Abstract
Key message
A comprehensive linkage atlas for seed yield in rapeseed.
Abstract
Most agronomic traits of interest for crop improvement (including seed yield) are highly complex quantitative traits controlled by numerous genetic loci, which brings challenges for comprehensively capturing associated markers/genes. We propose that multiple trait interactions underlie complex traits such as seed yield, and that considering these component traits and their interactions can dissect individual quantitative trait loci (QTL) effects more effectively and improve yield predictions. Using a segregating rapeseed (Brassica napus) population, we analyzed a large set of trait data generated in 19 independent experiments to investigate correlations between seed yield and other complex traits, and further identified QTL in this population with a SNP-based genetic bin map. A total of 1904 consensus QTL accounting for 22 traits, including 80 QTL directly affecting seed yield, were anchored to the B. napus reference sequence. Through trait association analysis and QTL meta-analysis, we identified a total of 525 indivisible QTL that either directly or indirectly contributed to seed yield, of which 295 QTL were detected across multiple environments. A majority (81.5%) of the 525 QTL were pleiotropic. By considering associations between traits, we identified 25 yield-related QTL previously ignored due to contrasting genetic effects, as well as 31 QTL with minor complementary effects. Implementation of the 525 QTL in genomic prediction models improved seed yield prediction accuracy. Dissecting the genetic and phenotypic interrelationships underlying complex quantitative traits using this method will provide valuable insights for genomics-based crop improvement.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-017-2911-7) contains supplementary material, which is available to authorized users.
Introduction
Crop production environments are subject to many abiotic and biotic factors affecting plant growth. In addition, networks of important agronomic traits interact with each other and with the environment in a very complex manner. Seed yield, one of the most important agronomic traits, is both extremely complex and strongly affected by the environment. Furthermore, yield performance depends on factors such as plant growth conditions, plant resistance to biotic and abiotic stresses, and interactions with other physiological and genetic processes through the entire crop life cycle. Yield itself is affected by many underlying traits and thousands of different genes (Bernardo 2008).
In the past few decades, many quantitative trait loci (QTL) analyses have been done using different statistical methods to dissect the genetic architecture of complex quantitative traits such as seed yield (Dargahi et al. 2014; Groos et al. 2003; Lacape et al. 2013; Li et al. 2014; Liu et al. 2015; Shi et al. 2009; Song et al. 2007; Xiao et al. 1996; Zhang et al. 2004). Nevertheless, understanding of such complex traits is still rudimentary, and this knowledge is insufficient to contribute substantially to further crop genetic improvement (Bernardo 2008). Theoretically, several hundred genes or QTL may account for differences in seed yield between two diverse parents, but it is difficult to detect such a large number of QTL in a single mapping study by conventional methods. Difficulties in detecting QTL arise for a number of reasons. Firstly, experimental variables such as soil fertility and time of crop emergence are difficult to control in standard field experiments. Secondly, genes with opposite contributions to seed yield may be located in the same QTL region, offsetting their contributions to yield. Thirdly, genotype-by-environment interactions are relatively poorly understood (Cowling and Balazs 2010; El-Soda et al. 2014). Finally, QTL mapping methods are often also statistically incapable of detecting genes with minor effects (Mackay et al. 2009; Würschum 2012).
More QTL can be detected by increasing the number of detectable recombination events in the segregating population. This usually involves enlarging the size of the population (and hence the number of recombinants) while maintaining genetic marker saturation such that all recombination events are detected (Mackay et al. 2009; Vales et al. 2005; Zeng 1994). However, larger population sizes and greater marker densities make experiments more difficult and costly, and often the trade-off is that fewer replicates are possible, hindering detection of field-scale environmental variation. To address this issue, a number of statistical or methodological approaches have been suggested to improve QTL detection efficiency. Long et al. (2007) proposed inclusion of minor-effect “micro-real” QTL, which do not meet QTL detection thresholds, but nevertheless are either detected repeatedly in different environments or which overlap with QTL for related traits. For the latter, where QTL overlap with loci for related traits, Shi et al. (2009) coined the term “indicator QTL”. In previous studies, indicator QTL with positive or negative effects on traits related to seed yield were often ignored. However, such loci may be very useful to further increase seed yield under selection in different genetic backgrounds and environments.
Multiple single trait interactions are likely to underlie all complex traits such as seed yield. We hypothesize that by considering these component traits and their interactions we can more effectively dissect individual QTL and more accurately predict the trait performance. To test this hypothesis, we investigated a large resource of phenotypic data generated in large-scale field trials: a total of 19 independent experiments in a doubled-haploid (DH) B. napus mapping population (BnaTNDH), with accompanying single nucleotide polymorphism (SNP) genotype data. Multi-environment phenotype data was available for seed yield and its components (Shi et al. 2009), flowering time (Long et al. 2007); seed quality traits (Jiang et al. 2014; Shi et al. 2009), glucosinolate content (Feng et al. 2012), nutrient use efficiency (Shi et al. 2013), tocopherol content (unpublished data), and disease resistance (unpublished data). Firstly, we analyzed multi-trait phenotypic and genetic correlations using this large trait data set in B. napus, and related this to expression of seed yield via path analysis. Secondly, we identified and integrated QTL regions associated with important traits (in particular seed yield) in B. napus, using a dense genetic map based on the Brassica Illumina Infinium 60 K SNP array. Thirdly, we analyzed the direct genetic effects and plasticity of QTL as well as indirect contributions to seed yield across environments, taking trait correlations into consideration. Finally, we tested the impact of yield-associated SNP markers on the accuracy of genomic prediction of seed yield.
Materials and methods
Plant material and available phenotype data
A doubled-haploid population (named “BnaTNDH”) derived from an F1 hybrid between “Tapidor” (a European winter-type rapeseed cultivar) and “Ningyou7” (a Chinese semi-winter type rapeseed cultivar) was used for this study (Qiu et al. 2006). From this population, 182 DH lines with genotyping data generated by the Brassica Illumina Infinium 60 K SNP array (Clarke et al. 2016) were recently used for genetic map construction (Zhang et al. 2016). Therefore, these 182 DH lines and this new map were used for QTL mapping in the present study with collected published phenotypic data (including seed yield and yield component traits, flowering time, glucosinolate presence in different tissues, seed erucic acid and oil content) and unpublished phenotype data (resistance to Leptosphaeria maculans, resistance to Sclerotinia sclerotiorum, and Vitamin E content) (see Tables 1, 2 for details of traits investigated and phenotyping locations).
Table 1.
Abbreviations and measurements of investigated traits in the BnaTNDH population
| Trait ID | Traits name | Trait abbreviation | Trait unit | Classification | Trait measurement |
|---|---|---|---|---|---|
| 1 | Seed yield per plant | SY | Gram | Yield and its components | Average dry weight of seeds of the harvested individuals in a plot |
| 2 | Seed number per pod | SN | NA | Average number of well-filled seeds from 100 well-developed pods, sampled from the primary branch in the middle of the harvested individuals in a plot | |
| 3 | Seed weight | SW | Gram | Average dry weight of 1000 well-filled seeds from three replicate samples, from the mixed seeds of the harvested individuals in a plot | |
| 4 | Pod number | PN | NA | Number of well-filled, normally developed pods on each harvested individual in a plot | |
| 5 | Seed yield per pod | PW | % | Dry weight of seed of each pod | |
| 6 | Biomass yield per plant | BY | Gram | Average total above-ground (except the seeds) dry weight of the harvested individuals in a plot | |
| 7 | Branch number | BN | NA | Developmental traits | Number of branches arising from the main shoot of each harvested individual in a plot |
| 8 | Development time of seeds | DT | Day | Interval between the date of flowering time and the date when harvest | |
| 9 | First branch height | FBH | cM | Measured from the base of the stem to the first primary branch | |
| 10 | Ratio of FBH to PH | RBH | NA | Relative first primary branch height | |
| 11 | Flowering time | FT | Day | Interval between the date of sowing and the date when the first flowers emerged on 50% of the plants in a plot | |
| 12 | Maturity time | MT | Day | Interval between the date of sowing and the date when pods on most of the plants in a plot were yellow | |
| 13 | Plant height | PH | cM | Measured from the base of the stem to the tip of the main shoot | |
| 14 | Pod number of main inflorescence | PNM | NA | Pod number of main inflorescence | |
| 15 | Resistance against Leptosphaeria Maculans (blackleg) | BR | Number | Resistance to stress | Symptoms were assessed at 16-18 days after inoculation using a 0–9 scale (0: no symptoms; 9: large grey lesions with pycnidia) (Koch et al. 1991); where 0-4 are classified as resistant, 5 as intermediate resistant and 6-9 as susceptible. The disease scores were used as quantitative data for QTL identification |
| 16 | Resistance against Sclerotinia sclerotiorum | SR | % | Radius of Sclerotinia sclrotiorum disease spots (oval) on the leaf (Trait abbriviation RA,RB,RC RD and RE represent the radius measured in different independent experiment) | |
| 17 | Erucic acid content | ERU | % | Seed quality | Erucic acid content of seed |
| 18 | Linolenic acid | LEN | % | Linolenic acid content | |
| 19 | Oil content | OC | % | Oil content of seed | |
| 20 | Protein content | PRO | % | Protein content in seeds | |
| 21 | Glucosinolates | ||||
| 1-Methoxy-3-indolylmethyl-GSL | Ind-NEO | μmol g−1 | Neoglucobrassicin content | ||
| 2-Hydroxy-3-butenyl-GSL | Ali-4C-PRO | μmol g−1 | Progoitrin content | ||
| 2-Hydroxy-4-pentenyl-GSL | Ali-5C-GNL | μmol g−1 | Gluconapoleiferin content | ||
| 3-Butenyl-GSL | Ali-4C-GNA | μmol g−1 | Gluconapin content | ||
| 3-Indolyl-methyl-GSL | Ind-GBS | μmol g−1 | Glucobrassicin content | ||
| 4C-Aliphatic glucosinolate | Ali-4C | μmol g−1 | Sum of Ali-4C-GRA,Ali-4C-GNA and Ali-4C-PRO | ||
| 4-Hydroxy-3-indolyl-methyl-GSL | Ind-4OH | μmol g−1 | 4-Hydroxyglucobrassicin content | ||
| 4-Methoxy-3-indolylmethyl-GSL | Ind-4ME | μmol g−1 | 4-Methoxyglucobrassicin content | ||
| 4-Methylsulfinylbutyl-GSL | Ali-4C-GRA | μmol g−1 | Glucoraphanin content | ||
| 4-Pentenyl-GSL | Ali-5C-GBN | μmol g−1 | Glucobrassicanapin content | ||
| 5C-Aliphatic glucosinolate | Ali-5C | μmol g−1 | Sum of Ali-5C-GAL,Ali-5C-GBN and Ali-5C-GNL | ||
| 5-Methylsulfinylamyl-GSL | Ali-5C-GAL | μmol g−1 | Glucoalyssin content | ||
| Aliphatic glucosinolate | Ali | μmol g−1 | Sum of Ali-4C and Ali-5C | ||
| Total Glucosinolate | TGS | μmol g−1 | Total glucosinolate content | ||
| Glucosinolate | GLU | μmol g−1 | Total glucosinolate content by NIRS | ||
| 2-Phenylethyl GSL | GST | μmol g−1 | Gluconasturtiin content | ||
| Indolyl glucosinolate | Ind | μmol g−1 | Sum of Ind-GBS,Ind-4OH,Ind-4ME and Ind-NEO | ||
| 22 | Vitamin E | ||||
| Tocopherol composition | TCO | NA | Ratio of the α-tocopherol content and γ-tocopherol content | ||
| Total tocopherol content | TTC | ppm | Total tocopherol content of the seed | ||
| α-Tocopherol content | αTC | ppm | α-Tocopherol content of the seed | ||
| γ-Tocopherol content | γTC | ppm | γ-Tocopherol content of the seed |
Table 2.
