Abstract
Leaf sheath hairiness is a morphological trait associated with various advantages, including tolerance to both abiotic and biotic stresses, thereby increasing yield. Understanding the genetic basis of this trait in barley can therefore improve the agronomic performance of this economically important crop. We scored leaf sheath hairiness in a two-year field trial in 1,420 BC1S3 lines from the wild barley nested association mapping (NAM) population HEB-25. Leaf sheath hairiness segregated in six out of 25 families with the reference parent Barke being glabrous. We detected the major hairy leaf sheath locus Hs (syn. Hsh) on chromosome 4H (111.3 cM) with high precision. The effects of the locus varied across the six different wild barley donors, with donor of HEB family 11 conferring the highest score of leaf sheath hairiness. Due to the high mapping resolution present in HEB-25, we were able to discuss physically linked pentatricopeptide repeat genes and subtilisin-like proteases as potential candidate genes underlying this locus. In this study, we proved that HEB-25 provides an appropriate tool to further understand the genetic control of leaf sheath hairiness in barley. Furthermore, our work represents a perfect starting position to clone the gene responsible for the 4H locus observed.
Introduction
Trichomes are specialized plant surface structures including hairs and exudate-secreting glands that originate and extend from the epidermis layer of aerial tissue [1]. They vary greatly in morphology, cellular composition, density, location, and function [1,2]. Non-glandular trichomes, through their reflectance properties, could play a role in temperature regulation of transpiring surfaces and the reduction of the heat load on leaves by reducing the absorption of radiant energy [3,4]. Interestingly, the wheat cultivar Pak 15800, which produced the largest number of leaf hair under water stress, was ranked as the most drought tolerant wheat accession in Hameed et al. [5]. Hence, hairiness could be a good selection criterion for drought tolerance, at least in wheat as well as barley as a temperate cereal [5]. Furthermore, hairiness could act as a deterrent to certain biotic stresses [6]. For instance, Kim et al. [7] reported a close association between trichome density and pepper mottle virus resistance in pepper, where the virus resistance could be due to the deterrence of the insect vectors of the virus by the hairs.
A number of studies on the molecular mechanism of the formation of trichomes have been conducted in Arabidopsis. Positive regulators of trichome formation include GLABROUS1 (GL1), an R2R3 MYB transcription factor; GLABRA3 (GL3), a basic helix-loop-helix transcription factor; Transparent Testa Glabra 1 (TTG1), a WD40 protein; and GLABRA2 (GL2), a homeodomain leucine zipper transcription factor. Triptychon (TYR) is an R3 MYB transcription factor and a negative regulator of trichome formation [8,9]. GL1 interacts with GL3, which interacts with TTG1. The GL1-GL3-TTG1 complex binds to the promotor of GL2 to initiate GL2 expression [10]. Epidermal cells expressing the GL2 protein are able to develop into trichome cells [9]. The GL1-GL3-TTG1 complex also binds to the promoter of TYR. The TYR protein expressed in trichome cells can move into neighboring cells and compete with GL1 for binding to GL3 to repress the GL2 expression [10].
In barley, leaf sheath hairiness is thought to follow a dominant single gene mode of inheritance [11,12]. The Hairy leaf sheath locus (Hs syn. Hsh) has initially been discovered by Pickering et al. [13]. The locus Hsb is derived from Hordeum bulbosum and is allelic to Hs [13]. Furthermore, it is homologous to Hairy peduncle Hp1, which governs leaf sheath and peduncle hairiness in rye and maps to the long arm of chromosome 5 [14]. The collinearity of the 5L rye chromosome to the distal part of the 4L chromosomes of other Triticeae members is due to a translocation event [15]. In addition, Hp1 and Hsb line up with Hairy leaf 1 (Hl1) (chromosome 4BL) that governs leaf hairiness in wheat [16]. Recently, Wang et al. [17] mapped a QTL controlling leaf sheath hairiness on chromosome 4HL in an association mapping diversity panel of 569 barley cultivars using 1,042 SNP markers from the Illumina GoldenGate oligonucleotide pool assay 1 (BOPA1). Furthermore, Wan et al. [18] detected a strong QTL on chromosome 4D associated with leaf sheath hairiness in wheat. The wild allele at this QTL originated from Aegilops tauschii, a wild progenitor of the hexaploid wheat, and was significantly associated with increased grain yield, grain weight and grain weight per spike [18]. The authors suggested that DNA markers for leaf sheath hairiness could be used for marker-assisted selection to increase yield in wheat [18].
In the present study, we used ‘Halle Exotic Barley 25’ (HEB-25), a recently developed nested association mapping (NAM) population, to study biodiversity of leaf sheath hairiness in wild barley. HEB-25 was developed by crossing 25 wild barley (Hordeum vulgare ssp. spontaneum, Hsp, and ssp. agriocrithon, Hag) accessions with the spring barley cultivar Barke (H. v. ssp. vulgare, Hv), ultimately resulting in 1,420 HEB lines in BC1S3 generation [19]. The fact that six wild HEB donors exhibit hairy leaf sheaths whereas Barke is glabrous makes HEB-25 an excellent tool (1) to identify QTL controlling hairiness in barley, (2) to locate the respective QTL at an increased genetic resolution and (3) to characterize the allelic variation at the respective QTL.
