Abstract
Background and Objectives:
Clostridium difficile is the leading cause of nosocomial diarrhea and pseudomembranous colitis. The prevalence of C. difficile infection differs in various geographical areas. The aim of this study was to determine the prevalence of C. difficile isolates and the prevalence of cdd3, tcdA and tcdB genes in beef samples in Kermanshah Province.
Materials and Methods:
One hundred ground beef samples were randomly collected from the butchers of Kermanshah province during March 2014 to March 2015. Following alcohol shock, minced meat samples were incubated in a specific culture medium for 5 to 7 days. The suspicious colonies were analyzed by biochemical tests and frequency of C. difficile and cdd3, tcdA and tcdB genes was assessed by PCR using specific primers.
Results:
In total, 30% samples were positive for C. difficile and all the isolates harbored Cdd3 gene. Combined dual-gene frequency of A+B+, A−B+ and A+B− strains in the positive were 0%, 3.3%, and 26.6% respectively, while 21 samples (70%) were non-toxigenic (A−B−).
Conclusion:
In this study, the presence of C. difficile in beef as a source of contamination was confirmed. It was also shown that the incidence of C. difficile in ground meat was relatively higher than many other studies.
Keywords: Clostridium difficile, Ground beef, PCR
INTRODUCTION
Clostridium difficile is a gram positive, spore-forming obligatory anaerobic bacterium (1). As the produced spores are highly resistant to harsh environments, C. difficile can sustain and live at various surfaces (2). C. difficile was isolated for the first time from stool of healthy newborns in 1935, while its virulence was confirmed in 1970s (3). The infection is a major global problem and is important in children and newborns. Nearly 25 to 80% of children and newborns are colonized without any symptoms and diarrhea (4) although it can cause colonic and extra-colonic infections. Colonization in the colon causes asymptomatic infection, pseudomembranous colitis, and fatal colitis with colonic perforation, peritonitis and eventually death (5). The prevalence of diarrhea associated with C. difficile led to a high mortality rate in Canada and America in 2003 (6). In a healthy person, the bacterial growth is controlled by the normal intestinal flora, but when a person is undergone to antibiotic treatment or when using gastric anti-acid drugs, the conditions become favorable for bacterial overgrowth. In addition to hospitals, C. difficile can be found in domestic animals, vegetables, meat and meat products, water, soil and fast foods (7–9).
This bacterium can be transmitted to humans through the fecal-oral route and from animals and animal products. Toxigenic C. difficile strains produces toxin A, toxin B and Binary Toxin (BT). Toxin A (tcdA) and toxin B (tcdB) genes are located close to each other on a Pathogencity Locus. Toxin A consists of 2710 amino acids (308 kDa) and toxin B consists of 2366 amino acids (270 kDa). These toxins are classified as lethal toxins among the group of large cytotoxic toxins (LCTS) (10) Toxin B causes more mucosal damage in colon comparing to toxin A, that’s why pathogenesis of strains lacking toxin B, is similar to non-toxigenic strains (11, 12).
There are not many reports about C. difficile infection in the Middle East, while the studies from rest of the world, such as Europe and North America show that infections caused by this bacterium are rapidly increasing (13).
According to the reports, ground meat can be a carrier for C. difficile and transmit the bacteria to humans. Moreover, there is little information about the prevalence of C. difficile in meat in Iran. Therefore, we analyzed the prevalence of C. difficile isolates and determined the prevalence of cdd3, tcdA and tcdB genes in beef samples in Kermanshah province.
MATERIALS AND METHODS
Specimens.
In this study, 100 random samples of ground beef were collected during March 2014 to March 2015. Samples were obtained from the butchers of Kermanshah and cities of West Islamabad, Sar-e Pol-e Zahab, Qasr-e Shirin and Dalahoo.
Culture and Primary Identification.
One gram of ground meat samples was slowly mixed with ethanol 96% and incubated for 2 minutes at room temperature. The resulting solution was cultured on Cycloserine, Cefoxitin, Fructose Agar F (CCFA) (Hi Media -India) enriched with 5% sheep blood and 250 μg /ml cycloserine and 8 μg / ml Cefoxitin antibiotics. Cultured Plates were incubated at 37°C under anaerobic atmosphere for 5–7 days.
