Abstract
Background:
MicroRNA deregulation may occur during hepatocellular carcinoma (HCC) genesis and progression stages. MicroRNA-34a (miR-34a) functions as a tumor suppressor and is down-regulated or silenced in a variety of human cancers, while heat shock proteins (Hsps) play important roles in assisting protein folding and preventing both protein aggregation and transport across membranes. The present study aimed to evaluating serum expression of miR-34a and its target Hsp70 for early detection of HCC in patients with liver cirrhosis (LC), focusing on correlations with clinicopathological features.
Methods:
A total of 180 patients were included: 120 with HCC on top of LC (60 with either early or late HCC) and 60 patients with HCV-related LC. In addition, 60 healthy individuals were considered as controls. Real-time polymerase chain reactions were performed for expression profiling of serum miR-34a and Hsp70 and for allelic discrimination of the promotor variant (rs2763979, C/T). In addition, in silico analysis was carried out.
Results:
All participants were heterozygote for the promotor polymorphism. miR-34a serum levels were significantly under-expressed in LC and especially HCC patients as compared to controls. Associations with a high Child-Turcotte-Pugh (CTP) score, advanced cancer stage, and number of masses were noted. In contrast the target Hsp70 was significantly overexpressed in cancer patients but not in LC group and inversely correlated with miR-34a levels.
Conclusion:
Utility of circulating miRNAs as biomarkers for early detection of HCC was raised. Future large-scale studies are warranted to confirm the current findings.
Keywords: Hepatocellular carcinoma, Hsp-70, liver cirrhosis, microRNA-34a, real-time polymerase chain reaction
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer in men and the seventh in women with high mortality and morbidity, especially in patients with liver cirrhosis (LC) (Wang et al., 2015). Monitoring of patients with LC for early detection of tumor transformation is of great importance. The incidence of HCC in the target population, the availability of efficient diagnostic tests, and effective treatment influence the success of monitoring (Antaki et al., 2010). Recently, early detection of HCC using new biogenetic markers has been valued.
MicroRNAs (miRNAs) are small non-coding ribonucleic acid molecules which target multiple messenger RNAs at their 3’ end and cause either translation repression or degradation (Toraih et al., 2016a). They play a key role in many essential biological processes and are stably secreted in extracellular spaces and body fluids (Toraih et al., 2015). The involvement of miRNAs in tumor development and progression is well established, as they can behave as tumor suppressors or potentiate oncogenesis depending on the cellular function of their targets (Otsuka et al., 2014). Deregulation of circulatory miRNAs and their targets was previously documented in various tumors (Afonso et al., 2016).
MicroRNA-34a (miR-34a) was found to be up-regulated in patients with nonalcoholic fatty liver disease (NAFLD) and hepatitis C virus (HCV) infection (Cermelli et al., 2011) and down-regulated in multiple tumors including hepatocellular carcinoma (Budhu et al., 2008), neuroblastoma (Welch et al., 2007), and non-small cell lung cancer (Bommer et al., 2007). MiR-34a regulates tumor suppression, controls the expression of a plethora of target proteins involved in cell cycle, differentiation and apoptosis, and antagonizes the necessary processes for basic cancer cell viability as well as cancer stemness, metastasis, and chemoresistance (van der Meer et al., 2012). The heat shock proteins (Hsps) are inducible chaperons produced in response to various physiologically and pathologically stressful conditions, including carcinogenesis (Mosser and Morimoto, 2004) The role of the Hsps in the tumorigenesis has been implicated in the regulation of cell cycle progression and apoptosis (van der Meer et al., 2012), in multidrug resistance (Ciocca et al., 1999), and as a modulator of p53 function (Levine et al., 1991). Hsp70; a major member in Hsp families, aids in stabilization of existing proteins against aggregation and mediates the folding of newly translated proteins in the cytosol and within organelles (Radons, 2016). Studies have linked Hsp70 to several cancer types, with overexpression levels to be associated with metastasis, poor prognosis, and therapeutic resistance (Radons, 2016).
Hsp70 chaperon was predicted to be targeted by miR-34a by computational tools. Understanding of the role and alterations of miRNA-mRNA complex in cellular functions and disease processes in the liver provides a strong basis for targeting miRNAs in treatment development. Most cases of HCC are associated with chronic liver diseases (hepatitis B or C viral infection) where the processes of chronic inflammation and fibrosis act as a stressful condition. Heat shock proteins induced in response to this stress condition may contribute to hepatocarcinogenesis. As there are few comprehensive studies of the expression of serum Hsp70 and correlation with miR-34a in HCC patients, the present study aimed at evaluating the role of serum miR-34a and Hs-70 in early detection of patients with HCC on top of liver cirrhosis, and evaluating their expression profiles according to the Child-Turcotte-Pugh (CTP) score, Barcelona-Clinic Liver Cancer (BCLC) staging system, and number of masses.
