Abstract
Background:
Head and neck squamous cell carcinomas (HNSCC) are a major health issue in many parts of the world. Recently, attention has focused on the human papilloma virus (HPV) as a potential causative agent for HNSCC. This study aimed to survey HPV occurrence in HNSCCs as part of a comprehensive molecular epidemiology approach.
Methods:
In this retrospective study, patients were recruited from hospitals affiliated to the Iran University of Medical Sciences, Tehran, Iran. Formalin-fixed paraffin-embedded (FFPE) blocks were subjected to DNA isolation by QIAamp® DNA FFPE Tissue Kit and nested PCR, HPV-16 specific conventional PCR, and extra INNO-LiPA HPV genotyping assays were subsequently performed. PCR products were purified with a High Pure PCR Product Purification Kit and sequenced with an ABI 3730 XL sequencer. CLC Main Workbench 5 and MEGA5 bioinformatics software was used to analyze the raw data and to create the phylogenetic tree. SPSS v.20 was applied for statistical analysis.
Results:
A total of 156 FFPE blocks were collected from 2011 to 2017. Total mean age (y) of participants was 60.5 ± 12.6; 77.6 % (121/156) being men and 22.4% (35/156) e women. Overall, 5/156 (3.2%) patients (3 females and 2 males) were found to be HPV positive using the three methods. HPV genotyping revealed HPV types 16, 2, 27, and 43 in these malignancies. Tumor location and lymph node involvement indicated significant differences between the sexes.
Conclusion:
Although high risk HPV genotypes have been associated with HNSCCs, our findings indicate a potential of low risk HPV types to also contribute to such malignancies.
Keywords: Human Papillomavirus, Head and Neck Squamous Cell Carcinoma, INNO-LiPA HPV genotyping
Introduction
Cancer is one of the major causes of human death worldwide. Iran as a developing country, reports several cases of cancers such as breast, prostate, stomach, colorectal, and lung cancers (Jalilvand et al., 2014; Salehi-Vaziri et al., 2015a; Ajdarkosh et al., 2016; Moradi et al., 2017; Safarnezhad Tameshkel et al., 2016). Cancer forms the third leading mortality factor after heart diseases and accidents (Emadzadeh et al., 2017; Mirzaei et al., 2016; Jalilvand et al., 2014).
Head and neck squamous cell carcinoma (HNSCC) is fifth most common cancer worldwide (Mirzaei et al., 2016; Siegel et al., 2017; Emadzadeh et al., 2017). This affects areas such as the oral cavity, nose, nasopharynx, oropharynx, hypopharynx, larynx, and salivary glands (Mirzaei et al., 2016; Emadzadeh et al., 2017; Siegel et al., 2017; Haratian et al., 2010). Annually, new cases of oral cavity cancers contribute to about 4% (63,030 cases) of the total cancer rate in the United States (Siegel et al., 2017; Knipe et al., 2013). Certain factors influencing the incidence of HNSCC include geographical districts, distinct lifestyles, tobacco and alcohol use, radiation exposure, vitamin deficiencies, dietary habits, human papilloma virus (HPV) cases, occupational exposures, and periodontal diseases (Haratian et al., 2010; Kermani et al., 2012; Mirzaei et al., 2016; Emadzadeh et al., 2017; Jalilvand et al., 2014).
The Papillomaviridae family comprises 39 genera and over 100 species; of these, 5 genera cause human infections and are termed as HPVs (Salehi-Vaziri et al., 2016; Javanmard et al., 2017; Dang et al., 2015b; Ghasemian et al., 2013). These viruses are about 60 nm in size and contain a single molecule of circular dsDNA. HPV as a major prevalent virus has the potential to establish neoplasms (Salehi-Vaziri et al., 2015a; Salehi-Vaziri et al., 2015b; Ghasemian et al., 2013). The role of HPV in certain malignancies such as vaginal, vulvar, ovarian, anal, penile, prostate, skin, urinary tract, and colorectal cancers has been explored (Salehi-Vaziri et al., 2015b; Salehi-Vaziri et al., 2016; Javanmard et al., 2017; Ghasemian et al., 2013). Other nonmalignant lesions such as recurrent respiratory papillomatosis (RRP) or genital warts are presumed to be a result of HPV infection; however, the association of HPV to these cancers remains controversial (Jalilvand et al., 2014; Knipe et al., 2013; Sadat Eftekhaar et al., 2017). HPV types categorized as high risk and low risk for cause malignant neoplasms. Some of the major high risk types include 16, 18, 31, 33, 35, 39, 45, 53 and low risk types include 6, 11, 40, 42, 43, 44, 54, 61, and 70 (Knipe et al., 2013; Javanmard et al., 2017; Salehi-Vaziri et al., 2016; Dang et al., 2015b). Several HPV types reported in the HNSCC patients till date include the high risk HPV types 16, 18, 31, 33, 52, and low risk HPV types 6, 11, 30, 35, 59 (Jalilvand et al., 2014; Haratian et al., 2010; Kermani et al., 2012; Lajer and Buchwald, 2010).
