Abstract
Glioblastoma (GBM) is the most common and deadly form of brain cancer. Interactions between GBM cells and vasculature in vivo contribute to poor clinical outcomes, with GBM-induced vessel co-option, regression, and subsequent angiogenesis strongly influencing GBM invasion. Here, elements of the GBM perivascular niche are incorporated into a methacrylamide-functionalized gelatin hydrogel as a means to examine GBM–vessel interactions. The complexity of 3D endothelial cell networks formed from human umbilical vein endothelial cells and normal human lung fibroblasts as a function of hydrogel properties and vascular endothelial growth factor (VEGF) presentation is presented. While overall length and branching of the endothelial cell networks decrease with increasing hydrogel stiffness and incorporation of brain-mimetic hyaluronic acid, it can be separately altered by changing the vascular cell seeding density. It is shown that covalent incorporation of VEGF supports network formation as robustly as continuously available soluble VEGF. The impact of U87-MG GBM cells on the endothelial cell networks is subsequently investigated. GBM cells localize in proximity to the endothelial cell networks and hasten network regression in vitro. Together, this in vitro platform recapitulates the close association between GBM cells and vessel structures as well as elements of vessel co-option and regression preceding angiogenesis in vivo.
Keywords: endothelial cells, glioblastoma, hyaluronic acid, hydrogels
1. Introduction
Glioblastoma (GBM) is one of the deadliest forms of brain cancer, with a median survival time of approximately one year.[1] In contrast to other forms of cancer, in which metastasis ultimately leads to patient mortality, the difficulty of treating GBM lies in the fact that cancer cells will invade diffusely into the surrounding brain parenchyma.[2] Clinical treatment prioritizes tumor resection followed by chemotherapy and radiotherapy. However, while the surgical margins for resection in the brain (debulking) are sharply defined, the physiological margins in the brain are diffuse, meaning cells that have already infiltrated remain within the patient. These cells are primarily responsible for the rapid recurrence (6.9 months post-resection) of GBM, with the majority of recurrence happening within 1–2 cm of the primary surgical site.[2]
Interactions between blood vessels and GBM cells contribute to the cancer’s aggressiveness. While metastatic cancers utilize blood vessels to intravasate and extravasate to secondary tumor sites, GBM cells utilize vessels as structural entities on which to crawl and invade into the surrounding brain parenchyma.[3–5] During invasion, GBM cells cluster around vessels through an act known as co-option. Co-option often leads to faulty vessel function and resulting vascular regression, and a region of necrosis develops around the compromised vessel.[6–8] The GBM cells surrounding the necrotic core form pseudo-palisades, or regions of densely packed and hypoxic cells. The cells within the pseudo-palisade send out proangioenic signals to stimulate sprouting angiogenesis at the tumor periphery, and then migrate toward these regions with new vessel formation at the margins.[9] It is also possible that the perivascular space serves as a niche for cancer stem cells, which are a highly aggressive and drug-resistant subpopulation that is suggested to contribute to recurrence.[10] An increased understanding of the interactions between GBM and vessel structures could therefore provide insight into novel methods of targeting GBM invasion.
Cancer biology studies and drug testing rely on models that accurately recapitulate elements of the in vivo tumor microenvironment. Traditional models include 2D cell culture, which do not reflect the 3D nature of the in vivo environment, and animal models, which are too complex to be used to establish relationships between microenvironmental cues and cancer behavior.[11–13] Researchers have therefore engineered 3D biomaterials that allow for controlled incorporation of specific microenvironmental properties to interrogate tumor response.[14,15] While these platforms have been used to study GBM cells alone,[16–19] few 3D in vitro models have investigated GBM behavior in the context of the perivascular niche. Cocultures of endothelial and GBM cells within biomaterials have demonstrated that endothelial cells form capillary sprouts in the presence of GBM, and that coculturing these cell types promotes differential proliferation and gene expression behavior compared to individual monocultures.[20–22] In GBM, however, the perivascular space includes not only endothelial cells, but also astrocytes and stromal cells in the form of pericytes.[23] Models for breast, prostate, lung, and colon cancers have utilized tricultures of tumor cells, endothelial cells, and stromal cells such as mesenchymal stem cells or fibroblasts within collagen, fibrin, and PEG-based biomaterials to study cancer cell cancer cell proliferation, malignant phenotype, and drug responsiveness within an in vitro vascular niche mimic.[24–27] These studies inspired our efforts to demonstrate a 3D in vitro perivascular niche environment that combines GBM cells with endothelial cells and stromal cells and uses a biomaterial that allows for the presentation of tunable biomolecular and biophysical (ligand content, mechanical properties, degradation sites) signals appropriate for those transitions observed across the GBM tumor microenvironment.
Our lab has previously developed a methacrylamide-functionalized gelatin (GelMA) platform with which to investigate GBM behavior in response to gradients of stiffness and hyaluronic acid (HA) presence as observed in the native microenvironment.[28–32] Hyaluronic acid is a predominant component of the extracellular matrix in the brain, and is additionally produced by glioblastoma cells.[28] Moreover, its receptor, CD44, is expressed at the tumor margins and is over-expressed in glioblastoma cells.[28] These studies found that the presence of matrix-bound HA significantly influenced malignant gene expression profiles and invasive phenotype of GBM specimens. In this study, we wanted to extend the use of our GelMA platform to incorporate elements of the perivascular niche of GBM. To do so, we first developed and characterized endothelial cell networks from cocultures of endothelial cells and fibroblasts within a series of HA-decorated GelMA hydrogels. While vascularization of GelMA has been shown previously,[33] we specifically evaluated endothelial cell network formation with respect to HA content and stiffness relevant to GBM phenotype. Furthermore, given that the availability of matrix-bound and soluble growth factors is known to strongly influence tumor progression[28,34,35] and angiogenesis,[36] we exploited acrylate chemistry to photoimmobilize vascular endothelial growth factor (VEGF) into the GelMA hydrogel to define the effect of soluble versus matrix-bound VEGF on endothelial cell network formation. Finally, we investigated the impact of direct incorporation of GBM cells on the stability of endothelial cell networks within the GelMA hydrogels as a means to examine processes associated with vessel co-option and regression.
2. Results
2.1. HAMA Content and Stiffness Decoupled in GelMA/HAMA Hydrogels
GelMA hydrogels were fabricated at 4 and 5 wt%, with and without 15% w/w methacrylate-functionalized hyaluronic acid (HAMA). Using a method recently described by our group to reduce the concentration of photoinitiator in hydrogels that incorporated HAMA,[32] we created a series of four hydrogels (4, 5 wt%; 0, 15% w/w HA) where the stiffness of the hydrogel increased from 3 to 5 kPa and depended only on total weight percent, not HAMA presence (Figure 1).
Figure 1.
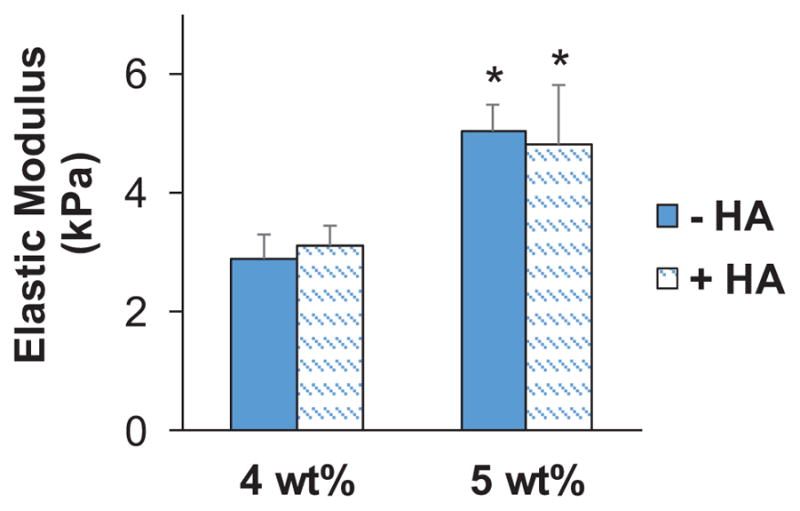
Elastic modulus of GelMA hydrogels. Data presented as mean±SD, n = 6, p-values calculated using one-way ANOVA with Tukey post hoc, *: significant compared to 4 wt% within same HA group (p < 0.05).
