Abstract
MicroRNAs (miRNAs) are ~22nt-long single-stranded RNA molecules that form a RNA-induced silencing complex with Argonaute (AGO) protein to post-transcriptionally downregulate their target messenger RNAs (mRNAs). To understand the regulatory mechanisms of miRNA, discovering the underlying functional rules for how miRNAs recognize and repress their target mRNAs is of utmost importance. To determine functional miRNA targeting rules, previous studies extensively utilized various methods including high-throughput biochemical assays and bioinformatics analyses. However, targeting rules reported in one study often fail to be reproduced in other studies and therefore the general rules for functional miRNA targeting remain elusive. In this review, we evaluate previously-reported miRNA targeting rules and discuss the biological impact of the functional miRNAs on gene-regulatory networks as well as the future direction of miRNA targeting research.
Keywords: Canonical site types, microRNA, miRNA targeting, miRNA, Noncanonical site types
CANONICAL SITE TYPES
It has been widely accepted that more than 60% of the entire human mRNAs are directly regulated by miRNAs (1–4). Accordingly, miRNAs participate in numerous biological processes, and their activity can lead to various human diseases (5–10). Although understanding the complete rules of how miRNAs recognize and regulate their target mRNAs is essential to learn the biological roles of miRNAs, comprehensive rules for functional miRNA targeting are yet to be determined.
miRNAs interact with their target mRNAs through Watson-Crick base pairing (WCP) at their 5′ ends (2, 11–15). Numerous empirical computational analyses have shown that perfect WCPs between the 2–7 nucleotide region at the 5′ end of the miRNA and its complementary target site on the mRNA are crucial for miRNA targeting (11, 13, 16). This 6nt region of miRNA is referred to as “seed”, and an additional base pairing at the 8nt position of miRNA or the existence of adenine on the mRNA side corresponding to the 1st nucleotide position of miRNA further improves the miRNA targeting efficacy. Based on these findings, four canonical site types (CSTs) were determined and are indicated as 8mer, 7mer-m8, 7mer-A1, and 6mer, respectively (14, 17).
To measure the impact of CSTs on the whole transcriptome, microarrays were utilized to monitor changes in the transcriptome after ectopic introduction of miRNAs. Accordingly, the widespread impact of the CSTs on the transcriptome was observed as a large number of mRNA targets were directly downregulated (3, 14). Also, whole proteomic analyses and ribosome profiling showed that miRNAs downregulate gene expression mainly through mRNA destabilization rather than translational repression (4, 15, 18). Lewis et al. (15) conducted comparative genomic analyses and found that many target sites of the CSTs were conserved across the species (11). Friedman et al. (1) used an extended list of vertebrate genomes to show that more than 60% of mammalian genes are conserved targets of miRNAs (1).
In terms of the molecular details of targeting mechanisms, a structural analysis by Schirle et al. (19) elucidated the functional mechanism of the CSTs by proposing a mechanistic model for seed pairing. This model includes a pocket to recognize adenine on the mRNA side, which explains why the adenine residue affects the miRNA targeting efficacy (19). The theory of the molecular mechanism for miRNA target recognition was reinforced by a single-molecule study that utilized a fluorescence resonance energy transfer (FRET) assay on human AGO2 (20). The researchers used their results to propose a stepwise model for miRNA target recognition that consists of the initial binding of AGO2 to a target site with WCPs for the miRNA 2–4nt region, which is referred to as the sub-seed recognition motif, and a subsequent step of lateral diffusion for the formation of complete seed pairing.
The broad impact of miRNA targeting and the conservation of miRNA target sites strongly indicate that the CSTs of miRNAs may play biologically important roles. Nonetheless, the response of the transcriptome cannot be fully explained only by the CSTs (12), implying that additional functional site types may exist in addition to the four CSTs.