Materials and Methods
Study participants
One hundred eighty patients (60 patients with HCV-related LC and 120 patients with HCC on top of LC) and 60 controls were enrolled in the study. Patients were recruited from the outpatient clinic of Tropical Medicine and Gastroenterology Department, Faculty of Medicine, Assuit, Egypt, during the period between November 2014 and August 2015. All patients had chronic hepatitis C infection with detectable anti-HCV for more than 6 months and/or HCV RNA in serum and had clinical evidence suggestive of LC and/or significant ultrasound findings. Patients positive for hepatitis B surface antigen (HBsAg) or serologic evidence of other chronic liver diseases as autoimmune hepatitis or metabolic diseases (as Hemochromatosis or Wilson disease) were excluded. The LC patients’ group had no evidence of cancer by initial screening and no history of alcohol intake or other potential hepatotoxic drug exposure. In HCC patient groups, HCC was diagnosed based on compatible clinical background (association with LC or viral hepatitis) with typical imaging findings and increase HCC-related tumor marker; alpha fetoprotein (AFP). Patients underwent thorough clinical evaluation and hepatic imaging (ultrasonography, computed tomography, arteriography, or magnetic resonance imaging). HCC patients were sorted into three groups according to the BCLC staging system (stage 0: very early stage, stage A/B: early to intermediate stage, stage C/D: advanced and end stage disease). All patients’ disease severity was scored according to the new CTP (Lee et al., 2014). The study was conducted according to the ethical guidelines of the 1964 Declaration of Helsinki (2008 revision), and approved by the Medical Research Ethics Committee of Assuit University. Written informed consent was obtained from all participants.
Specimen collection and biochemical analysis
Blood samples were collected from fasting individuals at the afternoon in Vacuette EDTA K3E blood bottles (Grenier Bio-one) and Vacutainer Serum Separator Tubes II (Becton Dickinson Plymouth). Whole blood was used for DNA extraction and Single nucleotide polymorphism (SNP) identification. The serum was separated and aliquoted into 1.5-mL Ribonuclease- free Eppendorf tubes and stored at -80ºC to be used for mRNA and miRNA expression analysis. Samples were allowed to undergo one freeze-thaw cycle only. Other laboratory tests done included: liver chemistry panel, complete blood count, prothrombin time, and alpha feto-protein.
MicroRNA-34a expression analysis
The total RNA, including small RNA, was isolated from serum using the Qiagen miRNeasy serum/plasma Kit (Qiagen, cat no 217184) following the protocol supplied by the manufacturer. Concentration of RNA was determined using the NanoDrop ND-1000 spectrophotometer (NanoDrop Tech., Inc. Wilmington, DE, USA) with the wavelength-dependent extinction coefficient ‘33’. The complementary DNA (cDNA) was prepared from total RNA (10 ng) using TaqManTM MicroRNA Reverse Transcription (RT) kit (P/N 4366596; Applied Biosystems, Foster City, CA, USA) and either the miR-34a-5p specific stem–loop primer (assay ID 000426) or the endogenous control RNU6B primer (assay ID 001093) according to the methodology described in Toraih et al.4 Reverse transcription was carried out in a T-Professional Basic, Biometra Polymerase Chain Reaction System (Biometra, Goettingen, Germeny) at 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min, then was held at 4°C. A negative control was included in each experiment, to ensure that PCR products were not due to contamination by genomic DNA. The PCR reactions were carried out in the final volumes of 20 µl, including 1.33 µl RT products, 2× TaqMan® Universal PCR Master Mix, 1 µl TaqMan® small RNA assay. No-template was included as a negative control. The PCR was performed on StepOne™ Real-Time PCR System (Applied Biosystems) and was incubated as follows: 95°C for 10 min followed by 40 cycles of 92°C for 15 seconds and 60°C for 1 minute.
Target gene heat shock protein-70 expression analysis
High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, P/N 4368814) was used for RT reaction. For each 20 µl RT reaction, 10 µl (10 ng) RNA sample was combined with 10 µl of 2x RT reaction mix containing 2 µl of 10x RT Buffer, 0.8 µl of 25x dNTP Mix (100 mM), 2 µl of 10x RT random primers, 1 µl of MultiScribe™ Reverse Transcriptase, 1 µl of RNase inhibitor, and 3.2 µl of nuclease-free water. Reverse transcription was carried out in a Mastercycler Gradient Thermocycler (Eppendorf, Hamburg, Germany) at 25 °C for 10 min, followed by 37 °C for 120 min, and finally 85 °C for 5 min, then hold at 4 °C. Appropriate positive and negative controls were included in each experiment. Expression of Hsp70 (HSPA1A and HSPA1B) was quantified using the TaqMan® assay (Applied Biosystems, assay ID Hs00271244_s1), TaqMan® Endogenous control assay for Glyceraldhyde 3 Phosphate dehydrogenase (GAPDH) (Applied Biosystems, catalogue no 402869), and Taqman® Universal PCR master mix II, No UNG (2×) (Applied Biosystems, P/N 4440043). The PCR reactions were carried out in the final volume of 20 µl, including 1.33 µl RT product, 2× TaqMan® Universal PCR Master Mix, 1 µl TaqMan®assays. The PCR profile was as follows: 95 °C for 10 min followed by 40 cycles of 92 °C for 15 seconds, and 60 °C for 1 min. All reactions were run in duplicate and included a no-template (with water instead of cDNA) and a no-reverse transcriptase controls.
