Abstract
Understanding what drives breast tumor progression is of utmost importance for blocking tumor metastasis; we have identified that semaphorin 7a is a potent driver of ductal carcinoma in situ (DCIS) progression. Semaphorin 7a is a GPI membrane anchored protein that promotes attachment and spreading in multiple cell types. Here we show that increased expression of SEMA7A occurs in a large percentage of breast cancers and is associated with decreased overall and distant metastasis free survival. In both in vitro and in vivo models, shRNA mediated silencing of SEMA7A reveals roles for semaphorin 7a in the promotion of DCIS growth, motility, and invasion as well as lymphangiogenesis in the tumor microenvironment. Our studies also uncover a relationship between COX-2 and semaphorin 7a expression and suggest that semaphorin 7a promotes tumor cell invasion on collagen and lymphangiogenesis via activation of β1-integrin receptor. Our results suggest that semaphorin 7a, may be novel target for blocking breast tumor progression.
Semaphorin 7a belongs to a large class of secreted and membrane bound proteins that were originally identified for their pleiotropic roles in neurite extension and migration(1–3). Unlike other semaphorins, which act to stunt axon outgrowth and inhibit angiogenesis, semaphorin 7a enhances axon migration and development(1–3) and angiogenesis(4–8). During neurogenesis, Sem7a promotes cell adhesion and motility through interaction with β1-integrin receptors. A similar role for Sem7a in monocyte and melanocyte migration has been described(9, 10). Furthermore, Sem7a also promotes differentiation in monocytes and osteoblasts(11, 12). In mice, Sem7a expression is observed at high levels in brain, spinal cord, lung, testis, bone and spleen as well as in immune cells; in human tissues, Sem7a expression has been observed in the nervous system, odontoblasts, fibroblasts, keritnocytes, endothelial cells, and melanocytes(3, 11, 13–15). While Sem7a has been primarily characterized for its role in development, re-activation of developmental programs often occurs during tumorigenesis and the pleiotropic nature of the semaphorins during development have led to investigation of semaphorins as multi-target therapies for cancer(16). Recently, in murine mammary tumor models, several studies demonstrated that semaphorin 7a expression promotes mouse mammary tumor cell growth, epithelial to mesenchymal transition, angiogenesis, and lung metastasis(5, 17, 18). However, a role for SEMA7A in human breast tumor progression remains largely unexplored.
To investigate whether Sem7a is a mediator of human breast tumor aggressiveness we silenced SEMA7A in MCF10DCIS cells. We chose this model based on its unique characteristics, which partially model human DCIS progression(19–21). Briefly, during establishment and outgrowth of the MCF10DCIS.com model, tumor cells create a non-invasive ductal carcinoma in situ-like structure that is similar to human DCIS. As early as a week after injection, myoepithelial cells at the radial edges of the DCIS-like structures are evident by expression of myoepithelial markers such as alpha-smooth muscle actin and p63, and deposition of a collagen type 4 positive basement membrane(20–22). Increased size of these DCIS-like lesions results from initial proliferation of tumor cells, which have the histologic appearance of micro-invasion, but then undergo a differentiation process that creates new DCIS-like neoplastic structures. In a period of approximately 4–8 weeks all tumors progress to IDC (SFigure 1A) that is characterized by tumor cell expression of EMT markers and fragmentation of the myoepithelial cell layer, underlying basement membranes and ‘cancerization’ of the associated stroma(20, 21). During this phase the neoplastic cells expand rapidly forming confluent sheets of tumor cells that invade directly into the stromal tissue. Within these sheets of invasive cancer, some areas of residual DCIS-like structures may be visible with maintained myoepithelial cells and basement membrane deposition(20, 21). Interestingly, p63 is initially expressed in peripheral cells of the DCIS-like structures that are morphologically consistent with myoepithelial cells; however, as the tumors in this model progress to an invasive phenotype p63 is expressed in the majority of the tumor cells(22, 23). Although this is a consistent feature of this model, it is noted this is not necessarily consistent with the natural progression of most human ductal carcinomas(24, 25). We examined the effects of Sem7a silencing on tumor cell growth, motility, and invasion in vitro and in vivo; we show that reducing Sem7a expression decreases human breast tumor cell growth, motility, invasion, and adhesion. Furthermore, we identify a novel role for Sem7a in promotion of lymphangiogenesis. We also show that Sem7a is regulated by COX-2 expression and that loss of Sem7a from tumor cells reduces tumor cell invasion and activation of β1-integrin receptor. Importantly, our analysis of patient datasets indicates that increased SEMA7A gene expression is observed in a high percentage of human breast tumors and is associated with poor prognosis. Cumulatively, our results suggest that further research into the role of Sem7a in breast cancer may lead to novel therapeutics for patients.