In general, the molecular epidemiology study of various cancers helps explore their potential risk factors, and may thus help in appropriate health care planning of general population; the policy makers could adapt their country’s health policy via research-based evidence (Banta, 1979; Samet, 2000; Emadzadeh et al., 2017). Thus, this study aimed to investigate the distribution of HPV types in Iranian HNSCC patients in order to use the data for further epidemiological and surveillance proposes.
Materials and Methods
Study design
In this retrospective study, archived formalin-fixed paraffin-embedded (FFPE) blocks were collected from HNSCC patients who were referred to hospitals affiliated to the Iran University of Medical Sciences, Tehran, Iran. The study was approved by the Ethics Committee of the university and is in compliance with the Helsinki Declaration. Patients were selected by sophisticated pathologists using pathological and clinical data that were retrieved from the clinical databases and patients’ health records from March 2011 to January 2017. According to the documentation of the tissue repository, blocks were fixed in 10% buffered formalin before paraffin embedment. Enrolled patients suffered from the HNSCC malignancies including pharynx (nasopharynx, oropharynx, and hypopharynx), larynx, tongue, salivary glands, and other head or oral cavity cancers. The paraffin-embedded tissue was used for further analyses.
DNA isolation
A thin 20-μm tissue section was removed from the blocks and was deparaffinized with xylene. Furthermore, series of distilled water and graded ethanol solutions were used for rehydration, according to the previous reports (Safarnezhad Tameshkel et al., 2016; Karbalaie Niya et al., 2016). QIAamp® DNA FFPE Tissue Kit (QIAGEN, Hilden, Germany) was used for DNA purification, according to the protocol (Safarnezhad Tameshkel et al., 2016; Karbalaie Niya et al., 2016). Quantitation and qualification of extracted DNA were performed via NanoDrop ND-1000® (Thermo Fisher Scientific Inc., Waltham, MA, USA) spectrophotometry. Isolated DNA was preserved at −20 °C until further use.
HPV L1 amplification
Specific MY and GP universal primers were used to amplify the HPV detected by nested PCR (nPCR) method, as described elsewhere (Salehi-Vaziri et al., 2016; Salehi-Vaziri et al., 2015b, Salehi-Vaziri et al., 2015a; Javanmard et al., 2017). MY09 and 11 primers were used as external primers and GP5+ and 6+ primers were used as internal specific primers, respectively. nPCR was performed for each sample and control for a total volume of 50 μl reaction mixture in every round containing 0.2–0.5 μM template or control, 0.5-mM of each of the four dNTPs into the dNTP mix (Fermentas GmbH, Germany), 0.5-μM of each primer, 1.5-μM MgCl2 solution (Fermentas GmbH, Germany), 5-units/μl of Taq DNA polymerase (Fermentas GmbH, Germany), and sterilized distilled water to achieve the total volume. CaSki cell line DNA extract was used as a positive control, and double-distilled water instead of sample DNA was included in the extraction and amplification procedures as negative control for HPV testing. PCR was carried out by Bio-Rad T100™ Thermal cycler using following protocol: 1 cycle, 95 °C for 5 min; 40 cycles, 95 °C for 1 min, 56 °C for 1 min (for external primers) or 52 °C for 30 s (for internal primers), and 72 °C for 1 min; 1 cycle, 72 °C for 10 min. Human β-globin sequence as an internal control used to validity assessment via described protocol (Salehi-Vaziri et al., 2015b, Javanmard et al., 2017). A gel electrophoresis system was used for visualizing the PCR products in each round via UV radiation emitted by E-Box Vilber version 15.03 (Vilber Lourmat, ZAC de Lamirault, France) gel documentation system. The PCR products were stained using ethidium bromide in 1.5% agarose gel.