Field experiment design and traits investigated in different environments
| Name of macro-environment | Name of agro-ecological area | Location and geographic feature | Rapeseed growing period | Code of experiment* | Investigated traits |
|---|---|---|---|---|---|
| Semi-winter | Middle valley of Yangtze River | Wuhan, Hubei, E114°21′/N30°37′/40 m | Oct 2002–May 2003 | S3 | BN, FT, MT, PH, PN, SN, SW, SY, OC, DT, ERU, GLU, PNM, PW |
| Wuhan, Hubei, E114°21′/N30°37′/40 m | Oct 2003–May 2004 | S4 | BN, FT, MT, PH, PN, SN, SW, SY, TTC, αTC, γTC, TCO, Ali-4C-GRA, Ali-4C-GNA, Ali-4C-PRO, Ali-5C-GAL, Ali-5C-GBN, Ali-5C-GNL, GST, Ind-4OH, Ind-4ME, Ind-NEO, Ali-4C, Ali-5C, Ali, Ind-GBS, Ind, TGS, OC, DT, ERU, GLU, PNM, PW | ||
| Wuhan, Hubei, E114°21′/N30°37′/40 m | Oct 2004–May 2005 | S5ʹ | Ali-4C-GNA, Ali-4C-PRO, Ali-5C-GAL, Ali-5C-GBN, Ali-5C-GNL, GST, Ind-4OH, Ind-4ME, Ali-4C, Ali-5C, Ali, Ali-4C-GRA, Ali-4C-GNA, Ali-4C-PRO, Ali-5C-GAL, Ali-5C-GBN, Ali-5C-GNL, GST, Ind-4OH, Ind-4ME, Ind-NEO, Ali-4C, Ali-5C, Ali, Ind-GBS, Ind, TGS, Ind-GBS, Ind, TGS | ||
| Wuhan, Hubei, E114°21′/N | Oct 2005–May 2006 | S6ʹ | Ali-4C-GNA, Ali-4C-PRO, Ali-5C-GAL, Ali-5C-GBN, Ali-5C-GNL, GST, Ind-4OH, Ind-4ME, Ind-NEO, Ali-4C, Ali-5C, Ali, Ali-4C-GRA, Ali-4C-GNA, Ali-4C-PRO, Ali-5C-GAL, Ali-5C-GBN, Ali-5C-GNL, GST, Ind-4OH, Ind-4ME, Ind-NEO, Ali-4C, Ali-5C, Ali, Ind-GBS, Ind, TGS, Ind-GBS, Ind, TGS, OC | ||
| 30°37′/40 m | |||||
| Wuhan, Hubei, E114°21′/N30°37′/40 m | Oct 2006–May 2007 | S7 | FT, MT, SW, SY, OC, SP, DT | ||
| Jiangling, Hubei, E113°25′/N30°30′/40 m | Oct 2004–May 2005 | S5 | BN, FT, MT, PH, PN, SN, SW, SY, TTC, αTC, γTC, TCO, OC, DT, ERU, GLU, PRO, PW | ||
| Daye, Hubei, E114°48′/N30°06′/100 m | Oct 2005–May 2006 | S6 | BN, BY, FT, MT, PH, PN, SN, SW, SY, Ali-4C-GRA, Ali-4C-GNA, Ali-4C-PRO, Ali-5C-GAL, Ali-5C-GBN, Ali-5C-GNL, GST, Ind-4OH, Ind-4ME, Ind-NEO, Ali-4C, Ali-5C, Ali, Ind-GBS, Ind, TGS, OC, DT, ERU, GLU, LEN, PRO, PW | ||
| Qichun, Hubei, E115°26′/N30°13′/30 m | Oct 2008–May 2009 | S9 | BN, PH, PN, SN, SW, SY, FBH, RBH under low and optimal phosphorus growth condition | ||
| Qichun, Hubei, E114°48′/N30°06′/30 m | Oct 2009–May 2010 | S10 | BN, PH, PN, SN, SW, SY, FBH, RBH under low and optimal phosphorus growth condition | ||
| Qichun, Hubei, E114°48′/N30°06′/30 m | Oct 2010–May 2011 | S11 | BN, PH, PN, SN, SW, SY, FBH, RBH under low and optimal phosphorus growth condition | ||
| Lower valley of Yangtze River | Hangzhou, Zhejiang, E120°12′/N30°16′/40 m | Oct 2006–May 2007 | E7 | FT, MT, SW, SY, OC | |
| Winter | The Loess Plateau | Dali, Shaanxi, E109°56′/N34°52′/800 m | Sep 2002–Jun 2003 | N3 | BN, FT, MT, PH, PN, SN, SW, SY, OC, DT, ERU, GLU, PNM, PW |
| Dali, Shaanxi, E109°56′/N34°52′/800 m | Sep 2003–Jun 2004 | N4 | BN, FT, MT, PH, PN, SN, SW, SY, DT, ERU, GLU, PNM, PW,TTC, αTC, γTC, TCO, OC | ||
| Dali, Shaanxi, E109°56′/N34°52′/800 m | Sep 2005–Jun 2006 | N6 | BN, BY, FT, MT, PH, PN, SN, SW, SY, OC, DT, ERU, GLU, LEN,PRO, PW, | ||
| Dali, Shaanxi, E109°56′/N34°52′/800 m | Sep 2006–Jun 2007 | N7 | FT, MT, SW, SY, OC, DT, MT | ||
| Spring | The Loess Plateau | Hezheng (35°36′ N, 103°11′ E, elev.2031 m) | Sep 2005–Jun 2006 | N6ʹ | OC |
| Green house | Wuhan, Hubei, E114°21′/N30°37′/40 m† | 2005 | G5 | SR (RA,RB) | |
| 2006 | G6 | SR (RC,RD,RE) | |||
| Britain, controlled environment | Jun 2009 | G9 | BR | ||
BN branch number, BY biomass yield, FT flowering time, MT maturity time, PH plant height, PN pod number, SN seed number, SW seed weight, SY seed yield
* The first letter represents the code of the agro-ecological area (middle valley of Yangtze River was in south China and coded as S; lower valley of Yangtze River was in east China and coded as E; the Loess Plateau was in north China and coded as N), the middle letter represents the year of harvest and the last letter represents the code of the population (D, DH population; R, RC-F2 population)
† Sclerotinia sclerotiorum inoculation was performed in the greenhouse
The field locations were classified into three macro-environments: semi-winter, winter and spring. The harvest year combined with the location was set as an “experiment” and all experiments used a randomized complete block design with three replications, as described in Shi et al. (2009). In addition, three experiments were conducted under greenhouse conditions (Table 2).
Collection of phenotype data on disease resistance
Resistance to Sclerotinia sclerotiorum (stem rot) was assessed at the seedling stage on the sixth leaf of each plant. The inoculation was performed in a plastic container, where mycelial agar disks of S. sclerotiorum were placed on the upper surfaces of leaves. Plants were kept in the dark for 16 h, then provided with 12 h photoperiod (12 h light at 32 °C/12 h darkness at 22 °C, light intensity at 210 µmol m−2 s−1). Diameters of S. sclerotiorum leaf lesions were recorded at three different time points to provide an index of resistance to S. sclerotiorum. This index was used as quantitative data for identification of QTL.
The phoma stem canker pathogen (Leptosphaeria maculans) isolate 99-79 was sourced from the AAFC Saskatoon Research Centre collection, kindly provided by Dr. H. R. Kutcher. This isolate produced distinct resistant (Tapidor) and susceptible (Ningyou7) phenotypes on parental lines and therefore was used to inoculate the BnaTNDH population lines to map the resistance gene.
Conidial suspension of isolate 99-79 was prepared from 12-day-old cultures on V8 agar and spore concentration was adjusted to 107 spores per ml. Seedlings of the 200 DH lines were grown in 40-well trays in a controlled environment room (20 °C day/20 °C night, 12 h light/12 h darkness, light intensity 210 µmol m−2 s−1). Cotyledons of 14-day-old seedlings were wounded and a 10 µl drop of conidial suspension was placed over each wound. Each seedling had four inoculation sites (one on each cotyledon lobe). Five seedlings of each DH line were inoculated (i.e. 20 inoculation sites). Symptoms were assessed at 16–18 days after inoculation using a 0–9 scale (0: no symptoms; 9: large gray lesions with pycnidia); where 0–4 were classified as a resistant response, 5 as an intermediate resistant response and 6–9 as a susceptible response. The disease scores were used as quantitative data for QTL identification.
