Abstract
Background
Peripheral inflammation is often associated with major depressive disorder (MDD), and immunological biomarkers of depression remain a focus of investigation.
Methods
We used microarray data on whole blood from two independent case-control studies of MDD: the GlaxoSmithKline–High-Throughput Disease-specific target Identification Program [GSK-HiTDiP] study (113 patients and 57 healthy control subjects) and the Janssen–Brain Resource Company study (94 patients and 100 control subjects). Genome-wide differential gene expression analysis (18,863 probes) resulted in a p value for each gene in each study. A Bayesian method identified the largest p-value threshold (q = .025) associated with twice the number of genes differentially expressed in both studies compared with the number of coincidental case-control differences expected by chance.
Results
A total of 165 genes were differentially expressed in both studies with concordant direction of fold change. The 90 genes overexpressed (or UP genes) in MDD were significantly enriched for immune response to infection, were concentrated in a module of the gene coexpression network associated with innate immunity, and included clusters of genes with correlated expression in monocytes, monocyte-derived dendritic cells, and neutrophils. In contrast, the 75 genes underexpressed (or DOWN genes) in MDD were associated with the adaptive immune response and included clusters of genes with correlated expression in T cells, natural killer cells, and erythroblasts. Consistently, the MDD patients with overexpression of UP genes also had underexpression of DOWN genes (correlation > .70 in both studies).
Conclusions
MDD was replicably associated with proinflammatory activation of the peripheral innate immune system, coupled with relative inactivation of the adaptive immune system, indicating the potential of transcriptional biomarkers for immunological stratification of patients with depression.
Keywords: Affymetrix, Bayesian, Biomarker, Inflammation, Systems, Transcriptome
Depression and inflammation are often associated with one another. Depressive symptoms in a large population sample were significantly related to blood concentrations of C-reactive protein (CRP; odds ratio ∼1.8 for depressive symptoms in people with CRP > 3 mg/L vs. CRP < 1 mg/L) (1). Multiple case-control studies of major depressive disorder (MDD) have reported increased peripheral blood concentrations of CRP (Cohen’s d ∼0.50) and proinflammatory cytokines such as interleukin 6 (d ∼0.50) and tumor necrosis factor (d ∼0.40) in MDD 2, 3. The prevalence of comorbid depression is increased in many nonpsychiatric inflammatory disorders (4).
There is growing evidence for a causal effect of inflammation on depression. Peripheral inflammation precedes the emergence of depressive symptoms in longitudinal epidemiological studies (5) and in about 30% of patients receiving proinflammatory interferon-α treatment for hepatitis C 6, 7. Experimental challenge with peripheral proinflammatory stimuli in animals robustly induces a syndrome of illness behavior and anhedonia that approximates depressive symptoms (8). Peripheral immune cells and cytokines are known to mediate signals across the blood-brain barrier by several mechanisms (9). Activation of microglia can locally amplify the effects of even a weak peripheral proinflammatory signal on neuronal function and behavior (10).
These observations suggest that pharmacological disruption of peripheral proinflammatory signals could be therapeutically effective, at least for a subgroup of patients with depression. However, it is most unlikely that any single anti-inflammatory drug will prove to be superior to all existing treatments for all patients (11). Only about a third of patients with depression have biological evidence for peripheral inflammation [e.g., CRP > 3 mg/L (12)], and an anti-inflammatory drug seems likely a priori to be most effective for an inflamed subgroup of patients with depression. There are many markers of the peripheral immune system that can be conveniently measured in venous blood samples from patients with depression, including cytokines, CRP, and other proteins; cell counts from flow cytometry; and gene transcription (13).
Transcriptional (messenger RNA [mRNA]) biomarkers have the potential advantages of assay stability (compared with cytokines) and target specificity (compared with cell counts). However, previous case-control studies of peripheral blood gene expression and MDD have been inconsistent 14, 15, 16, 17, 18 (Supplemental Table S1). To date, the Netherlands Study of Depression and Anxiety (NESDA) (14) is the largest single case-control study of the peripheral blood transcriptome in MDD (882 current patients with MDD, 635 remitted patients with MDD, and 331 control subjects). The NESDA reported statistically significant (false discovery rate [FDR] = 10%) differential expression of 129 genes enriched for interleukin 6 and natural killer (NK) cell signaling pathways (19).
We were primarily motivated by the hypothesis that MDD is associated with peripheral blood transcriptional markers of innate immune system activation 8, 20. We were also concerned to focus on results that were more likely to replicate across case-control studies of gene expression in MDD. We report Affymetrix microarray data on 18,863 probes from two independently designed and conducted case-control studies of MDD. We used a Bayesian method to identify genes that were differentially expressed in both studies. Focusing on a consensus set of 165 genes, we investigated the functional significance of the genes that were differentially (over- or under-) expressed in cases compared with controls. We also explored the secondary hypothesis that innate immune system activation is coupled to relative inactivation of the adaptive immune system in patients with MDD 21, 22, 23.
Methods and Materials
Samples
We analyzed data from two case-control studies of depression: the GlaxoSmithKline–High-Throughput Disease-specific target Identification Program (GSK–HiTDiP) study and the Janssen–Brain Resource Company (Janssen–BRC) study. Other aspects of these studies have been previously reported 24, 25, 26, 27, 28; demographic and clinical details on the samples are provided in Supplemental Table S2.
GSK–HiTDiP
This study was designed primarily to test for an association between genetic (DNA) variation and diagnosis of depression. Minimal sociodemographic and clinical data were collected, but microarray data were available for analysis (after quality control) from whole-blood samples stored for less than 6 years on a sample comprising 113 patients with MDD prospectively balanced for comorbid anxiety disorder [57 with generalized anxiety disorder diagnosed by the Mini-International Neuropsychiatric Interview (29) and 56 without anxiety disorder] and 57 healthy control subjects. All participants provided informed consent in writing. The study was approved by an independent ethics review board.
Janssen–BRC
This study was designed primarily for biomarker discovery. Microarray data were available for analysis (after quality control) from whole-blood samples stored for less than 1 year on a sample comprising 94 patients with MDD (40 with generalized anxiety disorder diagnosed post hoc by the Mini-International Neuropsychiatric Interview and 54 without anxiety disorder) and 100 healthy control subjects. Additional data on melancholic symptom severity, anxiety, substance use, and body mass index (BMI) were available for patients with MDD. All participants provided informed consent in writing. The study was approved by an independent ethics review board.
Whole-blood samples from both studies were analyzed using the Affymetrix Human Genome U133 plus 2.0 array. We applied identical quality control, normalizing, and annotation algorithms to the two datasets, resulting in the estimation of mRNA expression at each of 18,863 unique probes for each participant.