2.2. Endothelial Cell Network Formation in GelMA Is Modulated by HAMA Presence, Stiffness, and Cell Density
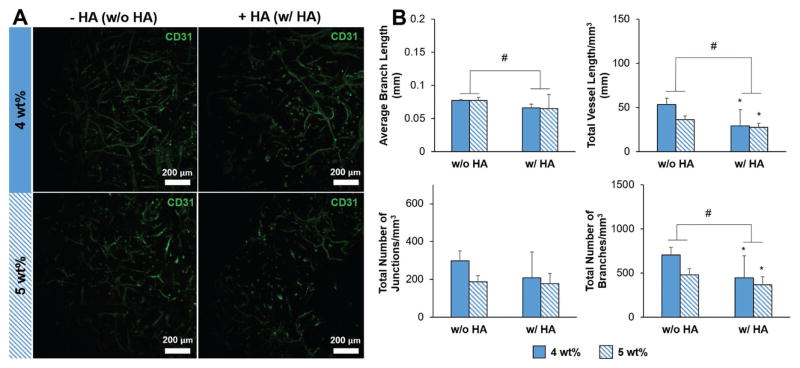
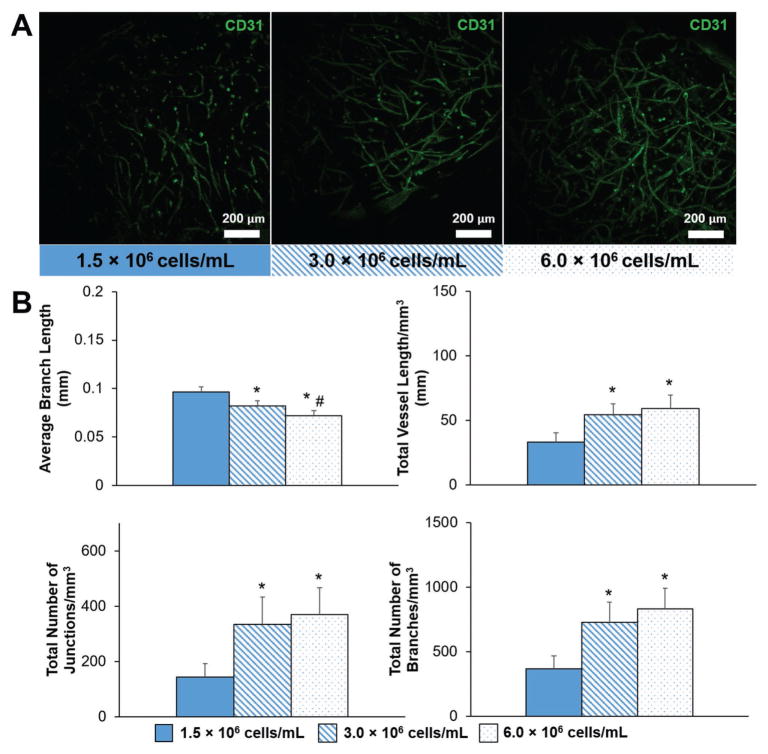
We next determined the impact of the inclusion of HAMA within the hydrogel and overall stiffness on endothelial cell network formation. We formed endothelial cell networks by culturing human umbilical vein endothelial cells (HUVECs) and normal human lung fibroblasts (NHLFs) in a 1:2 (HUVEC:NHLF) ratio. After 7 d of culture, staining for CD31 showed that endothelial cell network formation occurred in all hydrogel constructs (Figure 2A). We quantified the complexity of the endothelial cell networks using TubeAnalyst (IRB Bar-celona), an ImageJ macro. The macro generates 3D skeletons of the endothelial cell networks from z-stacks, and subsequently calculates metrics such as total vessel length, number of branches and junctions, and average branch length. These metrics, normalized to image volume, were used to define endothelial cell network complexity. Endothelial cell network complexity was reduced in the presence of matrix-immobilized HA, as measured by total vessel length mm−3 and total number of branches mm−3 (Figure 2B). Network formation also trended downward with increasing stiffness (p < 0.1). While increasing the initial cell seeding density (1.5–6 × 106 cells mL−1) significantly increased network formation, the positive effect of increasing cell density appeared to plateau at densities higher than 3.0 × 106 cells mL−1 (Figure 3).
Figure 2.
A) Representative maximum intensity projection images depicting CD31-labeled endothelial cell networks (green) within GelMA hydrogels after 7 d of culture. Scale bar: 200 μm. B) Characterization of endothelial cell network complexity: average branch length, total vessel length mm−3, total number of junctions mm−3, and total number of branches mm−3. Data presented as mean ± SD, n = 6, p-values calculated using one-way ANOVA with Tukey post hoc, #: significant main effect of HA (p < 0.05). The main effect considers only the effect of HA by averaging across 4 and 5 wt% constructs within an HA group. *: significant compared to 4 wt%, no HA GelMA hydrogel (p < 0.05).
Figure 3.
A) Representative maximum intensity projection images depicting endothelial cell network formation with varying initial HUVEC and NHLF density within GelMA hydrogels (4 wt%, no HA) after 7 d of culture. Endothelial cells are labeled with CD31. Scale bar: 200 μm. B) Quantitative comparison of endothelial cell network complexity with varying initial HUVEC and NHLF density. Data presented as mean ± SD, n = 6, p-values calculated using one-way ANOVA with Tukey post hoc, *: significant compared to 1.5 × 106 cells mL−1 (p < 0.05). #: significance between consecutive cell densities (p < 0.05).
2.3. Covalently Bound VEGF Maintains Endothelial Cell Network Formation within GelMA Hydrogel
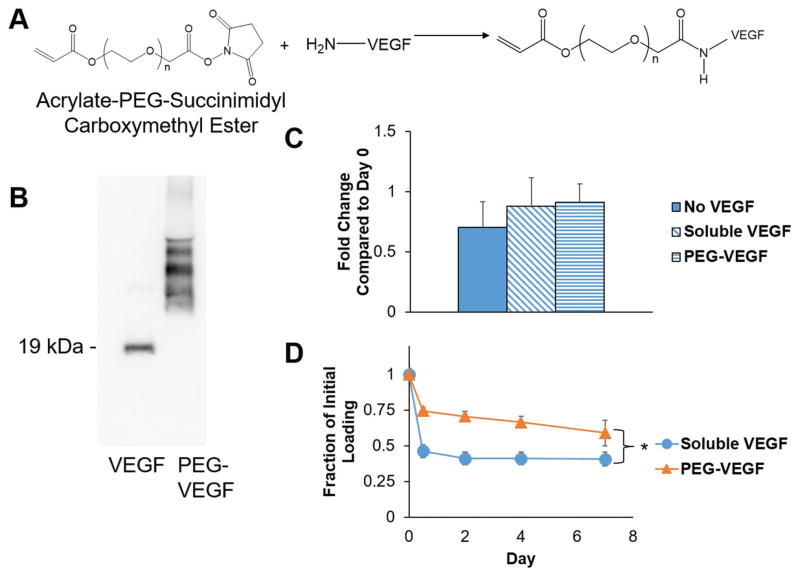
To investigate if covalent incorporation of VEGF into the hydrogel was sufficient to support endothelial network formation, we synthesized acrylate-PEG-VEGF to incorporate into the GelMA network during photopolymerization (Figure 4A). Acrylate-PEG-succinimidyl carboxymethyl ester was successfully conjugated to VEGF (Figure 4B). While unconjugated VEGF was observed via Western blot predominantly at 19 kDa for the monomer form, increased molecular mass was observed for acrylate-PEG-VEGF, with the width of the band suggesting multiple PEG molecules conjugated to each VEGF molecule. Acrylate-PEG-VEGF retained bioactivity, as HUVEC proliferation after 72 h was equivalent for EGM-2 media supplemented with soluble VEGF or acrylate-PEG-VEGF, while proliferation trended downward with VEGF-free EGM-2 media (Figure 4C). Finally, acrylate-PEG-VEGF was significantly better retained in the GelMA hydrogel after photopolymerization compared to soluble VEGF that was loaded into the prepolymer solution without tethering (Figure 4D).
