|---|
| M1 markers | ↓ | ↓ | ↓ |
| M2a markers | ↓ | ↓ | ↓ |
| Phagocytic M2c markers | ↑ | ↑ | — |
| Other M2c markers | — | ↓ | — |
| Angiogenesis markers | ↓ | ↓ | ↓ |
↑ (upregulated), ↓(downregulated), — (no change), compared to untreated controls.
ND-ODA, a relatively newly described form of ND,40 was selected as the optimal platform for the delivery of Dex because of its higher binding capacity and higher monolayer adsorption capacity in Dex’s solubility range, compared to ND-COOH. ND-ODA’s superior adsorption properties could be due to the fact that Dex and ND-ODA are both hydrophobic and may preferentially bond to each other over water molecules. While the Langmuir model predicted a higher theoretical monolayer adsorption capacity for ND-COOH over ND-ODA (Table 1), this effect could not be confirmed because it was only true for concentrations above Dex’s solubility. ND-ODA’s ability to adsorb up to 25 mg Dex per g ND-ODA allows for the delivery of clinically used concentrations of Dex. Depending on the severity of the inflammatory disease, up to 9 mg of Dex may be delivered intravenously to adult humans.57 If the maximum loading capacity is utilized, 9 mg Dex can be adsorbed onto 360 mg ND-ODA, which would correspond to 6 mg kg−1 for a 62 kg person. This dose is in line with other nanoparticle treatments used in clinical trials.58
Because of their foreign nature, nanoparticles and microparticles are immediately recognized by the immune system when introduced into the body. In fact, in order to avoid this, researchers have attempted to mask nanoparticles by coating them with polyethylene glycol, so that they are hydrophilic and minimize adsorption of macrophage-recognizing proteins in the blood,42 or even by coating with cell membranes to mimic red blood cells.59 However, the goal of immunomodulatory treatments is to directly target the cells of the immune system, especially macrophages, making nanoparticles and microparticles an ideal targeting strategy. While many different types of nanoparticles have been shown to be pro-inflammatory to macrophages,60–63 several studies have indicated that ND-COOH does not induce inflammation in murine macrophages. For example, Huang, H. et al.34 added ND-COOH to the murine RAW 264.7 cell line in concentrations ranging from 25 to 100 μg mL−1 and noted that there were no significant changes in pro-inflammatory Tnf, Il6, or Nos2 expression. Likewise, K.-J. Huang et al.35 noted ND-COOH did not affect expression of pro-inflammatory Tnf, Il1b, or Il6 in RAW 264.7 murine macrophages, and did not affect TNF-α levels in the blood of C57BL/6 mice. Finally, Thomas et al.38 showed that treatment of unactivated RAW 264.7 murine macrophages with ND-COOH of different sizes resulted in decreased expression of Tnf, Il1b, Vegfa, and Pdgfb. While the authors attributed these effects to the likelihood of cell death, we found no effects of ND-ODA treatment on macrophage cell viability in our study. Moreover, instead of a uniform decrease in gene expression, we observed differential effects across multiple macrophage phenotypes. Thus, our study showed that ND-ODA is anti-inflammatory, which was confirmed on the protein level with reduced secretion of TNF-α. It is possible that a major effect on IL-1β secretion was not detected because it is protected from extracellular release unless there is a large inflammatory stimulus.64 However, our results still strongly suggest that ND-ODA has distinct anti-inflammatory effects, which may be useful as a therapeutic agent in mediating inflammation.