Differential Expression Analysis
To determine differential gene expression between cases and controls, we adopted the same strategy for both studies; see Supplemental Figure S1 for a schematic overview of the data analysis strategy. For each gene i = 1, …, 18,863, we fitted a linear regression model that included group (coded Gr; two-level factor, case/control), batch (B; two-level factor, 1/2), gender (Ge; two-level factor, male/female), age (Ag; continuous), and presence/absence of anxiety (An; two-level factor, 0/1) as covariates. Denoting samples by j = 1, …, n and the design matrix by X, the model is
| (1) |
where εi ∼ N(0, σi2). For identifiability, we imposed the contrasts on model parameters. The model was fitted using the R package limma (30). Subsequently, we tested the null hypothesis Hi0: βi2Gr = 0; that is, there is no difference in expression of the ith gene between the two groups using the moderated t statistic (31). For each of the 18,863 probes in each study, the p value was generated using the asymptotic approximate distribution (and also nonparametrically by a permutation algorithm; see Supplemental Tables S3 and S4).
Combining p Values for Differential Gene Expression From Two Studies
To identify MDD-related genes that were replicated in both the GSK–HiTDiP and Janssen–BRC datasets, we set the p-value threshold for significance of differential expression of each gene in each study to optimize in some sense the number of genes that were differentially expressed in both studies compared with the number of coincidental differences expected by chance. To do this, we used the method of Blangiardo and Richardson (32), implemented in the R package sdef (33), and specified the p-value threshold as , which represents the largest (most lenient) p value < .05 for which there are at least twice as many significant case-control differences in common between the two studies as expected by chance.
Gene Coexpression Network Analysis
We used weighted gene correlation network analysis (34) to construct a normative coexpression network in which nodes represent genes and weighted edges represent correlations between the expression of pairs of genes in healthy control subjects (34). To maximize the amount of data available for this estimation, and to ensure that the resulting network was representative of both studies, we included the healthy control data from both the GSK–HiTDiP and Janssen–BRC studies (N = 157 in total). We used the consensus weighted gene correlation network analysis method to construct a weighted undirected graph that could be decomposed into a set of modules of strongly coexpressed genes (34). Eigengenes were used to test for case-control differences in the gene expression profile on average within each module of the normative weighted gene correlation network analysis transcriptome (Supplemental Table S11).
Gene Ontology Enrichment Analysis
We performed enrichment analysis for various gene lists such as the list of genes differentially expressed in both studies and the list of genes comprising each module of the coexpression network. We used the R package topGO (35) to determine whether these lists were significantly enriched for specific Gene Ontology (GO) terms (36) using a stringent Bonferroni correction (q < .05) for multiple comparisons across all 10,124 terms, resulting in a p-value threshold of 4.94 × 10−6.
Protein–Protein Interaction Mapping
We used the Search Tool for the Retrieval of Interacting Genes/Proteins [http://string-db.org (37)] to determine the network of known protein–protein interactions among the genes differentially expressed in both studies (38).
Cell Type–Specific Gene Expression Analysis
We investigated the genes differentially expressed in both studies in relation to a third independent microarray (Affymetrix) dataset designed to examine cell type–specific expression patterns (39). In particular, we focused on the expression of MDD-related genes across 37 sorted cell samples of the following immune cell types: erythroblasts (8 samples), monocytes (6 samples), monocyte-derived dendritic cells (5 samples), neutrophils (3 samples), B cells (4 samples), CD4+ T cells (5 samples), CD8+ T cells (5 samples), and NK cells (6 samples). We used the BioLayout Express3D software 40, 41 to visualize clusters or subgroups of genes that shared similar patterns of expression across different cell types.
Results
Combining Differential Expression Across Two Case-Control Studies of MDD
Separate differential expression analyses of the GSK–HiTDiP and Janssen–BRC datasets yielded two lists of p values for the same set of 18,863 gene expression probes (Supplemental Tables S3 and S4). The Bayesian method identified q2 = .0246 as the largest p value threshold associated with twice the number of genes differentially expressed in both studies compared with the number of coincidental case-control differences expected by chance. At this threshold, 173 genes were differentially expressed in both studies. We further refined this gene list by restricting attention to the 165 consensus genes (95%) that showed the same direction of overexpression (UP) or underexpression (DOWN) in the two case-control studies (Supplemental Table S5).
These findings were corroborated by Fisher’s chi-square test for combining p values, which identified 393 genes that were differentially expressed in both studies with FDR = 10%, of which 146 were included in the list of replicably, concordantly, differentially expressed genes defined by Bayesian analysis (Supplemental Table S6).
Furthermore, the set of 165 replicably concordant genes (henceforth the MDD-165 consensus set) was partially corroborated by the prior results of the NESDA case-control study (14). Considering the 15,830 genes that were measured by the microarrays used in all three studies (NESDA, Janssen–BRC, and GSK–HiTDiP), 150 of the MDD-165 consensus set were included, of which 7 genes (CD247, PRKCH, PGLYRP1, NFATC2, ST6GAL1, MAPK14, and MTSS1) were also differentially expressed, with the same sign of fold change, in the NESDA study at FDR = 5%. When the FDR threshold for the NESDA study was relaxed to 10% and 20%, 18 and 45 of the 150 genes differentially expressed in both the GSK–HiTDiP and Janssen–BRC studies, respectively, were also differentially expressed in the NESDA study (Supplemental Table S5). By comparison, we expect by chance to find 1.95, 8.30, and 30.40 genes in common with the NESDA study at FDR thresholds of 5%, 10%, and 20%, respectively. In contrast, there was less overlap between the MDD-165 consensus set and the top 29 most significantly differentially expressed genes reported in another large case-control study of MDD (16); only SRSF5 was underexpressed in both of these lists.
To assess the robustness of our results to key modeling assumptions, we conducted two sensitivity analyses. First, we repeated the analysis using a model for differential expression that did not include comorbid anxiety as an explanatory factor. Only 24 genes were identified by Bayesian analysis as significantly coexpressed; of these 24 genes, 21 were concordant for sign of fold change, and 15 of these 21 genes were also included in the MDD-165 consensus set (Supplement). The reduced sensitivity when not adjusting for anxiety likely reflects the importance of refining or homogenizing the clinical phenotype in the search for biomarkers. Second, we repeated the analysis using a model for differential expression that included BMI as a covariate. BMI was not measured in the GSK–HiTDiP study, so this analysis was restricted to 74 patients and 80 control subjects from the Janssen–BRC dataset. Inclusion of BMI in the model tended to increase the p values for MDD-related differences, and only 73 genes in the MDD-165 consensus set were differentially expressed at the probability threshold q2 = .0246 (Supplemental Figure S3). The reduced sensitivity of the analysis including BMI as a covariate likely reflects the known proinflammatory effect of obesity and the reduced sample size available.
Gene Set Enrichment Analyses Across GO Terms
We conducted a series of enrichment analyses to functionally characterize the MDD-165 consensus set. The top 10 most significantly enriched GO terms all were related to the immune response to infection (Bonferroni q < .05 for the top 9 terms) (Supplemental Table S7). Among the MDD-165 consensus set, 90 genes were overexpressed in cases compared with controls. The UP gene list was significantly enriched for GO terms related to the innate immune system and response to infection (Bonferroni q < .05 for the top 17 terms) (Table 1). In addition, 75 genes were significantly underexpressed in cases compared with controls. As we hypothesized based on prior literature (19), this gene list was enriched for GO terms related to the adaptive immune system (p ≤ 10−3 for top 20 GO terms; Table 1), but there was no significant enrichment of the DOWN genes at the more stringent Bonferroni threshold.