Hsp-70 SNP identification
Genomic DNA was extracted from blood leukocytes using QIAamp DNA Blood Mini (QIAGEN, cat. No. 51104) according to the manufacturer’s instructions. Concentration and purity of the extracted DNA were measured by NanoDrop ND-1000 (NanoDrop Tech., Inc. Wilmington, DE, USA). Extracted DNA was genotyped for the Hsp70 promotor polymorphism (rs2763979; C/T) using Real-Time PCR allelic discrimination technology. PCR was performed in a 25-µl reaction volume containing 12.5 μL (2x)Taqman® Universal PCR Master Mix (cat. No. 4440043), 1.25 µl 20 x TaqMan® SNP Genotyping Assay Mix (Applied Biosystems, Foster City, CA, USA, C_3052606_10), and 20 ng genomic DNA diluted to 11.25μL with Nuclease-free water (Toraih et al., 2016a). Real-time PCR amplification was performed on StepOne™ Real-Time PCR System (Applied Biosystems) using the following conditions: an initial hold (95°C for 10 min) followed by a 40-cycle two-step PCR (95°C denaturation for 15 s and annealing/extension 60°C for 1 min). Appropriate negative controls were used. Allelic discrimination was called by the SDS software version 1.3.1 (Applied Biosystems).
Identification of target genes and pathway enrichment analysis
The list of computationally-predicted gene targets for miR-34a-3p and 5p were obtained from DIANA-microT-CDS v5.0 database (http://www.microrna.gr/microT-CDS) and miRDB (http://mirdb.org), TargetScanHuman v6.2 (http://www.targetscan.org/), and miRNAMap v2.0 (Hsu et al., 2006). The predicted miRNA target genes were analyzed for gene ontology (GO) terms and Kyoto encyclopedia of genes and genomes (KEGG) enrichment pathways using DIANA-miRPath v2.0 web-server (Vlachos et al., 2012), to merge all the predicted and experimentally validated miRNA targets (in CDS, 3’UTR, or 5’UTR regions) into significantly selected KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways according to their biological function.
Statistical analysis
Data were managed using the “Statistical Package for the Social Sciences (SPSS) for windows” software (version 20.0). Categorical variables were compared using the chi-square or Fisher’s exact tests where appropriate, while Mann-Whitney U (MW) and Kruskal-Wallis (KW) tests were used to compare continuous variables. Correlation analysis using Spearman rank correlation (rs) and Pearson’s correlation coefficient were performed where appropriate. A two-tailed P-value of < 0.05 was considered statistically significant. The allele frequency within each group was determined as the number of occurrences of an individual allele divided by the total number of alleles (Toraih et al., 2016b). Hardy-Weinberg equilibrium (HWE) was evaluated using the Online Encyclopedia for Genetic Epidemiology (OEGE) software (http://www.oege.org/software/hwe-mr-calc.shtml) and tested by c2 test to compare the expected genotype frequencies among patient and control groups. Odds ratio (OR) with a 95% confidence interval (CI) was calculated to investigate the strength of the association between the SNP and the susceptibility to LC and HCC. Binary logistic regression analysis (enter method) was performed to adjust the mutual effect of miR-34a polymorphism and expression levels. The fold change of miRNA and mRNA expression in each patient relative to the normal controls was calculated via Livak method based on the threshold cycle (CT) value with the following equation: relative quantity = 2−ΔΔCT (Toraih et al., 2015).
Results
Baseline demographic characteristics of control and patient groups are shown in Table 1.
Table 1.
