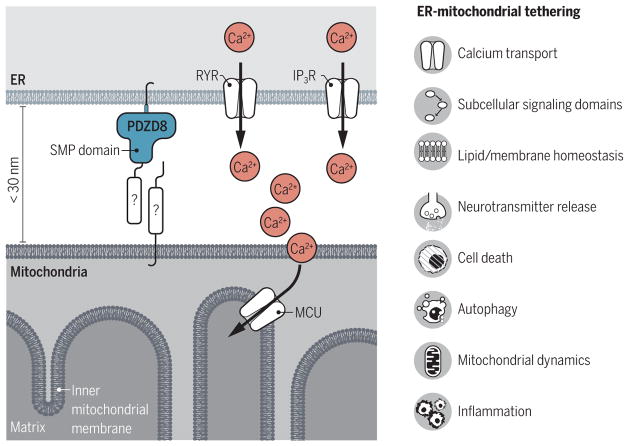
Cellular function and survival require the highly ordered subcellular localization of organelles. In particular, there is considerable interest in understanding the interconnectedness of the endoplasmic reticulum (ER) and mitochondria. The direct interaction of ER and mitochondrial membranes is referred to as tethering, typically defined as an interorganelle distance of less than 30 nm. Tethering has been implicated in numerous physiological processes, including lipid synthesis and transfer, coupling of Ca2+ transfer, autophagosome formation, inflammatory signaling, mitochondrial morphology, and mitochondrial DNA (mtDNA) synthesis and distribution (1–5). Although the functional relevance of ER-mitochondrial tethering is widely accepted, identification of the proteins that constitute these interconnected structures has remained elusive. On page 623 of this issue, Hirabayashi et al. (6) show that PDZ domain containing protein 8 (PDZD8) is required for the tethering of ER and mitochondrial membranes and is critical for Ca2+ transfer from ER to mitochondria.
The most well-defined ER-mitochondrial tethering complex is the yeast ER-mitochondria encounter structure (ERMES). The ERMES complex is composed of the ER-membrane–bound protein maintenance of mitochondrial morphology protein 1 (Mmm1), the cytosolic linker mitochondrial distribution and morphology protein 12 (Mdm12), and the outer mitochondrial membrane proteins Mdm34 and Mdm10. To date, no ERMES functional ortholog has been identified in metazoans (multicellular organisms). Using bioinformatics and structural modeling approaches, Hirabayashi et al. identified PDZD8 as a mammalian protein likely to contain an SMP (synaptotagmin-like mitochondrial lipid binding protein) domain, a functional feature synonymous with yeast ERMES proteins. Through a series of elegant experiments, they demonstrated that PDZD8 is an ortholog of the yeast ERMES complex protein Mmm1. MMM1-deficient yeast cells exhibit a dramatic phenotype attributed to loss of ER-mitochondrial contacts, including collapse of the mitochondrial network, inheritance defects, and complete loss of mtDNA (7). Expression of a yeast-MMM1–mouse-PDZD8 chimeric protein in MMM1-null yeast rescued the phenotype. These experiments prove that the SMP domain from mammalian PDZD8 is functionally equivalent to members of the yeast ERMES complex.
Next, they showed that PDZD8 in mammalian cell lines localized to ER membranes at sites in close proximity to mitochondria (see the figure). Further, the authors showed that mammalian cells lacking PDZD8 had significantly fewer ER-mitochondrial contact sites. Using three-dimensional serial electron microscopy, they showed that loss of PDZD8 markedly altered mitochondrial surface area in contact with the ER and that the reciprocal ER-mitochondrial interface was reduced. This dramatic difference (~80% reduction) in ER-mitochondrial contacts has never been shown for any of the candidate genes proposed to tether ER and mitochondria in mammalian systems. Furthermore, the difference in contact sites was not due to changes in either mitochondrial or ER morphology. This is of particular importance as alterations in mitochondrial structure have hampered the interpretation of other genes implicated in tethering (5). The alterations in tethering were causally linked to a decrease in ER-mitochondrial Ca2+ transfer. Importantly, loss of PDZD8 had no effect on Ca2+ release from the ER or ER-independent mitochondrial Ca2+ uptake, indicating that PDZD8 is specifically necessary for ER-dependent mitochondrial Ca2+ uptake.
Figure. Coupling ER and mitochondrial membranes.
The proteins that mediate the close coupling of ER and mitochondrial membranes (tethering) in mammalian cells have remained elusive. PDZD8 is an ER-bound protein that is critical for the tight association of ER and mitochondrial membranes. This will now allow the search for other possible binding partners and regulators that make up this newly identified tethering complex in mammalian cells. The close proximity of the ER and mitochondria is essential for several cellular processes. IP3R, inositol-1,4,5-trisphosphate receptor; MCU, mitochondrial calcium uniporter; RYR, ryanodine receptor.