Materials and methods
Plant material
In this study, 1,420 HEB lines from HEB-25 were grown in the field. HEB-25 is a barley NAM population resulting from crosses between the two-rowed German spring barley cultivar Barke and 25 wild donors. One of the wild donors is a Tibetan H. vulgare ssp. agriocrithon (Hag) accession and the other 24 accessions are H. vulgare ssp. spontaneum (Hsp) originating from the Fertile Crescent [19]. F1 plants were backcrossed to Barke and selfed three times through single seed descent. The lines used in this study were in BC1S3:6 in the field trial of 2013 and in BC1S3:8 in the field trial of 2015. Further details about the development of HEB-25 are available in Maurer et al. [19].
Field trials
A two-year field trial (2013 and 2015) was conducted at the ‘Kühnfeld Experimental Station’ of the Martin Luther University, Halle-Wittenberg (51°29′46.47″N; 11°59′41.81″E). A complete randomized block design was applied, where the 1,420 HEB lines were grown in two replicates (two blocks) per year. In 2013, the plots consisted of two rows (30 seeds each) with a length of 1.50 m, a distance of 0.20 m between rows and a spacing of 0.50 m between plots. In 2015, plots dimensions were identical, however sowing density was increased to 50 seeds per row. Barke was integrated as a check line in all trials. All field trials were sown in March (i.e. early spring), with fertilization and pest management carried out according to local practice.
Phenotyping of HEB-25
Hairiness of the leaf sheath was recorded after plants reached shooting stage (BBCH31 stage) and before flowering (BBCH49 stage, recorded as the number of days from sowing until 50% of the plants in the plot had their first awns visible) [20]. Fertilization occurs at this stage of spring barley development [21]. Based on visual assessment, the density of hairs covering the leaf sheath was scored. Therefore, a scale of 1–7 was used, where “1” is the score for glabrous (non-hairy) leaf sheaths, and “3”, “5” and “7” are the scores for leaf sheaths that are slightly hairy, medium hairy and very hairy, respectively. Photographs illustrating each hairiness score are given in S1 Fig. Detailed microscopic images were created for Barke and all 25 wild barley donors as well as for all lines of HEB family 23 using a VHX-500 F digital 3D microscope (Keyence, Osaka, Japan).
Genotyping of HEB-25
HEB-25 lines, Barke, and the 25 donors were genotyped with 7,864 SNPs using the barley Infinium iSelect 9k SNP chip [22]. In total, 5,709 SNPs, which were polymorphic in at least one HEB family, showed less than 10% missing data and less than 12.5% heterozygosity, were used for subsequent analyses. Based on parental genotype information, the exotic allele could be specified in each segregating family, where homozygous Barke genotypes, heterozygous genotypes and homozygous exotic genotypes were assigned scores of 0, 1 and 2 at each SNP locus, respectively. Score values, thus, represent the number of wild barley alleles at each SNP (genotype data is available in Additional file 5 in Maurer et al. [19]).
Phenotypic data analysis
We used SAS 9.4 software (SAS Institute Inc., Cary, NC, USA) to analyze the phenotypic data. Pearson correlation was used to show association of leaf sheath hairiness between years. A mixed linear model was fitted to calculate genotype, year and genotype-by-year interaction effects. These effects were assumed random to estimate variance components and determine broad-sense heritability (h2) across the whole population based on the entry means.
, where , , and represent the genotypic, genotype-by-year, and residual variance components; while y and r refer to the number of years and the number of replicates, respectively.
The genotype effect was considered fixed to derive the best linear unbiased estimates (BLUEs). BLUEs were used for the subsequent genome-wide association study (GWAS).
Association mapping analysis
We conducted a GWAS on the BLUEs of the trait using Model-A, as suggested in Liu et al. [23], where cofactors were included in addition to the tested SNP in the multiple linear regression model. Würschum et al. [24] showed that this model revealed the highest predictive power and detected the highest number of QTL in joint linkage association mapping. This model allows the post-hoc determination of family-specific QTL effects [25], which is explained further down. The analysis was conducted in SAS and consisted of two steps. In the first step, cofactors, i.e. SNPs controlling population structure and genetic background, were selected by stepwise forward-backward regression. Overfitting was prevented by minimizing the Schwarz Bayesian Criterion (SBC or BIC) [26]. In the second step, an F test compared a reduced model (without the tested SNP effect) with a full model (with the tested SNP effect included) for each of the 5,709 SNPs to identify SNPs associated with the phenotypic trait [27]. Resulting p-values were adjusted by Bonferroni–Holm procedure to reduce false positives resulting from multiple testing [28]. Only SNPs having an adjusted p-value <0.05 were considered significant. The R package qqman [29] was used to create the Manhattan plot. The amount of phenotypic variance explained by a single significant SNP, R2, was estimated by fitting only that SNP in a linear model and R2adj of all significant SNPs together was calculated according to Utz et al. [30]. In addition, prediction ability (R2val) was determined in a five-fold cross-validation scenario. For this, 100 subsets were extracted out of the total phenotypic data. Each subset consisted of 80% randomly chosen HEB lines per family. This set was used as the training set to define significant markers and to estimate their effects, while the remaining 20% of lines were used as the validation set. Thereby the phenotypes of the validation set lines (20%) were predicted based on marker effects estimated in the training set (80%). Prediction ability (R2val) was then calculated as the squared Pearson product-moment correlation between the observed and predicted phenotypes of the validation set. The average of all 100 runs was then taken as the final record for R2val.