Demographic, Clinical, and Biochemical Characteristics of the Study Groups
| Variables | Controls | LC | Early HCC | Late HCC |
|---|---|---|---|---|
| Number | 60 | 60 | 60 | 60 |
| Age | ||||
| Age, mean (SD) | 53.8 ± 6.6 | 54.9 ± 7.2 | 55.2 ± 5.8 | 48.2 ± 8.3 |
| Age groups (y) | ||||
| <55 y | 36 (60) | 30 (50) | 37 (61.7) | 57 (95) |
| ≥55 y | 24 (40) | 30 (50) | 23 (38.3) | 3 (5) |
| Gender | ||||
| Females | 24 (40) | 10 (16.7) | 8 (13.3) | 44 (73.3) |
| Males | 36 (60) | 50 (83.3) | 52 (86.7) | 16 (26.7) |
| CTP class | ||||
| Child A | ---- | ---- | 60 (100) | ---- |
| Child B | ---- | 16 (26.7) | ---- | ---- |
| Child C | ---- | 44 (73.3) | ---- | 60 (100) |
| Tumor stage | ||||
| Stage 0 | ---- | ---- | 20 (33.3) | ---- |
| Stage A | ---- | ---- | 40 (66.7) | ---- |
| Stage B | ---- | ---- | ---- | 8 (13.3) |
| Stage C | ---- | ---- | ---- | 20 (33.3) |
| Stage D | ---- | ---- | ---- | 32 (53.3) |
| Number of masses | ||||
| Solitary | ---- | ---- | 16 (26.7) | 4 (6.7) |
| Multiple | ---- | ---- | 44 (73.3) | 56 (93.3) |
| Lab investigations | ||||
| ALT (U/L) | 38.6 ± 4.9 | 77.3 ± 11.4 | 69.8 ± 13.4 | 70.4 ±10.8 |
| AST (U/L) | 37.3 ± 4.5 | 66.8 ± 16.8 | 75.8 ± 11.1 | 72.7 ±10.3 |
| ALP (U/L) | 124.9 ± 6.3 | 157.5 ± 27.3 | 123.1 ± 8.9 | 131.7 ±7.8 |
| Bilirubin (mg/dl) | 0.89 ± 0.9 | 2.33 ± 2.0 | 2.23 ± 5.7 | 8.22 ± 2.5 |
| Albumin (g/L) | 6.5 ± 0.2 | 4.26 ± 1.1 | 4.85 ± 1.4 | 1.31 ± 0.3 |
| PC (%) | 97.6 ± 3.4 | 57.8 ± 7.96 | 81.72 ± 9.1 | 51.73 ± 5.6 |
| PT (sec) | 11.4 ± 0.9 | 15.2 ± 2.0 | 12.7 ± 2.0 | 21.4 ± 2.8 |
| AFP (IU/ml) | 4.6 ± 2.5 | 80.0 ± 30.7 | 460.8 ± 194.1 | 1138 ± 1359 |
| HB (g/dL) | 12.08 ± 0.9 | 9.5 ± 1.3 | 10.16 ± 1.59 | 9.68 ± 1.4 |
| WBC (x109/L) | 5.86 ± 1.9 | 5.83 ± 1.9 | 3.37 ± 1.8 | 4.2 ± 2.2 |
| PLT (x109 /L) | 157.8 ± 27.8 | 91.26 ± 14.1 | 92.7 ± 12.5 | 85.6 ± 9.7 |
Data are shown as number (percentage) or mean ± SD); LC, liver cirrhosis; HCC, hepatocellular carcinoma patients; CTP, Child-Turcotte-Pugh classification; BCLC, Barcelona-Clinic Liver Cancer staging system for HCC patients; ALT, alanine transaminase; AST, aspartate transaminase; ALP, alkaline phosphatase; PC, prothrombin concentration; PT, prothrombin time; AFP, alpha fetoprotein; HB, hemoglobin count; WBC, while blood cells; PLT, platelet count.
MiR-34a and Hsp-70 levels
The serum miR-34a level was significantly under-expressed in LC and HCC patients compared to control group (P-value ≤ 0.001), with more down-regulation in late HCC patients [0.42 (0.34-0.93)] compared to early HCC group [0.88 (0.72-1.41)]. On the other hand, its target gene Hsp70 was significantly overexpressed in cancer patients but not in the LC group compared to controls. Expression levels in LC were 0.99 (0.78-2.07), whereas the median and quartile levels in early and late HCC patients were 1.49 (0.61-2.8) and 2.03 (0.57-3.44), respectively; Figure 1A-C. Stratified analysis by age and gender showed no differential effect of them. Analysis of Hsp70 promotor polymorphism (rs2763979; C/T) showed heterozygosity of all patients and controls in the current study population, thus did not render any effect on Hsp70 expression levels.
Figure 1.