PREVIOUSLY REPORTED NONCANONICAL SITE TYPES
Accumulated evidence from studies over the past decade have expanded the miRNA targeting rules and have led to the discovery of noncanonical site types (NSTs). Aside from CSTs, two NSTs were identified: centered site and offset 6mer (Table 1) (1, 21). Compared to CSTs, NSTs lack perfect WCPs between the seed site of miRNA and the target site of mRNAs and exhibit weaker but significant effects in downregulating target mRNAs.
Table 1.
Previously reported noncanonical site types
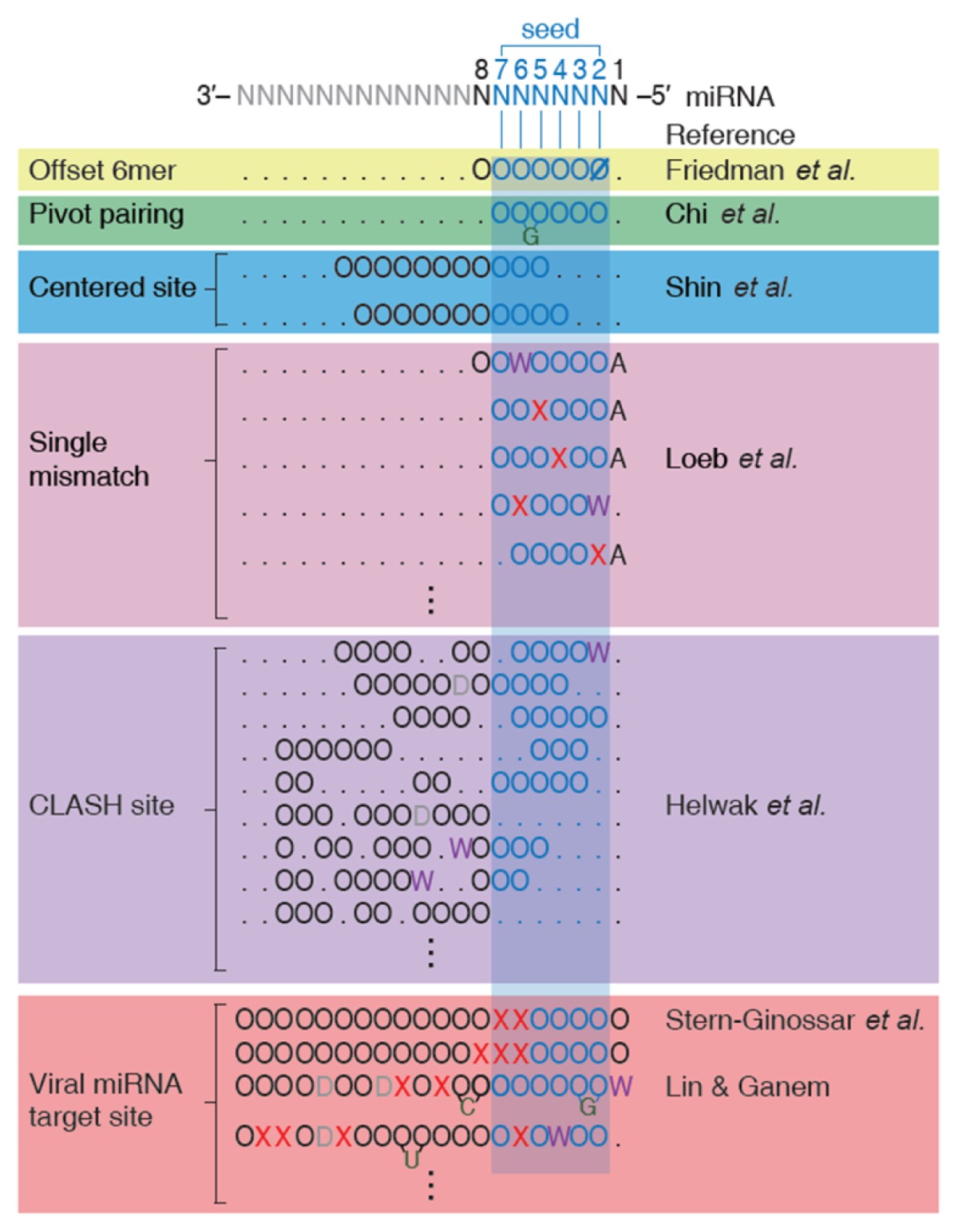
The residue ‘O’ indicates a Watson-Crick base pairing between miRNA and mRNA target and ‘Ø‘ represents all the other interactions other than a Watson-Crick base pairing. ‘W’ and ‘X’ indicate wobble pairing and mismatch, respectively. For the mRNA region responding to the 1st nucleotide position of miRNA, ‘A’ is assigned if adenine is in that position. A bulge on mRNA side is depicted as a protrusion of a nucleotide on the mRNA strand, and a bulge on miRNA side is represented as the residue ‘D’.