Linkage map and marker alignment to the B. napus “Darmor-bzh” reference genome
A genetic linkage map of the BnaTNDH population (182 lines) with a total of 2041 genetic bins was constructed from 12960 SNP markers detected with the Brassica 60 K Illumina Infinium array (Zhang et al. 2016; Supplementary Table 1). We aligned all the SNP markers included in the genetic loci of the BnaTNDH genetic linkage map to the reference B. napus genome of “Darmor-bzh 4.1” (Chalhoub et al. 2014) by BLAST (Altschul et al. 1990) as performed in Zou et al. (2016). For those representative bin markers with no hits or uncertain hits, we used the physical location of other markers in the same genetic bin. All information related to the physical alignment of the SNP markers in the genetic map to the reference genome is listed in Supplementary Table 1.
Phenotype and QTL analyses
Phenotypic correlation between all traits in a single environment was estimated using Spearman’s rank correlation coefficient (Myers et al. 2010). Genetic correlation between traits investigated in multi-environments was estimated using multivariate restricted maximum likelihood estimation (REML) in SAS Proc MIXED (Holland et al. 2002). Heritability was calculated using the equation: h 2 = σ2g/(σ2g + σ2ge/n + σ2e/nr), where σ2g is the genetic variance, σ2ge is the variance representing genotype-by-environment interactions and σ2e is the error variance, n is the number of environments and r is the number of replications. The variance of the components was estimated by SAS using a general linear model (GLM). Path analysis (Wright 1934) was also done for each environment to further investigate links between traits and their effects on seed yield. Seed yield (SY) was set as the dependent variable while other traits investigated in corresponding experiments were set as independent variables. The direct or indirect effects of the independent variables on the dependent variable were estimated. The variables were initially standardized and the following path analysis model was established:
where S i was the standard deviation of x, S y was the standard deviation of y, and α 1, α 2, …α m were the direct effects of independent variables on SY. A system of normal equations was used to estimate the direct and indirect effects of each explanatory variable on SY:
The indirect effect of independent variable x j on independent variable x i was calculated as r ij α jy. Statistical analysis was conducted in Microsoft Office Excel® and R (R Core Team 2015).
WinQTLcart v2.5 (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm) was used for QTL detection by composite interval mapping (Silva Lda et al. 2012; Wang et al. 2012). The number of control markers, window size, and walking speed were set to 5, 10, and 1 cM, respectively. Permutation analysis with 1000 repetitions was performed to obtain the LOD threshold (Churchill and Doerge 1994). A significance level of (P = 0.05) was adopted as the LOD threshold, and QTL with LOD scores greater than the LOD threshold were accepted as significant QTL. To avoid elimination of minor effect QTL with small LOD scores, a smaller LOD (P = 0.5) was adopted to determine suggestive QTL. Suggestive QTL that were repeatedly detected in multiple experiments were accepted for further analysis. Finally, significant QTL overlapping with suggestive QTL were admitted as authentic QTL and named as identified QTL according to the method reported by Long et al. (2007) and Shi et al. (2009). The identified QTL with R 2 ≥ 20% were accepted as major QTL and those with R 2 < 20% were accepted as minor QTL.
A meta-analysis algorithm (Goffinet and Gerber 2000) was developed to determine the number of “real” QTL within a region containing many independently identified QTL. The BioMercator v4.2 (http://moulon.inra.fr/index.php/fr/seminairedoc/cat_view/21-logiciels/101-abi-project-and-software/104-biomercator) was used to perform this meta-analysis: an adjusted Akaike criterion was adopted to determine the probable QTL numbers from the five given models (1-, 2-, 3-, 4- and N-QTL model, corresponding to the number of predicted true QTL). The software also provided a method to estimate the confidence interval of the predicted QTL position, where S 2i was the variance of the position of the QTLi and k was the number of selected QTL selected for the meta-analysis (Chardon et al. 2004; Khatkar et al. 2004; Shi et al. 2009).
Firstly, all QTL identified for each trait were integrated using meta-analysis to get consensus QTL, using the methods of Shi et al. (2009). Consensus QTL that could be detected in more than one experiment were classified as reproducible QTL, while others were classified as non-reproducible. As glucosinolate and tocopherol content include a large number of components, meta-analysis was done for these two traits and their components as described in Feng et al. (2012). Secondly, based on the phenotypic analysis and complex QTL distribution, a second round of meta-analysis was done for all consensus QTL for different traits to obtain indivisible QTL related to seed yield across the whole genome.
Accuracy of genomic prediction on seed yield
Ridge regression best linear unbiased prediction (RR-BLUP) was applied with different sets of markers for genomic prediction of seed yield using R software package “rrBLUP” (Endelman 2011). Considering that the seed yield and related trait phenotypes for the QTL detection were mostly detected in the semi-winter environments (9) rather than in the winter environments (4), phenotype data of seed yield from semi-winter environments was processed for testing the prediction. The environmental effects were estimated according to Zhao et al. (2013, 2015). In total, 100 runs of fivefold cross-validation were performed for the model, and the genomic selection accuracy was evaluated by the R 2 value divided by the square root of the heritability and predicted seed yield performance (Zhao et al. 2015; Zou et al. 2016).
Results
Correlations between agronomic traits and their contributions to seed yield in the BnaTNDH population
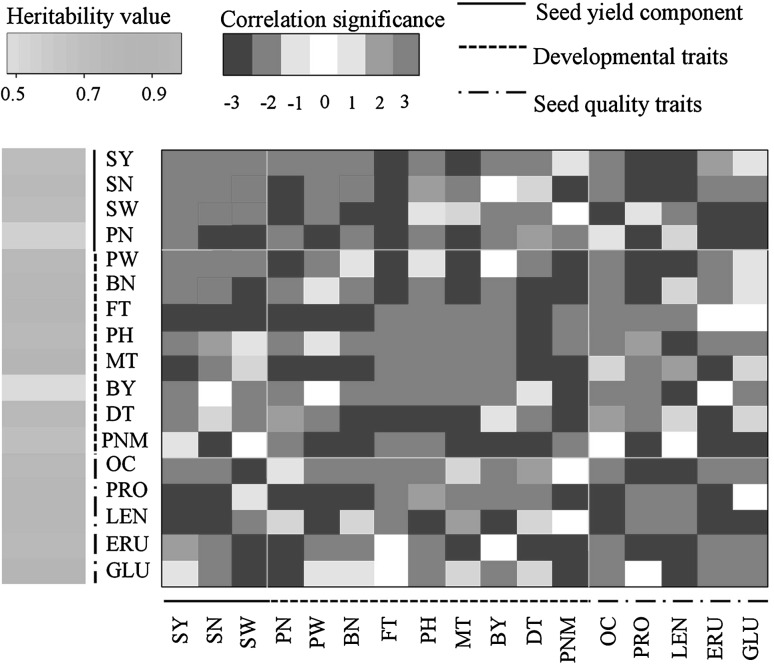
Phenotype data for a total of 22 traits in the BnaTNDH population were collected over eight growing seasons, eight locations, and three greenhouse experiments (Tables 1, 2). The glucosinolate and tocopherol content measurements included different components as shown in Table 1. We calculated the heritability of the 17 traits which had phenotype data collected over multiple environments (Fig. 1; Supplementary Table 2). Biomass yield (straw weight + seed yield) showed the lowest heritability (0.48), while heritability of all seed quality traits (erucic acid, oil content, glucosinolate content, linolenic acid) and developmental traits (flowering time, time to maturity) was >0.8 and seed yield showed heritability of 0.72.
Fig. 1.
The abbreviations SY, SN, SW, PN, PW, BN, FT, PH, MT, BY, DT, PNM, OC, PRO, LEN, ERU and GLU represent seed yield, seed number per pod, seed weight, pod number, pod weight, branch number, flowering time, plant height, maturity time, biomass yield, development time for seeds, pod number on main inflorescence, oil content, protein content, linolenic acid content, erucic acid content and glucosinolate content, respectively (Table 1). The values for correlation significance “0”, “1”, “2”, “3” represent the significance levels “no significant correlation”, “significant in a single environment (i.e., single experiment) with P < 0.01”, “significant in several environments (i.e., several experiments) with P < 0.05” and “significant in several environments with P < 0.01”, respectively
Genetic correlations and phenotypic correlations were calculated between the investigated traits across environments and within single environment. Significant correlations with seed yield were observed for all traits across the different environments with the exception of glucosinolate content (GLU) and pod number per main branch (PNM) (Fig. 1; Supplementary Table 3). These two traits were not significantly correlated with seed yield across all experiments, but showed significant correlations in a single experiment and hence were also considered as associated traits for the subsequent analyses. Disease resistance traits were investigated separately in different experiments, and hence only phenotypic correlation was calculated for these traits. However, disease resistance traits also showed significant correlation with yield in several experiments (Supplementary Table 3).
Table 3.