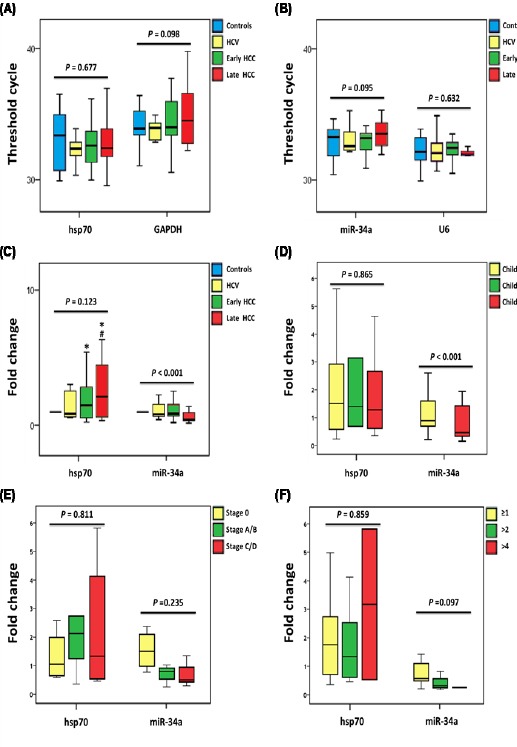
Serum MicroRNA-34a and hsp70 Expression Levels in Healthy Controls and Patients with Hepatitis C Virus and Hepatocellular Carcinoma. Values are represented as medians. The box defines upper and lower quartiles (25% and 75%, respectively) and the error bars indicate upper and lower adjacent limits. Kruskal-Wallis, One-way ANOVA followed by post hoc Bonferroni tests were used. (*) compared to controls, (#) compared to HCV group. (A) Cycle threshold values of Hsp70 and its endogenous control GAPDH. (B) Cycle threshold values of miR-34a and its endogenous control RNU6B. (C) Relative expression levels of Hsp70 and miR-34a in the study groups calculated by LIVAC method. (D) Fold change of Hsp70 and miR-34a in cancer patients according to Child-Turcotte-Pugh score. (E) Fold change of Hsp70 and miR-34a in cancer patients according to Barcelona-Clinic Liver Cancer staging system. (F) Fold change of Hsp70 and miR-34a in cancer patients, according to number of masses.
Association of miR-34a and Hsp70 with clinic-pathological characteristics
Association of miR-34a and its target Hsp70 with clinicopathological features are demonstrated in Table 2 and Figure 1. Most LC patients had Child score C while less than one third had B score. Comparing these two categories of patients revealed a significant lower expression of serum miR-34a in Child C compared to Child B patients (P-value < 0001). Regarding cancer groups, all early HCC patients had Child A score, while the late HCC group were classified as Child C. Our results showed differential expression of miR-34a among HCC patients with different Child scores (P-value < 0.001); Figure 1D. Serum miR-34a expression tended to decline with more advanced cancer stage (P-value = 0.001) and a higher number of liver masses (P-value = 0.001). On the contrary, its target gene Hsp70 expression showed significant up-regulation with cancer patients with a number of masses more than 4 (P-value = 0.011).
Table 2.
Association Analysis between Expression Levels of Serum Mir-34a and Hsp-70 and the Clinicopathological Features
| Variables | miR-34a | P value | Hsp70 | P value |
|---|---|---|---|---|
| CTP Score | ||||
| Child A | 0.88 (0.72-1.41) | < 0.001* | 1.49 (0.60-2.84) | 0.865 |
| Child B | 6.92 (6.02-6.99) | 1.38 (0.68-3.10) | ||
| Child C | 0.46 (0.33-1.42) | 1.26 (0.61-2.62) | ||
| Cancer stage | ||||
| Stage 0 | 1.59 (1.11-2.11) | < 0.001* | 1.04 (0.60-1.78) | 0.25 |
| Stage A/B | 0.77 (0.47-0.92) | 2.28 (1.26-2.98) | ||
| Stage C/D | 0.44 (0.35-0.95) | 1.36 (0.44-4.58) | ||
| Number of masses | ||||
| 1/2 | 0.92 (0.75-1.95) | 0.001* | 1.84 (0.64-2.98) | 0.011* |
| 3/4 | 0.42 (0.31-0.88) | 1.36 (0.57-2.58) | ||
| 5/6 | 0.35 (0.23-0.54) | 3.48 (0.43-6.52) | ||
Data are presented as median (quartiles). Kruskal-Wallis test was used.
Statistical significance at P < 0.05.
CTP, Child-Turcotte-Pugh; cancer stage in HCC patients according to Barcelona-Clinic Liver Cancer staging system.
MiR-34a-Hsp70 interaction
Our in silico analysis predicted that Hsp70 genes (HSPA1A and HSPA1B) to be one of the targets of miR-34a. Figure 2B showed direct interaction between conserved 8-mer miR-34a seed region and 3’ end of HSPA1A and HSPA1B with mirSVR score of -0.2131 and -1.1247, respectively, that combines alignment-based and energy-based algorithm (http://www.microRNA.org). In addition, Spearman’s rank correlation analysis revealed a moderate inverse relationship between miR-34a level and that of Hsp70 (r = -0.228, P = 0.003, R2 = 0.128); Figure 2C.
Figure 2.