INNO-LiPA HPV genotyping
The INNO-LiPA HPV genotyping extra assay was used for HPV genotyping via specific biotinylated consensus primers (SPF10). These cocktail primers amplified 65 bp of L1 variable region for typing 28 common HPVs (Salehi-Vaziri et al., 2015a, Salehi-Vaziri et al., 2015b, Salehi-Vaziri et al., 2016, Sadat Eftekhaar et al., 2017). A total volume of 50 μl reaction tube was used for PCR. Mastermix was included 37.7 μl of HPV Genotyping Extra Amp, 2.3 μl of HPV Genotyping Extra ENZ, and 5 μl of DEPC-treated water before 5 μl of sample DNA was added.
Genotyping was performed by hybridization of biotinylated PCR products on HPV type-specific oligonucleotide probes that were located on the nitrocellulose membranes. Moreover, colorimetric detection was carried out via an alkaline phosphatase–streptavidin conjugate. The Auto-LiPA 48 instrument was used post-PCR hybridization for colorimetric detection, according to protocol. The hybridization/stringent wash temperatures, timing, reagent addition, and aspiration were controlled by a preprogrammed test method. The Line Reader and Analysis software (LiRAS) was used for the strips scanning and evaluation. Trained operators confirmed the results by visual inspection. The result obtained by visualizing at least one of the defined type-specific banding patterns or one of the HPV control lines was considered positive.
HPV-16 PCR
One cycle of conventional PCR (cPCR) was performed via specific primers of HPV-16 E7 gene to detect the HPV-16 strains. The total volume of reaction mix and concentration of each PCR mix, the thermal cycler heating program, and the visualization procedure was same as described protocol in HPV L1 amplification section (except heating at 58 °C for 30 s for the annealing step). HPV-16 E7 forward primer was 5′-TGC AAC CAG AGA CAA CTG AT-3′ and reverse was 5′-TGT CTA CGT GTG TGC TTT GT-3′ that amplified 183 bp of HPV-16 E7 gene (Cheung et al., 2006).
Nucleotide sequencing and sequence analyses
High Pure PCR Product Purification Kit (Roche Diagnostic GmbH, Mannheim, Germany) was used to purify the HPV positive PCR products, according to instructions. A bidirectional sequencing with MY primers was performed using ABI 3730 XL sequencer (Thermo Fisher Scientific, Life Technologies Corporation, 5791 Van Allen Way, Carlsbad, California, USA). Bioinformatics software CLC Main Workbench 5 and MEGA5 were used to trim the raw data, comparison and analyses with HPV reference sequence (GenBank accession number NC_001348).
Statistical analysis
We reported the mean and related standard deviations for the continuous variables. Frequency and percentage was reported for the nominal and ordinal variables. To compare the mean age (with normal distribution), we performed the independent t-test. Furthermore, to determine the association between the nominal or ordinal variables, we performed chi square and Fisher’s exact tests. A p-value of 0.05 was considered as statistically significant for all analyses. All analyses were performed using SPSS version 20 software (SPSS Inc., Chicago, IL, USA).
Study population
A total of 156 HNSCC patients of mean age (y) 60.5 ± 12.6 and age range of 28–86 years were included. Of these participants, 77.6% (n = 121) were men and 22.4% were women (n = 35). The mean age was 60.7 ±11.9 years (range 30–83 years) for men and 59.7 ± 14.9 years (range 28–86 years) for women (p = 0.642). Table 1 displays the other basic characteristics of participants based on their sex. While the tumor location and lymph node involvement revealed significant difference between the two sexes, the other characteristics were not significantly different.
Table 1.