Suspected colonies with yellowish gray or white, round and slightly raised was identified with Gram staining.
Molecular Methods.
DNA of confirmed colonies was extracted by a kit (Sinaclon) and presence of tcdA, tcdB and cdd3 was investigated by PCR using specific primers (primers were designed based on sequences retrieved from Gen Bank). Isolates that were positive for cdd3 gene in PCR were tested for tcdA and tcdB. List of used primers are shown in Table 1.
Table 1.
Sequences of primers used for the amplification of cdd3, tcdA, and tcdB genes
| Primers | Sequence | Product bp | References | |
|---|---|---|---|---|
| cdd3 | F | TGA TAA CTC TAG GTA ATA AGA CC | 497 | This study |
| R | GTA ATC CAG ATA TGT TAG GTG G | |||
| tcdA | F | GTG TAG ATT CAC TTT CCA ATG | 404 | This study |
| R | TGA TAT TGA TGC TAA TCC TGG | |||
| tcdB | F | GATGTTGATAATGTTGTGAGAG | 653 | This study |
| R | TCA GCT ACT CCA CTT TCA TC | |||
A PCR reaction was performed in a total volume of 15 μl containing1X PCR buffer (Sinaclon, Iran), 0.2mM deoxynucleotide mix, 1.5 mM MgCl2, 0.6μM of primers, 1 unit of Taq DNA polymerase and 1 μl of DNA template. The program consisted of initial denaturation at 94°C for 5 minutes and 35 thermal cycles including denaturation at 94 for 30 seconds, annealing at 54°C for cdd3, tcdA and 53°C for tcdB genes, and a final extension at 72°C for 5 minutes. PCR products were isolated by 1% agarose gel electrophoresis and resulting bands were visualized by staining with Ethidium Bromide solution.
Statistical Analysis.
Data were analyzed using the SPSS 16 software. The qualitative data were analyzed using chi-square test, and in all cases p < 0.05 was considered as significant.
RESULTS
Suspected colonies were identified by Gram stain and the isolates were confirmed by molecular methods. From a total of 100 samples of ground beef, C. difficile was positive in 30 samples (30%).
The size of PCR products for cdd3, tcdA and tcdB genes were 497 bp (Fig. 1), 404 bp (Fig. 2) and 653 bp (Fig. 3), respectively. The frequency of tcdA and tcdB genes was 1% and 8%, respectively, while all the isolates harbored cdd3 gene (30%). Among the 30 isolates, the combined dual frequency of, A+B+, A−B+ and A+B− genes in cdd3 positive C. difficile strains were 0%, 3.3%, 26.6%, respectively. Interestingly, 21 isolates (70%) were non-toxigenic (A−B) (Tables 1 and 2).
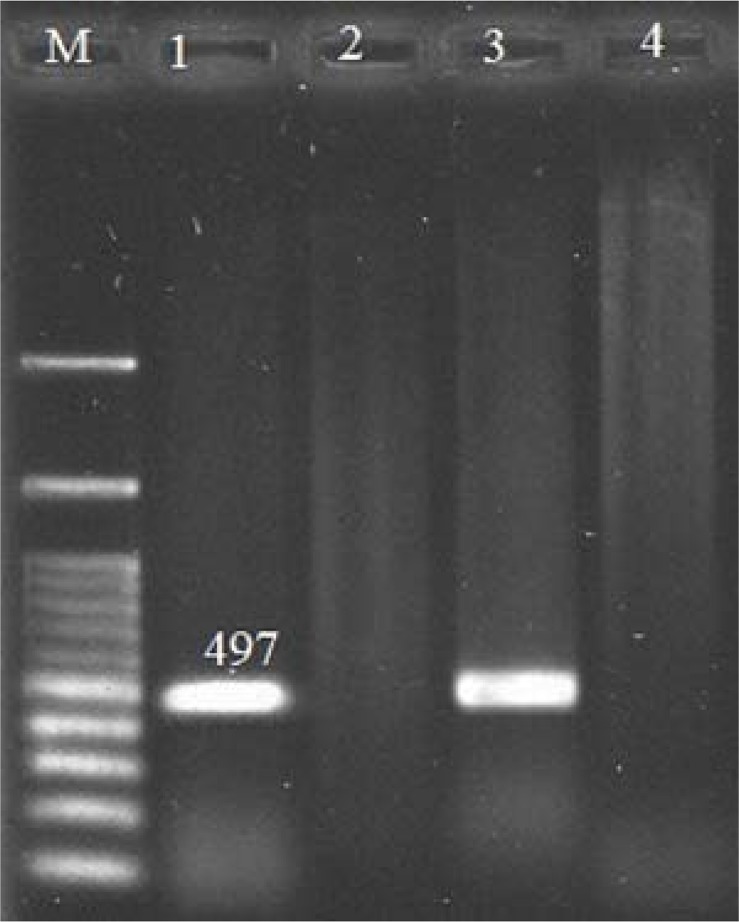
Fig. 1.