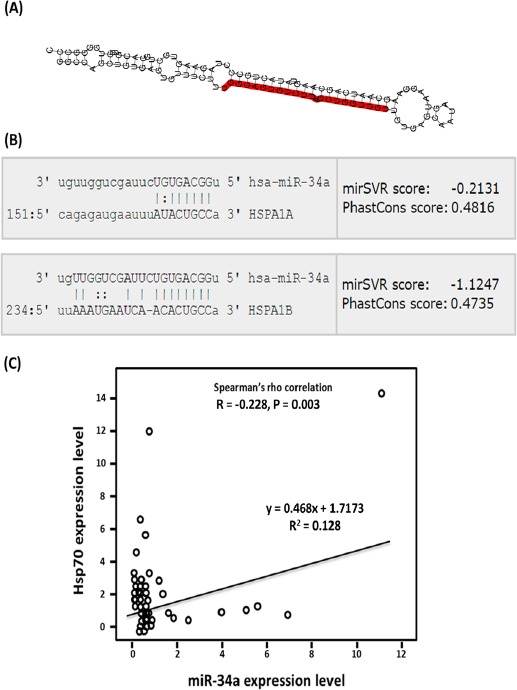
Correlation between Serum Expression Levels of miR-34a and Its Target Hsp70. (A) Secondary structure of pre-miR-34a. Both mature forms of miR-34a are illustrated (http://www.mirbase.org/). (B) Alignment of miR-34a and Hsp70 (HSPA1A and HSPA1B) interactions (http://www.microrna.org/). (C) Spearman’s rank correlation coefficient between the expression levels of miR-34a and Hsp70 in LC and cancer patients.
Discussion
Circulating miRNAs exhibit several advantages as potentially novel and ideal diagnostic and prognostic biomarkers (Morishita and Masaki, 2015); including stability in body fluids, resistance to endogenous RNase digestion, and easy analysis with sensitive methods as RT-PCR (Schwarzenbach et al., 2014; Arrese et al., 2015; Toraih et al., 2015). In the present study, we evaluated the role of the circulatory miR-34a and HSP-70 as biomarkers for early detection of HCC on top of LC and their relation to Child-Turcotte-Pugh score, Barcelona-Clinic Liver Cancer staging system, and number of masses. Our results revealed down-regulation of serum miR-34a in HCC patients compared to controls and LC patients, with more lower expression in late HCC than in an early HCC group. In addition, lower miR-34a expression levels were significantly associated with poor clinical outcome in cancer patients; higher Child score, advanced cancer stage, and more liver masses, reflecting the suppressive role of miRNA-34a on both development and progression of HCC.
Our in silico analysis demonstrated that miR-34a can regulate the expression of hundreds of genes and several essential cellular pathways including proliferation, apoptosis, and tumorigenesis. In prior studies, miR-34a was also down-regulated in various tumors (Li et al., 2011; Zhao et al., 2015). It was suggested to be a potent tumor suppressor gene. Ectopic overexpression of miR-34a in many tumor cell lines caused reactivation of apoptotic pathways and cell cycle arrest (Chen and Hu, 2012). Elevated expression of mir-34a in the study of Sun et al., (2008), induced a significant G1 cell cycle arrest via suppressing cyclin D1 (CCND1) and cyclin-dependent kinase 6 (CDK6) genes. Transfection of miR-34a in human glioma and medulloblastoma cells down-regulated multiple oncogenes as c-Met, Notch-1, Notch-2, and CDK6 (Li et al., 2009). Low expression levels of the tumor suppressor miR-34a renders activation of growth factor signaling transduction via ARAF (A-Raf Proto-Oncogene, serine/threonine kinase) and PIK3R2 (phosphoinositide-3-kinase, regulatory subunit 2), and regulates cell cycle progression by suppressing cyclins D3 and G2, E2F3 (eukaryotic transcription factor 3), SMAD4 [homologs of both the Drosophila protein, mothers against decapentaplegic (MAD) and the Caenorhabditis elegans protein SMA (for small body size)], MCM2 (minichromosome maintenance complex component 2) and MCM5 genes in cancer cell lines (Lal et al., 2011). In addition miR-34a was reported to target genes which inhibit apoptosis as BCL2 (B cell leukemia/lymphoma 2) (Bommer et al., 2007), enhance transcription of [MYB (proto-oncogene), HNF4A (hepatocyte nuclear factor 4 alpha) and FOXP1 (forkhead box P1)], (Rao et al., 2010; Takagi et al., 2010) and promote stem cell survival [WNT1 (Wingless-Type MMTV Integration Site Family, Member 1), DLL1 (Delta-like ligand 1), LEF1 (Lymphoid Enhancer Binding Factor 1), NOTCH1 (Notch homolog 1, translocation-associated), NOTCH2, JAG1 (Jagged 1), CD44 (Cluster of Differentiation 44)]. Moreover, growing evidence has shown that miR-34a is involved in metastasis and angiogenesis. MiR-34a causes silencing of ZO-1 (Zonula occludens-1), Occludin and Claudin-5 expression in glioma endothelial cells (Zhao et al., 2015).