Table 1.
Enrichment Analysis of the MDD-165 Consensus Set of Genes That Were Differentially Overexpressed (UP) or Underexpressed (DOWN) in Patients With MDD Compared With Healthy Control Subjects in Both GSK–HiTDiP and Janssen–BRC Studies
| Rank | GO:ID | Term | Annotated | Found | Expected | Fisher’s Test |
|---|---|---|---|---|---|---|
| Overexpressed UP (Innate Immunity) | ||||||
| 1 | GO:0009617 | Response to bacterium | 405 | 19 | 2.29 | 7.7e-13 |
| 2 | GO:0042742 | Defense response to bacterium | 164 | 13 | 0.93 | 7.0e-12 |
| 3 | GO:0043207 | Response to external biotic stimulus | 651 | 20 | 3.69 | 3.7e-10 |
| 4 | GO:0051707 | Response to other organism | 651 | 20 | 3.69 | 3.7e-10 |
| 5 | GO:0009607 | Response to biotic stimulus | 680 | 20 | 3.85 | 8.0e-10 |
| 6 | GO:0002376 | Immune system process | 2149 | 35 | 12.17 | 9.8e-10 |
| 7 | GO:0006955 | Immune response | 1310 | 26 | 7.42 | 6.3e-09 |
| 8 | GO:0098542 | Defense response to other organism | 351 | 14 | 1.99 | 9.1e-09 |
| 9 | GO:0051704 | Multi-organism process | 2176 | 32 | 12.32 | 9.2e-08 |
| 10 | GO:0050832 | Defense response to fungus | 21 | 5 | 0.12 | 9.7e-08 |
| 11 | GO:0032496 | Response to lipopolysaccharide | 253 | 11 | 1.43 | 1.7e-07 |
| 12 | GO:0002237 | Response to molecule of bacterial origin | 265 | 11 | 1.5 | 2.8e-07 |
| 13 | GO:0006952 | Defense response | 1372 | 24 | 7.77 | 3.2e-07 |
| 14 | GO:0009605 | Response to external stimulus | 1947 | 29 | 11.02 | 3.8e-07 |
| 15 | GO:0045087 | Innate immune response | 820 | 18 | 4.64 | 5.5e-07 |
| 16 | GO:0009620 | Response to fungus | 34 | 5 | 0.19 | 1.3e-06 |
| 17 | GO:0019731 | Antibacterial humoral response | 23 | 4 | 0.13 | 7.8e-06 |
| 18 | GO:0031640 | Killing of cells of other organism | 25 | 4 | 0.14 | 1.1e-05 |
| 19 | GO:0044364 | Disruption of cells of other organism | 25 | 4 | 0.14 | 1.1e-05 |
| 20 | GO:0019730 | Antimicrobial humoral response | 27 | 4 | 0.15 | 1.5e-05 |
| Underexpressed DOWN (Adaptive Immunity) | ||||||
| 1 | GO:0050851 | Antigen receptor-mediated signaling path | 133 | 5 | 0.59 | 0.00029 |
| 2 | GO:0046649 | Lymphocyte activation | 532 | 9 | 2.34 | 0.0005 |
| 3 | GO:0009059 | Macromolecule biosynthetic process | 4336 | 32 | 19.09 | 0.00052 |
| 4 | GO:0034645 | Cellular macromolecule biosynthetic proc | 4183 | 31 | 18.42 | 0.00064 |
| 5 | GO:0010557 | Positive regulation of macromolecule bio | 1361 | 15 | 5.99 | 0.00066 |
| 6 | GO:0032774 | RNA biosynthetic process | 3275 | 26 | 14.42 | 0.00086 |
| 7 | GO:0046631 | Alpha-beta T cell activation | 99 | 4 | 0.44 | 0.00094 |
| 8 | GO:0016070 | RNA metabolic process | 3873 | 29 | 17.06 | 0.00095 |
| 9 | GO:0050776 | Regulation of immune response | 721 | 10 | 3.18 | 0.00112 |
| 10 | GO:0006351 | Transcription, DNA-templated | 3161 | 25 | 13.92 | 0.00122 |
| 11 | GO:0097659 | Nucleic acid-templated transcription | 3177 | 25 | 13.99 | 0.00131 |
| 12 | GO:0042110 | T cell activation | 382 | 7 | 1.68 | 0.0014 |
| 13 | GO:0070489 | T cell aggregation | 382 | 7 | 1.68 | 0.0014 |
| 14 | GO:0071593 | Lymphocyte aggregation | 384 | 7 | 1.69 | 0.00144 |
| 15 | GO:0031328 | Positive regulation of cellular biosyn | 1470 | 15 | 6.47 | 0.00147 |
| 16 | GO:0002562 | Somatic diversification of immune recept | 52 | 3 | 0.23 | 0.00154 |
| 17 | GO:0016444 | Somatic cell DNA recombination | 52 | 3 | 0.23 | 0.00154 |
| 18 | GO:0070486 | Leukocyte aggregation | 390 | 7 | 1.72 | 0.00158 |
| 19 | GO:0045893 | Positive regulation of transcription, DN | 1178 | 13 | 5.19 | 0.00161 |
| 20 | GO:1903508 | Positive regulation of nucleic acid-temp | 1178 | 13 | 5.19 | 0.00161 |
The top 20 GO terms for biological processes are ranked according to their p values by Fisher’s exact test for significant enrichment. Bonferroni correction specifies a p-value threshold of p = 4.94 × 10−6 to achieve significance at q < 0.05. Such a correction is well known to be too stringent given that many of the GO terms are correlated; however, the top 17 terms were significantly enriched in the overexpressed UP gene set after Bonferroni correction.
GO, Gene Ontology; GSK–HiTDiP, GlaxoSmithKline–High-Throughput Disease-specific target Identification Program; Janssen–BRC, Janssen–Brain Resource Company; MDD, major depressive disorder; MDD-165 consensus set, set of 165 replicably concordant genes.
Protein Interaction and Whole-Genome Transcriptional Network Analyses
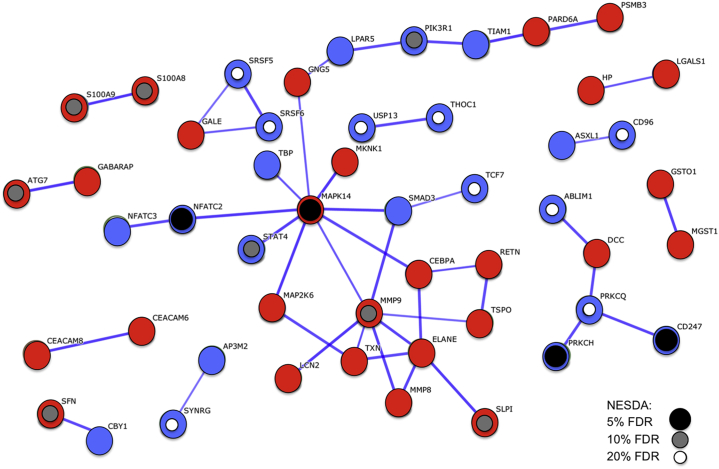
We used Search Tool for the Retrieval of Interacting Genes/Proteins analysis (37) to visualize the network of protein–protein interactions between the proteins coded by the MDD-165 consensus set. These genes were significantly enriched for protein–protein interactions (permutation test, p = 7 × 10−4) that were concentrated around mitogen-activated protein kinase 14 (MAPK14) and matrix metalloproteinase 9 (MMP9), which thus can be regarded as the most highly interactive hubs of this immune signaling network (Figure 1). Many of the proteins in this network are coded by genes that were independently identified by the NESDA study as differentially expressed in MDD; see Figure 1.