Results
Semaphorin 7a promotes tumor progression in breast cancer patients
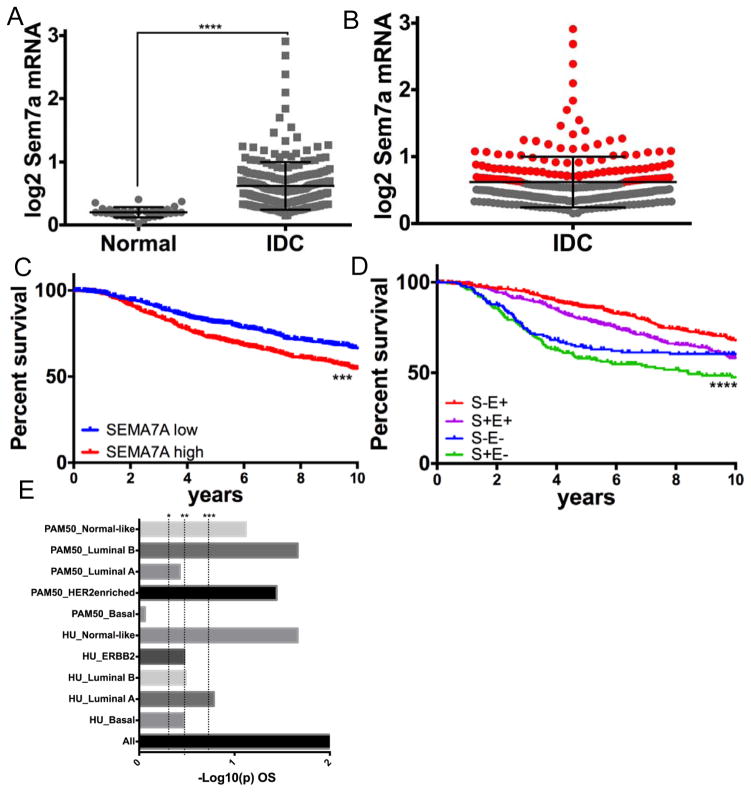
To determine whether SEMA7A expression correlates with disease progression in breast cancer patients we analyzed SEMA7A expression in normal breast tissue compared to invasive ductal carcinoma (IDC) in the TCGA dataset(26) using Oncomine(27). We find that SEMA7A expression is significantly increased in IDC when compared to normal tissue (Figure 1A). Furthermore, SEMA7A expression was above average in ~40% of IDC (red; Figure 1B). Additionally, SEMA7A was above average in >40% of cases in 3 additional datasets(18, 28, 29) (Table 1). We also observed association of SEMA7A with early recurrence, metastasis, and death in multiple publically available datasets (Table 2). We utilized Kaplan-Meier analysis to determine whether expression of SEMA7A is associated with poor prognosis in breast cancer patients in the METABRIC (Curtis)(28) breast tumor dataset, which is a large breast cancer dataset that is annotated for clinical characteristics and outcomes such as overall survival (OS) and ER status. Our analysis revealed decreased OS in patients with increased SEMA7A expression (Figure 1C). We also observed that SEMA7A was associated with decreased survival regardless of estrogen receptor status (Figure 1D and SFigure 1A&B). To further validate that Sem7a is associated with decreased overall survival, we utilized the gene expression-based outcome for breast cancer online (GOBO) tool(18) to analyze SEMA7A expression and outcomes in additional breast tumor data sets. In a dataset of 737 tumors, we utilized Kaplan-Meier analysis, shown here as –Log10(p) for OS, to show that high SEMA7A was most significantly associated with decreased OS in all breast cancer patients (Figure 1E). In addition, a higher proportion of patients with high Sem7a were grade 3 compared to those with low Sem7a (SFigure 1C). Taken together, these results implicate SEMA7A in the promotion of tumor metastasis in breast cancer patients, irrespective of the cancer biologic subtype.
Figure 1. SEMA7A is associated with poor prognosis in breast cancer patients.
A) Analysis of SEMA7A mRNA (log2 expression) in normal breast tissue from TCGA compared to invasive ductal carcinoma (IDC) reveals increased expression in IDC compared to normal. B) Cases of IDC expressing above average Sem7a in red. C) Kaplan-Meier 5 year survival analysis in the METABRIC (Curtis) dataset shows decreased overall survival (OS) with high expression of SEMA7A (n=607/1332). D) SEMA7A high patients have the worst survival regardless of whether they are ER+ (E+) or ER− (E−). E) –Log10(p) value of Kaplan-Meier survival analysis in reveals significantly decreased survival in all breast cancer patients with high expression of SEMA7A that does not appear to be driven by a single molecular subtype. *p<0.05, **P<0.01, ***P<0.001.
Table 1.
Increased SEMA7A is observed in a high percentage of breast cancers in multiple datasets.
| Dataset | %’OE | N |
|---|---|---|
| TCGA | 45% | 251 |
| NKI295 | 48% | 295 |
| GOBO | 45% | 737 |
| Curtis | 44% | 2,136 |
Table 2.
Increased SEMA7A (1.1–1.4 fold) is associated with increased early recurrence and decreased survival in multiple datasets.
| Dataset | N= | p value | Outcome |
|---|---|---|---|
| Esserman Breast | 130 | 0.041 | Recurrance at 3 years |
| Ma Breast 3 | 60 | 0.022 | Recurrance at 5 years |
| Sotiriou Breast 2 | 98 | 0.037 | Recurrance at 5 years |
| Bos Breast | 204 | 0.002 | Metastatic Event at 1 year |
| Minn Breast 2 | 121 | 0.044 | Metastatic Event at 3 years |
| vandeVijver Breast | 295 | 0.007 | Metastatic Event at 3 years |
| Hatzis Breast | 508 | 0.025 | Metastatic Event at 3 years |
| Symanns Breast 2 | 103 | 0.015 | Metastatic Event at 5 years |
| vandeVijver Breast | 295 | 0.008 | Dead at 3 years |
| Curtis Breast | 2,136 | 0.022 | Dead at 3 years |
| TCGA Breast | 593 | 0.021 | Dead at 3 years |
| TCGA Breast | 593 | 0.009 | Dead at 5 years |
| Curtis Breast | 2,136 | 6.81EJ04 | Dead at 5 years |
SEMA7A expression promotes human tumor cell growth and motility in vitro and in vivo
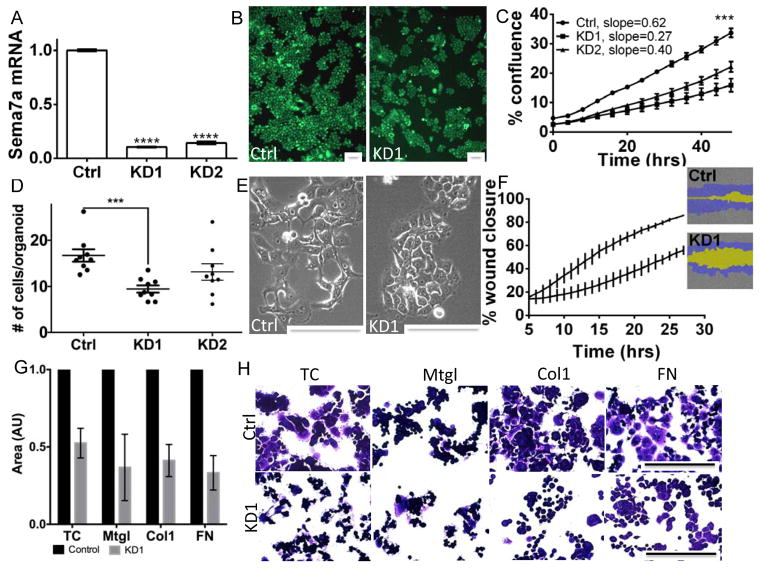
In order to determine mechanisms of SEMA7A promotion of breast cancer, we generated MCF10DCIS.com cells with stable silencing of SEMA7A (shSem7a). Decreased Sem7a protein expression was observed (Figure 2A; SFig 2A). In two independent lines, which harbor different shRNA sequences, we observed decreased tumor growth rate and overall growth when compared to control cells transfected with a non-targeting vector (Figure 2B&C; SFigure 2B). We also examined whether Sem7a was required for MCF10DCIS tumor cell growth in three dimensional (3D) culture using our previously described 3D culture system(23). We observed a decreased number of multicellular spheroids and number of cells/spheroid with shSem7a cells compared to controls (Figure 2D; SFig 2C). Furthermore, shSEM7a cells exhibited less mesenchymal morphology (Figure 2E) and decreased motility in wound closure assays compared to controls (Figure 2F). We also examined the ability of control and knockdown cells to adhere to different matrices and observed that both control and knockdown cells were most adherent and proliferative on fibronectin (SFig 2D&E) and that the shSem7a cells were less adherent on all matrices compared to control (Figure 2G&H). These in vitro results suggest that Sem7a promotes breast tumor progression by increasing tumor cell growth and motility.