In the current study, we identified Hsp70 to be one of the gene targets of miR-34a by different target prediction algorithms. This finding was supported in our lab by the inverse correlation between miR-34a and Hsp70 expression profiles. Serum Hsp70 levels was up-regulated among cancer patients but not in the LC group compared to controls, with the highest levels detected in late HCC patients and with those having large number of liver masses. In previous studies, Hsp70 were elevated in many types of cancer, including breast, colon, liver, prostate and esophageal carcinomas (Ciocca et al., 1999; Hwang et al., 2003; Joo et al., 2005; Alaiya et al., 2001; Tang et al., 2005; Sherman and Gabai, 2015). Similarly, Gehrmann et al., (2014), reported significantly higher expression of Hsp70 in the sera of HCC patients compared to healthy subjects and hepatitis patients. Wang et al., (2015) study revealed Hsp70 overexpression levels in HCC more than those of LC and controls with high sensitivity and specificity. Chuma et al., (2003), found Hsp-70 to be the most abundantly up-regulated gene in early HCC and a good sensitive biomarker that differentiate early malignancy from precancerous or non-cancerous liver. Shin et al., (2011), suggested Hsp70 to be an independent predictor of prognosis in malignancy. Hsp70 gene is normally expressed at low levels, and is induced under stress conditions to protect cells from proteotoxic effect associated with rapid cell proliferation during malignant transformation and to confer resistance to apoptotic cell death induced by chemo- and radiotherapy (Radons, 2016). In cancer cells, more Hsp70 molecules occur in the plasma membrane and are actively secreted into the blood circulation in lipid vesicles (Gehrmann et al., 2014). Based on these findings, our results support the evidence that Hsp70 have a key role in the development and progression of HCC thus could be a putative biomarker for early and late HCC.
The current study has some limitations, including that the number of cases was relatively small and the study explored the effect of only one member of miRNAs, which could reduce its accuracy for early detection of HCC and monitoring the progression. However, our study has the advantage to be the first one to explore the role of miRNA-34a/Hsp70 axis in HCC disease. We recommend a larger scale multi-center studies exploring a panel of miRNAs instead of a single one for a more sound conclusions and recommendations regarding surveillance for HCC. In conclusion, this study confirms that miRNA-34a along with Hsp-70 could be potential biomarkers for both development and progression of HCC among patients with HCV-related liver cirrhosis. Future prospective studies are required to confirm these findings.
References
- Afonso MB, Rodrigues PM, Simão AL, Castro RE. Aravalli RN, editor. Circulating microRNAs as potential biomarkers in Non-Alcoholic Fatty Liver Disease and Hepatocellular Carcinoma. J Clin Med. 2016;5:E30. doi: 10.3390/jcm5030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaiya AA, Oppermann M, Langridge J, et al. Identification of proteins in human prostate tumor material by two-dimensional gel electrophoresis and mass spectrometry. Cell Mol Life Sci. 2001;58:307–11. doi: 10.1007/PL00000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antaki N, Craxi A, Kamal S, et al. The neglected hepatitis C virus genotypes 4, 5, and 6:an international consensus report. Liver Int. 2010;30:342–55. doi: 10.1111/j.1478-3231.2009.02188.x. [DOI] [PubMed] [Google Scholar]
- Arrese M, Eguchi A, Feldstein AE. Circulating microRNAs:emerging biomarkers of liver disease. Semin Liver Dis. 2015;35:43–54. doi: 10.1055/s-0034-1397348. [DOI] [PubMed] [Google Scholar]
- Bommer GT, Gerin I, Feng Y, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Hu SJ. Effect of microRNA-34a in cell cycle, differentiation, and apoptosis:a review. J Biochem Mol Toxicol. 2012;26:79–86. doi: 10.1002/jbt.20412. [DOI] [PubMed] [Google Scholar]
- Chuma M, Sakamoto M, Yamazaki K, et al. Expression profiling in multistage hepatocarcinogenesis:identification of HSP70 as a molecular marker of early hepatocellular carcinoma. Hepatology. 2003;37:198–207. doi: 10.1053/jhep.2003.50022. [DOI] [PubMed] [Google Scholar]
- Ciocca DR, Clark GM, Tandon AK, et al. Heat shock protein Hsp70 in patients with axillary lymph node negative breast cancer:prognostic implications. J Natl Cancer Inst. 1999;85:570–4. doi: 10.1093/jnci/85.7.570. [DOI] [PubMed] [Google Scholar]
- Gehrmann M, Cervello M, Montalto G, et al. Heat shock protein 70 serum levels differ significantly in patients with chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Front Immunol. 2014;5:307. doi: 10.3389/fimmu.2014.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PW, Huang HD, Hsu SD, et al. miRNAMap:genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res. 2006;34:D135–9. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang TS, Han HS, Choi HK, et al. Differential, stage-dependent expression of Hsp70, Hsp110 and Bcl-2 in colorectal cancer. J Gastroenterol Hepatol. 2003;18:690–700. doi: 10.1046/j.1440-1746.2003.03011.x. [DOI] [PubMed] [Google Scholar]