Figure 1.
Protein–protein interaction network for proteins coded by the set of 165 replicably concordant genes differentially expressed in both case-control studies of major depressive disorder. The network is represented by an undirected graph where links correspond to known protein–protein interactions and weights are proportional to the Search Tool for the Retrieval of Interacting Genes/Proteins confidence score (38). Only high-confidence (>0.7) links are retained, and disconnected genes are not shown. Red (blue) nodes correspond to genes over- (under-) expressed in major depressive disorder in both the GlaxoSmithKline–High-Throughput Disease-specific target Identification Program and Janssen–Brain Resource Company datasets. Smaller inner circles highlight proteins that are coded by genes also differentially expressed and with the same sign of fold change in a third large independent case-control study of major depressive disorder (Netherlands Study of Depression and Anxiety [NESDA]) (14) thresholded to control false discovery rate (FDR) at 20% (white circles), 10% (gray circles), and 5% (black circles).
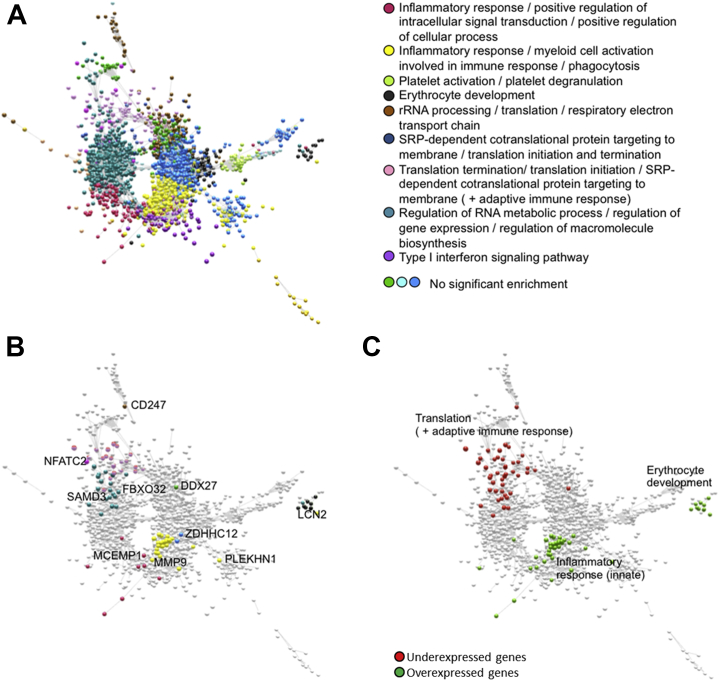
We constructed a graph representing significant coexpression of a pair of genes (nodes) as an edge drawn between them. As previously reported (42), this whole-genome transcriptional network or transcriptome had complex topological properties, including a community structure comprising modules of coexpressed genes enriched for specific GO terms (Figure 2). The UP genes were concentrated in the normative module (red) significantly enriched for innate immune response GO terms (e.g., myeloid cell activation involved in immune response, q = 5 × 10−5, Bonferroni corrected), whereas the DOWN genes were concentrated in a normative module (pink) specialized for translation-based terms (Figure 2). We confirmed in a hypothesis-driven analysis that the module associated with DOWN gene expression was also significantly enriched for adaptive immune response terms (e.g., T-cell differentiation, p = .0002). There were significant case-control differences in eigengene scores summarizing expression of all genes within modules enriched for innate immune response (red, p = .031; yellow, p = .015), translation (pink, p = .002), and one additional module with no significant enrichment terms (tan, p = .025); see Supplemental Table S11.
Figure 2.
Major depressive disorder (MDD)-related genes in the context of the normative whole-genome transcriptome. Overexpressed genes (or UP genes) in patients with MDD are concentrated in a module of the normative gene coexpression network specialized for innate immune response, whereas underexpressed genes (or DOWN genes) are concentrated in a module partially specialized for adaptive immune response. (A) The modules of the normative transcriptome are highlighted in different colors. (B) The MDD-related genes are colored according to their normative module affiliation, and representative genes are text-labeled. (C) The MDD-related genes are colored green for overexpressed (or UP) genes and are colored red for underexpressed (or DOWN) genes. The text labels highlight the functions of the corresponding modules of the normative transcriptome. rRNA, ribosomal RNA; SRP, signal recognition particle.
Cell Type–Specific Expression Patterns for the MDD-165 Consensus Set
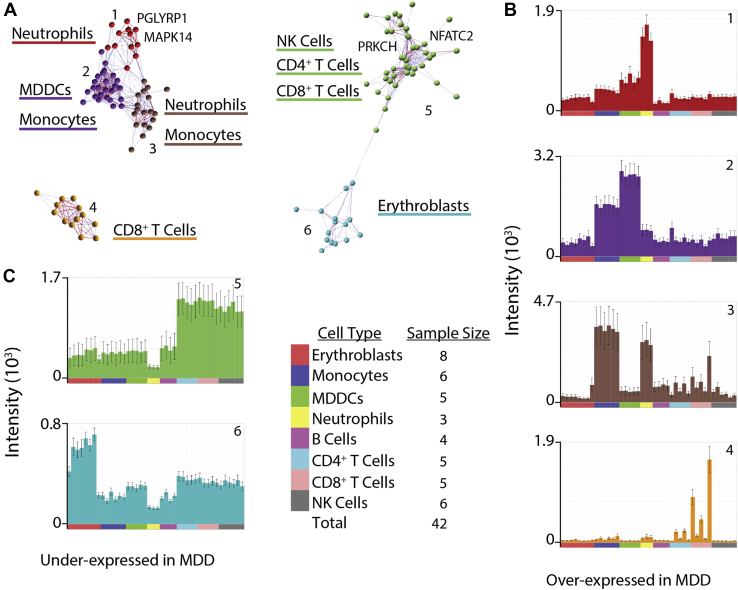
To assess the cellular specificity of the MDD-165 consensus set, we used microarray data from an independent study of cell type–specific gene expression across eight major classes of immune cells in healthy control subjects (39). Remarkably, 89 of the 90 UP genes formed four clusters of gene coexpression, three of which represented strong overexpression of a subset of UP genes in one or two myeloid cell classes (Figure 3). Likewise, 71 of the 75 DOWN genes formed two clusters of gene coexpression, each representing cell-specific overexpression of a subset of DOWN genes in one or two lymphoid or erythroblast classes (Figure 3).