Figure 4.
A) Schematic of acrylate-PEG-VEGF synthesis. B) Western blot depicting VEGF before and after conjugation to acrylate-PEG-succinimidyl carboxymethyl ester. C) Proliferation of HUVECs cultured in EGM-2 media supplemented with no VEGF, soluble VEGF, or acrylate-PEG-VEGF (72 h; normalized to the initial cell count on Day 0). D) Retention of soluble VEGF and acrylate-PEG-VEGF within GelMA hydrogels (4 wt%, no HA) over 7 d. Data presented as mean ± SD, n = 3, p-values calculated using one-way ANOVA with Tukey post hoc, *: significance between groups at a time point (p < 0.05).
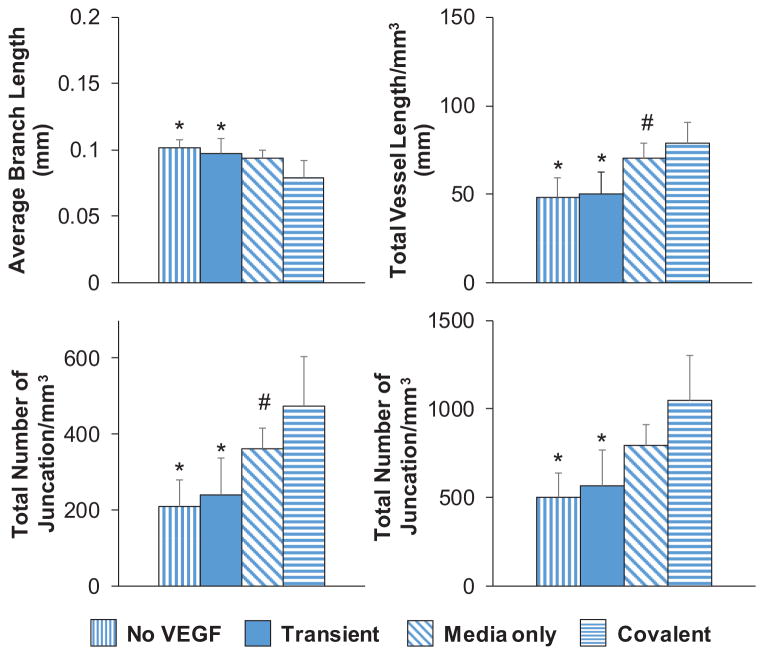
We subsequently showed that covalently bound VEGF within the GelMA hydrogel supported the development of endothelial cell networks in a manner comparable to conventional addition of soluble VEGF to the media (Figure 5). Covalently bound VEGF was as effective in promoting network formation as continuous supplementation of soluble VEGF in the cell culture media (Media only) or when soluble VEGF was included in the prepolymer solution and in the media throughout culture (Continuous) (Figure S1, Supporting Information). Network formation was significantly reduced when soluble VEGF was loaded into the hydrogel initially without additional supplementation over the culture period (Transient), or when soluble VEGF was absent from both the hydrogel and the media (No VEGF). Together, this shows a single dose of matrix-bound VEGF was as effective at maintaining endothelial cell networks as conventional methods that rely on continuous replenishment of VEGF in the media.
Figure 5.
Mode of VEGF presentation (soluble vs matrix-bound) affects endothelial network complexity in GelMA hydrogels (4 wt%, no HA) after 7 d of culture. No VEGF: Hydrogel cultured in VEGF-free EGM-2 media. Transient: Soluble VEGF (2 ng mL−1) was incorporated into the prepolymer solution prior to photopolymerization, but hydrogels were cultured in VEGF-free EGM-2 media. Media Only: No VEGF was added to the prepolymer solution prior to photopolymerization, but cell-seeded hydrogels were cultured in VEGF-containing EGM-2 media. Covalent: PEG-VEGF (2 ng mL−1) was incorporated in prepolymer solution to be covalently tethered to the hydrogel during photopolymerization, and hydrogels were subsequently cultured in VEGF-free EGM-2 media. Data presented as mean ± SD, n = 6, p-values calculated using one-way ANOVA with Tukey post hoc, *: significant compared to Covalent (p < 0.05). #: significant compared to No VEGF (p < 0.05).
2.4. Endothelial Cell Networks in GelMA Closely Associate with U87-MG and Alter U87-MG Cell Shape
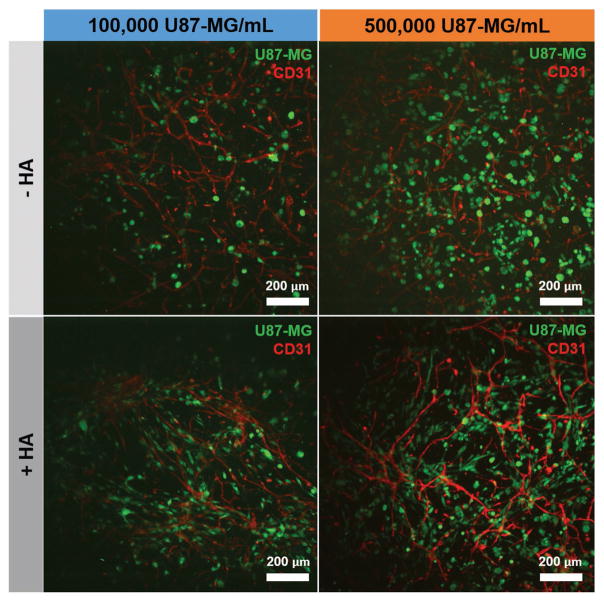
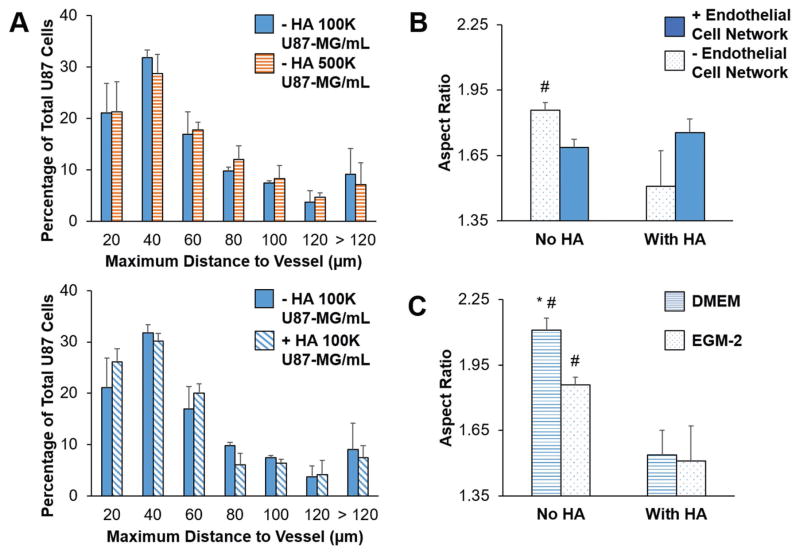
We subsequently investigated the impact of culturing U87-MG GBM cells along with HUVECs and NHLFs. U87-MG cells were labeled using a green CMFDA CellTracker dye and incorporated into the hydrogels at densities of 1 × 105 or 5 × 105 cells mL−1, while HUVECs and NHLFs were cultured at densities of 1 × 106 and 2 × 106 cells mL−1, respectively. Close association between CellTracker-labeled U87-MG cells and CD31+ endothelial networks was observed as rapidly as 3 d within the hydrogel (Figure 6). The majority of U87-MG cells were localized within 40 μm of an endothelial cord (Figure 7A). Interestingly, while U87-MG cells became more rounded (smaller aspect ratio) in the presence of an endothelial cell network in GelMA-only hydrogels, the trend was opposite in the presence of matrix-bound hyaluronic acid, with U87-MG cells becoming more elongated in the presence of an endothelial cell network (Figure 7B, Figure S2, Supporting Information). While all studies of tricultures were performed in EGM-2 media, we also examined the effect of choice of culture media (DMEM or EGM-2) on U87-MG-only experiments (Figure 7C); while U87-MG aspect ratio was not affected by media choice in HA-functionalized hydrogels, cells were more rounded when cultured in EGM-2 media in GelMA-only hydrogels.