Numbers of identified and consensus QTL for 22 traits investigated in the BnaTNDH rapeseed mapping population
| Trait classification | QTL type | A genome | C genome | Whole genome |
|---|---|---|---|---|
| Seed yield and its components (4)* | Identified QTL | |||
| Total | 241 | 125 | 366 | |
| R 2 | 0.07 | 0.06 | 0.07 | |
| Consensus QTL | ||||
| Total | 174 | 101 | 275 | |
| Developmental trait (10) | Identified QTL | |||
| Total | 376 | 324 | 700 | |
| R 2 | 0.06 | 0.06 | 0.07 | |
| Consensus QTL | ||||
| Total | 271 | 220 | 491 | |
| Seed quality trait (25) | Identified QTL | |||
| Total | 659 | 576 | 1235 | |
| R 2 | 0.07 | 0.06 | 0.08 | |
| Consensus QTL | ||||
| Total | 507 | 419 | 926 | |
| Stress resistance (2) | Identified QTL | |||
| Total | 176 | 92 | 268 | |
| R 2 | 0.08 | 0.06 | 0.07 | |
| Consensus QTL | ||||
| Total | 133 | 79 | 212 | |
| Total | Identified QTL | |||
| Total | 1452 | 1117 | 2569 | |
| R 2 | 0.08 | 0.07 | 0.08 | |
| Consensus QTL | ||||
| Total | 1085 | 819 | 1904 | |
* The number represents the number of traits classified into this category
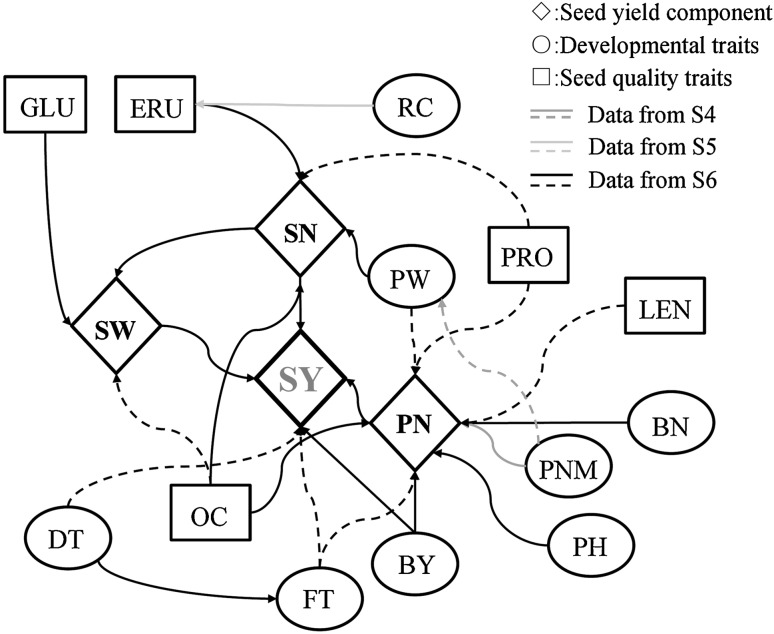
Path analysis was undertaken to identify complex connections between seed yield and other traits. The results demonstrated that although some specific traits did not affect seed yield directly, these traits affected seed yield indirectly via interactions with other traits (Fig. 2). For instance, in the N6 experiment, the direct path coefficient for plant height was −0.02, but its indirect path coefficient through pod number was 0.39. This suggests that a taller plant would usually have more pods, and therefore increased yield. In the S6 experiment, the direct path coefficient of seed development time was −0.1, but its indirect coefficients through flowering time and seed weight were 0.09 and 0.09, respectively, which explains its positive correlation of 0.15 with seed yield (Supplementary Table 4). Associated traits like oil content, seed weight, pod number and branch number showed a relatively consistent effect (positive correlation) on seed yield. Some traits also had differing impacts on seed yield in different environments. For example, flowering time showed a positive correlation with seed yield in winter environments, but was negatively correlated with seed yield in the semi-winter environments.
Fig. 2.
The abbreviations SY, SN, SW, PN, PW, BN, FT, PH, MT, BY, DT, PNM, OC, PRO, LEN, ERU and GLU represent seed yield, seed number per pod, seed weight, pod number, pod weight, branch number, flowering time, plant height, maturity time, biomass yield, development time for seeds, pod number on main inflorescence, oil content, protein content, linolenic acid content, erucic acid content and glucosinolate content, respectively (Table 1). Path analysis for the traits investigated in the environment S6 (experiment done in a semi-winter environment in 2006). Traits that have a relatively high path coefficient for seed yield are shown. For some traits not investigated in the environment S6, data from other environments (S4 and S5, experiments done in semi-winter in 2004 and 2005, respectively) were used and are indicated with blue and yellow lines. These traits include seed yield and its components (diamond), developmental traits (circle) and seed quality traits (square), linked by three different colors of lines. More results for the trait correlation and path analysis under other environments can be found in Supplementary Tables 3 and 4
Detection of consensus QTL accounting for all investigated traits using the high-density SNP genetic map of the BnaTNDH population
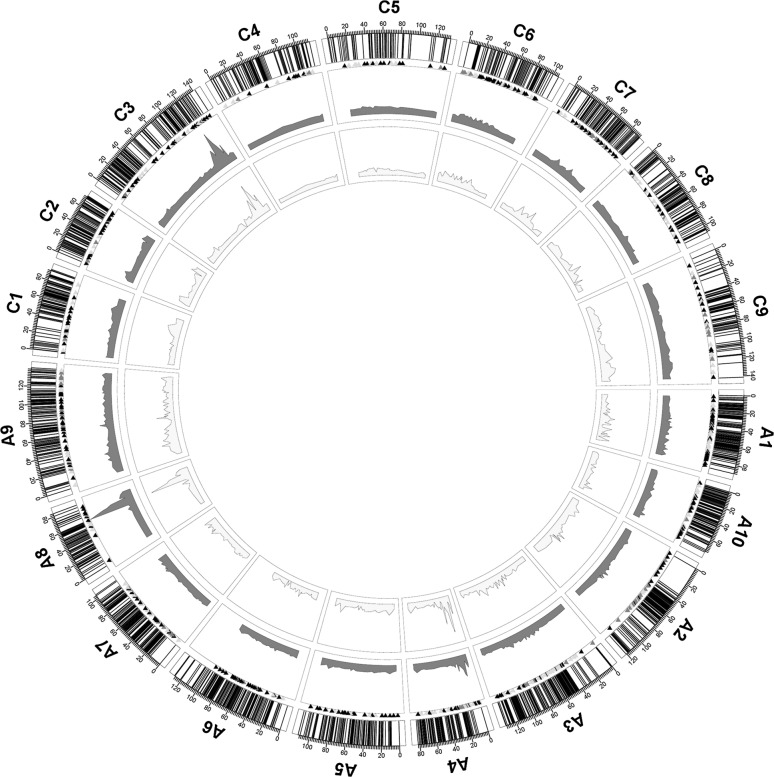
QTL accounting for seed yield and 21 additional traits collected from up to 19 experiments were analyzed using a newly constructed high-density SNP genetic map of the BnaTNDH population (Zhang et al. 2016). The newly constructed genetic map spanned a total of 2077.9 cM with an average of one locus per cM, covering 94.92% of the “Darmor-bzh” assembled reference genome as calculated by the physical position of the loci on each chromosome (Supplementary Tables 1, 5).
A total of 2569 identified QTL accounting for the 22 traits were detected with the new map: the average LOD value, R 2 and confidence interval were 4.92, 7.60% and 6.25 cM, respectively (Table 3; Supplementary Table 6). After integrating redundant QTL that were either repeatedly detected from different environments or re-counted by totals versus components of glucosinolate and tocopherol content traits, a total of 1904 consensus QTL were integrated for the 22 traits, with an average confidence interval reduced from 6.25 to 5.67 cM, in which there were 42 major QTL (R 2 > 20%), included seed yield, glucosinolate, erucic acid, disease resistance, seed number per pod, branch number, pod number, relative first primary branch height (Supplementary Tables 6, 7, 8). For those traits investigated in both semi-winter and winter macro-environments, a total of 155 QTL could be detected in both of the micro-environments. For the traits investigated in more than one experiment, 22.18% (420 out of 1894) of the QTL were repeatedly detected, and these were considered to be reproducible QTL. Furthermore, the proportion of reproducible QTL was significantly associated with the heritability of the traits (r = 0.63, P < 0.05).
After the meta-analysis, most consensus QTL for a single trait overlapped with QTL for other traits, illustrating a complex genetic regulatory network. The additive effects of some QTL also corresponded to the genetic correlations between traits. For example, qPN.A3-1 and qSY.A3-2 overlapping on linkage group A3 were responsible for the increase in pod number and seed yield, respectively (Supplementary Table 7), suggesting a pleiotropic effect or close linkage of the genes for the two traits. The two traits were also significantly correlated (r = 0.25, P < 0.001). A total of 12 QTL clusters distributed in linkage group A2, A3, A4, A7, A9, A10, C4, C5 and C6 were observed (Supplementary Table 7). For instance, in linkage group C6, there were 16 consensus QTL clustered in the region covering 13.9 cM, accounting for seed yield, branch number, pod number, plant height, flowering time, glucosinolates, and erucic acid, and these traits were significantly correlated (Supplementary Tables 3, 7).
Detection of “essential QTL” for seed yield with consideration of trait associations
A total of 80 consensus QTL responsible for seed yield were detected using the seed yield phenotype value: of these, 78 overlapped with QTL for other correlated traits. Correlation and path analysis also showed that seed yield was directly or indirectly correlated with almost every trait investigated. Therefore, meta-analysis was conducted to integrate the QTL for the traits correlated with seed yield (Supplementary Table 9). As the genetic correlation between seed yield and seed quality traits would be affected by the path correlation among the seed quality component traits, seed quality trait-related QTL that did not overlap with yield and yield-component trait QTL after meta-analysis were removed. As a result, 525 inseparable QTL involving 1266 markers across the whole genome were obtained, with an average QTL confidence interval of only 2.48 cM, and denoted as “essential QTL” contributing to the trait of seed yield (Table 4; Fig. 3; Supplementary Tables 9, 10; Supplementary Fig. 1). SNP alleles of the 1266 markers that positively contributed to seed yield are provided in Supplementary Table 10, and the candidate genes in the interval of the essential QTL are provided in Supplementary Table 11. Among the 525 essential QTL, 428 were integrated from QTL for multiple traits and were defined as pleiotropic, while the other 97 QTL were trait-specific. For the trait-specific essential QTL, two were seed yield-specific and 11 were seed yield component-specific. Most trait-specific essential QTL were responsible for developmental traits such as plant height, flowering time and branch number. There were also several seed quality trait QTL controlling, for example, glucosinolate components, seed oil and erucic acid content, which may affect seed yield in specific environments (Supplementary Table 9).
Table 4.