Ca2+ cross-talk between ER and mitochondria is arguably the most important function of ER-mitochondrial tethering. Ca2+ is an essential cellular signaling molecule, the effects of which are dependent on local changes in its concentration. Receptor-activated Ca2+ release from ER creates high-Ca2+ microdomains at discrete subcellular locations (8, 9). Indeed, it is estimated that ER Ca2+ release increases the Ca2+ concentration at ER-mitochondrial junctions more than 20-fold compared to the rest of the cytosol (8). Local elevations in Ca2+ regulate a vast array of effector molecules either directly through Ca2+-binding sites or indirectly by modulating Ca2+-dependent enzymatic processes (10). Thus, the spatiotemporal pattern of Ca2+ has profound effects on cellular function and survival, and ER-mitochondrial tethering is an important regulatory mechanism of Ca2+ dynamics and cellular physiology. The functional importance of subcellular Ca2+ domains is especially evident in the regulation of neurotransmitter release at neuronal synapses (for communication between neurons) (11, 12). When an action potential (electrical signal) reaches the presynaptic terminal, voltage-gated Ca2+ channels open, allowing for Ca2+ entry. The local rise in intracellular Ca2+ triggers fusion of neurotransmitter-filled vesicles with the presynaptic membrane (10). In addition, the duration and amplitude of Ca2+ spikes in dendrites (sites of postsynaptic neurotransmitter binding in neurons) is necessary for synaptic integrity and plasticity mechanisms that underlie memory allocation and storage (13, 14). Hirabayashi et al. show that PDZD8-dependent ER-mitochondrial tethering is also important for shaping Ca2+ microdomains in neuronal dendrites. Upon untethering mitochondria from ER, by decreasing PDZD8 expression, Ca2+ transfer was significantly reduced and cytosolic Ca2+ concentration was elevated, suggesting that PDZD8 is important for synaptic signaling. Future studies are likely already underway to determine the impact of the PDZD8-containing tethering complex on numerous neuronal functions.
Moving beyond cellular physiology, a better overall understanding of ER-mitochondrial associations may also shed light on mechanisms of disease. The ER-mitochondrial interface is the site of many biochemical processes that have been implicated in neurodegenerative diseases such as Ca2+ homeostasis, autophagy (the process of cellular organelle recycling), and mitochondrial dynamics (15). More importantly, it is known that ER-mitochondrial tethering is disturbed in Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis with associated frontotemporal dementia (15). However, the molecular mechanisms underlying ER-mitochondrial disruption are not fully understood. Although ER-mitochondrial contact sites represent a nexus for many signaling cascades and biochemical reactions, it is yet to be determined whether a disruption in tethering is causative in neurodegenerative disease initiation or represents a secondary alteration that occurs during disease progression. Clearly, this discovery will provide new tools to better understand the ER-mitochondrial axis with respect to physiology and disease across cell types.
Although several mammalian ER-mitochondrial tethering proteins have been proposed, most lack clear indisputable evidence, and the identification of bona fide ER-mitochondrial tethers has remained elusive. We now have the first description of a protein that appears to primarily function as a member of an ER-mitochondrial tethering complex that can be genetically manipulated without confounding alterations in ER or mitochondrial integrity. This exciting discovery will provide not only new molecular tools to begin to define the physiological functions of ER-mitochondrial connections but also stimulate the search for the mitochondrial interaction partner of PDZD8 and other potential yeast ERMES homologs in mammals.
References
- 1.Ah Young AP, et al. Proc Natl Acad Sci USA. 2015;112:E3179. [Google Scholar]
- 2.Friedman JR, et al. Science. 2011;334:358. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korobova F, Ramabhadran V, Higgs HN. Science. 2013;339:464. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garofalo T, et al. Autophagy. 2016;12:917. doi: 10.1080/15548627.2016.1160971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filadi R, Theurey P, Pizzo P. Cell Calcium. 2017;62:1. doi: 10.1016/j.ceca.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Hirabayashi Y, et al. Science. 2017;358:623. doi: 10.1126/science.aan6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kornmann B, et al. Science. 2009;325:477. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Csordás G, Thomas AP, Hajnóczky G. EMBO J. 1999;18:96. doi: 10.1093/emboj/18.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizzuto R, et al. Science. 1998;280:1763. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 10.Berridge MJ, Bootman MD, Roderick HL. Nat Rev Mol Cell Biol. 2003;4:517. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 11.Kwon S-K, et al. PLOS Biol. 2016;14:e1002516. doi: 10.1371/journal.pbio.1002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heidelberger R, Heinemann C, Neher E, Matthews G. Nature. 1994;371:513. doi: 10.1038/371513a0. [DOI] [PubMed] [Google Scholar]
- 13.Tran-Van-Minh A, Abrahamsson T, Cathala L, DiGregorio DA. Neuron. 2016;91:837. doi: 10.1016/j.neuron.2016.07.029. [DOI] [PubMed] [Google Scholar]
- 14.Tsay D, Dudman JT, Siegelbaum SA. Neuron. 2007;56:1076. doi: 10.1016/j.neuron.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paillusson S, et al. Trends Neurosci. 2016;39:146. doi: 10.1016/j.tins.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]