The effect of the wild allele of a significant SNP relative to the Barke allele was calculated by multiplying the regression coefficient (α effect) of that SNP by a factor of two, meaning the replacement of two Barke alleles with two wild barley alleles.
To estimate a family-specific QTL effect we cumulated significant SNP marker effects as described in Maurer et al. [25]. First, a peak marker for each expected donor-specific QTL was selected where the p-value was minimized. Each peak marker was placed centrally in a 26 cM interval (reflecting the mean introgression size in HEB-25) to look for further significant SNPs in this region. Effect estimates of all markers within this interval were then cumulated for each of the 25 donors. Since SNPs show different identity-by-state segregation patterns across the donors of HEB families, a different cumulated effect was obtained for each donor. In contrast to a haplotype-based approach, this method is more phenotypically orientated and is able to group family effects based on phenotypically-relevant SNP effects.
In order to identify potential candidate genes underlying the significant peak markers, we used the BARLEYMAP pipeline [31] with the recently released barley sequence [32]. We looked for genes within a window of 3 cM from the peak marker SCRI_RS_107762 and discussed the ones with a potential role in trichome formation and development based on literature review.
Results and discussion
Phenotypic data analysis
We phenotyped leaf sheath hairiness of 1,420 lines belonging to HEB-25, a wild barley nested association mapping population (S1 Table). Phenotyping was carried out for two years and the Pearson correlation between the years is equal to r = 0.82. The correlation is high, although lower than expected, possibly because the trait was recorded quantitatively (a score on a scale from 1–7) rather than scoring presence/absence. Also, different persons carried out the visual scoring in 2013 (BK) and 2015 (SS), adding another source of variation between years. Nevertheless, leaf sheath hairiness was found to be highly heritable with h2 = 80.4%, where the phenotypic variance in HEB-25 is predominantly controlled by genetic effects (Table 1).
Table 1. Variance partitioning and heritability of leaf sheath hairiness in HEB-25.
| Trait | Genotype | Year | Genotype-by-year | Residual | Broad-sense heritability (%) |
|---|---|---|---|---|---|
| Leaf sheath hairiness | 0.559 | 0.014 | 0.248 | 0.050 | 80.4 |
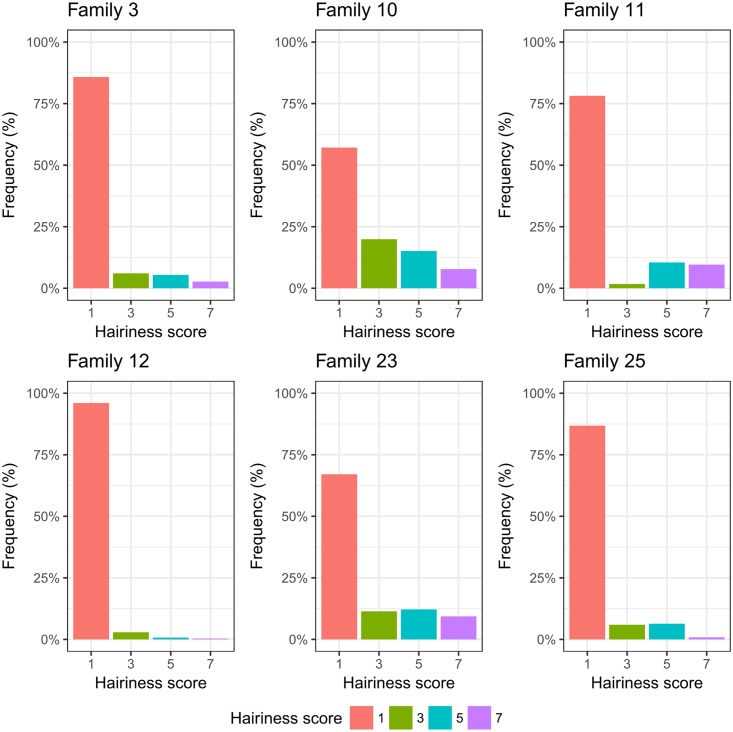
Out of 25 HEB families tested, leaf sheath hairiness segregated in six families (Table 2). Frequency distributions of the hairiness scores for each of the six hairiness HEB families are illustrated in Fig 1. In each family, the majority of HEB lines (>55%) revealed a non-hairy phenotype. The highest frequencies of hairy phenotypes were found in HEB families 10, 11 and 23. The observed hairness segregation suggests a monogenic or oligogenic inheritance for leaf sheath hairiness in the respective six segragting HEB families. The geographic origin of the HEB-25 wild barley donors are pinpointed in a map shown in S2 Fig. It occurs that most hairy donors (5 out of 6) originate from coastal regions of the Mediterranean Sea, whereas non-hairy donors predominantly originate from non-coastal regions of the Fertile Crescent. Possibly, there appears to be a selective advantage at coastal regions favoring the presence of leaf sheath hairiness, which might be associated with an increased coastal wind movement. In this regard, Grace and Russell [33] reported that the grass Festuca arundinacea, grown under windy field conditions, had more adaxial macro hairs than those grown in calm greenhouse conditions. In addition, we could postulate the association of leaf sheath hairiness with tolerance to higher temperatures since coastal regions tend to have lower elevations and higher temperatures than other regions.