Recently, a powerful biochemistry technique called AGO crosslinking immunoprecipitation and high-throughput sequencing (AGO CLIP-seq) was developed (22, 23). AGO CLIP-seq provides precise information of AGO protein binding regions on mRNAs. Using this technique, two additional NSTs were identified: the pivot pairing and single mismatch site types (STs) (24, 25). Pivot pairing ST contains a guanine bulge on the mRNA target site of the seed pairing region, and the single nucleotide mismatch ST includes a single nucleotide mismatch or a wobble pairing on the seed region. The detection accuracy was improved using photoactivatable-ribonucleoside-enhanced CLIP (PAR-CLIP), which incorporates photoreactive nucleoside analogs to facilitate cross-linking (23, 26–29).
Even though AGO CLIP-seq can effectively identify functional NSTs, the technique suffers from a few limitations. First, since this method detects the association among AGO, miRNAs, and mRNAs, it can only provide indirect evidence for the interactions between individual miRNAs and mRNAs. Second, although binding of AGO to the target mRNA is a necessary condition for downregulation of the target mRNA, it is not sufficient for detectable downregulation (22, 24, 29, 30). For instance, a portion of AGOs have the ability to bind to the ORF as well as 3′UTR (22, 25, 27, 31–33). Those ORF-bound AGOs can be detected by AGO CLIP-seq, but most of the mRNAs with ORF-bound AGOs would not be downregulated since the ORF-bound AGOs are likely to get bumped out by translating ribosomes (14, 34, 35). Therefore, the miRNA targets detected by AGO CLIP-seq tend to include a high rate of false positives.
To overcome the first limitation, an advanced technique, CLASH (CrossLinking, ligAtion, and Sequencing of Hybrids), that aims to identify direct interactions between AGO-bound miRNAs and mRNAs was developed (36, 37). CLASH includes an additional step of ligation for a miRNA and its target mRNAs, and it is thus able to provide a more direct profile of miRNA-mRNA interactions. Additional NSTs were proposed to be functional by analyzing improved binding information, generated by CLASH, among AGO protein, mRNA, and miRNA (Table 1) (37, 38). However, even with CLASH, the second limitation still remains unsolved.
Based on the interactions between viral miRNAs and mRNA targets, several additional NSTs that contain an imperfect seed pairing and additional complementary WCPs were reported (Table 1) (39, 40). These results imply the prevalence and potential contribution of the NSTs in functional miRNA targeting and thus emphasize the need to expand the miRNA targeting rules beyond the previously-accepted CSTs. Nonetheless, whether these proposed NSTs truly serve as general rules for functional miRNA targeting remains uncertain because of the aforementioned limitations.
SYSTEMATIC EVALUATION OF PREVIOUSLY REPORTED NSTS
In contrast with the prior results that indicate the widespread importance and functional roles of NSTs, a recent study claimed that almost all previously-identified NSTs are in fact not functional (41). In this study, researchers re-examined the efficacies of previously-reported NSTs by observing transcriptome changes after knocking out, knocking down, and ectopically expressing miRNAs. After careful and systematic evaluations of the transcriptome data, they concluded that even though NSTs were detected in AGO CLIP-seq studies, most of the NSTs except offset 6mer are non-functional and do not show any detectable downregulation of their target mRNAs (24, 36, 37), suggesting that these NSTs may be conditionally functional for a specific cell type or with specific miRNAs.