Figure 6.
Representative maximum intensity projection images depicting U87-MG cells (labeled with green CMFDA CellTracker) within endothelial cell networks formed within GelMA (−HA) or HA-functionalized GelMA (+HA) hydrogels (4 wt%) after 3 d. Endothelial cells (red) are labeled with CD31. Scale bar: 200 μm.
Figure 7.
A) Proximity of U87-MG cells to endothelial cell networks within GelMA hydrogels (4 wt%) after 3 d. B) Aspect ratio of U87-MG cells cultured with or without endothelial cell networks within GelMA hydrogels (4 wt%) in EGM-2 media after 3 d. #: significant compared to With HA within same group (p < 0.05). C) Aspect ratio of U87-MG cells cultured without endothelial cell networks within GelMA hydrogels (4 wt%) after 3 d. #: significant compared to With HA within same endothelial cell network group (p < 0.05). *: significant compared to EGM-2 within same HA group (p < 0.05). Data presented as mean ± SD, n = 3, p-values calculated using one-way ANOVA with Tukey post hoc.
2.5. Endothelial Cell Networks Regress in the Presence of U87-MG Cells
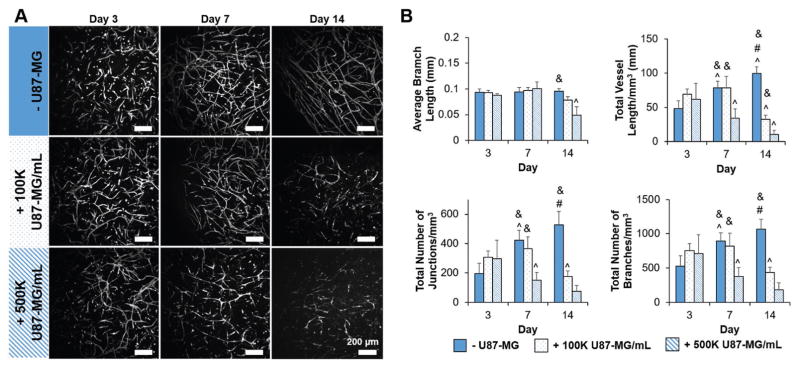
We subsequently traced the stability of the endothelial cell networks in the hydrogels over 14 d for the coculture (NHLF, HUVEC) and tumor triculture (NHLF, HUVEC, GBM) conditions that used 1 × 105 or 5 × 105 U87-MG mL−1 GBM cells (Figure 8). Without matrix-bound HA, network formation increased significantly in coculture (-U87-MG) conditions from Day 3 to Day 7 and persisted through Day 14. Interestingly, the addition of U87-MG cells led to regression of the endothelial cell networks over the two week culture in a GBM-density-dependent manner. With the incorporation of 1 × 105 U87-MG cells mL−1, the endothelial cell network persisted through Day 7, but showed signs of regression by Day 14 (reduced metrics of network complexity). With an increased density of GBM cells (5 × 105 U87-MG cells mL−1), the process of regression was already apparent by Day 7. These results were largely consistent in the presence of matrix-bound HA (Figure S3, Supporting Information). The endothelial cell network in the absence of GBM cells formed and persisted throughout the 14 d culture period. With the addition of 1 × 105 U87-MG cells mL−1 the endothelial cell network complexity increased from Day 3 to Day 7, before showing signs of regression by Day 14. Similarly, the endothelial cell network persisted for 7 d in the presence of 5 × 105 U87-MG cells mL−1, before regressing by Day 14.
Figure 8.
A) Representative maximum intensity projection images depicting endothelial cell network regression in the presence of U87-MG cells within GelMA hydrogels (4 wt%, no HA) over 14 d. Endothelial cells are labeled with CD31. Scale bar: 200 μm. B) Metrics for endothelial network complexity confirm endothelial cell network regression in the presence of U87-MG cells within GelMA hydrogels (4 wt%, no HA) over 14 d. Endothelial cell networks persist in the absence of U87-MG cells. Data presented as mean ± SD, n = 6, p-values calculated using one-way ANOVA with Tukey post hoc, #: significant compared to 100 K U87-MG mL−1 within time point (p < 0.05). &: significant compared to 500 K U87-MG mL−1 within time point (p < 0.05). ^: significant compared to prior time point within same U87-MG cell density (p < 0.05).
3. Discussion
Interactions between GBM cells and vessel structures within the brain are instrumental to the tumor’s progression. GBM cells crawl along vessels to invade into the parenchyma, and the cycle of vessel co-option, regression, and angiogenesis pushes forward the invasive front at the tumor margins.[3,6,7,9] Current models used to understand GBM–vessel interactions include in vivo models, 2D cell culture, and transwell migration assays.[4,5,37,38] The development of a 3D in vitro model would allow for controlled interrogation of signaling between GBM cells, endothelial cells, and associated stromal cells within a defined matrix environment. While various biomaterial constructs have been used to culture GBM cells, cocultures of GBM cells with endothelial cells have been investigated less extensively,[20–22] and tricultures that additionally incorporate a stromal cell type have not yet been reported in literature, suggesting an opportunity for developing new experimental tools. Inclusion of a stromal cell type is not only physiologically relevant, as the perivascular niche includes stromal cells along with endothelial cells and glioblastoma cells but also has been shown to improve and sustain vascularization of various bio-materials.[39,40] Chen et al. previously showed in GelMA that endothelial cells did not form capillary structures without the presence of stromal cells.[33]
Our lab has previously described a photopolymerizable GelMA system that allowed control over matrix stiffness and the capacity to selectively incorporate physiologically relevant biomolecular cues to investigate glioblastoma behavior.[29–32] Using this system, we have shown that varying matrix stiffness and the incorporation of matrix-bound HA informs changes in GBM cell phenotype (gene expression, signaling) related to invasion, and we have also described a dextran bead assay to quantify the influence of matrix-bound HA on cell invasion.[32] In this study, we demonstrated that this GelMA platform can be modified to investigate interactions between GBM cells and endothelial cell networks. While such networks are traditionally formed in alternative matrix systems (e.g., collagen, fibrin[41–44]), we report stable endothelial cell networks can be formed within the GelMA hydrogel system by coculturing endothelial cells and fibroblasts. Given the importance of tuning matrix stiffness and HA content in investigation of GBM cell activity,[19,29–32,45,46] we investigated the impact of these parameters on endothelial cell network formation. We subsequently investigated the mode of VEGF presentation as a material parameter. Finally, we demonstrated that the GelMA platform can support a triculture of GBM cells, endothelial cells, and fibroblasts, and that the presence of GBM correlates with differential endothelial cell network formation.