Table 2. Descriptive statistics for six HEB families segregating for leaf sheath hairiness.
| HEB family (n) (a) | Wild donor | Origin of donor | Lines with score 7 (%)(b) | Mean(c) | Min(d) | Max(d) | SD (e) | CV (in %) (f) |
|---|---|---|---|---|---|---|---|---|
| 3 (296) | HID_055 | Turkey | 2.7 | 1.50 | 1 | 7 | 1.36 | 90.36 |
| 10 (231) | HID_102 | Syria | 7.8 | 2.47 | 1 | 7 | 1.97 | 79.65 |
| 11 (229) | HID_109 | Syria | 9.6 | 2.03 | 1 | 7 | 2.04 | 100.46 |
| 12 (276) | HID_114 | Lebanon | 0.4 | 1.11 | 1 | 7 | 0.59 | 53.50 |
| 23 (246) | HID_359 | Israel | 9.3 | 2.28 | 1 | 7 | 2.04 | 89.60 |
| 25 (235) | HID_386 | Israel | 0.9 | 1.43 | 1 | 7 | 1.18 | 82.73 |
The 19 HEB families, not displayed in this table, did not reveal leaf sheath hairiness phenotypes.
(a) HEB family and number of observations (n) for the hairiness score.
(b) Percentage of lines (from n) with a leaf sheath hairiness score of 7 within each family.
(c) Mean of all lines of the respective family.
(d) Minimum and maximum scores for hairiness, where “1” is the score for glabrous (non-hairy) leaf sheaths and “7” is the score for leaf sheaths that are very hairy.
(e) Standard deviation.
(f) Coefficient of variation.
Fig 1. Frequency distribution (in %) of leaf sheath hairiness scores for six segregating HEB families.
When examined under the microscope, the hairs present in HEB-25 lines are enlarged at the base and pointy at the tip (S3 Fig). Further studies are needed to investigate hair length and density as well as to identify the exact morphology of these hairs including whether they are unicellular or multicellular. Interestingly, all 63 HEB lines investigated under the microscope, as well as all the 25 wild donors and Barke, carried a second type of hair (designated with the letter B in S3 Fig), in addition to the macroscopically visible type that is scored in this study. These additional hairs are microscopic and thorn-like (S3 Fig). Further molecular and genetic investigations are needed to elucidate a potential role of these thorn-like hairs in biotic and abiotic tolerance.
Association mapping analysis
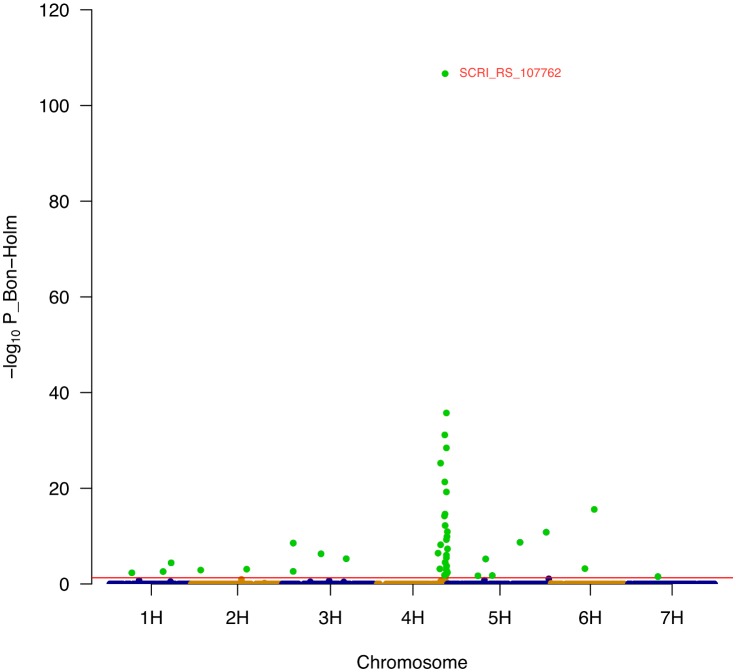
Based on the phenotypic data, we carried out a genome-wide association study on barley leaf sheath hairiness using 5,709 polymorphic SNP markers from the Infinium iSelect 9k SNP chip. In total, 44 significantly associated SNPs with P_Bonferroni-Holm<0.05 were detected (Table 3 and S2 Table). These SNPs were confounded to 16 QTL regions, present on all seven barley chromosomes (Fig 2 and S3 Table). The explained phenotypic variation (R2adj), calculated across all 44 significant SNPs, was equal to 0.84 and prediction ability (R2val) was 0.72 after cross-validation (Table 3). These findings indicate that the model used was able to explain an exceptionally large proportion of the phenotypic variation of leaf sheath hairiness in HEB-25.
Table 3. Summary of association mapping results for leaf sheath hairiness.
(a) Explained phenotypic variation based on linear regression of all 44 significant markers.
(b) Prediction ability obtained as an average of predicting 20% of lines in 100 cross-validation runs.