Figure 3.
Cell class–specific expression patterns for the set of 165 replicably concordant genes. We estimated the correlation between each possible pair of the 165 major depressive disorder (MDD)-related genes and identified clusters of genes with similar expression patterns in an independent microarray dataset on specific cell types. The set of 165 replicably concordant genes formed six clusters, with each cluster comprising a subset of genes that had strong mutual coexpression across a range of eight distinct cell classes: erythroblasts, monocytes, monocyte-derived dendritic cells (MDDCs), neutrophils, B cells, CD4+ T cells, CD8+ T cells, and natural killer (NK) cells. (A) The six clusters of genes with strongly correlated expression profiles. Clusters 1 to 4 (left) comprised genes that were overexpressed in MDD (or UP genes), and clusters 5 and 6 (right) comprised genes that were underexpressed in MDD (or DOWN genes). (B) Histograms of clustered gene expression across cell types for each of the four clusters of UP genes overexpressed in MDD (from top to bottom: clusters 1–4). The x-axis color legend codes for different cell types. (C) Histograms of clustered gene expression across cell types for each of the two clusters of DOWN genes underexpressed in MDD (from top to bottom: clusters 5 and 6). The x-axis color legend codes for different cell types.
Coupled UP and DOWN Gene Expression in Patients With MDD
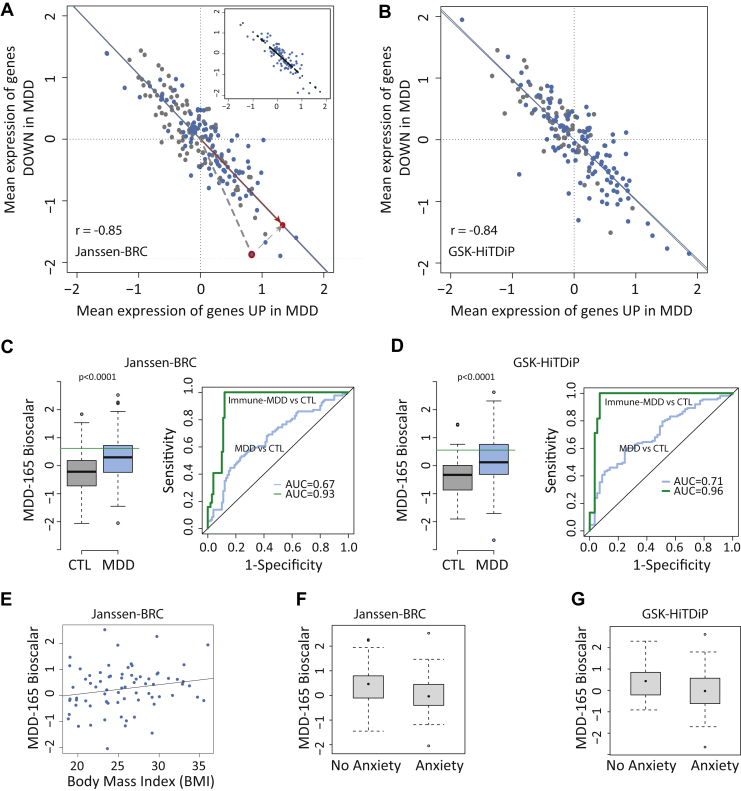
To explore the extent to which UP gene overexpression was related to DOWN gene underexpression, we estimated the mean UP gene expression (over all 90 differentially overexpressed genes) and the mean DOWN gene expression (over all 75 differentially underexpressed genes) for each of 113 patients with MDD in the GSK–HiTDiP study and each of 94 patients with MDD in the Janssen–BRC study. In both studies, UP and DOWN expression scores were strongly negatively correlated (Figure 4), indicating that these case-control differences were coupled at the level of individual patients (Janssen–BRC: r = −.82, p < 10−4; GSK–HiTDiP: r = −.74, p < 10−4).
Figure 4.
Opposing and coordinated expression of innate and adaptive immune transcripts in patients with major depressive disorder (MDD). (A, B) Scatter plots for the mean expression across the 75 underexpressed (DOWN) genes plotted against the mean expression of 90 overexpressed (UP) genes in the Janssen–Brain Resource Company (Janssen–BRC) (A) and GlaxoSmithKline–High-Throughput Disease-specific target Identification Program (GSK–HiTDiP) (B) datasets. Each point corresponds to a patient. Blue indicates case, and gray indicates control (CTL). Regression lines are shown in blue and gray, respectively. In (A), an individual patient’s data point (red outline) is projected onto the regression line (red circle). The distance from the origin to the point on the regression line is the bioscalar value for that patient. Inset illustrates the projection (black) of all individual patient data points (blue) onto the sample regression line. (C, D) Left panels: Box plot of MDD-165 bioscalar values in controls and cases. Green line indicates the threshold identifying the top third (tertile) of the MDD-165 bioscalar distribution as a subgroup of inflamed patients with MDD (designated immune-MDD). Right panels: Receiver operating characteristic curves were calculated for MDD vs. CTL and immune-MDD vs. CTL classifications. (E) Correlation of each depressed patient’s MDD-165 bioscalar with body mass index (BMI). (F, G) Box plots of MDD-165 bioscalar for subgroups defined by a comorbid diagnosis of anxiety disorder. AUC, area under the curve.
Given the strong linear relationship between UP and DOWN gene expression in patients with MDD, we mapped each patient’s data to a point on the fitted regression line, thereby characterizing each patient by a bioscalar that locates it on a single axis or dimension of coupled UP/DOWN gene expression; see Figure 4A (inset). As expected, there were significant case-control differences in the MDD-165 bioscalar in both studies (Cohen’s d = 0.68 for GSK–HiTDiP and 0.58 for Janssen–BRC, corresponding to a medium effect size; Figure 4C, D), with patients on average having more positive values, indicating a greater shift in the direction of coupled innate activation and adaptive inactivation. Receiver operating characteristic analysis indicated that cases and controls were classified with an area under the curve of 0.71 for the GSK–HiTDiP study and an area under the curve of 0.67 for the Janssen–BRC study (Figure 4C, D). This moderately accurate classification performance reflects the fact that the bioscalar distributions of cases and controls are overlapping. We also assessed the performance of the bioscalar not to discriminate cases from controls (which is ultimately a clinical diagnostic decision) but rather to identify the top third “most inflamed” patients with MDD. To do this, we defined a cutoff value for the bioscalar corresponding to the top tertile critical value of its distribution in each study (0.57). Participants with MDD-165 > 0.57 were classified correctly as belonging to such an inflamed MDD subgroup (sensitivity = 100% by definition) with a high specificity (93% and 88% in GSK–HiTDiP and Janssen–BRC studies, respectively).