Basic Characteristics of Participants Based on Sex
| Variables | Male (%) | Female (%) | Total (%) | P-value | |
|---|---|---|---|---|---|
| Location | Larynx (true & false vocal cord) | 60 (49.7) | 8 (22.8) | 68 (43.6) | 0.028* |
| Tongue | 16 (13.2) | 13 (37.1) | 29 (18.6) | ||
| Pharynx (naso-, hypo-, supra-, oro-) | 15 (12.4) | 6 (17.1) | 21 (13.5) | ||
| Glottis (epi-, supra-) | 15 (12.4) | 2 (5.7) | 17 (10.9) | ||
| Glands (parotid & tonsil) | 4 (3.3) | 2 (5.7) | 6 (3.8) | ||
| Lip | 3 (2.5) | 1 (2.9) | 4 (2.6) | ||
| Mandible | 3 (2.5) | 1 (2.9) | 4 (2.6) | ||
| Palatine | 2 (1.6) | 1 (2.9) | 3 (1.9) | ||
| Nose | 1 (0.8) | 1 (2.9%) | 2 (1.3) | ||
| Neck | 1 (0.8) | - | 1 (0.6) | ||
| Face | 1 (0.8) | - | 1 (0.6) | ||
| Tumor Differentiation | Well differentiated | 55 (45.5) | 21 (60) | 76 (48.7) | 0.516 |
| Moderately differentiated | 30 (24.8) | 7 (20) | 37 (23.7) | ||
| Poorly differentiated | 20 (16.5) | 4 (11.4) | 24 (15.4) | ||
| undifferentiated | 16 (13.2) | 3 (8.6) | 19 (12.2) | ||
| Lymph node involvement | Involved | 76 (62.8) | 13 (37.1) | 89 (57.0) | 0.007 |
| Not-involved | 45 (37.2) | 22 (62.9) | 67 (42.9) | ||
| Adjacent tissue Invasion | Invasive | 84 (69.4) | 23 (65.7) | 106 (67.9) | 0.677 |
| Not-invasive | 37 (30.6) | 12 (34.3) | 50 (32.1) |
Fisher exact test was used
HPV Genotyping
The nPCR was performed using MY and GP primers for all extracted DNAs. From a total of 156 specimens, only 4 (2.5%) were HPV positive (Figures 1 and 2). Of 4 HPV positives two were HPV-16 strains whom they were males by 54 and 77 years and others were HPV-2 and HPV-27 that include females by 65 and 68 years, respectively. The positive sample underwent duplicate testing for confirming the results. Each HPV type was detected in separate HNSCC locations. HPV-16 was found only in poorly differentiated tumor tissue and not invasive to adjacent tumor tissue types.
Figure 1.
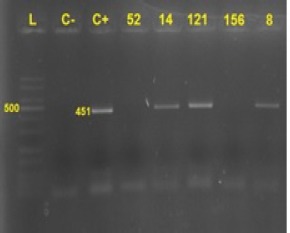
PCR Results by MY Primers. L,: Ladder; C-, control negative; C+, control positive; Numbers, tested samples No.; 14, 121 and 8 were positive. 451 bp band size was amplified by MY09 and 11 primers.
Figure 2.
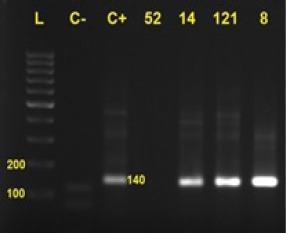
PCR Results by GP Primers. L, Ladder; C-, control negative; C+, control positive; Numbers, tested samples No.; 14, 121 and 8 were positive. 140 bp band size was amplified by GP5 and 6 primers.
Figure 3.
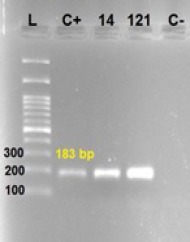
HPV-16 Confirmatory PCR by HPV-16 E7 Specific Primers. L, ladder 100-3000; C+, control positive; C-, control negative; 14 and 121: HPV-16 strains.
Results
The INNO-LiPA HPV genotyping assay detected two positive HPV type-16 isolates and one HPV-43 isolate that confirmed the PCR results. Total percentage of HPV positive subjects detected by INNO-LiPA method was 1.9% (3/156). HPV-43 was detected in a 50 year-old male.
Collectively, 5 (3.2%) HPV positive cases were detected with a mean age of 64.0 ± 9.3 years of which 3 (60%) were male and 2 (40%) were female. Interestingly, although the female participants were lesser than their male counterparts (35 females vs. 121 males), 5.7% (2/35) of females and 2.5% (2/121) of males were HPV positive. Result of the HPV investigation in detail is represented in Table 2. Although we used specific nPCR by general MY and GP primers and duplicated sampling, HPV-43 strain that detected by INNO-LiPA HPV genotyping assay was not confirmed by nPCR.
Table 2.
The Frequency of HPV Positive According to Tumor Characteristics
| Tumor characteristics | Categories | HPV positive | |
|---|---|---|---|
| N (%) | Type | ||
| Location | Larynx (true & false vocal cord) | 2 (2.9) | 27, 43 |
| Pharynx (naso-, hypo-, supra-, oro-) | 1 (4.7) | 2 | |
| Glands (parotid & tonsil) | 1 (16.6) | 16 | |
| Palatine | 1 (33.3) | 16 | |
| Tumor Differentiation | Well differentiated | 1 (1.3) | 27 |
| Moderate differentiated | 1 (2.7) | 43 | |
| Poorly differentiated | 2 (8.3) | 16, 16 | |
| undifferentiated | 1 (5.0) | 2 | |
| Lymph node involvement | Involved | 2 (2.2) | 16, 43 |
| Not-involved | 3 (3.9) | 16, 2, 27 | |
| Adjacent tissue Invasion | Invasive | 3 (2.8) | 2, 27, 43 |
| Not-invasive | 2 (4.0) | 16, 16 |
Another confirmatory test was carried out using HPV-16 E7 specific primers, and only 1.3% (2/156) patients were detected as HPV-16 positive. Results were shown in Figure 1. A specific183 bp band size was proposed HPV-16 positive in order to proper controls.