PCR Products Gel Electrophoresis for cdd3 Gene. M:100bp Plus DNA Ladder RTU (Ready-to-Use), lane1: positive control, lane 2: cdd3 Gene negative control, lane 3: cdd3 gene positive sample, lane 4: cdd3 gene negative sample.
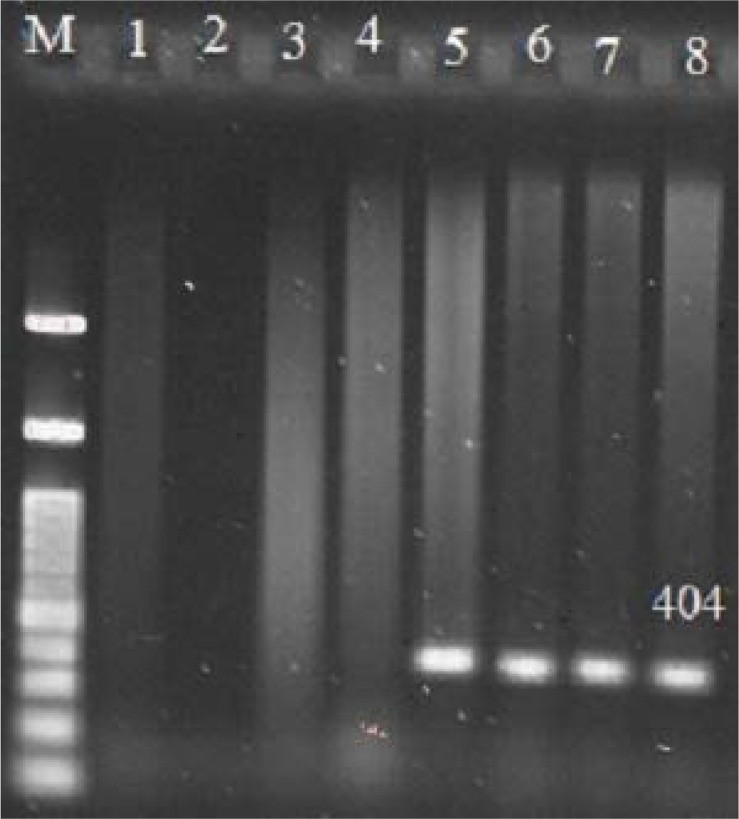
Fig. 2.
Agarose gel electrophoresis of PCR products for tcdA Gene. M:100bp Plus DNA Ladder RTU, lane1: positive control, lane 2, 3,4: tcdA Gene negative control, lane 5,6,7,8: tcdA gene positive sample.