Distribution of essential QTL for seed yield and informative markers in the BnaTNDH rapeseed mapping population
| Genome | No. essential QTL | Trait-specific | Pleiotropic | Average interval (cM) | Total covered distance (cM) | No. markers within confidence intervals |
|---|---|---|---|---|---|---|
| A01 | 41 | 11 | 30 | 1.54 | 63.06 | 103 |
| A02 | 34 | 3 | 31 | 2.49 | 84.64 | 59 |
| A03 | 43 | 4 | 39 | 1.73 | 74.41 | 102 |
| A04 | 29 | 7 | 22 | 2.2 | 63.8 | 81 |
| A05 | 24 | 5 | 19 | 3.24 | 77.73 | 69 |
| A06 | 25 | 9 | 16 | 2.35 | 58.64 | 78 |
| A07 | 29 | 5 | 24 | 2.37 | 68.69 | 92 |
| A08 | 14 | 6 | 8 | 2.57 | 35.98 | 35 |
| A09 | 45 | 7 | 38 | 1.83 | 82.24 | 100 |
| A10 | 23 | 2 | 21 | 1.88 | 43.35 | 53 |
| A genome | 307 | 59 | 248 | 2.22 | 681.23 | 772 |
| C01 | 21 | 5 | 16 | 2.52 | 52.85 | 56 |
| C02 | 19 | 5 | 14 | 2.21 | 42.06 | 48 |
| C03 | 33 | 6 | 27 | 2.83 | 93.31 | 80 |
| C04 | 14 | 4 | 10 | 4.8 | 67.22 | 42 |
| C05 | 20 | 3 | 17 | 4.12 | 82.3 | 40 |
| C06 | 34 | 3 | 31 | 2.63 | 89.47 | 60 |
| C07 | 23 | 3 | 20 | 2.96 | 68.16 | 52 |
| C08 | 26 | 5 | 21 | 3.04 | 79.07 | 70 |
| C09 | 28 | 4 | 24 | 2.73 | 76.5 | 46 |
| C genome | 218 | 38 | 180 | 3.09 | 674.49 | 494 |
| Total (A + C) | 525 | 97 | 428 | 2.48 | 1303.58 | 1266 |
Fig. 3.
The outer circle is the BnaTNDH linkage map, the short black lines indicate the positions of map markers. The red triangles, green triangles and black triangles under the linkage groups indicate the essential QTL for seed yield, seed yield components and other traits, respectively. The red wave in the middle circle indicates the LOD scores for corresponding composite QTL, while the yellow wave in the inner circle indicates the R 2 values for corresponding essential QTL. The details of LOD scores and the R 2 values for corresponding essential QTL are shown in Supplementary Table 9 (color figure online)
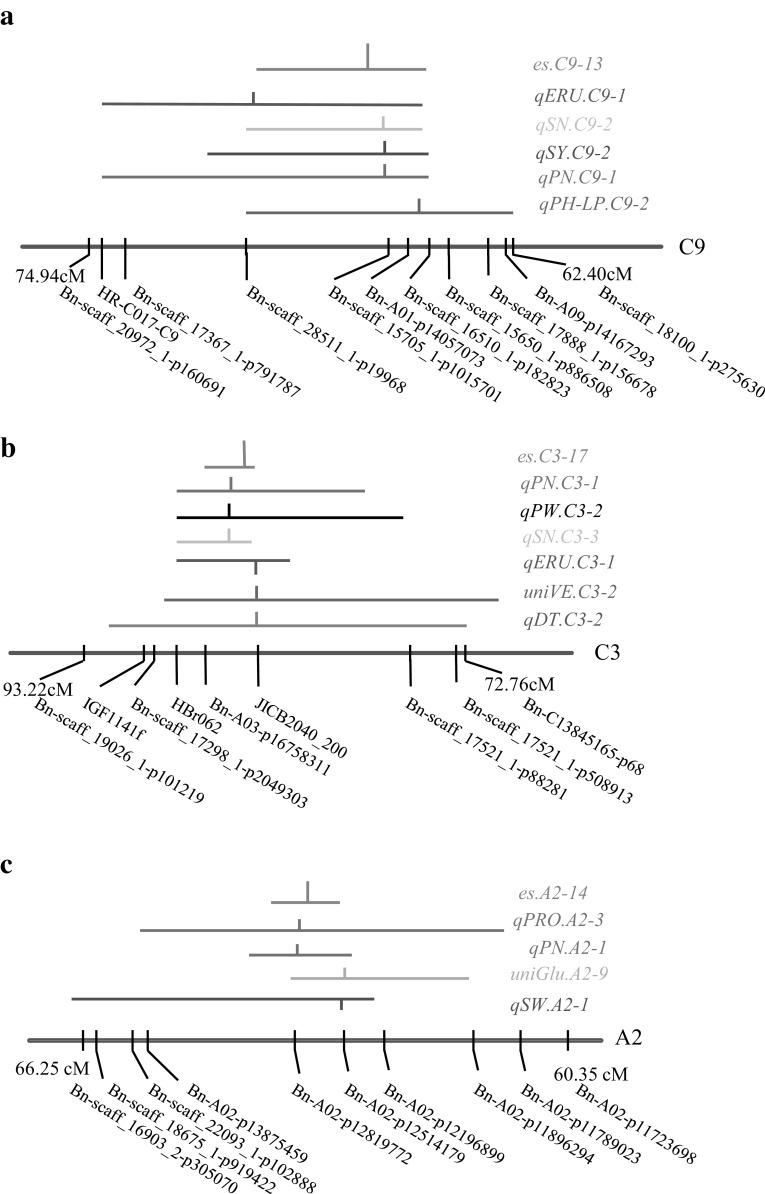
Some of the essential QTL (75) were directly comprised of consensus QTL for seed yield and other consensus QTL for yield-related traits. For example, seed yield QTL qSY.C9-2 on linkage group C9 overlapped with QTL for seed number (qSN.C9-2), pod number (qPN.C9-1), and plant height (qPH-LP.C9-2) (Supplementary Table 9, Fig. 4a). Some of the essential QTL (31) did not directly involve consensus QTL for seed yield, but contained at least two consensus QTL for seed yield-related traits with consistent effects (either positive or negative contribution to the traits). Although consensus QTL for seed yield could not be detected from such loci, the accumulated effects from these yield-component QTL could contribute substantially to seed yield. For instance, the allele of QTL qSN.C3-3 associated with increasing seed number overlapped with QTL qPN.C3-1 for increasing pod number (77–86 cM on linkage group C3; Fig. 4b). A number of the essential QTL (25) did not contain consensus QTL for seed yield, but contained consensus QTL for seed yield-related trait components with opposite phenotypic contributions (both positive and negative in relation to seed yield). For example, at 60.9–66.2 cM on linkage group A2, the Tapidor allele of QTL qSW.A2-1 was responsible for reducing seed weight, whilst the overlapping qPN.A2-1 was responsible for increasing pod number (Fig. 4c).
Fig. 4.
Horizontal lines indicate the QTL intervals while the short vertical lines indicate the positions of peaks. A downward peak position line indicates that the value of the genetic effect of the QTL is negative, and an upward line indicates that QTL has a positive genetic effect on the trait. a On the C9 linkage group, the seed yield QTL (qSY.C9-2) that overlapped with seed yield component QTL were integrated into es.C9-13; b On the C3 linkage group, QTL for seed yield components with positive contribution were integrated into es.C3-17; c On the A2 linkage group, QTL for seed yield components with opposite contributions were integrated via meta-analysis into one essential QTL named as es.A2-14. SN seed number per pod, SY seed yield, SW seed weight, PN pod number, ERU erucic acid content, PH-LP plant height under low phosphorus condition, PW seed yield per pod, VE vitamin E content, DT development time for seeds, PRO protein content, GLU glucosinolate content
For indirect QTL with consistent effects, 26 showed environment-specific contribution to seed yield, as they overlapped with several seed yield component QTL detected in the same macro-environment. Meanwhile, five indirect QTL affected the seed yield through different yield components in different macro-environments. For instance, es.C3-22 included QTL for pod number (qPN.C3-3, detected in the winter environment) and QTL for seed number (qSN.C3-5, detected in the semi-winter environment). This suggested that this essential QTL for seed yield could affect yield performance through different paths in different environments. Of the indirect QTL with mixed effects, 21 had opposite contributions to yield components, offsetting the yield improvement in the same macro-environment. However, four indirect QTL showed such a contribution in different macro-environments. These QTL showed an environment-specific potential for yield improvement, and mostly showed a positive contribution in the semi-winter environment. Only es.C9-17 showed a positive contribution to seed yield in the winter environment (through increased pod number).
Prediction accuracy for seed yield based on “essential QTL”
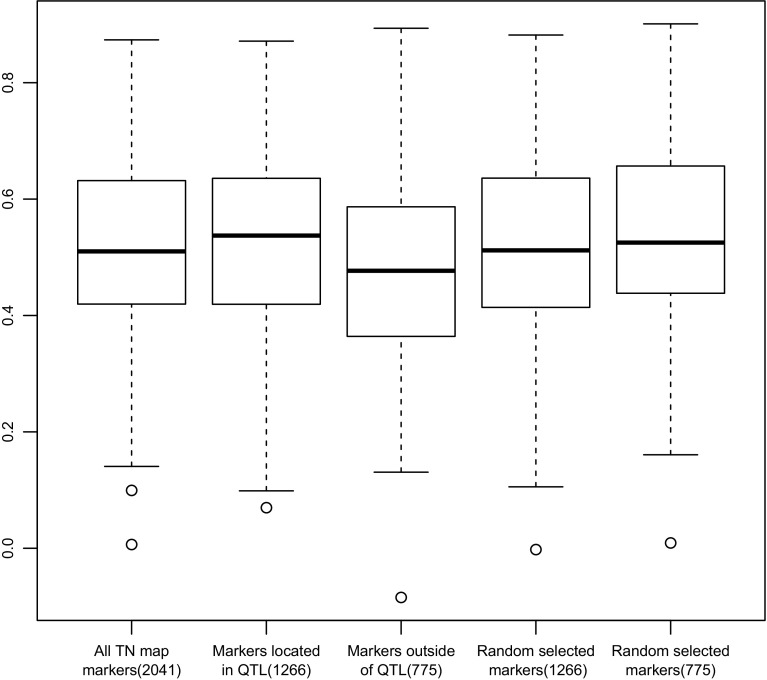
To test the prediction accuracy for seed yield based on predictions implementing essential QTL, we used the 2041 marker bins in the TN linkage map to perform prediction on seed yield in semi-winter environments using the prediction model RR-BLUP. Of these 2041 markers, 1266 markers were located within the confidence intervals of essential QTL (Supplementary Table 10) and 775 markers were not located within the confidence intervals of essential QTL. To avoid confounding effects from shared markers between the two datasets, two additional sets of 1266 and 775 markers were also randomly selected from the linkage map to assess prediction accuracy. The highest prediction accuracy of 0.5211 was achieved when the markers used only involved the 525 essential QTL. This prediction accuracy was superior to using the set of markers out of the QTL interval, using the same number of markers randomly chosen from the whole set of markers, or even to using the whole set of 2041 markers for prediction (accuracy of 0.5068) (Fig. 5).