Figure 2. Sem7a drives tumor cell growth and motility in vitro.
A) SEMA7A mRNA levels in MCF10DCISGFP cells stably transfected with vector only (Ctrl) or two different shRNA constructs (KD1 and KD2) shows significantly decreased SEMA7A expression. B) 10X images and C) % confluence and growth rate (slope) over 48 hours are decreased in shSem7a KD1 and KD2 cells. D) # of cells/structure in 3D matrigel cultures is decreased with Sem7a knockdown. E) 20X brightfield images of Ctrl and KD1 cells showing less mesenchymal morphology in shSem7a KD1 cells. F) % wound closure is decreased with shSem7a KD1. G&H) shSem7a cell adhesion is decreased on tissue culture treated plastic (TC), matrigel (Mtgl), collagen 1 (Col1), and fibronectin (FN) compared to control (set to 1). Scalebar=50um, *p<0.05, **P<0.01, ***P<0.001, ****p<0.0001
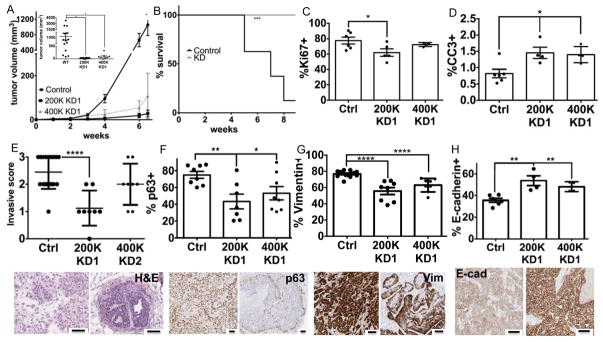
To determine whether Sem7a promotes breast tumor progression in vivo, we injected shSem7a and control tumor cells into intact nulliparous mouse mammary glands. We observed decreased growth with both KD1 and KD2 (SFigure 3A). In a larger study, with shSEM7a KD1 cells, we observed significantly decreased tumor growth (Figure 3A), which was maintained even when twice as many shSem7aDCIS KD1 cells were injected (Figure 3A). Furthermore, utilizing maximal tumor size (2cm3) as a surrogate for survival within groups, survival was reduced significantly in the control group compared to the knockdown group (Figure 3B). We confirmed that Sem7a protein was decreased in the knockdown group by IHC (SFigure 3B), but also observed that some of the tumor cells re-gained expression of Sem7a. Nevertheless, we observed decreased Ki67+ nuclei in the shSEM7a tumors suggestive of decreased tumor cell proliferation (Figure 3C;SFigure 3C). We also observed increased cleaved caspase 3 staining in the shSem7a tumors (Figure 3D) suggesting that Sem7a promotes tumor cell survival; however, this increase was observed at 2 weeks post-injection, but was not confirmed in our 8-week study where Ki67 was still decreased (SFig 3D). Whether this is due to a change in biology or can be attributed to re-expression of Sem7a in vivo is unanswered by our current studies.
Figure 3. Sem7a promotes tumor growth and invasion in vivo.
A) MCF10DCIS tumor cell growth is decreased with injection of the same number (200K) ofshSem7a KD1 cells as well as with twice as many (400K) KD1 cells (inset: all datapoints at study end). B) % survival, as measured by time to 2cm3 tumor volume, is significantly increased in the shSem7a KD1 group. C) % proliferation, measured by % Ki67+, is decreased in shSem7a KD1 tumors. D) % cleaved caspase 3 positive cells is increased in shSem7a KD1 tumors. E) Average invasive score (0=DCIS, 1=DCIS with microinvasion, 2=IDC with DCIS remnant, 3=IDC) is decreased in shSem7a KD1 tumors. F) % intratumor p63 and G) vimentin + is decreased in shSem7a KD1 tumors. H) % E-cadherin + is increased in Sem7a KD1. Scalebar=50um *p<0.05, **P<0.01, ***P<0.001, ****p<0.0001.
To determine invasiveness, lesions that contained only DCIS structures with clearly defined basement membranes and no evidence of micro-invasion were scored as 0, lesions that contained DCIS with identifiable micro-invasive foci were scored as 1, lesions that contained significant areas of sheet-like invasive tumor growth mixed with areas of DCIS were scored as 2, and lesions that contained entirely invasive tumor with rare to absent DCIS were scored as 3. While nearly all control tumors were comprised of either mixed DCIS and invasive carcinoma (2) or exclusively invasive carcinoma (3), shSem7a cells produced tumors that were either predominantly DCIS (0–1) or mixed in situ and invasive carcinoma (2) in a manner that appeared dependent on the number of tumor cells injected (Figure 3E). These results were compiled to create an average invasive score, which was significantly decreased when the same number (200K) of knockdown cells were injected compared to wild-type controls and trended towards significant when twice as many knockdown cells were injected. Furthermore, when all knockdown tumors were compared to wild-type, there was an overall significant decrease in the invasion score (SFigure 3E). Our initial studies examined invasion 4–6 weeks post-injection, however, we also examined invasion at a three weeks post-injection where we observed that 4/4 shSem7a tumors remained DCIS(0) (SFigure 3F) while 4/5 controls progressed to microinvasion(1). Therefore, decreased Sem7a expression appears to limit the in vivo invasive potential of the MCF10DCIS.com breast tumor cells. Additional evidence for lack of invasive/mesenchymal phenotype comes from our quantitation of intratumor p63, vimentin, and E-cadherin expression (Figure 3F–H); both p63 and vimentin were reduced and E-cadherin increased in the shSem7a tumors suggestive of a less mesenchymal cellular phenotype.