Primary HPV types were distinguished by the Basic Local Alignment Search Tool (BLAST) online software after analyzing the raw data. Phylogenetic analysis for 4 HPV positive strains was performed by MEGA5 software (Figure 4). Moreover, all L1 gene consensus sequences, after trimming, were submitted to the gene bank by BankIt online submission software and the released accession numbers were from MF134644 to MF134647.
Figure 4.
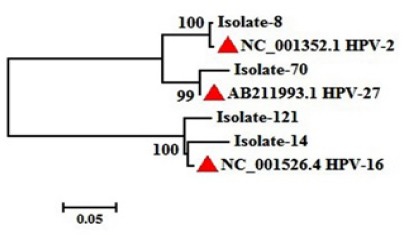
Neighbor-Joining Phylogenetic Tree of This Study HPV Strains. Red triangular showed reference sequences for each type. Isolates number showed our positive specimens.
Discussion
Briefly, in the present study, initially we investigated the HNSCC patients infected by HPVs and then HPV typing was carried out. Considerably, we used HNSCC patient’s archived FFPE blocks from the pathology section of hospitals affiliated to the Iran University of Medical Sciences, Tehran, Iran. After FFPE DNA extraction, a two-step nPCR was carried out to detect the HPV genome via specific MY and GP universal primers and four (4/156) positive products were sent for direct sequencing. Moreover, the trimmed sequences were analyzed by bioinformatics software and a phylogenetic tree was created. Four detected HPV positive specimens included one HPV-2, two HPV-16, and one HPV-27. INNO-LiPA HPV genotyping extra assay was used to evaluate the HPV types in all specimens, which detected two HPV-16 and one HPV-43 (3/156). Another confirmatory test by HPV-16 E7 specific primers enrolled and detected 1.3% (2/156) HPV-16 positive patients. In this study, the overall prevalence of HPV in the Iranian HNSCC patients was 3.2% (5/156) which 3 of detected HPV types (HPV-2, HPV-27, HPV-43) was not high risk for cancer involvement. Two HPV-16 and one HPV-43 strains were detected in males (60%), whereas HPV-2 and HPV-27 were detected in females (40%); moreover, both HPV-16 strains found in poorly differentiated tumor tissue were also present in the noninvasive tumor tissue.
Although this broad scale study comprises recent 5 years archived FFPE blocks and three methods (specific nPCR for L1, INNO-LiPAHPV Genotyping, and cPCR for HPV-16 E7) for each sample appropriately, the limited time for achieving healthy matched control group and high expenses of accurate tests such as Real-time PCR are likely to impact our results. The present study had certain limitations including low sample size, not using fresh tissue samples or frozen ones, matched healthy control group or adjacent tumor tissue, another confirmatory procedures such as Real-time PCR, different viral genome target except L1 due to loss of L1 in integrated form of HPV genome in cancerous cells and lack of data about patients’ immune system status.
Cancer is a major worldwide health problem with high mortality. Recently, HNSCC became a new concern of health services. Actually, epidemiological investigations could increase knowledge of health services to make up-to-date policy in each region (Samet, 2000; Emadzadeh et al., 2017). Furthermore, we set up a molecular epidemiology study to investigate HPV typing in Iranian HNSCC patients.
A study of Iranian HNSCC registry data from 2003 to 2009, in about 26,000 subjects, reported that the majority of patients were male (54%) and the highest incidence rate in both sexes was seen in patients of 80–84 years (Mirzaei et al., 2016). In the study of Kermani et al., (2012) the HNSCC patients mean age was 39.71 ± 6.7 years; female patients were 57.1% and male patients were 42.9%. In our study, the majority of our HNSCC patients were male (77.6%), and the mean age was 60.5 ± 12.6. It’s clear that broader sample size could define accurate results in age or male to female ratio and etc.