Fig 2. Manhattan plot for GWAS analysis of leaf sheath hairiness.
The y-axis shows the −log10 of Bonferroni-Holm-adjusted P-values. The horizontal red line (−log10 P_Bon-Holm = 1.3) indicates the significance threshhold of P_Bonferroni-Holm = 0.05. Green dots indicate signifcant SNPs exhibiting a Bonferroni-Holm corrected p-value <0.05.
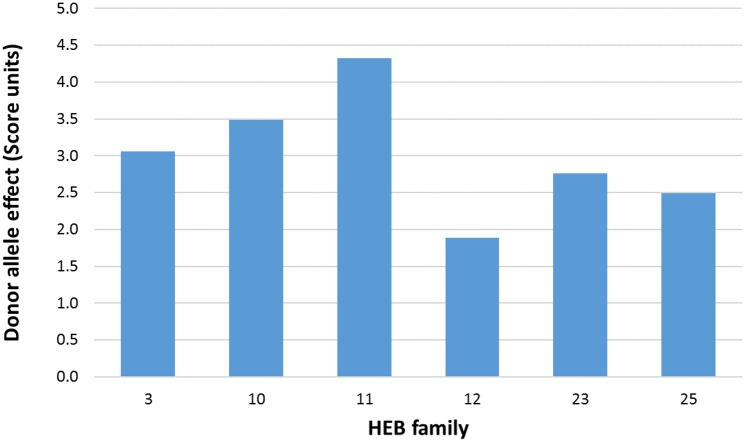
Among the 16 QTL, we detected a major QTL on chromosome 4H with high precision at position 111–113 cM. The most significant SNP, SCRI_RS_107762 at position 111.3 cM, revealed the highest Bonferroni-Holm adjusted -log10 P value of 107. The wild barley allele at this SNP had an increasing effect on leaf sheath hairiness equal to 2.54 score units relative to the Barke allele; in other words, the wild allele at this SNP increased leaf sheath hairiness score by 2.54 in comparison to the Barke leaf sheath hairiness score of 1. Clustering of a high number of 27 significant markers in this genomic region suggests the presence of family-specific effects [25]. Therefore, we calculated the cumulated wild barley donor-specific allele effect at the major QTL on chromosome 4H separately for each HEB family (Fig 3 and S3 Table). We found contrasting donor-specific effects ranging from 1.88 to 4.33 among the six polymorphic HEB families. The wild donor allele present in HEB family 11 conferred the highest effect on hairiness, where each line carrying a homozygous wild barley genotype is expected to have a hairiness score increased by 4.33 units compared to those lines carrying the homozygous Barke genotype.
Fig 3. Estimation of cumulated wild barley donor-specific allele effects at the major QTL on chromosome 4H (at 111.3 cM) for six leaf sheath hairness HEB families.
We used the BARLEYMAP pipeline [31] with the recently released barley sequence [32] to identify potential candidate genes, within a window of 3 cM from the peak marker SCRI_RS_107762, to explain the QTL. Here, we discuss the genes that might be candidates for leaf sheath hairiness based on their function and on literature reports linking the genes to hairiness. In the QTL region 4H-111.3 cM, pentatricopeptide repeat genes (HORVU4Hr1G085920.1) are present.
In plants, pentatricopeptide repeat (PPR) proteins constitute a large family of proteins characterized by a degenerate 35 amino acids motif in tandem repeats [34]. A large portion of these genes are targeted to mitochondria and chloroplasts [35] and are involved in organellar RNA processing [36–38]. Their roles also include seed development and growth [39], restoration of fertility in cytoplasmic male sterile plants[40], and response to abiotic stresses [41].
Interestingly, members of these gene families were found by Zeng et al. [42] when they fine mapped the glabrous leaf 6 (GL6) hairiness gene of Oryza sativa. In addition, Spyropoulou et al. [43] identified pentatricopeptides among the 30 most common InterPro entries in the RNAseq analysis of the Solanum lycopersicum trichomes.
Another candidate gene in that region is a subtilisin-like protease (HORVU4Hr1G085590). In fact, stomatal density and distribution 1–1 (SDD1) and abnormal leaf shape1 (ALE1) are two subtilisin-like proteases that were shown to be involved in the differentiation of epidermal cells in Arabidopsis: SDD1 protease is required in stomatal cell formation [44,45], while ALE1 plays a role in cuticle formation [46]. Furthermore, Liu et al. [47] and Luo et al. [48] showed that transgenic tobacco overexpressing Solaneum americanum SaPIN2b and SaPIN2a, which are serine protein inhibitors with SDD1 and ALE1 as possible targets, had significantly higher trichome density and increased branching. In addition, jasmonic acid-insensitive 1–1 (jai1-1) tomato mutants, which cannot produce PIN2, had significanly fewer glandular trichomes than wild type plants [49].
Both candidates lie within a 1 Mb interval in close proximity to the most significant SNP markers of the QTL region 4H-111.3 cM (Fig 4).
Fig 4. Physical map of the QTL region 4H-111.3cM including both candidate genes.
The position of the two most significant SNP markers is indicated as well as their wild allele effects on the hairiness score.