We provisionally explored correlations between the MDD-165 bioscalar and sociodemographic or clinical differences among the patients with MDD in the Janssen–BRC study. BMI (r72 = .20, p = .099), the CORE total score for melancholic symptom severity (r88 = .18, p = .097), and the presence or absence of comorbid substance abuse disorder (two-tailed t63 = 1.9, p = .07) were not significantly associated with bioscalar scores. There was a significant difference in bioscalar scores between subgroups of patients with MDD with or without comorbid anxiety disorder (t83 = −2.4, p = .02).
Discussion
We have reported the differential expression analysis of whole-genome microarray data from two independent case-control studies of patients with MDD compared with healthy control subjects. Using Bayesian methods, we identified a set of 165 consensus genes that were replicably associated with MDD, sharing the same direction of fold change in the cases compared with controls in both studies. The robustness of these results was further supported by comparison with the results of the largest prior case-control microarray study of depression, the NESDA study (14); several genes differentially expressed in the NESDA study were likewise differentially expressed in these data, for example, MAPK14 and MMP9. Both of these genes were consistently overexpressed in patients with MDD and both code proteins (MAPK14 and matrix metallopeptidase 9) that were hubs of a network of interactions between immune signaling proteins coded by many of the other differentially expressed genes. A MAPK14 inhibitor has been tried for treatment of major depression but did not demonstrate consistently significant effects on symptom rating scales compared with placebo at the single dose tested (43).
The consensus set of 165 genes associated with MDD was divided into two approximately equal-sized subsets; here, 90 so-called UP genes were overexpressed in patients and 75 so-called DOWN genes were underexpressed in patients. The overexpressed UP genes were significantly enriched for GO terms related to the response to infection and the innate immune system. These gene transcripts were not functionally unrelated to or independent of each other. Most UP genes were affiliated with the module of the normative gene transcriptional network specialized for innate immune response. Smaller clusters of highly correlated UP genes were typically enriched in neutrophils, monocytes, and monocyte-derived dendritic cells. These results are compatible with prior data implicating activation of the innate immune system and increased proinflammatory cytokine signaling in (some people with) depression 8, 20.
In contrast, we found that the underexpressed DOWN genes were enriched for GO terms related to T-cell function and adaptive immunity. The DOWN genes were affiliated with modules of the transcriptome partially specialized for adaptive immune function, and smaller clusters of strongly coexpressed DOWN genes were enriched in T cells, B cells, and NK cells. These results are compatible with prior data implicating relative suppression of the cellular immune system in (some people with) depression, including evidence for decreased NK cell cytotoxicity and decreased proliferation of lymphocytes challenged with mitogens in vitro (44), and with recent data suggesting that deficiencies of NK and T cells co-occur with inflammatory monocyte activation as related phenomena in the same patients with MDD 21, 22, 45, 46.
In both studies, we also observed a strong statistical association between mean UP gene overexpression and mean DOWN gene underexpression in each patient. This provided robust evidence that these complementary immunophenotypes are indeed coupled at the level of an individual patient. It also motivated the concept that each individual patient may be located by a single number (bioscalar) on a spectrum of coupled change in innate and adaptive immune system function. Patients with MDD had significantly higher scores on this bioscalar, indicating a mean shift to relatively increased UP gene expression and decreased DOWN gene expression. The subgroup of most-inflamed patients with MDD, defined as the top tertile of the MDD-165 bioscalar distribution, were identified with high sensitivity and specificity, suggesting that this may in the future prove to be a useful biomarker for defining an abnormally inflamed subgroup of patients with MDD.
So far, we have used the terms overexpression and underexpression simply to describe higher and lower levels of measured mRNA in cases compared with controls. At least three (mutually nonexclusive) explanatory factors are plausible: cellular, genetic, and environmental. First, case-control differences in expression could reflect differences in cell counts. For example, increased monocyte counts have been reported in MDD (47) and could cause apparent overexpression of innate immune system genes measured in whole-blood samples (and relative underexpression of adaptive immune genes). It will be important in the future to combine cytometry and transcriptional measurements in the same patients and to measure case-control expression differences in sorted cell samples. Second, mRNA changes could be quantitative traits determined by DNA variation at expression quantitative trait loci. This genetic explanation would require that (at least some) patients with MDD had a consistent profile of allelic variation in inflammation-related genes (48), but large genome-wide association studies have so far failed to identify genetic variants robustly linked to risk for major depression (49). The absence of significant genome-wide association study findings in MDD could be regarded as problematic for an expression quantitative trait locus interpretation of expression changes, or it could be discounted on the grounds that classically designed association studies were underpowered to detect DNA variations occurring in only a subgroup of immunologically dysfunctional patients. Third, gene expression changes could have been induced in patients by exposure to some shared environmental stimulus. The risk for depression is associated with adverse events in the biological and social environments such as infection, childhood abuse, and bereavement (50). In addition, psychosocial stress has been linked to peripheral immune state changes, such as increased proinflammatory cytokines and monocyte activation (51), that are compatible with overexpression of innate immune system genes.
It is a methodological limitation that the GSK–HiTDiP study, designed primarily to identify risk genes for MDD, did not provide data on severity of depressive symptoms or BMI. The case-control comparisons are not controlled for potentially confounding effects of cigarette smoking, race, comorbid medical disease, or socioeconomic status on peripheral immune status. Future studies of transcriptional biomarkers in MDD, with more detailed clinical and immunological phenotyping and more complete control of potential confounds, will be required to evaluate the generalizability of these results.
In short, we have reported replicable new evidence in support of peripheral immune gene expression markers for MDD. It may be fruitful in the future to investigate coupled overexpression of innate immune genes and underexpression of adaptive immune genes as a predictor of antidepressant response to novel immunotherapeutics.
Acknowledgments and Disclosures
This work was supported by a grant from the Medical Research Council to the Immunopsychiatry Consortium led by ETB (Grant No. MR/L014815/1). Data collection for the GSK–HiTDiP case-control study was funded by GlaxoSmithKline; data collection for the Janssen–BRC study was funded by Janssen. PEV is supported by a Medical Research Council Bioinformatics Research Fellowship (Grant No. MR/K020706/1). TCF is supported by an Institute Strategic Grant from the Biotechnology and Biological Sciences Research Council (Grant No. BB/JO1446X/1). CMP is supported by the National Institute for Health Research (NIHR) Mental Health Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. ETB is supported by the NIHR Cambridge Biomedical Research Centre.
We thank David Hume for comments on the manuscript; Fiona Kelly and Stewart Bates for supporting access to the GSK–HiTDiP dataset; Pieter Peeters, Wilhelmus Drinkenberg, and William Talloan for supporting access to the Janssen–BRC and HiTDiP datasets; and Evian Gordon from the BRC.
The microarray and clinical data from the Janssen-BRC study are accessible to scientists through the independent YODA Project process (http://yoda.yale.edu). The GSK-HiTDiP data are accessible on GEO (https://www.ncbi.nlm.nih.gov/geo) as GSE98793. For more information on combining the datasets please contact the YODA Project (yodap@yale.edu).