The role of HPV in HNSCC patients was initially described by Syrjanen et al., (1983). Studies from 1987 to 2010, with different method such as in situ hybridization (ISH), immunohistochemistry (IHC), cPCR, Real-time PCR (RT-PCR), and nested PCR (nPCR), reported distinct prevalence of 2–93% (Lajer and Buchwald, 2010). From 2011 to 2016, some studies (Deng et al., 2011; Bishop et al., 2012; Thibaudeau et al., 2013; Dang et al., 2015a; Picard et al., 2016) reported 17–30% of HPV incidence in HNSCC patients with HPV-16 being the common type. Another study of HPV related oropharyngeal cancer in Australia reported the increasing risk of 20–63% HPV infection in OSCC (Hong et al., 2016). Studies in Iranian HNSCC patients from 2005 to 2012 using nPCR and cPCR reported 20–62.5% HPV prevalence (Jalilvand et al., 2014). In the study by Haratian et al., (2010) in 2009, 82.5% and 62.5% of HPV infection was reported in Fanconi anemia (FA) cases and HNSCC without FA (control subjects), respectively. In a study by Tabatabai et al., (2015), HPV was detected in 43% of HNSCC subjects. These results slightly differ from our findings (3.2% incidence rate in our study), which could be due to the use of FFPE blocks and unexpected DNA fragmentation, low sample size, or method of detection; however, fewer studies used INNO-LiPA HPV genotyping extra assay, possibly due to its higher cost than other procedures. Moreover, in the present study, confirmatory test for HPV-16 strains was included.
In general, INNO-LiPA HPV genotyping extra assay used a hybridization method to detect the 28 HPV types including types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, and 82 as high risk types; HPV types 26, 53, and 66 as probable high risk types; HPV types 6, 11, 40, 43, 44, 54, and 70 as low risk types; and HPV types 69, 71, and 74 as unknown-risk types (Salehi-Vaziri et al., 2015a; Salehi-Vaziri et al., 2015b; Sadat Eftekhaar et al., 2017). In our study, we found two HPV-16 strains and one HPV-43 strain, detected by accurate INNO-LiPA HPV genotyping extra assay. Although the results of HPV-16 strains were confirmed by cPCR and nPCR (using general L1 and specific HPV-16 E7 primers), HPV-43 strain was not confirmed by the L1 consensus primers. This might be due to the low copy number of viral genome and may need an RT-PCR or other confirmatory method or specific primers for detection of HPV-43.
In a systematic review of HNSCC studies involving HPV reported that common HPV types were the high risk types 16, 18, 31, 33, and 52, and the low risk types 6, 11, 30, 35, and 59 (Lajer and Buchwald, 2010). The systematic review of Jalilv et al., (2014) reported that common HPV types in Iranian HNSCC included high risk types 16,18, 31, and 33, and the low risk types 6 and 11. In a study by Yahyapour et al., (2013) the Iranian ESCC by RT-PCR found 14 HPV types including 6, 11, 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 59. Haratian et al., (2010) found genotypes 16, 18, and 31 in HNSCC subjects. In a study by Kermani et al., (2012) HPV types 6, 16, and 18 were detected in 15 HNSCC patients. In the present study, we found high risk types 16 and low risk types 2, 27, and 43. Interestingly, we reported for the first time, HPV types 2, 27, and 43 in HNSCC patients. This elucidates some potential of the uncommon low risk HPV types to become or cause malignancy in the HNSCC patients, especially in Iran.
In general, HPV type 43 was found in rare surveys on cervical cancer (Salehi-Vaziri et al., 2016; Shafaghi et al., 2013; Beerens et al., 2005) and it is not reported in HNSCC patients up to now. Literature review did not report HPV-43 in HNSCC subjects. It might has sexual rout. HPV types 2 and 27 are generally found in common warts (Chan et al., 1997; Wang et al., 2007; Yan-Jun et al., 2009) and were not reported in HNSCC complication till date by any literature. This could emphasize the changing trends of low risk HPV types to cause malignancy, or alternatively, they could be noninvasive and have low or limited copy number in head and neck tissue after interventions. These patients underwent some procedures to suppress immune invasiveness that consequently could enhance the risk of opportunistic infections by other non-invasive organisms such as HPV types 2, 27 and 43.
HPV-16 accounted for 82% of HNSCC HPV positives (Pezzuto et al., 2015); in our study the prevalence was 40% (2/5). Tonsil SCC showed a high involvement (94%) by HPVs (Näsman et al., 2009); in our present study of 2 tonsil SCC patients, 1 (50%) was infected with HPV-16. These could be a result of low sample size or other confounding issues, as previously discussed.