The range of matrix biophysical properties tested in this project was informed by features of the native GBM microenvironment and our previous efforts examining GBM cell activity. Recent literature has shown that samples taken from the core or margins of GBM tissues have stiffness on the order of 1 kPa, while normal brain tissue has a stiffness on the order of 0.1 kPa.[47] We chose to examine network formation in hydrogels with elastic moduli on the order of 1–10 kPa, and by tuning the amount of photoinitiator used for UV photopolymerization we created hydrogels that decoupled stiffness from the incorporation of HA. To quantify the extent of vessel-like network formation within these hydrogels, we employed the TubeAnalyst macro to extract metrics of endothelial network complexity (total vessel length, number of branches, and number of junctions) from the 3D vascular skeleton reconstructed from z-stacks. While analysis of 2D maximum image projections of networks is more commonly used in the literature, the 3D analysis allows for a more accurate calculation of branching and junction complexity.[41,42,48,49] We found that endothelial cell network complexity decreased with the presence of matrix-bound HA. Camci-Unal et al. reported that HUVECs exhibited less cell spreading in hybrid GelMA-HA hydrogels, which could contribute to reduced network formation.[50] The size of HA has also been shown to impact angiogenesis. HA chains with less than 25 disaccharide units have been reported as proangiogenic, while larger HA chains are antiangiogenic.[51–54] In this study, we used 66–90 kDa HA, which induced differential GBM invasion in GelMA hydrogels in a recent study from our lab,[32] but whose size may also explain reduced network formation; these findings suggest an opportunity for future work to employ smaller HA fragments to enhance network formation. We also found that endothelial cell network formation trended downward with increasing material stiffness, which has been reported by others.[33] However, our capacity to modulate endothelial cell network formation via the initial cell density of endothelial cells and fibroblasts suggests the capacity to decouple hydrogel stiffness from the extent of endothelial cell network formation, and is therefore an important tool for developing biomimetic culture environments. Together, these results suggest that physiologically relevant matrix properties such as stiffness and HA presence impact endothelial cell network formation within our GelMA platform.
Biomolecular signals play a critical role within the tumor microenvironment, suggesting a need to regulate their mode of presentation and delivery in engineered tumor microenvironments. The mode of presentation of these bio-molecules is of physiological relevance, as ECM molecules are known to sequester and then release growth factors in a spatial and temporal manner.[35,55,56] The methacrylamide groups on the GelMA backbone, in addition to being essential for photopolymerization, provide a reaction site to covalently tether biomolecules to the GelMA hydrogel matrix. Here, we were motivated by the known significance of VEGF in GBM-associated angiogenesis[6,57] and the typical requirement that VEGF be available continuously to promote vascularization.[58] For extended culture times, continuous media supplementation can be expensive; further, it is difficult to control where in a hydrogel VEGF is available, a critical consideration in cases where nonuniform endothelial cell networks may be desirable. The capacity for matrix-bound VEGF to promote angiogenic processes[36] and regulate tumor progression[34,59,60] inspired us to covalently incorporate it into the GelMA hydrogel. We adapted an approach previously demonstrated by our lab to covalently immobilize acrylated stem cell factor (SCF) within the GelMA hydrogel to facilitate the culture of primary hematopoietic stem cells.[61] We found that covalently immobilized VEGF within the GelMA hydrogel (with no need for supplemental soluble VEGF in the media) promoted endothelial cell network formation as effectively as continuously provided soluble VEGF. Though statistically insignificant, there even appeared to be increased branching with covalently bound VEGF compared to continuously provided soluble VEGF. This observation correlates with work by Lee et al., which reported that vascular branching was favored by matrix-bound VEGF over soluble VEGF.[34] This differential response may be rooted in the ability of matrix-bound VEGF to initiate alternative signaling mechanisms compared to soluble VEGF.[36] While limited endothelial cell network formation still occurred when VEGF was absent or present only transiently, likely due to the presence of other angiogenic growth factors in the media such as epidermal growth factor (EGF),[62] endothelial cell network formation decreased significantly in these cases. Together, these results highlighted the adaptable nature of the GelMA system, allowing covalent immobilization of both brain-mimetic HA as well as VEGF to generate a culture platform that can support combined culture of endothelial networks and GBM specimens.
Finally, we wanted to demonstrate that a stable triculture of GBM cells, endothelial cells, and fibroblasts within GelMA can be used to generate a prototype of the perivascular niche within the GBM microenvironment. By 3 d of culture, we observed close association of GBM cells with developing endothelial cell networks within the hydrogel constructs, in agreement with previous in vivo and in vitro studies of GBM–endothelial cell interactions.[4,5,38] Interestingly, the presence of GBM cells additionally produced temporal changes in endothelial cell network formation not observed when GBM cells were lacking. While endothelial cell networks formed and persisted over a period of two weeks without the GBM cells, network regression occurred over the same time frame in the presence of GBM cells, and that this regression appears faster in the presence of larger densities of GBM cells. The initial GBM cell population was at most 15% of the total cell population, making these results likely not due to extreme cell densities. Instead, these observations are consistent with the phenomena of vessel co-option and regression typically seen in vivo.[7,63] GBM cells are known to displace astrocytes along vessel structures during invasion[4] as well as to compromise contractility of pericytes normally present in the perivascular space in the brain,[64] both resulting in compromised vessel function. Further, it has been shown that capillaries formed by endothelial cells regress in the absence of fibroblasts or mesenchymal stem cells, and that endothelial cells become apoptotic when in direct contact with tumor cells.[39,65,66] As a result, findings here motivate future application of this in vitro model to trace interactions between GBM cells with the fibroblasts and endothelial cells that compromise the stable endothelial cell networks. Ongoing efforts are employing gene expression studies to understand the mechanisms behind the association between GBM cells and endothelial cords, as well as network regression.
Taken together, we report an adaptable GelMA hydrogel model to create an in vitro culture environment that can integrate brain-mimetic HA, immobilized biomolecular cues, and the combined culture of GBM specimens and an endothelial cell network. Such platforms may be particularly appealing to examine mechanisms of GBM invasion and to test new therapeutic targets for GBM. We acknowledge that this platform is currently limited by the fact that endothelial cell networks develop concurrently with GBM culture, whereas in vivo GBM cells invade and interact with preexisting vessel structures. Thus, a critical step forward involves the development of temporal platforms in which endothelial cell networks or vessel structures can be formed before the addition of GBM cells to the material system. Ongoing efforts are employing micro-fluidic platforms[67–69] to first form a vascular network within the GelMA hydrogel prior to the addition of GBM specimens; this effort may also facilitate the formation of perfusable lumen within endothelial cell network.[70] The ability to determine how nutrient and oxygen supply impact GBM invasion would increase the complexity of our platform. Here, the use of patient-derived specimens cells along with brain microvascular endothelial cells and pericytes would increase the usefulness of our model for informing clinical outcomes and are the subject of current efforts building from this work.
4. Conclusion
Interactions between GBM cells and vascular networks in the brain contribute to the invasive spreading of GBM. Developing models to understand these interactions is critical in determining new methods of treating GBM. Here, we demonstrated the incorporation of endothelial cell networks within a GelMA hydrogel under development for studies of GBM using a coculture of endothelial cells and fibroblasts. The formation of endothelial cell networks is impacted by hyaluronic acid content and stiffness, both critical parameters of the GBM micro-environment, but can be corrected through manipulation of the starting concentration of endothelial cells and fibroblasts as well as importation of matrix-immobilized VEGF within the hydrogel. Finally, tricultures of GBM cells with endothelial cell networks lead to the onset of behavior reminiscent of vessel co-option and regression found in the GBM tumor microenvironment. Taken together, we show that this platform supports interactions between GBM cells and vascular networks, and can be further used to understand GBM behavior within a vascular microenvironment.
5. Experimental Section
Cell Culture
HUVECs and NHLFs were purchased from Lonza (Walkersville, MD). HUVECs were cultured in EGM-2, which contains 2% FBS, hEGF, hydrocortisone, VEGF, hFGF-B, R3-IGF-1, ascorbic acid, heparin, and gentamicin/amphotericin-B (Lonza, Walkersville, MD). NHLFs were cultured in FGM-2, which contains 2% FBS, hFGF-B, insulin, and gentamicin/amphotericin-B (Lonza, Walkersville, MD). U87-MG cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. All cells were cultured in a 5% CO2 environment at 37 °C. HUVECs were used before passage 6, while NHLFs were used before passage 8.