Semaphorin 7a mediates COX-2 dependent tumor cell invasion and promotion of lymphangiogenesis
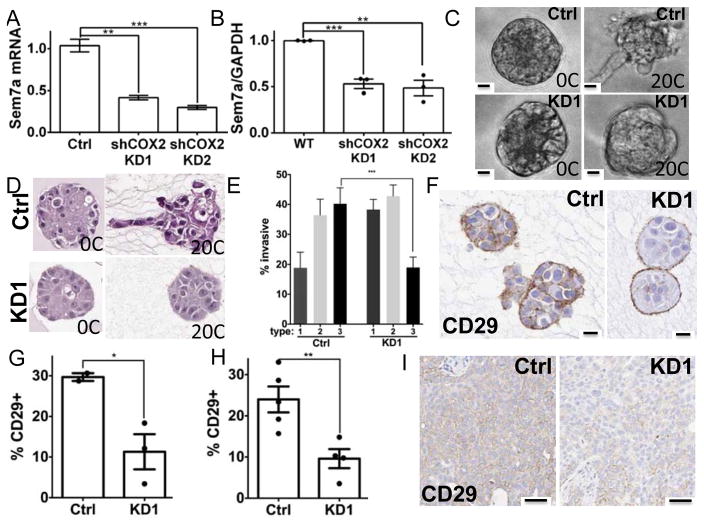
Increases in extracellular collagen 1 can promote COX-2 expression in DCIS.com tumor cells; COX-2 in turn promotes tumor cell growth, invasion, and lymphnagiogenesis(23, 30). To determine whether Sem7a may function downstream of COX-2 to promote growth and invasion, we examined Sem7a expression in shCOX-2 cell lines(23, 30, 31)(SFigure 4A). We observed significantly decreased expression of Sem7a in two independent shCOX-2 silenced MCF10DCIS.com cell lines (Figure 4A&B) suggesting that Sem7a expression is dependent upon COX-2 expression. To examine whether Sem7a was a potential mediator of COX-2 invasion on collagen we analyzed shSem7a cells in our 3D invasion model; as previously reported, we found that addition of collagen 1 resulted in increased invasion of the control MCF10DCIS.com cells(23, 32). In contrast, fewer invasive protrusions were observed with the shSem7a cells (Figure 4C&D). We quantitated % invasive by typing the structures as non-invasive (type 1), partially invasive (type 2), and fully invasive (type 3) (see SFigure 3B), similar to our published results with the shCOX-2 cells(23), and we observed that the majority of structures in the Ctrl group were types 2 and 3 while the majority of the structures in the shSem7a group were types 1 and 2 (Figure 4E). Because Sem7a promotes dendricity and spreading of neurons and melanocytes through β1-integrin receptors(3, 9), we next investigated whether activation of β1-integrin receptor was altered in 3D structures with shSem7a cells. Using an antibody that specifically recognizes the activated form of β1-integrin receptor to stain sections from our 3D invasion studies(32), we observed higher activation in control structures compared to shSem7a (Figure 4F&G). Furthermore, the localization of beta1integrin was restricted to the structure periphery in the shSem7a cells, but was observed throughout the structures in the controls (Figure 4F) suggesting that Sem7a mediates invasion on collagen through activation of β1-integrin receptors. Interestingly, when we block β1-integrin receptor in our 3D invasion assay we did not see decreases in COX-2 expression, but saw decreases in invasive structures(32). Thus, we propose that COX-2 upregulates Sem7a, which activates β1-integrin receptors on neighboring cells to promote tumor cell invasion. Finally, we also observe decreased activation of β1-integrin receptor shSem7a tumors (Figure 4H&I) suggesting that β1-integrin receptors mediate MCF10DCIS tumor cell invasion in vivo as well.
Figure 4. COX-2 drives Sem7a expression to promote tumor invasion and activation of β1-integrin.
A) qPCR and B) Western analyses reveal decreased SEMA7A/Sem7a in MCF10DCISGFP cells stably transfected with shRNAs for COX-2. C) Control and KD1 cells In live 3D cultures and D) H&E stained sections reveal that acquisition of invasive phenotype on 20% collagen is decreased with Sem7a knockdown. E) Quantitation of organoid invasive morphology on H&E stained sections. F) Activation of β1-integrin is significantly reduced in organoids with KD cells in 3D cultures on 20% collagen. G) Quantitation of %Cd29+ in organoid sections. H) CD29% + pixels are decreased in shSem7a KD1 tumors 2 weeks post-injection. I) Representative images depicting decreased staining for activated CD29 in xenografts from knockdown cells compared to controls. Scale bar=20um, *p<0.05, **p<0.01, ***p<0.001.
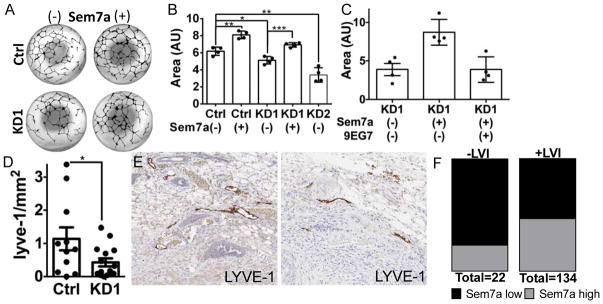
Having previously shown that COX-2 mediates lymphangiogenesis in the tumor microenvironment we also investigated the role of Sem7a in lymphangiogenesis. We added recombinant purified Sem7a to an in vitro lymphangiogenesis assay; PGE2 was utilized as a positive control for these studies as we have previously published PGE2 as a pro-lymphangiogenic factor(30). Compared to controls, we observed a significant increase in the size of in vitro lymphatic structures with addition of Sem7a to levels that were comparable to those observed with PGE2 (SFigure 3C). Having also shown that MCF10DCIS tumor cell conditioned media promotes lymphangiogenesis in vitro(30), we examined the effects of conditioned media from shSEMA7A cells on in vitro lymphangiogenesis. shSEMA7A cell conditioned media resulted in a significant reduction in lymphatic structure size compared to control, but addition of exogenous Sem7a rescued (Figure 5A&B). In addition, when we added a function blocking antibody for β1-integrin receptor, Sem7a was unable to rescue suggesting that Sem7a also mediates lymphangiogenesis via β1-integrin receptor activation (Figure 5C). Furthermore, in vivo tumor associated lymphangiogenesis was deceased with shSem7a tumor cells as evidenced by a significant decrease in lymphatic vessel density in the peritumor region of shSem7a tumors (Figure D&E; SFigure 3D). Finally, in the METABRIC dataset high Sem7a was associated with increased risk for lymphatic invasion (OR: 2.93, 95% CI: 1.021–8.397, p 0.0387), which we have previously shown to also be COX-2 dependent(30) (Figure 5F). Collectively, these in vivo, in vitro, and patient based results suggest that Sem7a exerts pleotropic effects, similar to those induced by COX-2, on tumor cells and the tumor microenvironment to promote breast tumor progression.