While our future studies will focus on systematically investigating the underlying mechanisms, we used protein secretion analysis to preliminarily investigate two potential mechanisms behind ND-ODA’s anti-inflammatory effects: (1) its recognition as apoptotic cells or cell debris, thus mimicking the process of efferocytosis; or (2) recognition of adsorbed haemoglobin via CD163 receptors, thus triggering increased secretion of the anti-inflammatory cytokine IL-10. One of macrophages’ key roles in wound resolution includes clearing apoptotic cells from the body, in a process called efferocytosis. During this process, macrophages phagocytose the apoptotic cells, triggering a switch to an “M2-like” phenotype characterized by secretion of anti-inflammatory cytokines such as interleukin-10 (IL-10) and transforming growth factor-β1 (TGF-β1).65 Therefore, impaired clearance of apoptotic cells leads to increased pro-inflammatory cytokine levels, and, consequently, chronic inflammation.66,67 Although the ND-ODA aggregates are micron-sized, ranging from to 1 to 7 μm, which falls directly in the size range of human cells of 1–10 μm, and display hydrocarbon chains that may resemble the apoptotic cell marker phosphatidylserine (PS),68 the macrophages did not significantly increase secretion of IL-10 or TGF-β1 when incubated with ND-ODA, suggesting that ND-ODA does not trigger a process resembling efferocytosis. Another possibility is that the hydrophobic hydrocarbon chains on ND-ODA increased adsorption of the relatively hydrophobic serum proteins, including haemoglobin, the target of the CD163 receptor. Although the concentration of haemoglobin in the human serum used in the cell culture media was three orders lower than the concentration in human blood,69 it is possible that haemoglobin adsorbed to ND-ODA surface, thus triggering uptake through the CD163 receptor, which has a strong affinity for haemoglobin-haptoglobin complexes in the blood as well as haemoglobin.70 Upon recognizing these haemoglobin-ND-ODA complexes, CD163 expression may have been upregulated,71 resulting in increased IL-10 production72 as well as the suppression of pro-inflammatory genes, such as TNF and IL1B.73 Because IL-10 secretion by macrophages was only moderately increased by ND-ODA treatment, the results do not strongly support this theory either. Thus, future experiments will focus on investigating the mechanisms by which ND-ODA exerts anti-inflammatory effects on human macrophages.
Surprisingly, compared to Dex, ND-ODA-Dex did not further increase the expression of either M2c-associated phagocytic genes compared to untreated controls. This major difference may be due to a decrease in bioactivity of Dex following adsorption to ND-ODA. In this case, the interactions between Dex and the glucocorticoid receptors would be dependent on Dex release from the ND-ODA-Dex complexes in the cytoplasm. It is possible that only very low amounts of Dex were released from the ND-ODA-Dex complexes in the timeframe of this study, resulting in a reduced effect. Additionally, compared to ND-ODA, ND-ODA-Dex has a smaller aggregate size (~1–2 μm) and different surface chemistry (due to the adsorption) compared to ND-ODA. Because of these differences between ND-ODA and ND-ODA-Dex, it is possible that different plasma proteins were adsorbed, causing macrophage recognition and uptake through different pathways (e.g. CD64-mediated uptake, in which macrophages recognize and bind to immunoglobulin-G;74 or scavenger receptor A-mediated uptake, which macrophages primarily use to recognize low-density lipoproteins75), thus resulting in differences in phenotype. Although the results strongly suggest that ND-ODA has anti-inflammatory properties, there were several limitations to this study. First, we chose to analyse a limited set of 14 genes and 4 proteins to characterize changes in macrophage behaviour. These selected genes and proteins represent a small subset of the hundreds to thousands of changes that can occur with changes in macrophage phenotype. In future studies, a larger panel of genes and proteins will be analysed and should include those that provide insight into the potential mechanisms of the anti-inflammatory action. In addition, we were unable to control ND-ODA particle size, since deaggregation methods typically result in low yields or high concentrations of contaminants.76,77 To gain a better understanding of ND–macrophage interactions, future studies should focus on discerning the effect of aggregate size, as well as surface chemistry, on macrophage uptake and inflammatory behaviour. Nonetheless, this study provides proof of concept that ND-ODA can directly cause anti-inflammatory effects on primary human macrophages, in addition to acting as a drug delivery platform to shuttle drugs directly to the inside of macrophages.
Conclusions
We have characterized changes in the gene expression of three macrophage phenotypes (M0, M1, and M2a) in response to Dex, ND-ODA, and ND-ODA-Dex, and found that ND-ODA had strong anti-inflammatory and pro-phagocytic effects on macrophage gene expression. The anti-inflammatory activity of ND-ODA, even in the absence of Dex, was confirmed on the protein level. These findings suggest that ND-ODA has the potential to be used as an anti-inflammatory therapeutic that can target macrophages by phagocytosis. Furthermore, use of inherently anti-inflammatory biomaterials would reduce the negative effects caused by many anti-inflammatory drugs, including immunosuppression and increased drug resistance. This knowledge, along with determination of the mechanism through which ND-ODA has anti-inflammatory effects on macrophages, holds potential to inform next generation biomaterial design strategies to mediate inflammation in chronic inflammatory diseases and the foreign body response.