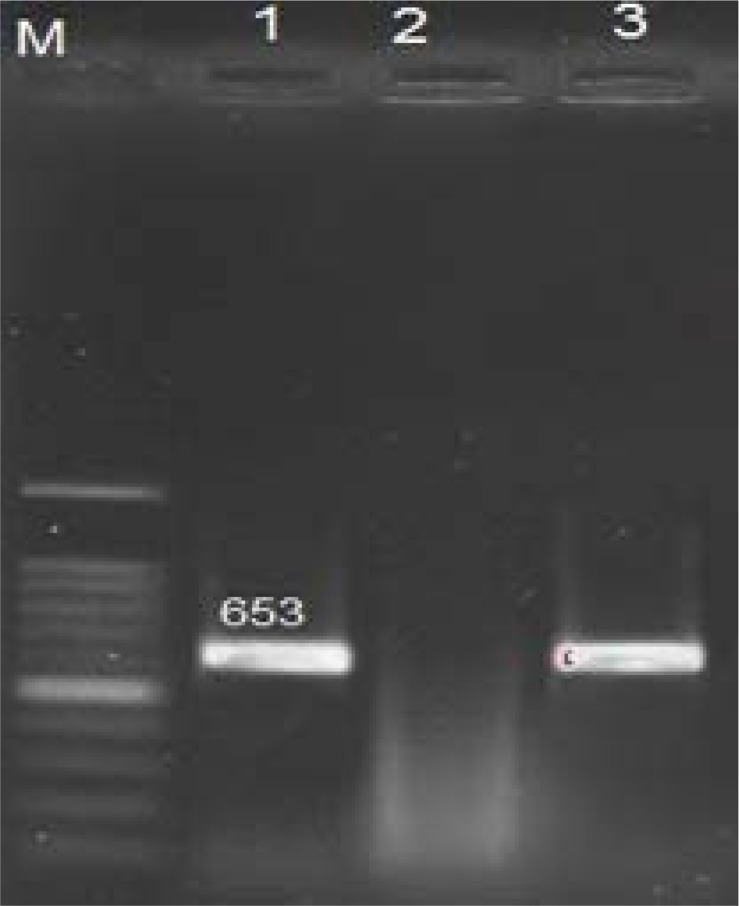
Fig. 3.
Agarose gel electrophoresis of PCR products for tcdB Gene. M: 100bp Plus DNA Ladder RTU, lane1: positive control, lane 2: tcdB Gene negative control, lane 3: tcdB gene positive sample.
Table 2.
The frequency of cultures, and cdd3, tcdA and tcdB genes in the collected samples obtained from Kermanshah province.
| Sample obtaining location | Culture | cdd3 | tcdA | tcdB |
|---|---|---|---|---|
| Kermanshah | 31 | 13 | 1 | 0 |
| West Islam Abad | 47 | 16 | 7 | 1 |
| Dlahoo | 11 | 1 | 0 | 0 |
| Qasre-Shirin | 6 | 0 | 0 | 0 |
| Sar-e Pol-e Zahab | 5 | 0 | 0 | 0 |
DISCUSSION
C. difficile is an important cause of nosocomial diarrhea. Spores of C. difficile are quite widespread in the environment and foods (14). It has been reported that the bacterium is a bovine pathogen which may be a serious threat to human health (15, 16). In various studies, the prevalence of infections with C. difficile has different values. As there are few studies concerning the prevalence of C. difficile in Iran, we aimed to assess the prevalence of the bacterium in ground meat samples in Kermanshah province.
Among samples of ground beef in several areas of Kermanshah Province, the frequency of A+B+, A−B+ and A+B− were obtained 0, 3.3, and 26.6%, respectively. In this study, 21 (70%) non-toxigenic isolates were also isolated from minced meat samples which show that the rate of non-toxigenic strains is relatively high. It was believed that all toxigenic isolates of C. difficile produce both toxins (A+B+ strains) and the toxin B is capable of inducing CDAD without the presence of toxin A (17). But, later, severe cases of CDAD and pseudo-membranous colitis were observed that were arisen from the strains harboring toxin B and not having toxin A (strain A−B+).
Killgor reported that the frequency of C. difficile was 42% and also C. difficile were 42.2% 41.3% and 44.4% in beef, pork and turkey meat samples (18).
The prevalence of the bacteria in ground beef in studies from America and France was lower comparing to our data, even though in some studies no C. difficile was isolated from ground beef (19).