Figure 5. Sem7a promotes tumor-associated lymphangiogenesis and lymph vessel invasion.
A) Representative images of entire wells from lymphangiogenesis assay quantitation for B. B) HDLEC tube formation is significantly reduced with conditioned media from Sem7a knockdown cells, but can be rescued with addition of 5 mg/ml exogenous Sem7a. C) HDLEC tube formation is significantly reduced with conditioned media from Sem7a knockdown cells and rescue with exogenous Sem7a is blocked by inhibition of β1-integrin activation with functional blocking mAb 9EG7. D) Peritumor LVD is decreased with shSem7a KD1 tumors. E) Representative images depicting decreased Lyve-1 staining at the tumor periphery of knockdown tumors compared to control. F) A higher proportion of patients in the METABRIC dataset for who are positive for lymph vessel invasion (LVI) exhibit higher than average Sem7a expression, OR: 2.928, 95% CI: 1.021–8.397, p=0.0387. Scale bar=20um, *p<0.05, **p<0.01, ***p<0.001.
Discussion
Sem7a is emerging as an important mediator of melanoma and mammary tumor progression in murine models(5, 18); here we characterize semaphorin 7a as a novel mediator of breast cancer progression using pre-clinical models of human breast cancer and patient genomic datasets. The results of our studies suggest that Sem7a promotes breast tumor progression via pleiotropic effects on the tumor and tumor-associated lymphatics. Specifically, we demonstrate a novel role for Sem7a in promotion of lymphangiogenesis via activation of β1-integrin receptor on lymphatic endothelial cells. Furthermore, we identify a role for of Sem7a in human tumor growth and invasion by proposing a novel mechanism whereby COX-2 expression drives expression of semaphorin 7a. In turn, semaphorin 7a expressing breast tumor cells promote activation of β1-integrin receptors on neighboring cells to promote invasion (Figure 5). This mechanism is similar to semphorin 7a promotion of melanocyte migration and neurite extension(1, 3). Furthermore, while semaphorin 7a has been primarily characterized as a membrane bound semaphorin, a soluble isoform has been demonstrated(33) and our results also suggest that semaphorin 7a secretion by tumor cells acts directly on the lymphatic endothelium in the tumor microenvironment. In support of this, secretome analysis of glioblastoma cells revealed significantly increased secretion of semaphorin 7a in invasive tumor cells(34).
Importantly, our analyses of multiple tumor datasets suggests that high Sem7a is associated with decreased survival and increased metastasis in breast cancer patients. Furthermore, in 3 independent datasets we observe increased expression of SEMA7A in nearly 40% of breast tumors. Thus, a targeted therapy designed to shut down the actions of Sem7a may benefit the large percentage of breast cancer patients predicted by our studies to exhibit increased tumor associated expression of Sem7a. The question remains of how to properly target Sem7a. Our results revealing that expression is low in normal mammary tissues compared to tumor, along with those of our colleagues showing that Sem7a-null mice are viable(3), suggest that targeting Sem7a directly may be a therapeutic option. To date, compounds that specifically inhibit Sem7a are unavailable; however, multiple downstream targets of Sem7a have been identified, including FAK and Erk, which are current targets in clinical trials. In addition, our published data showing that extracellular collagen deposition induced upregulation of COX-2 expression can be inhibited with COX-2 inhibitors(23), coupled with our current observations suggesting that COX-2 expression is necessary for Sem7a expression, make it plausible that COX-2 inhibition may also decrease Sem7a expression. COX-2 inhibitors are relatively safe and readily available. Thus, in the absence of targeted drugs for Sem7a, combination therapies that target downstream effectors and COX-2 could be tested in clinical trials to determine whether tumor progression can be blocked in breast cancer patients with high Sem7a expression. Our results are the first step toward proposing such therapies for breast cancer patients.
Materials and Methods
Analysis of publicly available datasets
Oncomine analysis: Using SEMA7A as input we queried all breast tumor datasets. For normal versus tumor analysis analysis (TCGA) we downloaded the data then utilized GraphPad prism to determine differential expression. For %OE analysis we determined average expression over the entire dataset and then determined the percentage of the total that were above average. For Metabric (Curtis) dataset analysis we downloaded the data and cleaned the dataset to include only invasive ductal carcinoma (IDC) cases that were sufficiently annotated for survival data and estrogen receptor (ER) status (n=1332). We assessed SEMA7A expression in all subjects and divided them into high or low expression (above or below mean expression in each dataset). We also divided subjects based on ER status and high or low SEMA7A expression. Kaplan Meier analyses were performed in Graph Pad Prism. Similar analyses were performed for NKI295 analysis. For GOBO analysis: This dataset is compiled from 11 independent, publicly available Affymetrix U133A microarray analyses(18) of human breast tumors; 10 year OS data and Sem7a expression data were available for 737 breast tumors.