GMW and WCD are employees and stockholders of Janssen Research & Development of Johnson & Johnson. SK and RH are employees and stockholders of GlaxoSmithKline (GSK). ETB is employed half-time by GSK and half-time by the University of Cambridge; he holds stock in GSK. ETB is also a shareholder in the BRC. CMP has received research funding from Johnson & Johnson and speaker fees from Lundbeck. The remaining authors all report no biomedical financial interests or potential conflicts of interest.
Footnotes
Immunopsychiatry Consortium: Edward Bullmore, Petra Vertes, Rudolf Cardinal (University of Cambridge); Sylvia Richardson, Gwenael Leday (Medical Research Council Biostatistics Unit); Tom Freeman, Tim Regan, David Hume, Zhaozong Wu (University of Edinburgh); Carmine Pariante, Annamaria Cattaneo, Patricia Zunszain, Alessandra Borsini, Robert Stewart, David Chandran (King’s College London); Livia Carvalho, Joshua Bell, Luis Souza-Teodoro (University College London); Hugh Perry (University of Southampton); Neil Harrison (University of Sussex); Wayne Drevets, Gayle Wittenberg, Declan Jones (Janssen Research & Development); Edward Bullmore, Shahid Khan, Annie Stylianou, Robbie Henderson (GlaxoSmithKline).
GGRL and PEV contributed equally to this work.
GMW and ETB contributed equally to this work.
Supplementary material cited in this article is available online at http://dx.doi.org/10.1016/j.biopsych.2017.01.021.
Contributor Information
Edward T. Bullmore, Email: etb23@cam.ac.uk.
MRC Immunopsychiatry Consortium:
Edward Bullmore, Petra Vertes, Rudolf Cardinal, Sylvia Richardson, Gwenael Leday, Tom Freeman, Tim Regan, David Hume, Zhaozong Wu, Carmine Pariante, Annamaria Cattaneo, Patricia Zunszain, Alessandra Borsini, Robert Stewart, David Chandran, Livia Carvalho, Joshua Bell, Luis Souza-Teodoro, Hugh Perry, Neil Harrison, Wayne Drevets, Gayle Wittenberg, Declan Jones, Edward Bullmore, Shahid Khan, Annie Stylianou, and Robbie Henderson
Supplementary Material
References
- 1.Wium-Andersen M.K., Ørsted D.D., Nielsen S.F., Nordestgaard B.G. Elevated C-reactive protein levels, psychological distress, and depression in 73,131 individuals. JAMA Psychiatry. 2013;70:176–184. doi: 10.1001/2013.jamapsychiatry.102. [DOI] [PubMed] [Google Scholar]
- 2.Valkanova V., Ebmeier K.P., Allan C.L. CRP, IL-6 and depression: A systematic review and meta-analysis of longitudinal studies. J Affect Disord. 2013;150:736–744. doi: 10.1016/j.jad.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Haapakoski R., Mathieu J., Ebmeier K.P., Alenius H., Kivimaki M. Cumulative meta-analysis of interleukins 6 and 1β, tumour necrosis factor α and C-reactive protein in patients with major depressive disorder. Brain Behav Immun. 2015;49:206–215. doi: 10.1016/j.bbi.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matcham F., Rayner L., Steer S., Hotopf M. The prevalence of depression in rheumatoid arthritis: A systematic review and meta-analysis. Rheumatology. 2013;52:2136–2148. doi: 10.1093/rheumatology/ket169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khandaker G.M., Pearson R.M., Zammit S., Lewis G., Jones P.B. Association of serum interleukin 6 and C-reactive protein in childhood with depression and psychosis in young adult life: A population-based longitudinal study. JAMA Psychiatry. 2014;71:1121–1128. doi: 10.1001/jamapsychiatry.2014.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bull S.J., Huezo-Diaz P., Binder E.B., Cubells J.F., Ranjith G., Maddock C. Functional polymorphisms in the interleukin-6 and serotonin transporter genes, and depression and fatigue induced by interferon-α and ribavirin treatment. Mol Psychiatry. 2009;14:1095–1104. doi: 10.1038/mp.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hepgul N., Cattaneo A., Agarwal K., Baraldi S., Borsini A., Bufalino C. Transcriptomics in interferon-α-treated patients identifies inflammation-, neuroplasticity- and oxidative stress-related signatures as predictors and correlates of depression. Neuropsychopharmacology. 2016;41:2502–2511. doi: 10.1038/npp.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dantzer R., O’Connor J., Freund G., Johnson R., Kelley K. From inflammation to sickness and depression: When the immune system subjugates the brain. Nat Rev Neurosci. 2008;9:46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banks W.A., Kastin A.J., Gutierrez E.G. Penetration of interleukin-6 across the murine blood-brain barrier. Neurosci Lett. 1994;179:53–56. doi: 10.1016/0304-3940(94)90933-4. [DOI] [PubMed] [Google Scholar]
- 10.Perry V.H., Cunningham C., Holmes C. Systemic infections and inflammation affect chronic neurodegeneration. Nat Rev Immunol. 2007;7:161–167. doi: 10.1038/nri2015. [DOI] [PubMed] [Google Scholar]
- 11.Miller A.H., Raison C.L. Are anti-inflammatory therapies viable treatments for psychiatric disorders? Where the rubber meets the road. JAMA Psychiatry. 2015;72:527–528. doi: 10.1001/jamapsychiatry.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raison C., Capuron L., Miller A. Cytokines sing the blues: Inflammation and the pathogenesis of depression. Trends Immunol. 2006;27:24–31. doi: 10.1016/j.it.2005.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin E., Tsai S.J. Genome-wide microarray analysis of gene expression profiling in major depression and antidepressant therapy. Prog Neuropsychopharmacol Biol Psychiatry. 2016;64:334–340. doi: 10.1016/j.pnpbp.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Jansen R., Penninx B.W.J.H., Madar V., Xia K., Milaneschi Y., Hottenga J.J. Gene expression in major depressive disorder. Mol Psychiatry. 2016;21:444. doi: 10.1038/mp.2015.94. [DOI] [PubMed] [Google Scholar]
- 15.Garbett K.A., Vereczkei A., Kálmán S., Brown J.A., Taylor W.D., Faludi G. Coordinated messenger RNA/microRNA changes in fibroblasts of patients with major depression. Biol Psychiatry. 2015;77:256–265. doi: 10.1016/j.biopsych.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mostafavi S., Battle A., Zhu X., Potash J.B., Weissman M.M., Shi J. Type I interferon signaling genes in recurrent major depression: Increased expression detected by whole-blood RNA sequencing. Mol Psychiatry. 2014;19:1267–1274. doi: 10.1038/mp.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glahn D.C., Curran J.E., Winkler A.M., Carless M.A., Kent J.W., Charlesworth J.C. High dimensional endophenotype ranking in the search for major depression risk genes. Biol Psychiatry. 2012;71:6–14. doi: 10.1016/j.biopsych.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shelton R.C., Claiborne J., Sidoryk-Wegrzynowicz M., Reddy R., Aschner M., Lewis D.A. Altered expression of genes involved in inflammation and apoptosis in frontal cortex in major depression. Mol Psychiatry. 2011;16:751–762. doi: 10.1038/mp.2010.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blume J., Douglas S.D., Evans D.L. Immune suppression and immune activation in depression. Brain Behav Immun. 2011;25:221–229. doi: 10.1016/j.bbi.