However, the prevalence of C. difficile in samples of ground beef in studies from America, Canada and the present study was 42%, 20.8 % and 30%, respectively. The different prevalence of C. difficile in ground beef samples in various studies can be attributed to the difference in sample size, geographical and regional conditions, and development of diagnostic test for C. difficile (18, 20).
There are some reports about the prevalence of toxigenic strains of C.difficile Iran. Rahimi et al reported a 1.65% frequency of C. difficile among 121 samples of beef, while the isolates produce both TcdA and TcdB toxins tcdA and tcdB genes (21). Moreover, in another study on chopped and ground meat carried out in Isfahan, the prevalence of C. difficile was 4% and 20% respectively which is close to our results. But with the difference that the number of samples examined in our study is much more than the study in Isfahan (22).
In study of Spain (2005), the prevalence of tcdA+B+ and tcdA−B+ encoding strains were 4.5%, 5% and 25.7%, 56.9% (23). The prevalence of A−B+isolates in Europe, China and Japan, was about 6.2%, 26.6% and 6.3% (24, 25). A−B+ strains are very rare and there are a few reports (26) that only one study that was conducted in one case of a total of 159 isolates. While in our study A+B− strain was observed in 8 isolates (26.6%) (27). According to the aforementioned findings, we can conclude that most toxigenic isolates of the C. difficile in Iran are those variants. This could affect the expression of toxins and affect the clinical manifestation of the disease.
Based on the results of current study, there is a high rate of C. difficile of ground beef in Kermanshah province. According to studies in this field, ground meat can be a carrier for of C. difficile and transmit the bacteria to humans. Thus, control and supervision by the health organization and hygiene and sanitation and proper cooking can reduce food contamination with C. difficile. It seems that identifying virulence factors and the prevalence of these bacteria can be effective in the prevention and control of the disease (28, 29).
Furthermore, more studies must be done in different parts of the country to determine the prevalence of the C. difficile on food products would be helpful in future.
REFERENCES
- 1.Paredes-Sabja D, Shen A, Sorg JA. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol 2014; 22:406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pelleschi ME. Clostridium difficile-associated disease: diagnosis, prevention, treatment, and nursing care. Crit Care Nurse 2008; 28: 27–35. [PubMed] [Google Scholar]
- 3.Hall I, O’Toole E. Intestinal microflora in newborn infants with a description of a new pathogenic anaerobe, Bacillus difficilis. Am J Dis Child 1935; 49: 390–402. [Google Scholar]
- 4.Kelly CP, Pothoulakis C, LaMont JT. Clostridium difficile colitis. N Engl J Med 1994; 330: 257–262. [DOI] [PubMed] [Google Scholar]
- 5.Rubin MS, Bodenstein LE, Kent KC. Severe Clostridium difficile colitis. Dis Colon Rectum 1995; 38(4): 350–354. [DOI] [PubMed] [Google Scholar]
- 6.Dial S, Alrasadi K, Manoukian C, Huang A, Menzies D. Risk of Clostridium difficile diarrhea among hospital inpatients prescribed proton pump inhibitors: cohort and case-control studies. Can Med Assoc J 2004;171:33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato H, Kato N, Katow S, Maegawa T, Nakamura S, Lyerly DM. Deletions in the repeating sequences of the toxin A gene of toxin A-negative, toxin B-positive Clostridium difficile strains. FEMS Microbiol Lett 1999;175: 197–203. [DOI] [PubMed] [Google Scholar]
- 8.Keel K, Brazier JS, Post KW, Weese JS, Songer JG. Prevalence of PCR ribotypes among Clostridium difficile isolates from pigs, calves, and other species. J Clin Microbiol 2007;45:1963–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rupnik M. Is Clostridium difficile-associated infection a potentially zoonotic and foodborne disease? Clin Microbiol Infect 2007;13:457–459. [DOI] [PubMed] [Google Scholar]