Cell culture
MCF10DCIS.com-GFP cells (and COX-shRNA derivatives) were obtained from K. Polyak and A.Marusyk (Harvard). Cells were validated by the DNA sequencing core at the University of Colorado Anschutz Medical Campus and identified to be a pure population of MCF10DCIS cells. Monthly mycoplasma testing confirmed they were negative throughout the studies. Cells were sub-cultured as previously described (23, 31, 35). Human Dermal Lymphatic Endothelial Cells (HDLECs) were obtained from PromoCell and maintained according to manufacturer’s protocol with the following modifications: for routine culturing, plates were coated with 10 μg/mL Matrigel 30 minutes prior to plating. 3 dimensional (3D) cultures were performed as previously described(23).
RNA isolation
RNA was isolated using the RNeasy Plus Mini Kit (Qiagen). Concentration and purity were assessed using absorbance at 280 nm, 260 nm, and 230 nm. cDNA was synthesized from 1 μg RNA using the Bio-Rad iScript cDNA Synthesis Kit in the following conditions: 25°C for 5 minutes, 42°C for 30 minutes, 85°C for 5 minutes.
shRNA knockdown
Four SureSilencing shRNA plasmids targeting Sema7a and 1 negative control shRNA (SABiosciences) were amplified in E. coli. Plasmid DNA was isolated by Plasmid Maxi-Prep (Qiagen). MCF10DCIS.com-GFP cells were grown overnight in a 24-well dish to 80% confluence. Each Sema7a specific shRNA plasmid and the negative control shRNA plasmid was diluted to 1 μg/μL in Transfectagro Reduced-Serum Medium (Corning), and 100 μL of this dilution was incubated for 15 minutes with 4 μL of X-treme Gene HP DNA transfection reagent (Roche). This mixture was added dropwise to the cells. Transfected cells were selected for by hygromycin resistance and stable knockdown was confirmed by qPCR. The negative control cells were transfected with a scrambled artificial sequence not matching human, mouse, or rat according to the product specification sheet provided by SABiosciences.
qPCR analysis
qPCR of cDNA samples was performed using the Bio-Rad iTaq Universal SYBR Green Supermix with primers for SEMA7A (forward: CTCAGCCGTCTGTGTGTATT, reverse: CTCAGGTAGTAGCGAGAGTTTG) and COX-2 (BioRad;PrimePCR). The conditions were as follows: 95°C for 3 minutes, then 40 cycles of 95°C for 15 seconds and 60°C for 1 minute, 95°C for 30 seconds, and 55°C for 1 minute. The fidelity of our primers was assessed after each qPCR reaction using a 95°C melt curve. mRNA quantification was normalized using the geometric average of two reference genes, GAPDH (forward: CAAGAGCACAAGAGGAAGAGAG, reverse: CTACATGGCAACTGTGAGGAG) and RPS18 (forward: GCGAGTACTCAACACCAACA, reverse: GCTAGGACCTGGCTGTATTT).
Western blot analysis
Sem7a western blots were performed by loading equal amounts of protein into each lane. Protein concentration in lysates was determined by Bradford assay. Blots were probed with anti-Sema7a (Atlas) or anti-COX2 (Cayman) primary antibodies, and goat anti-mouse or goat anti-rabbit secondary antibodies (LICOR). Blots were imaged and quantitated on an Odyssey CLx Imager (LICOR).
In vitro growth, proliferation rate, and motility assays
For proliferation assays, cells were plated at 2,000 cells per well of a 96-well dish. Four pictures of different regions in each well were taken every four hours for 48 hours using an IncuCyte ZOOM (Essen BioScience). Percent confluence was determined using the IncuCyte ZOOM software (Essen BioScience). For motility assays, wells of a 96-well dish were coated with 200 mg/mL matrigel (Corning) and allowed to gel for 2 hours at 37°C. Liquid was then aspirated and replaced with 2 mg/mL bovine serum albumin and blocked for 1 hour at 37°C. Wells were then washed once with PBS and cells were plated to produce a uniform monolayer 24 hours later. A WoundMaker (Essen BioScience) was used to make scratches across each well. Scratches were cleaned by washing wells two times with PBS. Pictures were taken at the center of the scratch every hour for 30 hours, and the IncucyteZOOM software was used to measure percent wound closure over time. For adhesion assays cells were plated at 5,000 cells/well in a 96-well dish. 6 hours post-plating the plates were washed with PBS and the cells were fixed in 10% neutral buffered formalin before being stained with 10% crystal violet. Images of each well were taken for analysis of positive pixels using Image J.
In vitro lymphangiogenesis assays
15,000 human dermal lymphatic endothelial cells (HDLECs) were seeded on 4 mg/mL matrigel pads on glass coverslips or in 96-well format and incubated with 50% experimental medias and 50% endothelial cell media with no additives. Experimental medias were made with either 5 μg/mL recombinant Sem7a (Origene), 0.082nM PGE2 (Sigma), or 9EG7. Experiments were photographed after 24 hours. Surface area of cellular structures was calculated using ImageJ software.
Animal Model
200,000 DCIS-GFP or shSEMA7A derivatives in 50 μl PBS were injected into right and left #4 mammary gland fat pads of female SCID Hairless Outbred mice from Charles River (SHO; Crl:SHO-PrkdcscidHrhr) at one day post-weaning or age-matched nulliparous group mice; for shSEMA7A all mice were 6–8 weeks of age and nulliparous. Mice were sacrificed at 3, 4, 6, or 8 weeks post-injection for tumor cell population isolation, fixed tissue, and/or RNA. Each study was replicated 2–3 times. Tumor size was calculated by GFP signal using Illumatool technology. All animal experiments were conducted in accordance to an institutional IUCAC approved protocol.
Histologic Analysis
Tumors were harvested at 3 or between between 4.5 and 7 weeks. Hematoxylin and eosin stained sections of tumors were examined by a board certified anatomic pathologist (AN) blinded to the experimental group. Interpretations of in situ versus invasion were made primarily on the grounds of morphologic phenotype observed on H&E sections, with p63 IHC used to clarify and confirm these conclusions in challenging cases. Those lesions which contained only well-developed DCIS structures with clearly defined basement membranes and no evidence of micro-invasion were scored as 0. Lesions that contained extensive DCIS with easily identifiable micro-invasive foci were scored as 1. Lesions that contained significant areas of sheet-like invasive tumor growth admixed with areas of DCIS were scored as 2. Finally, lesions that contained entirely invasive tumor with rare to absent DCIS remnants were scored as 3.