Supplementary Material
Acknowledgments
This work was supported in part by NHLBI R01 HL130037 to KLS. AEP is grateful for a National Science Foundation Graduate Research Fellowship. We would also like to thank Taron Makaryan for performing FTIR analysis on the samples and Dr Sergey Balashov at the Genomic Lab Core of Drexel University College of Medicine’s Clinical and Translational Research Institute for technical assistance with the Agilent 2100 Bioanalyzer as well as Nanostring nCounter Analysis System.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c7bm00294g
Conflicts of interest
There are no conflicts of interest to declare.
References
- 1.Lucas T, Waisman A, Ranjan R, Roes J, Krieg T, Müller W, Roers A, Eming SA. J Immunol. 2010;184:3964–3977. doi: 10.4049/jimmunol.0903356. [DOI] [PubMed] [Google Scholar]
- 2.Arnold L, Henry A, Poron F, Baba-Amer Y, Van Rooijen N, Plonquet A, Gherardi RK, Chazaud B. J Exp Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song E, Ouyang N, Hörbelt M, Antus B, Wang M, Exton MS. Cell Immunol. 2000;204:19–28. doi: 10.1006/cimm.2000.1687. [DOI] [PubMed] [Google Scholar]
- 4.Zizzo G, Hilliard BA, Monestier M, Cohen PL. J Immunol. 2012;189:3508–3520. doi: 10.4049/jimmunol.1200662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lurier EB, Dalton D, Dampier W, Raman P, Nassiri S, Ferraro NM, Rajagopalan R, Sarmady M, Spiller KL. Immunobiology. 2017;222(7):847–856. doi: 10.1016/j.imbio.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nassiri S, Zakeri I, Weingarten MS, Spiller KL. J Invest Dermatol. 2015;135:1700. doi: 10.1038/jid.2015.30. [DOI] [PubMed] [Google Scholar]
- 7.Ferraro NM, Dampier W, Weingarten MS, Spiller KL. Integr Biol. 2017;9(4):328–338. doi: 10.1039/c7ib00018a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma Y, Pope RM. Curr Pharm Des. 2005;11:569–580. doi: 10.2174/1381612053381927. [DOI] [PubMed] [Google Scholar]
- 9.Sindrilaru A, Peters T, Wieschalka S, Baican C, Baican A, Peter H, Hainzl A, Schatz S, Qi Y, Schlecht A. J Clin Invest. 2011;121:985–997. doi: 10.1172/JCI44490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan X, Koo SJ, Wen J. J Immunol. 2016;196:124. [Google Scholar]
- 11.Venneri MA, Giannetta E, Panio G, De Gaetano R, Gianfrilli D, Pofi R, Masciarelli S, Fazi F, Pellegrini M, Lenzi A. PLoS One. 2015;10:e0126580. doi: 10.1371/journal.pone.0126580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu T, Wang W, Nassiri S, Kwan T, Dang C, Liu W, Spiller KL. J Biomater Sci Polym Ed. 2016;27:721–742. doi: 10.1080/09205063.2016.1155881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milde R, Ritter J, Tennent GA, Loesch A, Martinez FO, Gordon S, Pepys MB, Verschoor A, Helming L. Cell Rep. 2015;13:1937–1948. doi: 10.1016/j.celrep.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson JM, Gristina A, Hanson S, Harker L, Johnson R, Merritt K, Naylor P, Schoen F. Biomater Sci. 1996:165–214. [Google Scholar]
- 15.Ju YM, Yu B, West L, Moussy Y, Moussy F. J Biomed Mater Res, Part A. 2010;93:200–210. doi: 10.1002/jbm.a.32512. [DOI] [PubMed] [Google Scholar]