- Joo M, Chi JG, Lee H. Expressions of HSP70and HSP27 in hepatocellular carcinoma. J Korean Med Sci. 2005;20:829–34. doi: 10.3346/jkms.2005.20.5.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Thomas MP, Altschuler G, et al. Capture of MicroRNA–bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 2011;7:1002363. doi: 10.1371/journal.pgen.1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-H, Hsu C-Y, Chu C-W, et al. A new child-turcotte-pugh class 0 for patients with hepatocellular carcinoma:determinants, prognostic impact and ability to improve the current staging systems. PLoS One. 2014;9:99115. doi: 10.1371/journal.pone.0099115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351:453–6. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- Li WB, Ma MW, Dong LJ, et al. MicroRNA-34a targets notch1 and inhibits cell proliferation in glioblastoma multiforme. Cancer Biol Ther. 2011;12:477–83. doi: 10.4161/cbt.12.6.16300. [DOI] [PubMed] [Google Scholar]
- Li Y, Guessous F, Zhang Y, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69:7569–76. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita A, Masaki T. miRNA in hepatocellular carcinoma. Hepatol Res. 2015;45:128–41. doi: 10.1111/hepr.12386. [DOI] [PubMed] [Google Scholar]
- Mosser DD, Morimoto RI. Molecular chaperones and the stress of oncogenesis. Oncogene. 2004;23:2907–18. doi: 10.1038/sj.onc.1207529. [DOI] [PubMed] [Google Scholar]
- Otsuka M, Kishikawa T, Yoshikawa T, et al. The role of microRNAs in hepatocarcinogenesis:Current knowledge and future prospects. J Gastroenterol. 2014;49:173–84. doi: 10.1007/s00535-013-0909-8. [DOI] [PubMed] [Google Scholar]
- Radons J. The human HSP70 family of chaperones:where do we stand? Cell Stress Chaperones. 2016;21:379–404. doi: 10.1007/s12192-016-0676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao DS, O'Connell RM, Chaudhuri AA, et al. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33:48–59. doi: 10.1016/j.immuni.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11:145–56. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- Sherman MY, Gabai VL. Hsp70 in cancer:back to the future. Oncogene. 2015;34:4153–61. doi: 10.1038/onc.2014.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin E, Ryu HS, Kim SH, et al. The clinicopathological significance of heat shock protein 70and glutamine synthetase expression in hepatocellular carcinoma. J Hepatobiliary Pancreat Sci. 2011;18:544–50. doi: 10.1007/s00534-010-0367-0. [DOI] [PubMed] [Google Scholar]
- Sun F, Fu H, Liu Q, et al. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582:1564–8. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- Takagi S, Nakajima M, Kida K, et al. MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem. 2010;285:4415–22. doi: 10.1074/jbc.M109.085431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Khaleque MA, Jones EL, et al. Expression of heat shock proteins and heat shock protein messenger ribonucleic acid in human prostate carcinoma in vitro and in tumors in vivo. Cell Stress Chaperone. 2005;10:46–58. doi: 10.1379/CSC-44R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toraih EA, Fawzy MS, Elgazzaz MG, et al. Combined genotype analyses of precursor miRNA196a2 and 499a variants with hepatic and renal cancer susceptibility- a preliminary study. Asian Pac J Cancer Prev. 2016a;17:3369–75. [PubMed] [Google Scholar]
- Toraih EA, Ismail NM, Toraih AA, Hussein MH, Fawzy MS. Precursor miR-499a variant but not miR-196a2 is associated with rheumatoid arthritis susceptibility in an Egyptian population. Mol Diagn Ther. 2016b;20:279–95. doi: 10.1007/s40291-016-0194-3. [DOI] [PubMed] [Google Scholar]
- Toraih EA, Mohammed EA, Farrag S, Ramsis N, Hosny S. Pilot study of serum microRNA-21 as a diagnostic and prognostic biomarker in Egyptian breast cancer patients. Mol Diagn Ther. 2015;19:179–90. doi: 10.1007/s40291-015-0143-6. [DOI] [PubMed] [Google Scholar]
- van der Meer AJ, Veldt BJ, Feld JJ, et al. Association between sustained virological response and all-cause mortality among patients with chronic hepatitis C and advanced hepatic fibrosis. JAMA. 2012;308:2584–93. doi: 10.1001/jama.2012.144878. [DOI] [PubMed] [Google Scholar]
- Vlachos IS, Kostoulas N, Vergoulis T, et al. DIANA miRPath v.2.0:investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012;40:498–504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gou W, Liu M, et al. Expression of P53 and HSP70 in chronic hepatitis, liver cirrhosis, and early and advanced hepatocellular carcinoma tissues and their diagnostic value in hepatocellular carcinoma:An immunohistochemical study. Med Sci Monit. 2015;21:3209–15. doi: 10.12659/MSM.895592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–22. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- Zhao W, Wang P, Ma J, et al. MiR-34a regulates blood–tumor barrier function by targeting protein kinase C. Mol Biol Cell. 2015;26:1786–96. doi: 10.1091/mbc.E14-10-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]


