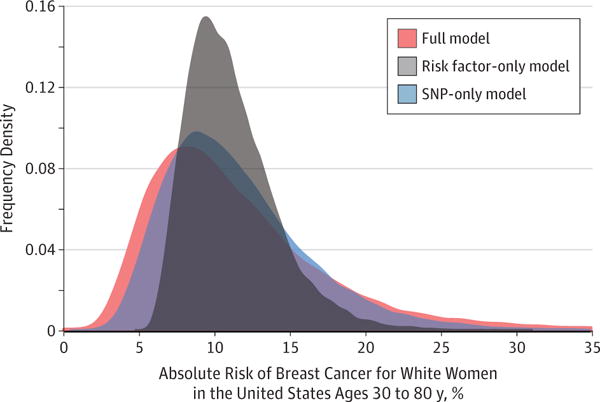
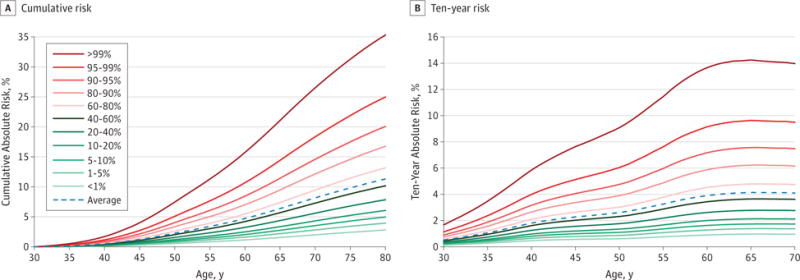
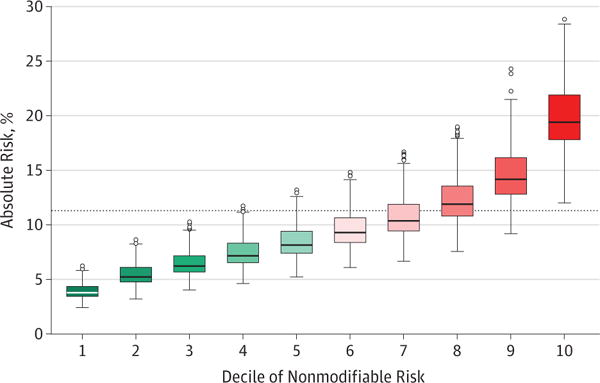
Paige Maas
Paige Maas, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Myrto Barrdahl
Myrto Barrdahl, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Amit D Joshi
Amit D Joshi, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Paul L Auer
Paul L Auer, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Mia M Gaudet
Mia M Gaudet, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Roger L Milne
Roger L Milne, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Fredrick R Schumacher
Fredrick R Schumacher, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
William F Anderson
William F Anderson, MD, MPH
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
David Check
David Check, BS
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Subham Chattopadhyay
Subham Chattopadhyay, BS
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Laura Baglietto
Laura Baglietto, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Christine D Berg
Christine D Berg, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Stephen J Chanock
Stephen J Chanock, MD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
David G Cox
David G Cox, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Jonine D Figueroa
Jonine D Figueroa, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Mitchell H Gail
Mitchell H Gail, MD, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Barry I Graubard
Barry I Graubard, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Christopher A Haiman
Christopher A Haiman, ScD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Susan E Hankinson
Susan E Hankinson, ScD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Robert N Hoover
Robert N Hoover, MD, ScD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Claudine Isaacs
Claudine Isaacs, MD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Laurence N Kolonel
Laurence N Kolonel, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Loic Le Marchand
Loic Le Marchand, MD, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
I-Min Lee
I-Min Lee, ScD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Sara Lindström
Sara Lindström, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Kim Overvad
Kim Overvad, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Isabelle Romieu
Isabelle Romieu, MD, MPH, ScD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Maria-Jose Sanchez
Maria-Jose Sanchez, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Melissa C Southey
Melissa C Southey, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Daniel O Stram
Daniel O Stram, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Rosario Tumino
Rosario Tumino, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Tyler J VanderWeele
Tyler J VanderWeele, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Walter C Willett
Walter C Willett, MD, DrPh
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Shumin Zhang
Shumin Zhang, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Julie E Buring
Julie E Buring, ScD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Federico Canzian
Federico Canzian, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Susan M Gapstur
Susan M Gapstur, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Brian E Henderson
Brian E Henderson, MD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
David J Hunter
David J Hunter, MBBS, ScD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Graham G Giles
Graham G Giles, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Ross L Prentice
Ross L Prentice, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Regina G Ziegler
Regina G Ziegler, PhD, MPH
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Peter Kraft
Peter Kraft, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Montse Garcia-Closas
Montse Garcia-Closas, MPH, DrPH
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1,
Nilanjan Chatterjee
Nilanjan Chatterjee, PhD
1Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland (Maas, Anderson, Check, Chattopadhyay, Chanock, Figueroa, Gail, Graubard, Hoover, Ziegler, Garcia-Closas, Chatterjee); Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany (Barrdahl); Program in Genetic Epidemiology and Statistical Genetics, Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Joshi, Lindström, Hunter, Kraft); Fred Hutchinson Cancer Research Center, Seattle, Washington (Auer, Prentice); School of Public Health, University of Wisconsin-Milwaukee, Milwaukee (Auer); Epidemiology Research Program, American Cancer Society, Atlanta Georgia (Gaudet, Gapstur); Cancer Epidemiology Centre, Cancer Council Victoria, Melbourne, Australia (Milne, Baglietto, Giles); Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Victoria, Australia (Milne, Baglietto, Giles); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles (Schumacher, Haiman, Stram, Henderson); Division of Cancer Prevention, National Cancer Institute, Bethesda, Maryland (Berg); INSERM U1052 – Cancer Research Center of Lyon, Centre Léon Bérard, Lyon, France (Cox); Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, England (Cox); Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst (Hankinson); Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Hankinson); Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC (Isaacs); Epidemiology Program, Cancer Research Center, University of Hawaii, Honolulu (Kolonel); University of Hawaii Cancer Center, Honolulu (Le Marchand); Division of Preventive Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts (Lee, Zhang, Buring); Department of Public Health, Aarhus University, Aarhus, Denmark (Overvad); Nutrition and Metabolism Section, International Agency for Research on Cancer, Lyon, France (Romieu); Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA. Hospitales Universitarios de Granada/Universidad de Granada, Granada, Spain (Sanchez); CIBER de Epidemiología y Salud Pública (CIBERESP), Spain (Sanchez); Genetic Epidemiology Laboratory, Department of Pathology, The University of Melbourne, Melbourne, Australia (Southey); Cancer Registry and Histopathology Unit, “Civic-M.P.Arezzo” Hospital, ASP Ragusa, Italy (Tumino); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Biostatistics, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (VanderWeele); Department of Nutrition, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Willett); German Cancer Research Center (DKFZ), Heidelberg, Germany (Canzian); Department of Epidemiology and Preventive Medicine, Monash University, Melbourne, Victoria, Australia (Giles); University of Washington, School of Public Health and Community Medicine, Seattle (Prentice); Breakthrough Breast Cancer Research Centre, Division of Genetics and Epidemiology, The Institute of Cancer Research, London, England (Garcia-Closas); Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland (Chatterjee); Department of Oncology, School of Medicine, Johns Hopkins University, Baltimore, Maryland (Chatterjee)
1