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller A., Maletic V., Raison C. Inflammation and its discontents: The role of cytokines in the pathophysiology of major depression. Biol Psychiatry. 2009;65:732–741. doi: 10.1016/j.biopsych.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grosse L., Hoogenboezem T., Ambrée O., Bellingrath S., Jörgens S., de Wit H.J. Deficiencies of the T and natural killer cell system in major depressive disorder: T regulatory cell defects are associated with inflammatory monocyte activation. Brain Behav Immun. 2016;54:38–44. doi: 10.1016/j.bbi.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Snijders G., Schiweck C., Mesman E., Grosse L., de Wit H.J., Nolen W.A. A dynamic course of T cell defects in individuals at risk for mood disorders. Brain Behav Immun. 2016;58:11–17. doi: 10.1016/j.bbi.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Grosse L., Carvalho L.A., Wijkhuijs A.M., Bellingrath S., Ruland T., Ambree O. Clinical characteristics of inflammation-associated depression: Monocyte gene expression is age-related in major depressive disorder. Brain Behav Immun. 2015;44:48–56. doi: 10.1016/j.bbi.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Domenici E., Willé D.R., Tozzi F., Prokopenko I., Miller S., McKeown A. Plasma protein biomarkers for depression and schizophrenia by multi analyte profiling of case-control collections. PLoS One. 2010;5:e9166. doi: 10.1371/journal.pone.0009166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muglia P., Tozzi F., Galwey N.W., Francks C., Upmanyu R., Kong X.Q. Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Mol Psychiatry. 2010;15:589–601. doi: 10.1038/mp.2008.131. [DOI] [PubMed] [Google Scholar]
- 26.Tilahun A., Lin D., Shkedy Z., Geys H., Alonso A., Peeters P. Genomic biomarkers for depression: Feature-specific and joint biomarkers. Stat Biopharm Res. 2010;2:419–434. [Google Scholar]
- 27.Zoon H., Veth C., Arns M., Drinkenburg W., Talloen W., Peeters P. EEG alpha power as an intermediate measure between brain-derived neurotrophic factor Val66Met and depression severity in patients with major depressive disorder. J Clin Neurophys. 2013;30:261–267. doi: 10.1097/WNP.0b013e3182933d6e. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y., Yieh L., Yang T., Drinkenburg W., Peeters P., Steckler T. Metabolomic biosignature differentiates melancholic depressive patients from healthy controls. BMC Genomics. 2016;17:669. doi: 10.1186/s12864-016-2953-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheehan D.V., Lecrubier Y., Sheehan K.H., Amorim P., Janavs J., Weiller E. The Mini-International Neuropsychiatric Interview (MINI): The development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry. 1998;59:22–33. [PubMed] [Google Scholar]
- 30.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smyth G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 32.Blangiardo M., Richardson S. Statistical tools for synthesizing lists of differentially expressed features in related experiments. Genome Biol. 2007;8:R54. doi: 10.1186/gb-2007-8-4-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blangiardo M., Cassese A., Richardson S. sdef: An R package to synthesize lists of significant features in related experiments. BMC Bioinformatics. 2010;11:270. doi: 10.1186/1471-2105-11-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang B., Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 35.Alexa A, Rahnenfuhrer J (2010): topGO: Enrichment analysis for gene ontology. R package, version 2.0.
- 36.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M. Gene Ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A. STRING v9: 1. Protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mabbott N.A., Baillie J.K., Brown H., Freeman T.C., Hume D.A. An expression atlas of human primary cells: Inference of gene function from coexpression networks. BMC Genomics. 2013;14:632. doi: 10.1186/1471-2164-14-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freeman T.C., Goldovsky L., Brosch M., Van Dongen S., Mazière P., Grocock R.J. Construction, visualisation, and clustering of transcription networks from microarray expression data. PLoS Comput Biol. 2007;3:e206. doi: 10.1371/journal.pcbi.0030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Theocharidis A., van Dongen S., Enright A., Freeman T. Network visualization and analysis of gene expression data using BioLayout Express3D. Nat Protoc. 2009;4:1535–1550. doi: 10.1038/nprot.2009.177. [DOI] [PubMed] [Google Scholar]
- 42.Langfelder P., Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inamdar A., Merlo-Pich E., Gee M., Makumi C., Mistry P., Robertson J. Evaluation of antidepressant properties of the p38 MAP kinase inhibitor losmapimod (GW856553) in major depressive disorder: Results from two randomised, placebo-controlled, double-blind, multicentre studies using a Bayesian approach. J Psychopharmacol. 2014;28:570–581. doi: 10.1177/0269881114529377. [DOI] [PubMed] [Google Scholar]
- 44.Zorrilla E.P., Luborsky L., McKay J.R., Rosenthal R., Houldin A., Tax A. The relationship of depression and stressors to immunological assays: A meta-analytic review. Brain Behav Immun. 2001;15:199–226. doi: 10.1006/brbi.2000.0597. [DOI] [PubMed] [Google Scholar]
- 45.Carvalho L.A., Bergink V., Sumaski L., Wijkhuijs J., Hoogendijk W.J., Birkenhager T.K. Inflammatory activation is associated with a reduced glucocorticoid receptor alpha/beta expression ratio in monocytes of inpatients with melancholic major depressive disorder. Transl Psychiatry. 2014;4:e344. doi: 10.1038/tp.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weigelt K., Carvalho L.A., Drexhage R.C., Wijkhuijs A., de Wit H., van Beveren N.J. TREM-1 and DAP12 expression in monocytes of patients with severe psychiatric disorders: EGR3, ATF3 and PU.1 as important transcription factors. Brain Behav Immun. 2011;25:1162–1169. doi: 10.1016/j.bbi.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 47.Maes M., Lambrechts J., Bosmans E., Jacobs J., Suy E., Vandervorst C. Evidence for a systemic immune activation during depression: Results of leukocyte enumeration by flow cytometry in conjunction with monoclonal antibody staining. Psychol Med. 1992;22:45–53. doi: 10.1017/s0033291700032712. [DOI] [PubMed] [Google Scholar]
- 48.Wong M.L., Dong C., Maestre-Mesa J., Licinio J. Polymorphisms in inflammation-related genes are associated with susceptibility to major depression and antidepressant response. Mol Psychiatry. 2008;13:800–812. doi: 10.1038/mp.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium. Ripke S., Wray N.R., Lewis C.M., Hamilton S.P., Weissman M.M. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Danese A., Pariante C.M., Caspi A., Taylor A., Poulton R. Childhood maltreatment predicts adult inflammation in a life-course study. Proc Natl Acad Sci U S A. 2007;104:1319–1324. doi: 10.1073/pnas.0610362104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rohleder N. Stimulation of systemic low-grade inflammation by psychosocial stress. Psychosom Med. 2014;76:181–189. doi: 10.1097/PSY.0000000000000049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.