Hydrogel Fabrication and Mechanical Characterization
GelMA and HAMA were synthesized as previously described.[29] GelMA was characterized by 1H NMR to have a 50% degree of functionalization. 4 and 5 total wt% hydrogels were formed from either GelMA alone or GelMA combined with 15% w/w HAMA. Hydrogels were created by dissolving the polymers in PBS at 65 °C, along with lithium acylphosphinate (LAP) as a photoinitiator. 0.1% w/v LAP was used for GelMA-only gels, and 0.02% w/v LAP was used for GelMA/HAMA gels. The prepolymer solution was pipetted into circular Teflon molds with a diameter of 5 mm and a thickness of 1 mm, and exposed to UV light (λ = 365 nm, 5.69 mW cm−2) for 30 s. Teflon molds were prepared by the university machine shop from 2.2 cm × 2.8 cm × 1 mm rectangular pieces of Teflon, in which circles with a diameter of 5 mm were punched out. Acellular hydrogels were maintained in PBS at pH = 7.4.
Hydrated hydrogels were tested in unconfined compression using an Instron 5943 mechanical tester (Instron, Norwood, MA) with a preload of 0.005 N and a strain rate of 0.1 mm min−1. The elastic modulus was determined by plotting stress versus strain and calculating the slope of the linear regime (≈0–40% strain).
Endothelial Cell and Fibroblast Coculture; GBM Triculture Platforms
Endothelial cell networks were formed within the hydrogels by coculturing HUVECs and NHLFs in a 1:2 ratio (HUVEC:NHLF) with cell densities ranging from 1.5 × 106 to 6 × 106 total cells mL−1. Briefly, cells were suspended in the hydrogel prepolymer solution, pipetted into Teflon molds (5 mm diameter, 1 mm thick), and photopolymerized as previously described. Tricultures were formed by suspending U87-MG cells at densities of 1 × 105 or 5 × 105 cells mL−1 with HUVECs and NHLFs. For experiments in which U87-MG cells were visualized, U87-MG was incubated with 10 × 10−6 M green CMFDA CellTracker (Thermo Fisher Scientific, Waltham, MA) before incorporation into the hydrogels. Hydrogels were maintained in 48-well plates in EGM-2 media for 3–14 d, with daily media changes.
Immunofluorescent Staining
Hydrogels were fixed in formalin (Sigma Aldrich, St. Louis, MO) and washed with PBS. Encapsulated cells were permeabilized with PBS containing 0.5% Tween 20 (Thermo Fisher Scientific, Waltham, MA), and then blocked in PBS with 2% BSA (Sigma Aldrich, St. Louis, MO) and 0.1% Tween 20. Endothelial cells were stained with mouse anti-human CD31 (Dako, Denmark) overnight at 4 °C, followed by goat anti-mouse Alexa Fluor 488 or 561 (Thermo Fisher Scientific, Waltham, MA) overnight at 4 °C. PBS with 0.1% Tween was used for all wash steps following permeabilization.
Image Acquisition and Analysis
Endothelial cell networks were imaged using a DMi8 Yokogawa W1 spinning disk confocal microscope outfitted with a Hamamatsu EM-CCD digital camera (Leica Microsystems, Buffalo Grove, IL). Images were taken as z-stacks with a depth of 200 μm and a step size of 5 μm. Six regions were imaged per hydrogel. Endothelial cell networks were quantified using metrics such as total length, number of branches and junctions, and average branch length using TubeAnalyst (IRB Barcelona), which is available as an ImageJ (NIH, Bethesda, MD) macro. The TubeAnalyst macro was employed to quantify vessel-like structures from 3D z-stacks. This method, compared to analysis of 2D images, allowed for accurate determination of the number of branches and junctions within the endothelial cell network.[49] The algorithm identifies vessel-like structures, then segments and skeletonizes the image to quantify total vessel length, total number of junctions, and number and length of subsequent branches extending from each junction. Metrics were subsequently normalized to image volume. TubeAnalyst results were visualized using Volume Viewer in ImageJ and manually compared to 2D maximum intensity projection images to eliminate regions of the skeleton that corresponded to endothelial cells forming sheets instead of cords.
PEG-VEGF Synthesis and Characterization
PEG-VEGF was synthesized using methods previously described by our group and West and co-workers.,[61,71,72] by reacting human recombinant VEGF165 (R&D Systems, Minneapolis, MN) with 3500 MW acrylate-PEG-succinimidyl carboxymethyl ester (JenKem Technology USA, Plano, TX) in a 200:1 molar ratio (acrylate-PEG-succinimidyl carboxymethyl ester: VEGF) in PBS (pH = 8.0) for 2 h at 4 °C. The reaction mixture was dialyzed overnight using a 10 K Slide-A-Lyzer G2 dialysis cassette (Thermo Fisher Scientific, Waltham, MA) to remove unreacted acrylate-PEG-succinimidyl carboxymethyl ester, and then lyophilized to obtain the final PEG-VEGF product. A VEGF Quantikine ELISA kit was used to determine the concentration of PEG-VEGF (R&D Systems, Minneapolis, MN). To confirm the conjugation of acrylate-PEG-succinimidyl carboxymethyl ester to VEGF, Western blot was performed under reducing conditions using a 4–20% Mini-PROTEAN TGX precast polyacrylamide gel (Bio-Rad, Hercules CA), with goat anti-human VEGF and horseradish peroxidase-conjugated anti-goat secondary antibody (R&D Systems, Minneapolis, MN). The SuperSignal West Pico Chemiluminescent Substrate kit (Thermo Fisher Scientific, Waltham, MA) was used for detection with an ImageQuant LAS 4000 (GE Healthcare Bio-Sciences, Pittsburgh, PA).
Release Profile of VEGF from GelMA
PEG-VEGF or soluble VEGF was added into the GelMA prepolymer solution at 2 ng mL−1, which is the concentration of VEGF in the EGM-2 media. Hydrogels were photopolymerized and maintained in PBS (pH = 7.4) at 37 °C. At 12 h, and days 2, 4, and 7, PBS was collected and frozen at −80 °C until analysis. The VEGF Quantikine ELISA kit was used to quantify the amount of VEGF released from the hydrogel (R&D Systems, Minneapolis, MN).
Confirming VEGF Bioactivity
HUVECs were seeded in six-well plates and fed with EGM-2 media, VEGF-free EGM-2 media, or VEGF-free media supplemented with 2 ng mL−1 acrylate-PEG-VEGF. Proliferation was determined via a Hoescht assay (after 72 h) normalized to the starting cell population (Day 0).
Comparison of Soluble versus Covalently Bound VEGF on Endothelial Cell and Fibroblast Coculture
HUVECs and NHLFs were cocultured as previously described in 4 wt% GelMA hydrogels. Hydrogels were maintained for 7 d before fixation, immunofluorescent staining, and imaging. VEGF was incorporated into the hydrogels in one of the following manners: No VEGF: Hydrogel was cultured in VEGF-free EGM-2 media. Transient: Soluble VEGF (2 ng mL−1) was incorporated into the prepolymer solution during fabrication, but hydrogels were cultured in VEGF-free EGM-2 media. Media Only: No VEGF was added to the prepolymer solution during fabrication, but cell-seeded hydrogels were cultured in VEGF-containing EGM-2 media. Continuous: Soluble VEGF (2 ng mL−1) was incorporated into the prepolymer solution, and hydrogels were cultured in VEGF-containing EGM-2 media. Covalent: acrylate-PEG-VEGF (2 ng mL−1) was incorporated in prepolymer solution to be covalently tethered to the hydrogel, and hydrogels were subsequently cultured in VEGF-free EGM-2 media. Metrics of vessel complexity were gathered as described earlier for all five conditions.