Although almost all previously-reported NSTs were found to be non-functional, one critical question still remains unexplored: are there any additional functional NSTs? Compared to the astronomical number of interactions that can possibly occur between miRNAs and mRNAs, previous studies have evaluated only a tiny fraction of possible STs (42). The limited scope of the examination could be the reason for past failures in detecting functional STs, calling for a systematic and exhaustive evaluation of all possible interactions between miRNAs and mRNAs based on direct evidence and the discovery of comprehensive rules for functional miRNA targeting.
COMPREHENSIVE EVALUATION OF FUNCTIONAL SITE TYPES
Kim et al. (42) systematically determined all possible interactions that can occur between miRNAs and the target mRNAs to expand the number of evaluated STs and utilized large-scale microarray data that measured the transcriptome response when miRNAs are ectopically introduced to evaluate whether these interactions are functional (42). The authors statistically evaluated whether each of the > 2 billion STs is enriched in genes that are highly downregulated when miRNAs are overexpressed. Since the approach the authors adopted was to examine an astronomical number of STs based on direct evidence of actual transcriptome response to miRNA overexpression, their research is free from the limitations of studies based on AGO CLIP-seq.
Through a massive-scale bioinformatics search, the authors discovered three functional NSTs in addition to the CSTs. The newly discovered NSTs consist of previously-identified offset 6mer ST, a novel NST termed as offset 7mer, and another novel NST termed as 6mer-A1 (42). Offset 7mer contains an additional WCP compared to the offset 6mer ST and 6mer-A1 is similar to canonical 7mer-A1 ST with an exception that it is one nucleotide shorter. Kim et al. (42) observed that local contexts that are known to affect miRNA targeting, such as local AU content of the surrounding region of the target site, 3′UTR length of the target mRNA, the target site abundance, and the thermodynamic pairing stability between miRNA and mRNA (11, 14, 17, 43–48), also have significant impact on the proficiency of the three newly-discovered NSTs. The authors searched for additional