- 16.Patil SD, Papadimitrakopoulos F, Burgess DJ. Diabetes Technol Ther. 2004;6:887–897. doi: 10.1089/dia.2004.6.887. [DOI] [PubMed] [Google Scholar]
- 17.Graversen JH, Svendsen P, Dagnæs-Hansen F, Dal J, Anton G, Etzerodt A, Petersen MD, Christensen PA, Møller HJ, Moestrup SK. Mol Ther. 2012;20:1550–1558. doi: 10.1038/mt.2012.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Cousin JM, Hughes J, Van Damme J, Seckl JR, Haslett C, Dransfield I, Savill J, Rossi AG. J Immunol. 1999;162:3639–3646. [PubMed] [Google Scholar]
- 19.Lim HY, Müller N, Herold MJ, Van Den Brandt J, Reichardt HM. Immunology. 2007;122:47–53. doi: 10.1111/j.1365-2567.2007.02611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Goes A, Hoekstra K, van den Berg TK, Dijkstra CD. J Leukocyte Biol. 2000;67:801–807. doi: 10.1002/jlb.67.6.801. [DOI] [PubMed] [Google Scholar]
- 21.Kunicka JE, Talle MA, Denhardt GH, Brown M, Prince LA, Goldstein G. Cell Immunol. 1993;149:39–49. doi: 10.1006/cimm.1993.1134. [DOI] [PubMed] [Google Scholar]
- 22.Verhoef CM, van Roon JA, Vianen ME, Lafeber FP, Bijlsma JW. Ann Rheum Dis. 1999;58:49–54. doi: 10.1136/ard.58.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pentecost AE, Lurier EB, Spiller KL. Microscale Technologies for Cell Engineering. ch. 14. Springer; 2016. pp. 291–304. [Google Scholar]
- 24.Mochalin VN, Shenderova O, Ho D, Gogotsi Y. Nat Nanotechnol. 2012;7:11–23. doi: 10.1038/nnano.2011.209. [DOI] [PubMed] [Google Scholar]
- 25.Liu KK, Cheng CL, Chang CC, Chao JI. Nanotechnology. 2007;18:325102. [Google Scholar]
- 26.Liu KK, Zheng WW, Wang CC, Chiu YC, Cheng CL, Lo YS, Chen C, Chao JI. Nanotechnology. 2010;21:315106. doi: 10.1088/0957-4484/21/31/315106. [DOI] [PubMed] [Google Scholar]
- 27.Schrand AM, Huang H, Carlson C, Schlager JJ, Ōsawa E, Hussain SM, Dai L. J Phys Chem B. 2007;111:2–7. doi: 10.1021/jp066387v. [DOI] [PubMed] [Google Scholar]
- 28.Neitzel I, Mochalin V, Gogotsi Y. In: Ultananocrystalline Diamond. 2. Gruen DM, editor. William Andrew Publishing; Oxford: 2012. pp. 421–456. [Google Scholar]
- 29.Zhang Q, Mochalin VN, Neitzel I, Hazeli K, Niu J, Kontsos A, Zhou JG, Lelkes PI, Gogotsi Y. Biomaterials. 2012;33:5067–5075. doi: 10.1016/j.biomaterials.2012.03.063. [DOI] [PubMed] [Google Scholar]
- 30.Mochalin VN, Pentecost A, Li XM, Neitzel I, Nelson M, Wei C, He T, Guo F, Gogotsi Y. Mol Pharm. 2013;10:3728–3735. doi: 10.1021/mp400213z. [DOI] [PubMed] [Google Scholar]
- 31.Giammarco J, Mochalin VN, Haeckel J, Gogotsi Y. J Colloid Interface Sci. 2016;468:253–261. doi: 10.1016/j.jcis.2016.01.062. [DOI] [PubMed] [Google Scholar]
- 32.Chen M, Pierstorff ED, Lam R, Li SY, Huang H, Osawa E, Ho D. ACS Nano. 2009;3:2016–2022. doi: 10.1021/nn900480m. [DOI] [PubMed] [Google Scholar]
- 33.Knapinska AM, Tokmina-Roszyk D, Amar S, Tokmina-Roszyk M, Mochalin VN, Gogotsi Y, Cosme P, Terentis AC, Fields GB. Biopolymers. 2015;104:186–195. doi: 10.1002/bip.22636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang H, Pierstorff E, Osawa E, Ho D. Nano Lett. 2007;7:3305–3314. doi: 10.1021/nl071521o. [DOI] [PubMed] [Google Scholar]
- 35.Huang K-J, Lee CY, Lin YC, Lin CY, Perevedentseva E, Hung SF, Cheng CL. J Biophotonics. 2017 doi: 10.1002/jbio.201600202. [DOI] [PubMed] [Google Scholar]