In conclusion, these date slightly bright the future trends of research on HNSCC patients infected by HPVs by a higher attention to other low risk types and define their roles in these malignancies since they are likely an emerging or super-infecting these patients to suffer more pain additionally to overwhelming radiotherapy, chemotherapy or surgery. On the other hand, if the low risk types were entered by contaminated procedures or lower immune system protection to these injured organs after interventions our health system should be improved to eliminate these contaminations. More data needs to comprehend various aspects of HPV low risk types roles by case-control or cohort studies in a broad scale sample size and high precise detection methods.
Conflict of interest
None.
Acknowledgments
We are thankful from Keyvan laboratory, Tehran, Iran personnel for unaccountable technical supports and assistance. This project was fund by Iran University of Medical Sciences, Tehran, Iran in order to a grant number 92215402.
References
- Ajdarkosh H, Safarnezhad Tameshkel F, et al. Association of Helicobacter Pylori infection with colon polyp and colorectal cancer. British J Med and Med Res. 2016;16:1–6. [Google Scholar]
- Banta JE. Epidemiology in health care planning. Am J Trop Med Hyg. 1979;28:1077–8. [Google Scholar]
- Beerens E, Van Renterghem L, Praet M, et al. Human papillomavirus DNA detection in women with primary abnormal cytology of the cervix:prevalence and distribution of HPV genotypes. Cytopathology. 2005;16:199–205. doi: 10.1111/j.1365-2303.2005.00266.x. [DOI] [PubMed] [Google Scholar]
- Bishop JA, Ma X-J, Wang H, et al. Detection of transcriptionally active high risk HPV in patients with head and neck squamous cell carcinoma as visualized by a novel E6/E7 mRNA in situ hybridization method. Am J Surg Pathol. 2012;36:1874. doi: 10.1097/PAS.0b013e318265fb2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S-Y, Chew S-H, Egawa K, et al. Phylogenetic analysis of the human papillomavirus type 2 (HPV-2), HPV-27, and HPV-57 group, which is associated with common warts. Virol J. 1997;239:296–302. doi: 10.1006/viro.1997.8896. [DOI] [PubMed] [Google Scholar]
- Cheung JL, Lo KW, Cheung TH, Tang JW, Chan PK. Viral load, E2 gene disruption status, and lineage of human papillomavirus type 16 infection in cervical neoplasia. J Infect Dis. 2006;194:1706–12. doi: 10.1086/509622. [DOI] [PubMed] [Google Scholar]
- Dang J, Feng Q, Eaton KD, Jang H, Kiviat NB. Detection of HPV in oral rinse samples from OPSCC and non-OPSCC patients. BMC Oral Health. 2015a;15:126. doi: 10.1186/s12903-015-0111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang J, Manrique H, Veron D, Feng Q. Oral human papillomavirus (HPV) infection in healthy individuals and patients with head and neck squamous cell carcinoma (HNSCC) Epidemiology (sunnyvale) 2015b;5:1161–5. [Google Scholar]
- Deng Z, Hasegawa M, Matayoshi S, et al. Prevalence and clinical features of human papillomavirus in head and neck squamous cell carcinoma in Okinawa, southern Japan. Eur Arch Otorhinolaryngol. 2011;268:1625–31. doi: 10.1007/s00405-011-1515-0. [DOI] [PubMed] [Google Scholar]
- Emadzadeh M, Shahidsales S, Mohammadian Bajgiran A, et al. Head and neck cancers in North-East Iran:A 25 year survey. Iran J Otorhinolaryngol. 2017;29:137–45. [PMC free article] [PubMed] [Google Scholar]
- Ghasemian E, Monavari S, Irajian GR, et al. Evaluation of human papillomavirus infections in prostatic disease:a cross-sectional study in Iran. Asian Pac J Cancer Prev. 2013;14:3305–8. doi: 10.7314/apjcp.2013.14.5.3305. [DOI] [PubMed] [Google Scholar]
- Haratian K, Mohseni Meybodi A, Zari Moradi S, Vosough P. Detection of high risk human papillomavirus dna sequences in head and neck squamous cell carcinoma in iranian fanconi anemia patients. Yakhteh Med J. 2010;12:43–50. [Google Scholar]