STs whose target sites with good contexts exhibit detectable downregulation and identified four additional functional NSTs. They named these NSTs context-dependent noncanonical site types (CDNSTs). When compared to CSTs, the seven newly-discovered NSTs and CDNSTs elicit weaker-but-still-significant target repression (42). Also, NSTs and CDNSTs have more target sites than CSTs, indicating they may exert considerable influence on the regulation of the transcriptome (Table 2).
Table 2.
Comprehensive rules for functional microRNA targeting
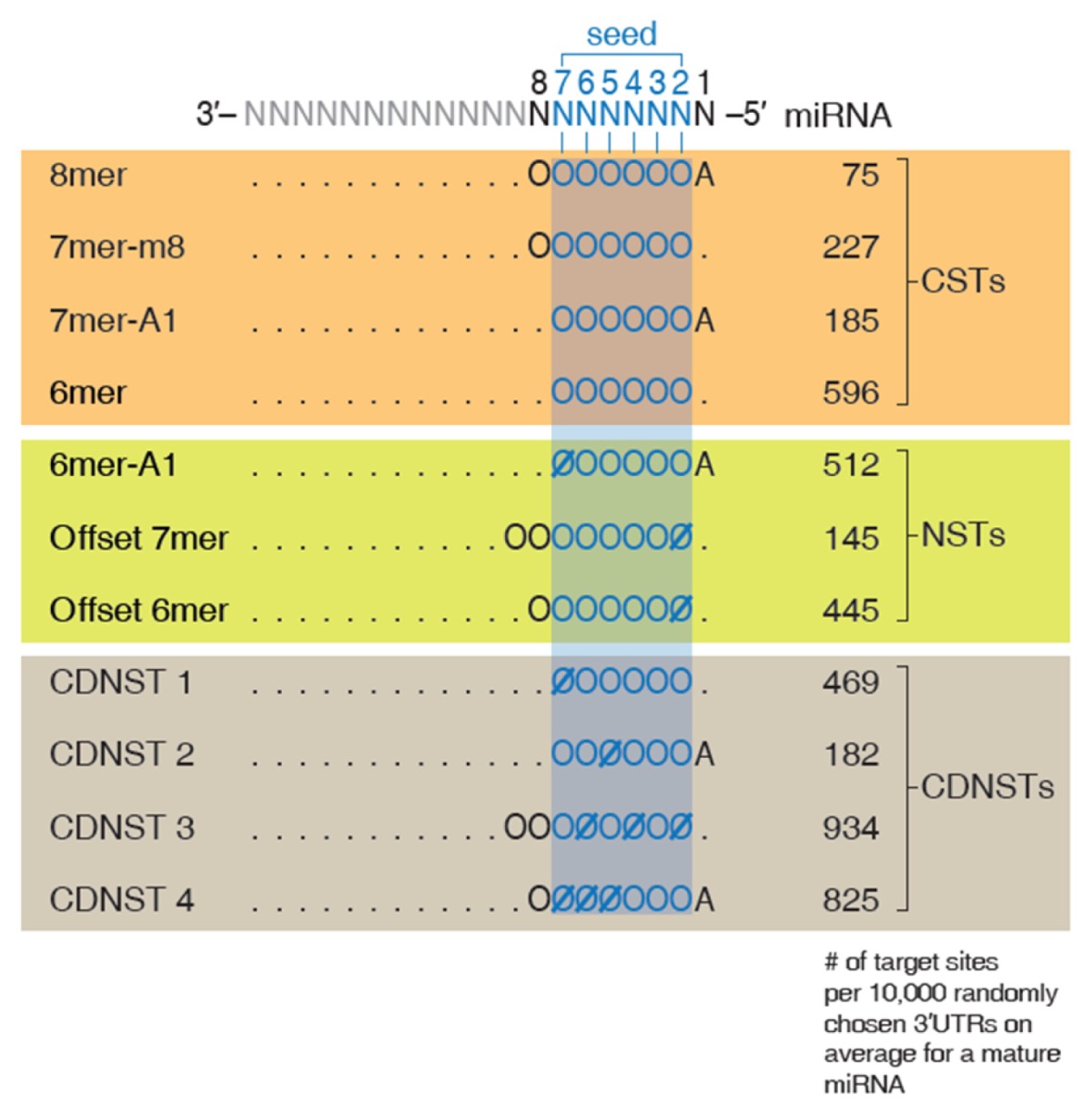
An expanded view of functional miRNA targeting (modified from Fig. 3B of Kim et al.). The normalized numbers of targets for each site type are shown in right side and the representation of interactions follows notations described in Table 1.
The newly-discovered NSTs were thoroughly validated through various experiments and computational analyses. In the luciferase assay, an overall 70% of the target mRNAs of NSTs and CDNSTs exhibited significant repression, which confirms their functionality in vivo (42). Also, independent microarray data obtained from various human cell lines was evaluated by monitoring the transcriptome response against the overexpressed miRNA to further demonstrate that these NSTs and CDNSTs are generally functional. In addition, the biological significance of the NSTs and CDNSTs was validated by analyzing the miRNA knock-out and knock-down microarray data, which strongly indicates they effectively downregulate their target mRNAs in an endogenous environment.
The impact of the NSTs and CDNSTs on the transcriptome was assessed by estimating the overall amount of mRNA repression mediated by CSTs, NSTs, and CDNSTs. The analysis showed that even though the individual impact of NSTs and CDNSTs was relatively weak, when added together, the overall impact of the NSTs and CDNSTs on the transcriptome was comparable to that of CSTs (42). Moreover, a comparative genomics analysis confirmed that the target sites in 10 out of 11 functional STs are evolutionarily conserved across the vertebrate genome (42). Therefore, novel NSTs and CDNSTs may have physiologically important functions, and the influence of NSTs and CDNSTs should be carefully considered when identifying miRNA targets.