Patterson [12] and Taketa and Takeda [11] described leaf sheath hairiness in barley as a monogenic trait and also Cockram et al. [50] specified that hairiness of leaf sheath is associated with a single locus. However, Wang et al. [17] and our study revealed additional minor QTL besides the major 4H QTL (Fig 2 and S2 Table). The genes underlying these additional QTL could be involved in fine tuning rather than in initiation of the development of epidermal hairs.
Conclusions
This work showed that that the cumulation method [25] reliably identified QTL effects of hairy donors and that allelic series seem to exist in HEB-25. HEB-25 can, thus, be used as a tool for further analyses of leaf sheath hairiness in barley and for fine-mapping and, ultimately, cloning of the causative gene/s. In the future, functional characterization of hair development in HEB-25 and their potential to increase barley yields under harsh environmental conditions will be investigated. The high accuracy of QTL mapping due to the NAM population design and the Infinium iSelect 9k SNP chip already allowed narrowing down the position of the major QTL and defining potentially plausible candidate genes. As a strategy to clone the 4H-111.3 cM QTL in HEB-25 we propose to apply cloning by sequencing as outlined by Jost et al. [51]. For this, we will select HEB lines, which, in generation BC1S3, proved to be heterozygous at the most significant SNP, SCRI_RS_107762, and homozygous in the remaining genome background. After propagating these HEB lines to the current generation (BC1S3:9), two pools of sister lines could be selected, which are expected to be fixed in the genomic background and segregate 1:1 for leaf sheath hairiness. The resulting two sister pools will be used to select recombination events close to the SCRI_RS_107762 locus. Subsequently, these recombinants will be used for whole exome-capture resequencing to validate candidate genes [52]. The final identity of the true candidate gene, causing the 4H-111.3cM QTL effect on leaf sheath hairiness, could be achieved through a CRISP/Cas9-based targeted knock out of the candidate gene [53,54].
Supporting information
The value of “1” refers to non-hairy leaf sheaths, whereas “3”, “5” and “7” indicate leaf sheaths, which are scored as slightly hairy, medium hairy, and very hairy, respectively.
(PDF)
Red and white dots indicate origins of hairy and non-hairy donors of HEB-25, respectively.
(PDF)
The elongated hair, designated with letter (A), is the type of hair segregating in HEB-25 and phenotyped in this study. The two thorn-like hairs, designated with letter (B), indicate a second type of hair, which was found in all investigated HEB lines and donors.
(PDF)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
HEB-25 was genotyped with the Infinium iSelect 9k SNP chip due to the efforts of TraitGenetics GmbH, Gatersleben, Germany. We are furthermore grateful to Maria Umann for 3D microscopy work.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This project was funded by German Research Foundation (DFG) via Grants Pi339/7-1 and Pi339/8-1 (AM): http://www.dfg.de/; Interdisciplinary Centre for Crop Plant Research (IZN), Halle (VD): http://www.uni-halle.de/izn/; King Abdullah University of Science and Technology (KAUST) (SS): https://www.kaust.edu.sa/en; Ege University Erasmus Programme (BK): http://ebys.ege.edu.tr/ogrenci/ebp/llp-erasmus.htm.
References
- 1.Levin DA. Role of Trichomes in Plant Defense. Q Rev Biol. 1973;48: 3–15. [Google Scholar]
- 2.Werker E. Trichome diversity and development. Adv Bot Res. 2000;31: 1–35. [Google Scholar]
- 3.Johnson HB. Plant pubescence: An ecological perspective. Bot Rev. 1975;41: 233–58. [Google Scholar]
- 4.Ehleringer JR, Mooney HA. Leaf hairs: Effects on physiological activity and adaptive value to a desert shrub. Oecologia. 1978;37: 183–200. doi: 10.1007/BF00344990 [DOI] [PubMed] [Google Scholar]
- 5.Hameed M, Mansoor U, Ashraf M, Rao A. Variation in leaf anatomy in wheat germplasm from varying drought-hit habitats. Int J Agri Biol. 2002;4: 12–6. [Google Scholar]
- 6.Graham J, Smith K, Tierney I, MacKenzie K, Hackett CA. Mapping gene H controlling cane pubescence in raspberry and its association with resistance to cane botrytis and spur blight, rust and cane spot. Theor Appl Genet. 2006;112: 818–31. doi: 10.1007/s00122-005-0184-z [DOI] [PubMed] [Google Scholar]
- 7.Kim HJ, Han JH, Kim S, Lee HR, Shin JS, Kim JH, et al. Trichome density of main stem is tightly linked to PepMoV resistance in chili pepper (Capsicum annuum L.). Theor Appl Genet. 2011;122: 1051–8. doi: 10.1007/s00122-010-1510-7 [DOI] [PubMed] [Google Scholar]
- 8.Larkin JC, Oppenheimer DG, Lloyd AM, Paparozzi ET, Marks MD. Roles of the GLABROUS1 and TRANSPARENT TESTA GLABRA genes in Arabidopsis trichome development. Plant Cell. 1994;6: 1065–76. doi: 10.1105/tpc.6.8.1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marks MD. Molecular genetic analysis of trichome development in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol. 1997;48: 137–63. doi: 10.1146/annurev.arplant.48.1.137 [DOI] [PubMed] [Google Scholar]