Fig. 5.
Prediction of genomic selection accuracy was done using 2041 representative bin markers from the BnaTNDH genetic map, 1266 bin markers located within QTL confidence intervals, 775 bin markers located outside QTL confidence intervals, and by comparison 1266 randomly selected bin markers and 775 randomly selected bin markers, respectively. The top and bottom of each box represent the prediction values of the upper 25% and the lower 25% of the markers; the bold line within each box represents the median prediction value. The upper and lower lines outside the box represent the largest and smallest prediction values
Discussion
Most agronomic traits of interest in crop breeding are complex quantitative traits, controlled by many genes with minor effects (Chesler et al. 2005). Detecting these minor-effect genes and biologically and statistically validating their effects on crop phenotypes is challenging. As a result of crop domestication, a considerable number of traits are associated, either by co-selection, or through co-regulation of underlying genes (Balasubramanian et al. 2007; Bernardo 2008). When estimating and attempting to improve seed yield, breeders always consider not only the trait of seed yield itself, but also traits associated with seed yield, such as plant height, branching, flowering time and pod number. However, these interrelationships between traits are rarely considered in QTL prediction. This may limit prediction accuracy as well as our understanding of how different phenotypes and underlying genetic loci interact to control complex traits. In this study, we propose the concept of “essential QTL”: combined multi-trait or multi-environment QTL that contribute either directly or indirectly (via interactions between related traits) to seed yield. Our results provide a model for predicting and understanding complex traits in crops.
Our analysis of correlations between traits and QTL locations in the large BnaTNDH data set (Figs. 1, 2, 3; Supplementary Table 8) revealed a complex genetic architecture for seed yield. A total of 525 essential QTL for seed yield were identified, spanning 2.48 cM on average (about 731.56 kb; Table 4), anchored by at least one unique SNP-bin marker. Most of the essential QTL showed pleiotropic effects, as well as environment effects. Finally, a total of 1266 essential QTL markers (or representative markers for genetic bins) were identified for reference in future studies (Supplementary Table 10).
Using essential QTL significantly improved prediction accuracy for seed yield under a genomic selection model relative to other marker datasets used in our study (Fig. 5). Although there are a few reports on the prediction accuracy for seed yield with GS models as 0.45 (Jan et al. 2016; Würschum et al. 2014), little research related to prediction accuracy of seed yield has been done in rapeseed. In this study, the genomic prediction accuracy for seed yield in semi-winter environment using the whole marker dataset was 0.5068, but the accuracy was improved (to 0.5211) by using markers located in QTL regions (Fig. 5), which highlights the potential use of these markers in increasing prediction accuracy. With the implementation of essential QTL on the whole genome selection model for seed yield, the prediction accuracy would be further improved. Therefore, combining previously acquired QTL information with our proposed prediction strategy could facilitate the process of whole genome selection for complex traits in future.
Of the essential QTL we have identified, nearly one-fifth had a direct effect on seed yield, but the majority had an indirect effect on seed yield through the contribution of yield-related traits. These latter essential QTL had either a direct effect (all QTL positive or all negative with respect to effect on seed yield) or contained a mixture of positive and negative QTL affecting seed yield in the essential QTL region. In common practice, only direct QTL are detected via the final value of seed yield (Chen et al. 2010; Maccaferri et al. 2008; Moncada et al. 2001; Peng et al. 2011; Radoev et al. 2008). Using the essential QTL approach, we detected an additional 56 QTL that influenced seed yield indirectly through control of other traits. Nearly half (25) of these essential QTL were composed of QTL with opposite effects, offsetting each other in their final contribution to seed yield. The effect of co-localized QTL has also been reported in other studies (Sun et al. 2012; Zhao et al. 2016). In rice, gene OsPTR9 was found to increase seed yield by 16.7–24.5% compared with wild-type lines by improving the nitrogen utilization efficiency (Fang et al. 2013). Recently, Fan et al. (2016) also identified a protein controlling nitrogen uptake efficiency which could increase rice yield by fifty percent. These results provide evidence that yield-related traits such as nutrient uptake efficiency can affect seed yield. The detection of “essential QTL” provides a means of identifying the genetic basis of complex traits under a dynamic growing system with sophisticated trait associations. Our analysis of seed yield comprises an effective approach to dissect complex traits, which would be also used for similar investigation of other traits in other crops.
Comprehensive QTL analysis to discover trait-associated markers is necessary for molecular marker-based selection in rapeseed pre-breeding and breeding. The high-density genetic map newly constructed with the assistance of the Illumina Infinium Brassica 60 K SNP array (Zhang et al. 2016), combined with eight years of phenotype data across different environments for up to 22 traits (including seed yield and yield components, plant development, seed quality, and biotic stresses), allowed us detect a total of 1904 consensus QTL accounting for 22 traits with an average confidence interval of 5.67 cM (Supplementary Table 7). Furthermore, the available “Darmor-bzh” B. napus reference genome sequence (Chalhoub et al. 2014) allowed us to align 60 K SNP markers to physical positions genome-widely.
From the QTL analysis of BnaTNDH population across multiple-environments, a majority of the QTL were environment-specific, presenting in one macro-environment or single experiment. It indicated significant environment effects on complex traits, and these QTL would provide reference for understanding the environment interactions with genes (Li et al. 2003; Yang et al. 2007). While there were also considerable QTL detected in both macro-environments (155 QTL) and multiple experiments (420), introgression of the favorable alleles of these QTL would be useful for improving the adaptation of B. napus. Quite a lot of QTL clusters and major QTL were also identified (Supplementary Table 7). These QTL clusters would also provide references for further understanding of the co-selection or linkage drags in rapeseed for trait improvement. The major QTL as well as the minor QTL would also provide references for gene cloning. For example, the resistance QTL qBR.A10-1 against L. maculans identified on A10 was found to be the major resistance gene Rlm2, which has now been cloned (Larkan et al. 2014). Two small QTL for resistance against L. maculans were also detected on A4 and C3 of the BnaTNDH population, which suggests that these two QTL may contribute to quantitative resistance. Previous studies also provided evidence that there were several QTL on A4 and C3 contributing to resistance against L. maculans (Jestin et al. 2015; Huang et al. 2016). Candidate genes FLOWERING LOCUS T, FAE1, Rlm2 for flower time, erucic acid content, resistance against L. maculans, were consistently detected in the QTL regions for these traits (Supplementary Table 11).
The BnaTNDH genetic mapping population of B. napus was derived from the cross between the typical European winter-type cultivar “Tapidor” and typical Chinese semi-winter cultivar “Ningyou7”, and hence contains genomic diversity between the two major gene pools of B. napus growing in Europe and Asia (Qiu et al. 2006). More QTL were discovered in the A genome than in the C genome in our study (Table 3), supporting previous results (Shi et al. 2009). This may have been due to richer allelic variation in the A genome than in the C genome in the rapeseed germplasm pool, and this may particularly be the case for the two parents of the BnaTNDH mapping population, as the parent cultivar Ningyou7 is known to carry introgressions from the more diverse A-genome progenitor species B. rapa (Bancroft et al. 2011).
In future, as data will be continually obtained through QTL mapping and genome wide association analysis (e.g. Chen et al. 2007; Huang et al. 2016; Lee et al. 2015; Raman et al. 2014; Ramchiary et al. 2015; Xu et al. 2009), it would be possible to integrate the data from this study into further meta-analyses using this approach with consideration of trait association to provide better understanding of complex traits for crop improvement. These data and the integration of genetic and physical maps will provide an important theoretical and applied resource for studies in rapeseed genetics and genomics.
Author contribution statement
ZLL and MW performed the research, analyzed the data and wrote the manuscript; YJH and BF provided the unpublished phenotypic data for resistance against Leptosphaeria maculans and revised the manuscript; JLM, CYZ and XL provided the unpublished phenotypic data for resistance against Sclerotinia sclerotiorum and data for tocopherol components; JXX, YL, LS and JLM provided the SNP genotypes, genetic map and the previous published phenotypic data of BnaTNDH population; ASM, RS, PFL and JLM contributed to the methods; JZ designed the research, analyzed the data, and wrote the manuscript. All authors contributed, read, revised and approved the final manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1. Distribution of 525 essential QTL on each linkage group in the BnaTNDH rapeseed mapping population (TIFF 967 kb)
Supplementary material 2 (XLSX 1447 kb)
Supplementary material 7 (XLSX 346 kb)
Supplementary material 8 (XLSX 404 kb)
Supplementary material 10 (XLSX 230 kb)
Supplementary material 11 (XLSX 127 kb)
Supplementary material 12 (XLSX 6379 kb)
Acknowledgments
The authors gratefully thank the previous members (Dr. Ruiyuan Li, Dr. Jiaqin Shi, Dr. Ji Feng, Dr. Congcong Jiang, Dr. Dan Qiu, etc.) from the research group at Huazhong Agricultural University for their previous work on the BnaTNDH population. We thank Dr H. R. Kutcher for providing the Leptosphaeria maculans isolate. We thank Prof. Jiana Li of Southwest University for help with path analysis. This work was supported by the National Basic Research Program of China (Grant No. 2015CB150200, 2011CB109306), National Key Research and Development Program of China (No. 2016YFD0100305, 2016YFD0101300) and the UK Biotechnology and Biological Sciences Research Council (BB/E001610/1 & BB/I017585/1). ASM is supported by the DFG Emmy Noether Grant MA6473/1-1.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Ziliang Luo and Meng Wang contributed equally.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-017-2911-7) contains supplementary material, which is available to authorized users.