Immunohistochemistry
Mammary glands (tumors intact) were prepared for H&E and IHC as described(23). Antibody information is in Supplemental Table 1. Quantification of total tumor area (necrotic and stromal areas removed) and percent positive stain was performed using Image Aperio Analysis software. Briefly, entire histological sections were imaged with Aperio ScanCope T3 scanner at 0.47 microns/pixel. The images were then down-sampled to a resolution of 1.5 microns per pixel to facilitate subsequent image manipulation. Areas for quantification were annotated using Aperio analysis tools and percent weak, medium, and strong determined using the color-deconvolution tool. For Ki67 and p63+ the denominator is total nuclei, for vimentin and E-cadherin the denominator is total tumor area.
Experimental replicates, sample size, and statistical analyses
For all in vitro studies individual experiments were performed in triplicate and each experiment replicated three times. Representative data from a single experiment is shown for all in vitro studies. For animal studies, we chose the number of mice/cell line/group/time-point, based on power calculations from previous and pilot studies to achieve 80% or more power (β), with α = 0.05 (two-sided). For all animal studies each tumor is represented by a dot on the graph to indicate the exact number of tumors per group. All animal studies were replicated twice with representative or pooled data shown. Aside from instances of unexpected death or cell contamination, no samples or animals were excluded from our analysis. Unpaired t-test (two-sided), Chi-square, Log-rank (Mantel-Cox), Gehan-Breslow-Wilcoxon, and Pearson correlations were performed in GraphPad Prism, assuming independent samples and normal distributions. Only p-values less than 0.05 were considered significant.
Supplementary Material
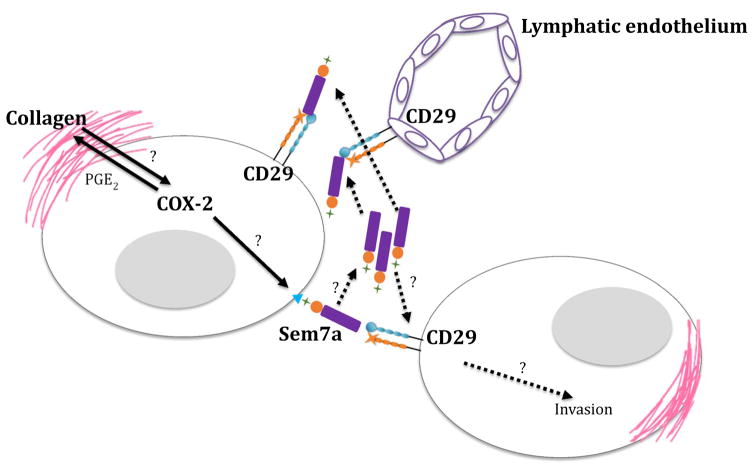
Figure 6. A model depicting COX-2 mediated Sem7a signaling.
We have previously shown that collagen 1 promotes upregulation of COX-2 through an unknown mechanism. Our current data suggests that COX-2 promotes expression of Sem7a through an unknown mechanism. Sem7a, in turn, promotes tumor cell invasion and lymphnangiogenesis via activation of β1-integrin receptors on neighboring tumor and lymphatic endothelial cells.
Acknowledgments
We gratefully acknowledge K. Polyak and A. Marusyk (Harvard Medical School) for providing MCF10DCIS parental cells and shCOX-2 derivatives; AC Tan, K. Hunter (NCI), J. Richer, T. Rogers, and M. Gordon for advice on dataset analysis; A. Elder, M. Kobritz, J. Pruitt, A. Pham, S. Tarullo, and V. Wessells for technical support; V. Borges and P. Schedin for critical review of the manuscript and crucial advice. This work was supported by 1KL2TR001080-01 CCTSI Mentored Career Development Award, CCTSI CO PILOT M-13-161, and 1R21CA185226-01 NIH/NCI to TL.
Footnotes
Conflict of interest disclosure: The authors declare that there are no conflicts of interest.
References
- 1.Pasterkamp RJ, Kolk SM, Hellemons AJ, Kolodkin AL. Expression patterns of semaphorin7A and plexinC1 during rat neural development suggest roles in axon guidance and neuronal migration. BMC Dev Biol. 2007;7:98. doi: 10.1186/1471-213X-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pasterkamp RJ, Kolodkin AL. Semaphorin junction: making tracks toward neural connectivity. Current opinion in neurobiology. 2003 Feb;13(1):79–89. doi: 10.1016/s0959-4388(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 3.Pasterkamp RJ, Peschon JJ, Spriggs MK, Kolodkin AL. Semaphorin 7A promotes axon outgrowth through integrins and MAPKs. Nature. 2003 Jul 24;424(6947):398–405. doi: 10.1038/nature01790. [DOI] [PubMed] [Google Scholar]
- 4.Adams RH, Eichmann A. Axon guidance molecules in vascular patterning. Cold Spring Harb Perspect Biol. 2010 May;2(5):a001875. doi: 10.1101/cshperspect.a001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Areas R, Libreros S, Amat S, Keating P, Carrio R, Robinson P, et al. Semaphorin7A promotes tumor growth and exerts a pro-angiogenic effect in macrophages of mammary tumor-bearing mice. Frontiers in physiology. 2014;5:17. doi: 10.3389/fphys.2014.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gelfand MV, Hong S, Gu C. Guidance from above: common cues direct distinct signaling outcomes in vascular and neural patterning. Trends in cell biology. 2009 Mar;19(3):99–110. doi: 10.1016/j.tcb.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu C, Giraudo E. The role of semaphorins and their receptors in vascular development and cancer. Exp Cell Res. 2013 Feb 17; doi: 10.1016/j.yexcr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghanem RC, Han KY, Rojas J, Ozturk O, Kim DJ, Jain S, et al. Semaphorin 7A promotes angiogenesis in an experimental corneal neovascularization model. Current eye research. 2011 Nov;36(11):989–96. doi: 10.3109/02713683.2011.593730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott GA, McClelland LA, Fricke AF. Semaphorin 7a promotes spreading and dendricity in human melanocytes through beta1-integrins. The Journal of investigative dermatology. 2008 Jan;128(1):151–61. doi: 10.1038/sj.jid.5700974. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki K, Okuno T, Yamamoto M, Pasterkamp RJ, Takegahara N, Takamatsu H, et al. Semaphorin 7A initiates T-cell-mediated inflammatory responses through alpha1beta1 integrin. Nature. 2007 Apr 5;446(7136):680–4. doi: 10.1038/nature05652. [DOI] [PubMed] [Google Scholar]
- 11.Delorme G, Saltel F, Bonnelye E, Jurdic P, Machuca-Gayet I. Expression and function of semaphorin 7A in bone cells. Biology of the cell/under the auspices of the European Cell Biology Organization. 2005 Jul;97(7):589–97. doi: 10.1042/BC20040103. [DOI] [PubMed] [Google Scholar]