In summary, a massive-scale computational search revealed seven novel functional noncanonical interactions that were validated by multiple lines of strong evidence, suggesting that these NSTs and CDNSTs may serve in important roles in the miRNA-mRNA regulatory network.
DISCUSSION
Since the discovery of miRNA, numerous scientists have attempted to understand miRNA in terms of its biogenesis, functions, and significance. In 2005, Lewis et al. discovered CSTs and verified that they are functional in vivo (11). This discovery was a scientific breakthrough because CSTs not only exert substantial influence on the whole transcriptome and proteome, but are also evolutionarily conserved, suggesting their biological significance (1, 3, 15). The accumulation of genome-wide data and the development of advanced technologies, such as AGO CLIP-seq and CLASH, have led to a discovery of additional NSTs involved in miRNA targeting (1, 21, 24, 25, 37, 39, 40). Although there are large numbers of previously reported NSTs, these NSTs are not fully accepted as a part of general miRNA targeting rules due to inconsistent results found in various studies (41). Therefore, a recent study made an attempt to systematically and comprehensively evaluate miRNA-target interactions by employing a massive-scale bioinformatics approach (42). In this study, seven potentially functional NSTs and CDNSTs were discovered. Validations via luciferase assays and analyses of independent data suggest that most of these NSTs and CDNSTs may be functional, and the evolutionary conservation and estimated regulatory effect on the transcriptome of NSTs and CDNSTs clearly indicate that expanded miRNA targeting rules could potentially play biologically relevant roles.
A deeper understanding of miRNA targeting rules raises important issues. One major issue is the lack of research on RNA-binding proteins (RBPs) that act as determinants of miRNA targeting and the mechanisms through which these RBPs regulate miRNA targeting proficiencies. Several unique cases were reported in which RBPs influence the proficiency of repression of miRNA target mRNAs (49, 50), but a comprehensive model depicting the interplay between RBPs and miRNA targeting remains to be evaluated. Another issue is the lack of complete understanding of the biological consequences of miRNA targeting on translational regulation. Guo et al. (2010) showed that miRNA-mediated gene silencing in a steady state is mainly mediated by mRNA destabilization and that translational repression contributes little to the overall downregulation (18). However, in a transient state, the translational control appears to be a major mechanism of miRNA targeting (51), and even in the steady state, translational control may play more prominent roles for specific miRNAs (52–54). Hence, discovering miRNA targeting determinants associated with translational repression would provide valuable knowledge to understand miRNA targeting mechanisms more completely.
An expanded repertoire of functional miRNA targets implies that miRNA-target mRNA interactions and their regulatory networks are far more intricate than are currently understood. The comprehensive rules of miRNA targeting revealed in recent studies may lead to a deeper understanding of the complex gene-regulatory network controlled by miRNAs, reduction in the off-targeting effects when designing siRNA/shRNA libraries, and an improvement in the accuracy of miRNA target prediction algorithms.