- 10.Wang S, Chen JG. Arabidopsis transient expression analysis reveals that activation of GLABRA2 may require concurrent binding of GLABRA1 and GLABRA3 to the promoter of GLABRA2. Plant Cell Physiol. 2008;49: 1792–804. doi: 10.1093/pcp/pcn159 [DOI] [PubMed] [Google Scholar]
- 11.Taketa S, Takeda K. Expression of dominant marker genes of barley in wheat-barley hybrids. Genes Genet Syst. 1997;72: 101–6. [Google Scholar]
- 12.Patterson F. Independent inheritance of four characters in barley. Agron J. 1957;49: 218–9. [Google Scholar]
- 13.Pickering RA, Hill AM, Kynast RG. Characterization by RFLP analysis and genomic in situ hybridization of a recombinant and a monosomic substitution plant derived from Hordeum vulgare L. x Hordeum bulbosum L. crosses. Genome. 1997;40: 195–200. [DOI] [PubMed] [Google Scholar]
- 14.Korzun V, Malyshev S, Pickering RA, Börner A. RFLP mapping of a gene for hairy leaf sheath using a recombinant line from Hordeum vulgare L. x Hordeum bulbosum L. cross. Genome. 1999;42: 960–3. [Google Scholar]
- 15.Devos KM, Atkinson MD, Chinoy CN, Francis HA, Harcourt RL, Koebner RMD, et al. Chromosomal rearrangements in the rye genome relative to that of wheat. Theor Appl Genet. 1993;85: 673–80. doi: 10.1007/BF00225004 [DOI] [PubMed] [Google Scholar]
- 16.Dobrovolskaya O, Pshenichnikova TA, Arbuzova VS, Lohwasser U, Röder MS, Börner A. Molecular mapping of genes determining hairy leaf character in common wheat with respect to other species of the Triticeae. Euphytica. 2007;155: 285–93. [Google Scholar]
- 17.Wang M, Jiang N, Jia T, Leach L, Cockram J, Comadran J, et al. Genome-wide association mapping of agronomic and morphologic traits in highly structured populations of barley cultivars. Theor Appl Genet. 2012;124: 233–46. doi: 10.1007/s00122-011-1697-2 [DOI] [PubMed] [Google Scholar]
- 18.Wan HS, Yang YM, Li J, Zhang ZF, Yang WY. Mapping a major QTL for hairy leaf sheath introgressed from Aegilops tauschii and its association with enhanced grain yield in bread wheat. Euphytica. 2015;205: 275–85. [Google Scholar]
- 19.Maurer A, Draba V, Jiang Y, Schnaithmann F, Sharma R, Schumann E, et al. Modelling the genetic architecture of flowering time control in barley through nested association mapping. BMC Genomics. 2015;16: 290 doi: 10.1186/s12864-015-1459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lancashire PD, Bleiholder H, Vandenboom T, Langeluddeke P, Stauss R, Weber E, et al. A uniform decimal code for growth-stages of crops and weeds. Ann Appl Biol. 1991;119: 561–601. [Google Scholar]
- 21.Alqudah AM, Schnurbusch T. Heading date is not flowering time in spring barley. Front Plant Sci. 2017;8: 896 doi: 10.3389/fpls.2017.00896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Comadran J, Kilian B, Russell J, Ramsay L, Stein N, Ganal M, et al. Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat Genet. 2012;44: 1388–92. doi: 10.1038/ng.2447 [DOI] [PubMed] [Google Scholar]
- 23.Liu W, Gowda M, Steinhoff J, Maurer HP, Wurschum T, Longin CF, et al. Association mapping in an elite maize breeding population. Theor Appl Genet. 2011;123: 847–58. doi: 10.1007/s00122-011-1631-7 [DOI] [PubMed] [Google Scholar]
- 24.Würschum T, Liu W, Gowda M, Maurer HP, Fischer S, Schechert A, et al. Comparison of biometrical models for joint linkage association mapping. Heredity (Edinb). 2012;108: 332–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maurer A, Sannemann W, Léon J, Pillen K. Estimating parent-specific QTL effects through cumulating linked identity-by-state SNP effects in multiparental populations. Heredity (Edinb). 2017;118: 477–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarz G. Estimating Dimension of a Model. Ann Stat. 1978;6: 461–4. [Google Scholar]
- 27.Reif JC, Liu W, Gowda M, Maurer HP, Möhring J, Fischer S, et al. Genetic basis of agronomically important traits in sugar beet (Beta vulgaris L.) investigated with joint linkage association mapping. Theor Appl Genet. 2010;121: 1489–99. doi: 10.1007/s00122-010-1405-7 [DOI] [PubMed] [Google Scholar]
- 28.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6: 65–70. [Google Scholar]
- 29.Turner SD. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv. 2014; http://dx.doi.org/10.1101/005165. [Google Scholar]
- 30.Utz HF, Melchinger AE, Schon CC. Bias and sampling error of the estimated proportion of genotypic variance explained by quantitative trait loci determined from experimental data in maize using cross validation and validation with independent samples. Genetics. 2000;154: 1839–49. [PMC free article] [PubMed] [Google Scholar]
- 31.Cantalapiedra CP, Boudiar R, Casas AM, Igartua E, Contreras-Moreira B. BARLEYMAP: physical and genetic mapping of nucleotide sequences and annotation of surrounding loci in barley. Mol Breeding. 2015;35: [Google Scholar]
- 32.Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, et al. A chromosome conformation capture ordered sequence of the barley genome. Nature. 2017;544: 427–33. doi: 10.1038/nature22043 [DOI] [PubMed] [Google Scholar]