- 36.Shkurupy V, Arkhipov S, Neshchadim D, Akhramenko E, Troitskii A. Bull Exp Biol Med. 2015;158:500. doi: 10.1007/s10517-015-2794-z. [DOI] [PubMed] [Google Scholar]
- 37.Huang H, Pierstorff E, Osawa E, Ho D. ACS Nano. 2008;2:203–212. doi: 10.1021/nn7000867. [DOI] [PubMed] [Google Scholar]
- 38.Thomas V, Halloran BA, Ambalavanan N, Catledge SA, Vohra YK. Acta Biomater. 2012;8:1939–1947. doi: 10.1016/j.actbio.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spiller KL, Wrona EA, Romero-Torres S, Pallotta I, Graney PL, Witherel CE, Panicker LM, Feldman RA, Urbanska AM, Santambrogio L. Exp Cell Res. 2016;347:1–13. doi: 10.1016/j.yexcr.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 40.Mochalin VN, Gogotsi Y. J Am Chem Soc. 2009;131:4594–4595. doi: 10.1021/ja9004514. [DOI] [PubMed] [Google Scholar]
- 41.Zahr AS, Davis CA, Pishko MV. Langmuir. 2006;22:8178–8185. doi: 10.1021/la060951b. [DOI] [PubMed] [Google Scholar]
- 42.Mosqueira VCF, Legrand P, Gulik A, Bourdon O, Gref R, Labarre D, Barratt G. Biomaterials. 2001;22:2967–2979. doi: 10.1016/s0142-9612(01)00043-6. [DOI] [PubMed] [Google Scholar]
- 43.Getts DR, Terry RL, Getts MT, Deffrasnes C, Müller M, van Vreden C, Ashhurst TM, Chami B, McCarthy D, Wu H. Sci Transl Med. 2014;6:219ra217. doi: 10.1126/scitranslmed.3007563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez FO, Gordon S, Locati M, Mantovani A. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 45.Spiller KL, Anfang RR, Spiller KJ, Ng J, Nakazawa KR, Daulton JW, Vunjak-Novakovic G. Biomaterials. 2014;35:4477–4488. doi: 10.1016/j.biomaterials.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 47.Jones JA, McNally AK, Chang DT, Qin LA, Meyerson H, Colton E, Kwon I, Matsuda T, Anderson JM. J Biomed Mater Res Part A. 2008;84:158–166. doi: 10.1002/jbm.a.31220. [DOI] [PubMed] [Google Scholar]
- 48.Newby AC. Cardiovasc Res. 2006;69:614–624. doi: 10.1016/j.cardiores.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 49.Osswald S, Yushin G, Mochalin V, Kucheyev SO, Gogotsi Y. J Am Chem Soc. 2006;128:11635–11642. doi: 10.1021/ja063303n. [DOI] [PubMed] [Google Scholar]
- 50.Langmuir I. J Am Chem Soc. 1918;40:1361–1403. [Google Scholar]
- 51.Freundlich H. Kapillarchemie, eine Darstellung der Chemie der Kolloide und verwandter Gebiete, von Dr. Herbert Freundlich. akademische Verlagsgesellschaft; 1909. [Google Scholar]
- 52.Champion JA, Walker A, Mitragotri S. Pharm Res. 2008;25:1815–1821. doi: 10.1007/s11095-008-9562-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Witherel CE, Graney PL, Freytes DO, Weingarten MS, Spiller KL. Wound Repair Regen. 2016;24:514–524. doi: 10.1111/wrr.12423. [DOI] [PubMed] [Google Scholar]
- 54.Strober W. Curr Protoc Immunol. 2001:A3.B.1–A3.B.3. doi: 10.1002/0471142735.ima03bs111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chikanza I, Kozaci D. Rheumatology. 2004;43:1337–1345. doi: 10.1093/rheumatology/keh333. [DOI] [PubMed] [Google Scholar]
- 56.Paleolog EM. Arthritis Res. 2002;4:S81. doi: 10.1186/ar575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.A. M. Association. AMA drug evaluations. 4. John Wiley and Sons; New York: 1980. [Google Scholar]