- 10.Lyerly DM, Krivan HC, Wilkins TD. Clostridium Difficile: its Diseases and Toxins. Clin Microbiol Rev 1998; 1:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuehne SA, Cartman ST. Both, Toxin A and Toxin B are Important in Clostridium difficile Infection. Gut Microbes 2011; 2(4):252–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riggs MM, Sethi AK, Zabarsky TF, Eckstein EC, Jump RL, Donskey CJ. Asymptomatic Carriers are a Potential Source for Transmission of Epidemic and Non Epidemic Clostridium difficile Strains among long - Term Care Facility Residents. Clin Infect Dis 2007; 45:992–998. [DOI] [PubMed] [Google Scholar]
- 13.Yoo J, Lightner AL. Clostridium difficile Infections: What Every Clinician Should Know. Perm J 2010; 14: 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers MA, Greene MT, Saint S, Chenoweth CE, Malani PN, Trivedi I, et al. Higher rates of Clostridium difficile infection among smokers. PLoS One 2012; 7(7): e42091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniel A, Rapose A. The evaluation of Clostridium difficile infection (CDI) in a community hospital. J Infect Public Health 2015; 8:155–160. [DOI] [PubMed] [Google Scholar]
- 16.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in empidemiology and pathogenesis. Nat Rev Microbiol 2009; 7: 526–536. [DOI] [PubMed] [Google Scholar]
- 17.Guilbault C, Labbe AC, Poirier L, Busque L, Beliveau C, Laverdiere M. Development and evaluation of a PCR method for detection of the Clostridium difficile toxin B gene in stool specimens. J Clin Microbiol 2002; 40: 2288–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Songer JG, Trinh HT, Killgore GE, Thompson AD, McDonald LC, Limbago BM. Clostridium difficile in Retail Meat Products, USA, 2007. Emerg Infect Dis 2009; 15:819–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouttier S, Barc MC, Felix B, Lambert S, Collignon A, Barbut F. Clostridium difficile in ground meat, France. Emerg Infect Dis 2010; 16:733–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguz-Palacios A, Staemfli H, Duffield T, Weese JS. Clostridium difficile in retail Ground Meat, Canada. Emerg Infect Dis 2007; 13:485–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahimi E, Jalali M, Weese JS. Prevalence of Clostridium difficile in raw beef, cow, sheep, goat, camel and buffalo meat in Iran. BMC Public Health 2014; 14:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esfandiari Z, Jalali M, Ezzatpanah H, Weese JS, Chamani M. Prevalence and Characterization of Clostridium difficile in Beef and Mutton Meats of Isfahan Region, Iran. Jundishapur J Microbiol 2014; 7(8): e16771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alonso R, Martin A, Pelaez T, Marin M, Rodriguez-Creixems M, Bouza E. Toxigenic status of Clostridium difficile in a large Spanish teaching hospital. J Med Microbiol 2005; 54:159–162. [DOI] [PubMed] [Google Scholar]
- 24.Cobeta-Garcia JC, Domingo-Morera JA, Gracia P. Clostridium difficile-associated diarrhea in a patient with rheumatoid arthritis: comment on the article by Ramos et al. Arthritis Rheum 1998;41:2083–2085. [DOI] [PubMed] [Google Scholar]
- 25.Kikkawa H, Hitomi S, Watanabe M. Prevalence of toxin A-nonproducing/toxin-B-producing Clostridium difficile in the Tsukuba-Tsuchiura district, Japan. J Infect Chemother 2007;13:35–38. [DOI] [PubMed] [Google Scholar]
- 26.Persson S, Torpdahl M, Olsen KE. New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection. Clin Microbiol Infect 2008; 14: 1057–1064. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez-Palacios A, Reid-Smith R, Staempfli HR, Weese JS. Colstridium difficile survives minimal temperature recommended for cooking ground meats. Anaerobe 2010; 16: 540–542. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez-Palacios A, Lejeune JT. Moist-Heat resistance, spore aging, and superdormancy in Clostridium difficile. Appl Environ Microbiol 2011;77:3085–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemee L, Dhalluin A, Testelin S, Mattrat M. A, Maillard K, Lemeland JF, et al. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (toxin A), and tcdB (toxin B) genes for toxigenic culture of Clostridium difficile. J Clin Microbiol 2004;42:5710–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]