ACKNOWLEDGEMENTS
This work was supported by the Institute for Basic Science (Grant number IBS-R008-D1), and by the National Research Foundation, funded by the Ministry of Science, ICT & Future Planning, Republic of Korea (Grant numbers 2014M3C9A 3063541 and 2012M3A9D1054622).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 4.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 5.Kasinski AL, Slack FJ. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melo SA, Kalluri R. miR-29b moulds the tumour microenvironment to repress metastasis. Nat Cell Biol. 2013;15:139–140. doi: 10.1038/ncb2684. [DOI] [PubMed] [Google Scholar]
- 7.Babar IA, Cheng CJ, Booth CJ, et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Nat Acad Sci U S A. 2012;109:E1695–1704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 9.Cheng CJ, Bahal R, Babar IA, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costinean S, Zanesi N, Pekarsky Y, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Nat Acad Sci U S A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 12.Farh KK, Grimson A, Jan C, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 13.Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 14.Grimson A, Farh KKH, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Gene regulation. Structural basis for microRNA targeting. Science. 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandradoss SD, Schirle NT, Szczepaniak M, MacRae IJ, Joo C. A dynamic search process underlies microRNA targeting. Cell. 2015;162:96–107. doi: 10.1016/j.cell.2015.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011;8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 24.Chi SW, Hannon GJ, Darnell RB. An alternative mode of microRNA target recognition. Nat Struct Mol Biol. 2012;19:321–327. doi: 10.1038/nsmb.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loeb GB, Khan AA, Canner D, et al. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell. 2012;48:760–770. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hafner M, Landthaler M, Burger L, et al. PAR-CliP--a method to identify transcriptome-wide the binding sites of RNA binding proteins. Journal of visualized experiments : JoVE J Vis Exp. 2010:e2034. doi: 10.3791/2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corcoran DL, Georgiev S, Mukherjee N, et al. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011;12:R79. doi: 10.1186/gb-2011-12-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majoros WH, Lekprasert P, Mukherjee N, et al. MicroRNA target site identification by integrating sequence and binding information. Nat Methods. 2013;10:630–633. doi: 10.1038/nmeth.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konig J, Zarnack K, Luscombe NM, Ule J. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet. 2011;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 31.Gottwein E, Corcoran DL, Mukherjee N, et al. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe. 2011;10:515–526. doi: 10.1016/j.chom.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leung AK, Young AG, Bhutkar A, et al. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol. 2011;18:237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skalsky RL, Corcoran DL, Gottwein E, et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012;8:e1002484. doi: 10.1371/journal.ppat.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 35.Gu S, Jin L, Zhang FJ, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc Nat Acad Sci U S A. 2011;108:10010–10015. doi: 10.1073/pnas.1017386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khorshid M, Hausser J, Zavolan M, van Nimwegen E. A biophysical miRNA-mRNA interaction model infers canonical and noncanonical targets. Nat Methods. 2013;10:253–255. doi: 10.1038/nmeth.2341. [DOI] [PubMed] [Google Scholar]
- 39.Stern-Ginossar N, Elefant N, Zimmermann A, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin HR, Ganem D. Viral microRNA target allows insight into the role of translation in governing microRNA target accessibility. Proc Nat Acad Sci U S A. 2011;108:5148–5153. doi: 10.1073/pnas.1102033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim D, Sung YM, Park J, et al. General rules for functional microRNA targeting. Nat Genetics. 2016;48:1517–1526. doi: 10.1038/ng.3694. [DOI] [PubMed] [Google Scholar]
- 43.Ui-Tei K, Naito Y, Nishi K, Juni A, Saigo K. Thermodynamic stability and Watson-Crick base pairing in the seed duplex are major determinants of the efficiency of the siRNA-based off-target effect. Nucleic Acids Res. 2008;36:7100–7109. doi: 10.1093/nar/gkn902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arvey A, Larsson E, Sander C, Leslie CS, Marks DS. Target mRNA abundance dilutes microRNA and siRNA activity. Mol Syst Biol. 2010;6:363. doi: 10.1038/msb.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 47.Xia T, SantaLucia J, Jr, Burkard ME, et al. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 48.Kim D, Kim J, Baek D. Global and Local Competition between Exogenously Introduced microRNAs and Endogenously Expressed microRNAs. Mol Cells. 2014;37:412–417. doi: 10.14348/molcells.2014.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 50.Triboulet R, Gregory RI. Pumilio turns on microRNA function. Nat Cell Biol. 2010;12:928–929. doi: 10.1038/ncb1010-928. [DOI] [PubMed] [Google Scholar]
- 51.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meijer HA, Kong YW, Lu WT, et al. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013;340:82–85. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- 53.Jangra RK, Yi M, Lemon SM. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol. 2010;84:6615–6625. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moretti F, Thermann R, Hentze MW. Mechanism of translational regulation by miR-2 from sites in the 5′ untranslated region or the open reading frame. RNA. 2010;16:2493–2502. doi: 10.1261/rna.2384610. [DOI] [PMC free article] [PubMed] [Google Scholar]