- 58.Zuckerman JE, Davis ME. Nat Rev Drug Discovery. 2015;14:843–856. doi: 10.1038/nrd4685. [DOI] [PubMed] [Google Scholar]
- 59.Doshi N, Zahr AS, Bhaskar S, Lahann J, Mitragotri S. Proc Natl Acad Sci U S A. 2009;106:21495–21499. doi: 10.1073/pnas.0907127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park MV, Neigh AM, Vermeulen JP, de la Fonteyne LJ, Verharen HW, Briedé JJ, van Loveren H, de Jong WH. Biomaterials. 2011;32:9810–9817. doi: 10.1016/j.biomaterials.2011.08.085. [DOI] [PubMed] [Google Scholar]
- 61.Park EJ, Park K. Toxicol Lett. 2009;184:18–25. doi: 10.1016/j.toxlet.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 62.Brown DM, Kinloch IA, Bangert U, Windle A, Walter D, Walker G, Scotchford C, Donaldson K, Stone V. Carbon. 2007;45:1743–1756. [Google Scholar]
- 63.Nicolete R, dos Santos DF, Faccioli LH. Int Immunopharmacol. 2011;11:1557–1563. doi: 10.1016/j.intimp.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 64.Lopez-Castejon G, Brough D. Cytokine Growth Factor Rev. 2011;22:189–195. doi: 10.1016/j.cytogfr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Voll RE, Herrmann M, Roth EA, Stach C, Kalden JR, Girkontaite I. Nature. 1997;390:350–351. doi: 10.1038/37022. [DOI] [PubMed] [Google Scholar]
- 66.Gaipl US, Munoz LE, Grossmayer G, Lauber K, Franz S, Sarter K, Voll RE, Winkler T, Kuhn A, Kalden J. J Autoimmun. 2007;28:114–121. doi: 10.1016/j.jaut.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 67.Koh JS, Wang Z, Levine JS. J Immunol. 2000;165:4190–4201. doi: 10.4049/jimmunol.165.8.4190. [DOI] [PubMed] [Google Scholar]
- 68.Verhoven B, Schlegel R, Williamson P. J Exp Med. 1995;182:1597–1601. doi: 10.1084/jem.182.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boehringer PA, Darden IL. Blood. 2006;108:777–778. doi: 10.1182/blood-2006-03-008789. [DOI] [PubMed] [Google Scholar]
- 70.Schaer DJ, Schaer CA, Buehler PW, Boykins RA, Schoedon G, Alayash AI, Schaffner A. Blood. 2006;107:373–380. doi: 10.1182/blood-2005-03-1014. [DOI] [PubMed] [Google Scholar]
- 71.Ugocsai P, Barlage S, Dada A, Schmitz G. Cytometry Part A. 2006;69:203–205. doi: 10.1002/cyto.a.20235. [DOI] [PubMed] [Google Scholar]
- 72.Philippidis P, Mason J, Evans B, Nadra I, Taylor K, Haskard D, Landis R. Circ Res. 2004;94:119–126. doi: 10.1161/01.RES.0000109414.78907.F9. [DOI] [PubMed] [Google Scholar]
- 73.Van den Heuvel MM, Tensen CP, van As JH, Van den Berg TK, Fluitsma DM, Dijkstra CD, Döpp E, Droste A, Van Gaalen FA, Sorg C. J Leukocyte Biol. 1999;66:858–866. doi: 10.1002/jlb.66.5.858. [DOI] [PubMed] [Google Scholar]
- 74.Hulett MD, Hogarth PM. Mol Immunol. 1998;35:989–996. doi: 10.1016/s0161-5890(98)00069-8. [DOI] [PubMed] [Google Scholar]
- 75.Goldstein JL, Ho Y, Basu SK, Brown MS. Proc Natl Acad Sci U S A. 1979;76:333–337. doi: 10.1073/pnas.76.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ōsawa E. Pure Appl Chem. 2008;80:1365–1379. [Google Scholar]
- 77.Krüger A, Kataoka F, Ozawa Maa, Fujino T, Suzuki Y, Aleksenskii A, Vul AY, Ōsawa E. Carbon. 2005;43:1722–1730. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.