A correction to this article is available online at https://doi.org/10.1007/s00122-017-3005-2.
Contributor Information
Ziliang Luo, Email: ziliangluo@webmail.hzau.edu.cn.
Meng Wang, Email: wm17@webmail.hzau.edu.cn.
Yan Long, Email: longyan@caas.cn.
Yongju Huang, Email: y.huang8@herts.ac.uk.
Lei Shi, Email: leish@mail.hzau.edu.cn.
Chunyu Zhang, Email: zhchy@mail.hzau.edu.cn.
Xiang Liu, Email: xiang_liu@hotmail.com.
Bruce D. L. Fitt, Email: b.fitt@herts.ac.uk
Jinxia Xiang, Email: 1421906806@qq.com.
Annaliese S. Mason, Email: annaliese.mason@agrar.uni-giessen.de
Rod J. Snowdon, Email: rod.snowdon@agrar.uni-giessen.de
Peifa Liu, Email: liupeifa@webmail.hzau.edu.cn.
Jinling Meng, Email: jmeng@mail.hzau.edu.cn.
Jun Zou, Phone: 86 27 8728 8183, Email: zoujun@mail.hzau.edu.cn.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Balasubramanian V, Sie M, Hijmans RJ, Otsuka K (2007) Increasing rice production in Sub-Saharan Africa: Challenges and opportunities. In: Sparks DL (ed) Advances in Agronomy, Vol 94, pp 55–133. doi:10.1016/S0065-2113(06)94002-4
- Bancroft I, Morgan C, Fraser F, Higgins J, Wells R, Clissold L, Baker D, Long Y, Meng J, Wang X, Liu S, Trick M. Dissecting the genome of the polyploid crop oilseed rape by transcriptome sequencing. Nat Biotechnol. 2011;29(8):762–766. doi: 10.1038/nbt.1926. [DOI] [PubMed] [Google Scholar]
- Bernardo R. Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Sci. 2008;48:1649–1664. doi: 10.2135/cropsci2008.03.0131. [DOI] [Google Scholar]
- Chalhoub B, Denoeud F, Liu S, Parkin IAP, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, Correa M, Da Silva C, Just J, Falentin C, Koh CS, Le Clainche I, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Arnaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao M, Edger PP, Chelaifa H, Tack D, Lassalle G, Mestiri I, Schnel N, Le Paslier M-C, Fan G, Renault V, Bayer PE, Golicz AA, Manoli S, Lee T-H, Vinh Ha Dinh T, Chalabi S, Hu Q, Fan C, Tollenaere R, Lu Y, Battail C, Shen J, Sidebottom CHD, Wang X, Canaguier A, Chauveau A, Berard A, Deniot G, Guan M, Liu Z, Sun F, Lim YP, Lyons E, Town CD, Bancroft I, Wang X, Meng J, Ma J, Pires JC, King GJ, Brunel D, Delourme R, Renard M, Aury J-M, Adams KL, Batley J, Snowdon RJ, Tost J, Edwards D, Zhou Y, Hua W, Sharpe AG, Paterson AH, Guan C, Wincker P. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345:950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- Chardon F, Virlon B, Moreau L, Falque M, Joets J, Decousset L, Murigneux A, Charcosset A. Genetic architecture of flowering time in maize as inferred from quantitative trait loci meta-analysis and synteny conservation with the rice genome. Genetics. 2004;168:2169–2185. doi: 10.1534/genetics.104.032375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Faris JD, Hu J, Stack RW, Adhikari T, Elias EM, Kianian SF, Cai X. Saturation and comparative mapping of a major Fusarium head blight resistance QTL in tetraploid wheat. Mol Breed. 2007;19:113–124. doi: 10.1007/s11032-006-9049-7. [DOI] [Google Scholar]
- Chen G, Geng J, Rahman M, Liu X, Tu J, Fu T, Li G, McVetty PBE, Tahir M. Identification of QTL for oil content, seed yield, and flowering time in oilseed rape (Brassica napus) Euphytica. 2010;175:161–174. doi: 10.1007/s10681-010-0144-9. [DOI] [Google Scholar]
- Chesler EJ, Lu L, Shou SM, Qu YH, Gu J, Wang JT, Hsu HC, Mountz JD, Baldwin NE, Langston MA, Threadgill DW, Manly KF, Williams RW. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke WE, Higgins EE, Plieske J, Wieseke R, Sidebottom C, Khedikar Y, Batley J, Edwards D, Meng J, Li R, Lawler CT, Bouquet J, Laga B, Chung W, Iniguez-Luy F, Dyrszka E, Rae S, Stich B, Snowdon RJ, Sharpe AG, Ganal MW, Parkin IAP. A high-density SNP genotyping array for Brassica napus and its ancestral diploid species based on optimised selection of single-locus markers in the allotetraploid genome. Theor Appl Genet. 2016;129:1887–1899. doi: 10.1007/s00122-016-2746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core Team R. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2015. [Google Scholar]
- Cowling WA, Balazs E. Prospects and challenges for genome-wide association and genomic selection in oilseed Brassica species. Genome. 2010;53:1024–1028. doi: 10.1139/G10-087. [DOI] [PubMed] [Google Scholar]
- Dargahi H, Tanya P, Somta P, Abe J, Srinives P. Mapping quantitative trait loci for yield-related traits in soybean (Glycine max L.) Breed Sci. 2014;64:282–290. doi: 10.1270/jsbbs.64.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Soda M, Malosetti M, Zwaan BJ, Koornneef M, Aarts MG. Genotype x environment interaction QTL mapping in plants: lessons from Arabidopsis. Trends Plant Sci. 2014;19:390–398. doi: 10.1016/j.tplants.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Endelman JB. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome. 2011;4:250–255. doi: 10.3835/plantgenome2011.08.0024. [DOI] [Google Scholar]
- Fan X, Tang Z, Tan Y, Zhang Y, Luo B, Yang M, Lian X, Shen Q, Miller AJ, Xu G. Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc Natl Acad Sci USA. 2016;113(26):7118–7123. doi: 10.1073/pnas.1525184113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z, Xia K, Yang X, Grotemeyer MS, Meier S, Rentsch D, Xu X, Zhang M. Altered expression of the PTR/NRT1 homologue OsPTR9 affects nitrogen utilization efficiency, growth and grain yield in rice. Plant Biotechnol J. 2013;11:446–458. doi: 10.1111/pbi.12031. [DOI] [PubMed] [Google Scholar]
- Feng J, Long Y, Shi L, Shi J, Barker G, Meng J. Characterization of metabolite quantitative trait loci and metabolic networks that control glucosinolate concentration in the seeds and leaves of Brassica napus. New Phytol. 2012;193:96–108. doi: 10.1111/j.1469-8137.2011.03890.x. [DOI] [PubMed] [Google Scholar]
- Goffinet B, Gerber S. Quantitative trait loci: a meta-analysis. Genetics. 2000;155:463–473. doi: 10.1093/genetics/155.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groos C, Robert N, Bervas E, Charmet G. Genetic analysis of grain protein-content, grain yield and thousand-kernel weight in bread wheat. Theor Appl Genet. 2003;106:1032–1040. doi: 10.1007/s00122-002-1111-1. [DOI] [PubMed] [Google Scholar]
- Holland JB, Portyanko VA, Hoffman DL, Lee M. Genomic regions controlling vernalization and photoperiod responses in oat. Theor Appl Genet. 2002;105:113–126. doi: 10.1007/s00122-001-0845-5. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Jestin C, Welham SJ, King GJ, Manzanares-Dauleux MJ, Fitt BDL, Delourme R. Identification of environmentally stable QTL for resistance against Leptosphaeria maculans in oilseed rape (Brassica napus) Theor Appl Genet. 2016;129:169–180. doi: 10.1007/s00122-015-2620-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan HU, Abbadi A, Lücke S, Nichols RA, Snowdon RJ. Genomic prediction of testcross performance in canola (Brassica napus L.) PLoS ONE. 2016;11:e0147769. doi: 10.1371/journal.pone.0147769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jestin C, Bardol N, Lodé M, Duffé P, Domin C, Vallée P, Mangin B, Manzanares-Dauleux MJ, Delourme R. Connected populations for detecting quantitative resistance factors to phoma stem canker in oilseed rape (Brassica napus L.) Mol Breed. 2015;35:167. doi: 10.1007/s11032-015-0356-8. [DOI] [Google Scholar]
- Jiang C, Shi J, Li R, Long Y, Wang H, Li D, Zhao J, Meng J. Quantitative trait loci that control the oil content variation of rapeseed (Brassica napus L.) Theor Appl Genet. 2014;127:957–968. doi: 10.1007/s00122-014-2271-5. [DOI] [PubMed] [Google Scholar]
- Khatkar MS, Thomson PC, Tammen I, Raadsma HW. Quantitative trait loci mapping in dairy cattle: review and meta-analysis. Genet Sel Evol. 2004;36:163–190. doi: 10.1186/1297-9686-36-2-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacape JM, Gawrysiak G, Cao TV, Viot C, Llewellyn D, Liu SM, Jacobs J, Becker D, Barroso PAV, de Assuncao JH, Palai O, Georges S, Jean J, Giband M. Mapping QTLs for traits related to phenology, morphology and yield components in an inter-specific Gossypium hirsutum x G. barbadense cotton RIL population. Field Crop Res. 2013;144:256–267. doi: 10.1016/j.fcr.2013.01.001. [DOI] [Google Scholar]
- Larkan NJ, Lydiate DJ, Yu F, Rimmer SR, Borhan MH. Co-localisation of the blackleg resistance genes Rlm2 and LepR3 on Brassica napus chromosome A10. BMC Plant Biol. 2014;14:387. doi: 10.1186/s12870-014-0387-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Izzah NK, Jayakodi M, Perumal S, Joh HJ, Lee HJ, Lee S-C, Park JY, Yang K-W, Nou I-S, Seo J, Yoo J, Suh Y, Ahn K, Lee JH, Choi GJ, Yu Y, Kim H, Yang T-J. Genome-wide SNP identification and QTL mapping for black rot resistance in cabbage. BMC Plant Biol. 2015;15:32. doi: 10.1186/s12870-015-0424-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZK, Yu SB, Lafitte HR, Huang N, Courtois B, Hittalmani S, Vijayakumar CH, Liu GF, Wang GC, Shashidhar HE, Zhuang JY, Zheng KL, Singh VP, Sidhu JS, Srivantaneeyakul S, Khush GS. QTL x environment interactions in rice. I. heading date and plant height. Theor Appl Genet. 2003;108(1):141–153. doi: 10.1007/s00122-003-1401-2. [DOI] [PubMed] [Google Scholar]