- 12.Narod SA, Iqbal J, Giannakeas V, Sopik V, Sun P. Breast Cancer Mortality After a Diagnosis of Ductal Carcinoma In Situ. JAMA oncology. 2015 Aug 20; doi: 10.1001/jamaoncol.2015.2510. [DOI] [PubMed] [Google Scholar]
- 13.Maurin JC, Delorme G, Machuca-Gayet I, Couble ML, Magloire H, Jurdic P, et al. Odontoblast expression of semaphorin 7A during innervation of human dentin. Matrix biology: journal of the International Society for Matrix Biology. 2005 May;24(3):232–8. doi: 10.1016/j.matbio.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Czopik AK, Bynoe MS, Palm N, Raine CS, Medzhitov R. Semaphorin 7A is a negative regulator of T cell responses. Immunity. 2006 May;24(5):591–600. doi: 10.1016/j.immuni.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 15.Koh JM, Oh B, Lee JY, Lee JK, Kimm K, Kim GS, et al. Association study of semaphorin 7a (sema7a) polymorphisms with bone mineral density and fracture risk in postmenopausal Korean women. Journal of human genetics. 2006;51(2):112–7. doi: 10.1007/s10038-005-0331-z. [DOI] [PubMed] [Google Scholar]
- 16.Moserle L, Casanovas O. Exploiting pleiotropic activities of semaphorins as multi-target therapies for cancer. EMBO molecular medicine. 2012 Mar;4(3):168–70. doi: 10.1002/emmm.201200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allegra M, Zaragkoulias A, Vorgia E, Ioannou M, Litos G, Beug H, et al. Semaphorin-7a reverses the ERF-induced inhibition of EMT in Ras-dependent mouse mammary epithelial cells. Molecular biology of the cell. 2012 Oct;23(19):3873–81. doi: 10.1091/mbc.E12-04-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ringner M, Fredlund E, Hakkinen J, Borg A, Staaf J. GOBO: gene expression-based outcome for breast cancer online. PloS one. 2011;6(3):e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Behbod F, Kittrell FS, Lamarca H, Edwards D, Kerbawy S, Heestand JC, et al. An intra-ductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast cancer research: BCR. 2009 Sep 7;11(5):R66. doi: 10.1186/bcr2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller FR, Santner SJ, Tait L, Dawson PJ. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. Journal of the National Cancer Institute. 2000 Jul 19;92(14):1185–6. doi: 10.1093/jnci/92.14.1185a. [DOI] [PubMed] [Google Scholar]
- 21.Tait LR, Pauley RJ, Santner SJ, Heppner GH, Heng HH, Rak JW, et al. Dynamic stromal-epithelial interactions during progression of MCF10DCIS.com xenografts. International journal of cancer Journal international du cancer. 2007 May 15;120(10):2127–34. doi: 10.1002/ijc.22572. [DOI] [PubMed] [Google Scholar]
- 22.Hu M, Yao J, Carroll DK, Weremowicz S, Chen H, Carrasco D, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008 May;13(5):394–406. doi: 10.1016/j.ccr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyons TR, O’Brien J, Borges VF, Conklin MW, Keely PJ, Eliceiri KW, et al. Postpartum mammary gland involution drives progression of ductal carcinoma in situ through collagen and COX-2. Nature medicine. 2011 Sep;17(9):1109–15. doi: 10.1038/nm.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stefanou D, Batistatou A, Nonni A, Arkoumani E, Agnantis NJ. p63 expression in benign and malignant breast lesions. Histology and histopathology. 2004 Apr;19(2):465–71. doi: 10.14670/HH-19.465. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Mori I, Tang W, Nakamura M, Nakamura Y, Sato M, et al. p63 expression in normal, hyperplastic and malignant breast tissues. Breast cancer. 2002;9(3):216–9. doi: 10.1007/BF02967592. [DOI] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Research N. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nature genetics. 2013 Oct;45(10):1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004 Jan-Feb;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012 Jun 21;486(7403):346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002 Dec 19;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 30.Lyons TR, Borges VF, Betts CB, Guo Q, Kapoor P, Martinson HA, et al. Cyclooxygenase-2-dependent lymphangiogenesis promotes nodal metastasis of postpartum breast cancer. The Journal of clinical investigation. 2014 Sep 2;124(9):3901–12. doi: 10.1172/JCI73777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu M, Peluffo G, Chen H, Gelman R, Schnitt S, Polyak K. Role of COX-2 in epithelial-stromal cell interactions and progression of ductal carcinoma in situ of the breast. Proceedings of the National Academy of Sciences of the United States of America. 2009 Mar 3;106(9):3372–7. doi: 10.1073/pnas.0813306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maller O, Hansen KC, Lyons TR, Acerbi I, Weaver VM, Prekeris R, et al. Collagen architecture in pregnancy-induced protection from breast cancer. Journal of cell science. 2013 Sep 15;126(Pt 18):4108–10. doi: 10.1242/jcs.121590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esserman L, Yau C. Rethinking the Standard for Ductal Carcinoma In Situ Treatment. JAMA oncology. 2015 Aug 20; doi: 10.1001/jamaoncol.2015.2607. [DOI] [PubMed] [Google Scholar]
- 34.Formolo CA, Williams R, Gordish-Dressman H, MacDonald TJ, Lee NH, Hathout Y. Secretome signature of invasive glioblastoma multiforme. Journal of proteome research. 2011 Jul 1;10(7):3149–59. doi: 10.1021/pr200210w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aslakson CJ, Rak JW, Miller BE, Miller FR. Differential influence of organ site on three subpopulations of a single mouse mammary tumor at two distinct steps in metastasis. International journal of cancer Journal international du cancer. 1991 Feb 1;47(3):466–72. doi: 10.1002/ijc.2910470327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.