Measurement of U87-MG Cell Shape and Localization Relative to the Endothelial Cell Networks
Tricultures formed via adding green CMFDA CellTracker-stained U87-MG cells at densities of 1 × 105 or 5 × 105 cells mL−1 into the hydrogels with HUVECs and NHLFs were cultured for 3 d before fixation and staining for CD31. z-stacks were obtained as previously described to show GBM cells in the presence of endothelial cell networks. From each z-stack, 50 random U87-MG cells were identified, and the perpendicular distance between the center of the cell to the nearest endothelial cord was measured using ImageJ (≈900 measurements per condition). Cell aspect ratio was quantified by manually thresholding maximum intensity projection images in ImageJ to define U87-MG cells as particles, then using “Analyze Particles” in ImageJ to determine aspect ratio. The aspect ratio of U87-MG cells was compared as a function of the presence of endothelial cell networks and the presence of matrix-bound HA within the hydrogel.
Statistical Analysis
Statistical analysis was performed using one-way analysis of variance (ANOVA) tests with Tukey post hoc in OriginPro (OriginLab, Northampton, MA). Mechanical characterization and experiments that characterized vessel network formation used n = 6 hydrogel samples (6 image volumes per hydrogel) per group. Experiments defining acrylate-PEG-VEGF bioactivity and release, as well as those to measure U87-MG cell aspect ratio and proximity to endothelial cell networks used n = 3 hydrogel samples (6 image volumes per hydrogel) per group. Significance was set at p < 0.05. Data are presented as mean ± SD.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Eli Karvelis and Christine Hunter for assistance with immunofluorescent staining. Research reported in this publication was also supported by the National Cancer Institute of the National Institutes of Health under Award No. R01 CA197488. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors are grateful for the funding for this study provided by the NSF Graduate Research Fellowship DGE-1144245 (M.T.N.). The authors are also grateful for additional funding provided by the Department of Chemical and Biomolecular Engineering and the Carl R. Woese Institute for Genomic Biology at the University of Illinois at Urbana-Champaign.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Mai T. Ngo, 193 Roger Adams Laboratory, 600 S. Mathews Ave, Urbana, IL 61801, USA
Prof. Brendan A. Harley, 110 Roger Adams Laboratory, 600 S. Mathews Ave, Urbana, IL 61801, USA
References
- 1.DeAngelis LM. N Engl J Med. 2001;344:114. doi: 10.1056/NEJM200101113440207. [DOI] [PubMed] [Google Scholar]
- 2.Paw I, Carpenter RC, Watabe K, Debinski W, Lo HW. Cancer Lett. 2015;362:1. doi: 10.1016/j.canlet.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farin A, Suzuki SO, Weiker M, Goldman JE, Bruce JN, Canoll P. Glia. 2006;53:799. doi: 10.1002/glia.20334. [DOI] [PubMed] [Google Scholar]
- 4.Watkins S, Robel S, Kimbrough IF, Robert SM, Ellis-Davies G, Sontheimer H. Nat Commun. 2014;5:4196. doi: 10.1038/ncomms5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winkler F, Kienast Y, Fuhrmann M, Von Baumgarten L, Burgold S, Mitteregger G, Kretzschmar H, Herms J. Glia. 2009;57:1306. doi: 10.1002/glia.20850. [DOI] [PubMed] [Google Scholar]
- 6.Hardee ME, Zagzag D. Am J Pathol. 2012;181:1126. doi: 10.1016/j.ajpath.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holash J, Maisonpierre PC, Compton D, Boland P, Alexander CR, Zagzag D, Yancopoulos GD, Wiegand SJ. Science. 1999;284:1994. doi: 10.1126/science.284.5422.1994. [DOI] [PubMed] [Google Scholar]
- 8.Vartanian A, Singh SK, Agnihotri S, Jalali S, Burrell K, Aldape KD, Zadeh G. Neuro-Oncology. 2014;16:1167. doi: 10.1093/neuonc/nou035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rong Y, Durden DL, Van Meir EG, Brat DJ. J Neuropathol Exp Neurol. 2006;65:529. doi: 10.1097/00005072-200606000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Lathia JD, Heddleston JM, Venere M, Rich JN. Cell Stem Cell. 2011;8:482. doi: 10.1016/j.stem.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chwalek K, Bray LJ, Werner C. Adv Drug Delivery Rev. 2014;79:30. doi: 10.1016/j.addr.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Katt ME, Placone AL, Wong AD, Xu ZS, Searson PC. Front Bioeng Biotechnol. 2016;4:12. doi: 10.3389/fbioe.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song HH, Park KM, Gerecht S. Adv Drug Delivery Rev. 2014;79:19. doi: 10.1016/j.addr.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fong EL, Harrington DA, Farach-Carson MC, Yu H. Biomaterials. 2016;108:197. doi: 10.1016/j.biomaterials.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu L, Mooney DJ. Nat Rev Cancer. 2016;16:56. doi: 10.1038/nrc.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Kievit FM, Erickson AE, Silber JR, Ellenbogen RG, Zhang M. Adv Healthcare Mater. 2016;5:3173. doi: 10.1002/adhm.201600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Tong X, Yang F. Mol Pharm. 2014;11:2115. doi: 10.1021/mp5000828. [DOI] [PubMed] [Google Scholar]
- 18.Rao SS, Dejesus J, Short AR, Otero JJ, Sarkar A, Winter JO. ACS Appl Mater Interfaces. 2013;5:9276. doi: 10.1021/am402097j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heffernan JM, Overstreet DJ, Le LD, Vernon BL, Sirianni RW. Ann Biomed Eng. 2015;43:1965. doi: 10.1007/s10439-014-1223-1. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen DT, Fan YT, Akay YM, Akay M. IEEE Trans Nanobiosci. 2016;15:289. doi: 10.1109/TNB.2016.2528170. [DOI] [PubMed] [Google Scholar]
- 21.Chen Z, Htay A, Dos Santos W, Gillies GT, Fillmore HL, Sholley MM, Broaddus WC. J Neurooncol. 2009;92:121. doi: 10.1007/s11060-008-9742-y. [DOI] [PubMed] [Google Scholar]
- 22.Avci NG, Fan YT, Dragomir A, Akay YM, Akay M. IEEE Trans Nanobiosci. 2015;14:790. doi: 10.1109/TNB.2015.2477818. [DOI] [PubMed] [Google Scholar]
- 23.Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H. Glia. 2011;59:1169. doi: 10.1002/glia.21136. [DOI] [PubMed] [Google Scholar]
- 24.Bray LJ, Binner M, Holzheu A, Friedrichs J, Freudenberg U, Hutmacher DW, Werner C. Biomaterials. 2015;53:609. doi: 10.1016/j.biomaterials.2015.02.124. [DOI] [PubMed] [Google Scholar]
- 25.de Sampaio PC, Auslaender D, Krubasik D, Failla AV, Skepper JN, Murphy G, English WR. PLoS One. 2012;7:e30753. doi: 10.1371/journal.pone.0030753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Del Bufalo F, Manzo T, Hoyos V, Yagyu S, Caruana I, Jacot J, Benavides O, Rosen D, Brenner MK. Biomaterials. 2016;84:76. doi: 10.1016/j.biomaterials.2016.01.030. [DOI] [PubMed] [Google Scholar]
- 27.Nyga A, Loizidou M, Emberton M, Cheema U. Acta Biomater. 2013;9:7917. doi: 10.1016/j.actbio.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiranowska M, Rojiani MV. In: Glioma – Exploring its Biology and Practical Relevance. Ghosh A, editor. Ch. 12 InTech; Rijeka, Croatia: 2011. [Google Scholar]
- 29.Pedron S, Becka E, Harley BA. Biomaterials. 2013;34:7408. doi: 10.1016/j.biomaterials.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 30.Pedron S, Becka E, Harley BAC. Adv Mater. 2015;27:1567. doi: 10.1002/adma.201404896. [DOI] [PubMed] [Google Scholar]