- Li N, Shi J, Wang X, Liu G, Wang H. A combined linkage and regional association mapping validation and fine mapping of two major pleiotropic QTLs for seed weight and silique length in rapeseed (Brassica napus L.) BMC Plant Biol. 2014;14:114. doi: 10.1186/1471-2229-14-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hua W, Hu Z, Yang H, Zhang L, Li R, Deng L, Sun X, Wang X, Wang H. Natural variation in ARF18 gene simultaneously affects seed weight and silique length in polyploid rapeseed. Proc Natl Acad Sci USA. 2015;112:E5123–E5132. doi: 10.1073/pnas.1502160112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, Shi J, Qiu D, Li R, Zhang C, Wang J, Hou J, Zhao J, Shi L, Park B-S, Choi SR, Lim YP, Meng J. Flowering time quantitative trait loci analysis of oilseed Brassica in multiple environments and genomewide alignment with Arabidopsis. Genetics. 2007;177:2433–2444. doi: 10.1534/genetics.107.080705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccaferri M, Sanguineti MC, Corneti S, Ortega JLA, Ben Salem M, Bort J, DeAmbrogio E, Garcia Fernando, del Moral L, Demontis A, El-Ahmed A, Maalouf F, Machlab H, Martos V, Moragues M, Motawaj J, Nachit M, Nserallah N, Ouabbou H, Royo C, Slama A, Tuberosa R. Quantitative trait loci for grain yield and adaptation of durum wheat (Triticum durum Desf.) across a wide range of water availability. Genetics. 2008;178:489–511. doi: 10.1534/genetics.107.077297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nat Rev Genet. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- Moncada P, Martinez CP, Borrero J, Chatel M, Gauch H, Jr, Guimaraes E, Tohme J, McCouch SR. Quantitative trait loci for yield and yield components in an Oryza sativa X Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor Appl Genet. 2001;102:41–52. doi: 10.1007/s001220051616. [DOI] [Google Scholar]
- Myers JL, Well A, Lorch RF (2010) Research design and statistical analysis. Routledge
- Peng B, Li Y, Wang Y, Liu C, Liu Z, Tan W, Zhang Y, Wang D, Shi Y, Sun B, Song Y, Wang T, Li Y. QTL analysis for yield components and kernel-related traits in maize across multi-environments. Theor Appl Genet. 2011;122:1305–1320. doi: 10.1007/s00122-011-1532-9. [DOI] [PubMed] [Google Scholar]
- Qiu D, Morgan C, Shi J, Long Y, Liu J, Li R, Zhuang X, Wang Y, Tan X, Dietrich E, Weihmann T, Everett C, Vanstraelen S, Beckett P, Fraser F, Trick M, Barnes S, Wilmer J, Schmidt R, Li J, Li D, Meng J, Bancroft I. A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor Appl Genet. 2006;114:67–80. doi: 10.1007/s00122-006-0411-2. [DOI] [PubMed] [Google Scholar]
- Radoev M, Becker HC, Ecke W. Genetic analysis of heterosis for yield and yield components in rapeseed (Brassica napus L.) by quantitative trait locus mapping. Genetics. 2008;179:1547–1558. doi: 10.1534/genetics.108.089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman H, Dalton-Morgan J, Diffey S, Raman R, Alamery S, Edwards D, Batley J. SNP markers-based map construction and genome-wide linkage analysis in Brassica napus. Plant Biotechnol J. 2014;12:851–860. doi: 10.1111/pbi.12186. [DOI] [PubMed] [Google Scholar]
- Ramchiary N, Pang W, Nguyen VD, Li X, Choi SR, Kumar A, Kwon M, Song HY, Begum S, Kehie M, Yoon M-K, Na J, Kim H, Lim YP. Quantitative trait loci mapping of partial resistance to Diamondback moth in cabbage (Brassica oleracea L) Theor Appl Genet. 2015;128:1209–1218. doi: 10.1007/s00122-015-2501-5. [DOI] [PubMed] [Google Scholar]
- Shi J, Li R, Qiu D, Jiang C, Long Y, Morgan C, Bancroft I, Zhao J, Meng J. Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics. 2009;182:851–861. doi: 10.1534/genetics.109.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Li R, Zhao Z, Ding G, Long Y, Meng J, Xu F, Shi L. QTL for yield traits and their association with functional genes in response to phosphorus deficiency in Brassica napus. PLoS One. 2013;8:e54559. doi: 10.1371/journal.pone.0054559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva Lda C, Wang S, Zeng ZB. Composite interval mapping and multiple interval mapping: procedures and guidelines for using Windows QTL Cartographer. Methods Mol Biol (Clifton, NJ) 2012;871:75–119. doi: 10.1007/978-1-61779-785-9_6. [DOI] [PubMed] [Google Scholar]
- Song XJ, Huang W, Shi M, Zhu MZ, Lin HX. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- Sun F, Liu P, Ye J, Lo LC, Cao S, Li L, Yue GH, Wang CM. An approach for jatropha improvement using pleiotropic QTLs regulating plant growth and seed yield. Biotechnol Biofuels. 2012;5:42. doi: 10.1186/1754-6834-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vales MI, Schön CC, Capettini F, Chen XM, Corey AE, Mather DE, Mundt CC, Richardson KL, Sandoval-Islas JS, Utz HF, Hayes PM. Effect of population size on the estimation of QTL: a test using resistance to barley stripe rust. Theor Appl Genet. 2005;111:1260–1270. doi: 10.1007/s00122-005-0043-y. [DOI] [PubMed] [Google Scholar]
- Wang S, Basten CJ, Zeng Z-B (2012) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm). Accessed 12 Nov 2015
- Wright S. The method of path coefficients. Ann Math Stat. 1934;5(3):161–215. doi: 10.1214/aoms/1177732676. [DOI] [Google Scholar]
- Würschum T. Mapping QTL for agronomic traits in breeding populations. Theor Appl Genet. 2012;125:201–210. doi: 10.1007/s00122-012-1887-6. [DOI] [PubMed] [Google Scholar]
- Würschum T, Abel S, Zhao Y. Potential of genomic selection in rapeseed (Brassica napus L.) breeding. Plant Breed. 2014;133:45–51. doi: 10.1111/pbr.12137. [DOI] [Google Scholar]
- Xiao J, Li J, Yuan L, Tanksley SD. Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor Appl Genet. 1996;92:230–244. doi: 10.1007/BF00223380. [DOI] [PubMed] [Google Scholar]
- Xu Y, This D, Pausch RC, Vonhof WM, Coburn JR, Comstock JP, McCouch SR. Leaf-level water use efficiency determined by carbon isotope discrimination in rice seedlings: genetic variation associated with population structure and QTL mapping. Theor Appl Genet. 2009;118:1065–1081. doi: 10.1007/s00122-009-0963-z. [DOI] [PubMed] [Google Scholar]
- Yang DL, Jing RL, Chang XP, Li W. Identification of quantitative trait loci and environmental interactions for accumulation and remobilization of water-soluble carbohydrates in wheat (Triticum aestivum L.) stems. Genetics. 2007;176(1):571–584. doi: 10.1534/genetics.106.068361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WK, Wang YJ, Luo GZ, Zhang JS, He CY, Wu XL, Gai JY, Chen SY. QTL mapping of ten agronomic traits on the soybean (Glycine max L. Merr.) genetic map and their association with EST markers. Theor Appl Genet. 2004;108:1131–1139. doi: 10.1007/s00122-003-1527-2. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Thomas CL, Xiang J, Long Y, Wang X, Zou J, Luo Z, Ding G, Cai H, Graham NS, Hammond JP, King GJ, White PJ, Xu F, Broadley MR, Shi L, Meng J. QTL meta-analysis of root traits in Brassica napus under contrasting phosphorus supply in two growth systems. Sci Rep. 2016;6:33113. doi: 10.1038/srep33113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zeng J, Fernando R, Reif JC. Genomic prediction of hybrid wheat performance. Crop Sci. 2013;53:802–810. doi: 10.2135/cropsci2012.08.0463. [DOI] [Google Scholar]
- Zhao Y, Mette MF, Reif JC. Genomic selection in hybrid breeding. Plant Breed. 2015;134:1–10. doi: 10.1111/pbr.12231. [DOI] [Google Scholar]
- Zhao W, Wang X, Wang H, Tian J, Li B, Chen L, Chao H, Long Y, Xiang J, Gan J, Liang W, Li M. Genome-wide identification of QTL for seed yield and yield-related traits and construction of a high-density consensus map for QTL comparison in Brassica napus. Front Plant Sci. 2016;7:17. doi: 10.3389/fpls.2016.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Zhao Y, Liu P, Shi L, Wang X, Wang M, Meng J, Reif JC. Seed quality traits can be predicted with high accuracy in Brassica napus using genomic data. PLoS One. 2016;11(11):e0166624. doi: 10.1371/journal.pone.0166624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Distribution of 525 essential QTL on each linkage group in the BnaTNDH rapeseed mapping population (TIFF 967 kb)
Supplementary material 2 (XLSX 1447 kb)
Supplementary material 7 (XLSX 346 kb)
Supplementary material 8 (XLSX 404 kb)
Supplementary material 10 (XLSX 230 kb)
Supplementary material 11 (XLSX 127 kb)
Supplementary material 12 (XLSX 6379 kb)