- 33.Grace J, Russell G. Effect of Wind on Grasses: III. Influence of Continuous Drought or Wind on Anatomy and Water Relations in Festuca arundinacea Schreb. J Exp Bot. 1977;28: 268–78. [Google Scholar]
- 34.Saha D, Prasad AM, Srinivasan R. Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiol Bioch. 2007;45: 521–34. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura T, Schuster G, Sugiura M, Sugita M. Chloroplast RNA-binding and pentatricopeptide repeat proteins. Biochem Soc Trans. 2004;32: 571–4. doi: 10.1042/BST0320571 [DOI] [PubMed] [Google Scholar]
- 36.Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16: 2089–103. doi: 10.1105/tpc.104.022236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Small ID, Peeters N. The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25: 46–7. [DOI] [PubMed] [Google Scholar]
- 38.Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15: 1480–95. doi: 10.1105/tpc.010397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutierrez-Marcos JF, Dal Pra M, Giulini A, Costa LM, Gavazzi G, Cordelier S, et al. empty pericarp4 encodes a mitochondrion-targeted pentatricopeptide repeat protein necessary for seed development and plant growth in maize. Plant Cell. 2007;19: 196–210. doi: 10.1105/tpc.105.039594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bentolila S, Alfonso AA, Hanson MR. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci U S A. 2002;99: 10887–92. doi: 10.1073/pnas.102301599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma S, Gong Q, Bohnert HJ. Dissecting salt stress pathways. J Exp Bot. 2006;57: 1097–107. doi: 10.1093/jxb/erj098 [DOI] [PubMed] [Google Scholar]
- 42.Zeng YH, Zhu YS, Lian L, Xie HG, Zhang JF, Xie HA. Genetic analysis and fine mapping of the pubescence gene GL6 in rice (Oryza sativa L.). Chinese Sci Bull. 2013;58: 2992–9. [Google Scholar]
- 43.Spyropoulou EA, Haring MA, Schuurink RC. RNA sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genomics. 2014;15: 402 doi: 10.1186/1471-2164-15-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berger D, Altmann T. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 2000;14: 1119–31. [PMC free article] [PubMed] [Google Scholar]
- 45.Von Groll U, Berger D, Altmann T. The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell. 2002;14: 1527–39. doi: 10.1105/tpc.001016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka H, Onouchi H, Kondo M, Hara-Nishimura I, Nishimura M, Machida C, et al. A subtilisin-like serine protease is required for epidermal surface formation in Arabidopsis embryos and juvenile plants. Development. 2001;128: 4681–9. [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Xia KF, Zhu JC, Deng YG, Huang XL, Hu BL, et al. The nightshade proteinase inhibitor IIb gene is constitutively expressed in glandular trichomes. Plant Cell Physiol. 2006;47: 1274–84. doi: 10.1093/pcp/pcj097 [DOI] [PubMed] [Google Scholar]
- 48.Luo M, Wang Z, Li H, Xia KF, Cai Y, Xu ZF. Overexpression of a weed (Solanum americanum) proteinase inhibitor in transgenic tobacco results in increased glandular trichome density and enhanced resistance to Helicoverpa armigera and Spodoptera litura. Int J Mol Sci. 2009;10: 1896–910. doi: 10.3390/ijms10041896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, et al. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell. 2004;16: 126–43. doi: 10.1105/tpc.017954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cockram J, White J, Zuluaga DL, Smith D, Comadran J, Macaulay M, et al. Genome-wide association mapping to candidate polymorphism resolution in the unsequenced barley genome. Proc Natl Acad Sci U S A. 2010;107: 21611–6. doi: 10.1073/pnas.1010179107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jost M, Taketa S, Mascher M, Himmelbach A, Yuo T, Shahinnia F, et al. A homolog of Blade-On-Petiole 1 and 2 (BOP1/2) controls internode length and homeotic changes of the barley inflorescence. Plant Physiol. 2016;171: 1113–27. doi: 10.1104/pp.16.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mascher M, Jost M, Kuon JE, Himmelbach A, Assfalg A, Beier S, et al. Mapping-by-sequencing accelerates forward genetics in barley. Genome Biol. 2014;15: R78 doi: 10.1186/gb-2014-15-6-r78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41: e188 doi: 10.1093/nar/gkt780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li M, Li X, Zhou Z, Wu P, Fang M, Pan X, et al. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci. 2016;7: 377 doi: 10.3389/fpls.2016.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The value of “1” refers to non-hairy leaf sheaths, whereas “3”, “5” and “7” indicate leaf sheaths, which are scored as slightly hairy, medium hairy, and very hairy, respectively.
(PDF)
Red and white dots indicate origins of hairy and non-hairy donors of HEB-25, respectively.
(PDF)
The elongated hair, designated with letter (A), is the type of hair segregating in HEB-25 and phenotyped in this study. The two thorn-like hairs, designated with letter (B), indicate a second type of hair, which was found in all investigated HEB lines and donors.
(PDF)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.