- 31.Pedron S, Harley BAC. J Biomed Mater Res, Part A. 2013;101:3405. doi: 10.1002/jbm.a.34637. [DOI] [PubMed] [Google Scholar]
- 32.Chen JW, Pedron S, Harley BAC. Macromol Biosci. 2017;17:1700018. doi: 10.1002/mabi.201700018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen YC, Lin RZ, Qi H, Yang YZ, Bae HJ, Melero-Martin JM, Khademhosseini A. Adv Funct Mater. 2012;22:2027. doi: 10.1002/adfm.201101662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S, Jilani SM, Nikolova GV, Carpizo D, Iruela-Arispe ML. J Cell Biol. 2005;169:681. doi: 10.1083/jcb.200409115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park JE, Keller GA, Ferrara N. Mol Biol Cell. 1993;4:1317. doi: 10.1091/mbc.4.12.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen TT, Luque A, Lee S, Anderson SM, Segura T, Iruela-Arispe ML. J Cell Biol. 2010;188:595. doi: 10.1083/jcb.200906044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khodarev NN, Yu J, Labay E, Darga T, Brown CK, Mauceri HJ, Yassari R, Gupta N, Weichselbaum RR. J Cell Sci. 2003;116:1013. doi: 10.1242/jcs.00281. [DOI] [PubMed] [Google Scholar]
- 38.Rao S, Sengupta R, Choe EJ, Woerner BM, Jackson E, Sun T, Leonard J, Piwnica-Worms D, Rubin JB. PLoS One. 2012;7:e33005. doi: 10.1371/journal.pone.0033005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newman AC, Nakatsu MN, Chou W, Gershon PD, Hughes CC. Mol Biol Cell. 2011;22:3791. doi: 10.1091/mbc.E11-05-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakatsu MN, Sainson RC, Aoto JN, Taylor KL, Aitkenhead M, Perez-del-Pulgar S, Carpenter PM, Hughes CC. Microvasc Res. 2003;66:102. doi: 10.1016/s0026-2862(03)00045-1. [DOI] [PubMed] [Google Scholar]
- 41.Chen X, Aledia AS, Ghajar CM, Griffith CK, Putnam AJ, Hughes CC, George SC. Tissue Eng, Part A. 2009;15:1363. doi: 10.1089/ten.tea.2008.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X, Aledia AS, Popson SA, Him L, Hughes CC, George SC. Tissue Eng, Part A. 2010;16:585. doi: 10.1089/ten.tea.2009.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan EC, Kuo SM, Kong AM, Morrison WA, Dusting GJ, Mitchell GM, Lim SY, Liu GS. PLoS One. 2016;11:e0149799. doi: 10.1371/journal.pone.0149799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cross VL, Zheng Y, Choi NW, Verbridge SS, Sutermaster BA, Bonassar LJ, Fischbach C, Stroock AD. Biomaterials. 2010;31:8596. doi: 10.1016/j.biomaterials.2010.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rape AD, Kumar S. Biomaterials. 2014;35:8846. doi: 10.1016/j.biomaterials.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim Y, Kumar S. Mol Cancer Res. 2014;12:1416. doi: 10.1158/1541-7786.MCR-13-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miroshnikova YA, Mouw JK, Barnes JM, Pickup MW, Lakins JN, Kim Y, Lobo K, Persson AI, Reis GF, McKnight TR, Holland EC, Phillips JJ, Weaver VM. Nat Cell Biol. 2016;18:1336. doi: 10.1038/ncb3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghajar CM, Kachgal S, Kniazeva E, Mori H, Costes SV, George SC, Putnam AJ. Exp Cell Res. 2010;316:813. doi: 10.1016/j.yexcr.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeon JS, Bersini S, Whisler JA, Chen MB, Dubini G, Charest JL, Moretti M, Kamm RD. Integr Biol. 2014;6:555. doi: 10.1039/c3ib40267c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Camci-Unal G, Cuttica D, Annabi N, Demarchi D, Khademhosseini A. Biomacromolecules. 2013;14:1085. doi: 10.1021/bm3019856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.West DC, Hampson IN, Arnold F, Kumar S. Science. 1985;228:1324. doi: 10.1126/science.2408340. [DOI] [PubMed] [Google Scholar]
- 52.Lam J, Truong NF, Segura T. Acta Biomater. 2014;10:1571. doi: 10.1016/j.actbio.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perng CK, Wang YJ, Tsi CH, Ma H. J Surg Res. 2011;168:9. doi: 10.1016/j.jss.2009.09.052. [DOI] [PubMed] [Google Scholar]
- 54.Deed R, Rooney P, Kumar P, Norton JD, Smith J, Freemont AJ, Kumar S. Int J Cancer. 1997;71:251. doi: 10.1002/(sici)1097-0215(19970410)71:2<251::aid-ijc21>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 55.Schultz GS, Wysocki A. Wound Repair Regener. 2009;17:153. doi: 10.1111/j.1524-475X.2009.00466.x. [DOI] [PubMed] [Google Scholar]
- 56.Hortensius RA, Harley BA. Biomaterials. 2013;34:7645. doi: 10.1016/j.biomaterials.2013.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tate MC, Aghi MK. Neurotherapeutics. 2009;6:447. doi: 10.1016/j.nurt.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foster GA, Headen DM, Gonzalez-Garcia C, Salmeron-Sanchez M, Shirwan H, Garcia AJ. Biomaterials. 2017;113:170. doi: 10.1016/j.biomaterials.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bergers G, Brekken R, McMahon G, Vu TH, Itoh T, Tamaki K, Tanzawa K, Thorpe P, Itohara S, Werb Z, Hanahan D. Nat Cell Biol. 2000;2:737. doi: 10.1038/35036374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Belotti D, Paganoni P, Manenti L, Garofalo A, Marchini S, Taraboletti G, Giavazzi R. Cancer Res. 2003;63:5224. [PubMed] [Google Scholar]
- 61.Mahadik BP, Pedron Haba S, Skertich LJ, Harley BA. Biomaterials. 2015;67:297. doi: 10.1016/j.biomaterials.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seano G, Chiaverina G, Gagliardi PA, di Blasio L, Sessa R, Bussolino F, Primo L. Blood. 2013;121:e129. doi: 10.1182/blood-2012-08-452292. [DOI] [PubMed] [Google Scholar]
- 63.Zagzag D, Amirnovin R, Greco MA, Yee H, Holash J, Wiegand SJ, Zabski S, Yancopoulos GD, Grumet M. Lab Invest. 2000;80:837. doi: 10.1038/labinvest.3780088. [DOI] [PubMed] [Google Scholar]
- 64.Caspani EM, Crossley PH, Redondo-Garcia C, Martinez S. PLoS One. 2014;9:e101402. doi: 10.1371/journal.pone.0101402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duffy GP, Ahsan T, O’Brien T, Barry F, Nerem RM. Tissue Eng, Part A. 2009;15:2459. doi: 10.1089/ten.TEA.2008.0341. [DOI] [PubMed] [Google Scholar]
- 66.Kebers F, Lewalle JM, Desreux J, Munaut C, Devy L, Foidart JM, Noel A. Exp Cell Res. 1998;240:197. doi: 10.1006/excr.1998.3935. [DOI] [PubMed] [Google Scholar]
- 67.Chen MB, Whisler JA, Jeon JS, Kamm RD. Integr Biol. 2013;5:1262. doi: 10.1039/c3ib40149a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whisler JA, Chen MB, Kamm RD. Tissue Eng, Part C. 2014;20:543. doi: 10.1089/ten.tec.2013.0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bersini S, Jeon JS, Dubini G, Arrigoni C, Chung S, Charest JL, Moretti M, Kamm RD. Biomaterials. 2014;35:2454. doi: 10.1016/j.biomaterials.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jeon JS, Bersini S, Gilardi M, Dubini G, Charest JL, Moretti M, Kamm RD. Proc Natl Acad Sci USA. 2015;112:214. doi: 10.1073/pnas.1417115112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leslie-Barbick JE, Moon JJ, West JL. J Biomater Sci, Polym Ed. 2009;20:1763. doi: 10.1163/156856208X386381. [DOI] [PubMed] [Google Scholar]
- 72.Cuchiara ML, Horter KL, Banda OA, West JL. Acta Biomater. 2013;9:9258. doi: 10.1016/j.actbio.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.