- Hong A, Lee CS, Jones D, et al. Rising prevalence of human papillomavirus–related oropharyngeal cancer in Australia over the last 2 decades. Head Neck. 2016;38:743–50. doi: 10.1002/hed.23942. [DOI] [PubMed] [Google Scholar]
- Jalilvand S, Shoja Z, Hamkar R. Human papillomavirus burden in different cancers in Iran:a systematic assessment. Asian Pac J Cancer Prev. 2014;15:7029–35. doi: 10.7314/apjcp.2014.15.17.7029. [DOI] [PubMed] [Google Scholar]
- Javanmard D, Namaei MH, Haghighi F, et al. The frequency and typing of human papilloma virus among women with normal and abnormal cytology in southern Khorasan, Eastern Iran. Jundishapur J Microbiol. 2017;10:43213. [Google Scholar]
- Karbalaie Niya M, Basi A, Koochak A, et al. Sensitive high-resolution melting analysis for screening of KRAS and BRAF mutations in Iranian human metastatic colorectal cancers. Asian Pac J Cancer Prev. 2016;17:5147–52. doi: 10.22034/APJCP.2016.17.12.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermani IA, Seifi S, Dolatkhah R, et al. Human papilloma virus in head and neck squamous cell cancer. Iran J Cancer Prev. 2012;5:21. [PMC free article] [PubMed] [Google Scholar]
- Knipe D, Howley P, Griffin D, et al. Fields virology. I+II. Lippincott, Williams and Wilkins; 2013. pp. 1662–703. [Google Scholar]
- Lajer CB, Buchwald CV. The role of human papillomavirus in head and neck cancer. APMIS. 2010;118:510–9. doi: 10.1111/j.1600-0463.2010.02624.x. [DOI] [PubMed] [Google Scholar]
- Mirzaei M, Hosseini S-A, Ghoncheh M, et al. Epidemiology and trend of head and neck cancers in Iran. Global J Health Sci. 2016;8:189. doi: 10.5539/gjhs.v8n1p189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradi P, Keyvani H, Mousavi S-A J, et al. Investigation of viral infection in idiopathic pulmonary fibrosis among Iranian patients in Tehran. Microb Pathog. 2017;104:171–4. doi: 10.1016/j.micpath.2017.01.030. [DOI] [PubMed] [Google Scholar]
- Näsman A, Attner P, Hammarstedt L, et al. Incidence of human papillomavirus (HPV) positive tonsillar carcinoma in Stockholm, Sweden:An epidemic of viral-induced carcinoma? Int J Cancer. 2009;125:362–6. doi: 10.1002/ijc.24339. [DOI] [PubMed] [Google Scholar]
- Pezzuto F, Buonaguro L, Caponigro F, et al. Update on head and neck cancer:current knowledge on epidemiology, risk factors, molecular features and novel therapies. Oncology. 2015;89:125–36. doi: 10.1159/000381717. [DOI] [PubMed] [Google Scholar]
- Picard A, Badoual C, Hourseau M, et al. Human papilloma virus prevalence in HIV patients with head and neck squamous cell carcinoma. Aids. 2016;30:1257–66. doi: 10.1097/QAD.0000000000001030. [DOI] [PubMed] [Google Scholar]
- Sadat Eftekhaar N, Karbalaie Niya MH, Izadi F, Teaghinezhad-S S, Keyvani H. Human papillomavirus (HPV) genotype distribution in patients with recurrent respiratory papillomatosis (RRP) in Iran. Asian Pac J Cancer Prev. 2017;18:1973–6. doi: 10.22034/APJCP.2017.18.7.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safarnezhad Tameshkel F, Sohrabi MR, Babaee MR, et al. Mutation analysis of KRAS and BRAF genes in metastatic colorectal cancer:a First large scale study from Iran. Asian Pac J Cancer Prev. 2016;17:603–8. doi: 10.7314/apjcp.2016.17.2.603. [DOI] [PubMed] [Google Scholar]
- Salehi-Vaziri M, Sadeghi F, Alamsi-Hashiani A, et al. Merkel cell polyomavirus and human papillomavirus infections in cervical disease in Iranian women. Arch Virol. 2015a;160:1181–7. doi: 10.1007/s00705-015-2368-4. [DOI] [PubMed] [Google Scholar]
- Salehi-Vaziri M, Sadeghi F, Bokharaei-Salim F, et al. The prevalence and genotype distribution of human papillomavirus in the genital tract of males in Iran. Jundishapur J Microbiol. 2015b;8:21912. doi: 10.5812/jjm.21912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi-Vaziri M, Sadeghi F, Hashemi FS, et al. Distribution of human papillomavirus genotypes in Iranian women according to the severity of the cervical lesion. Iran Red Crescent Med J. 2016;18:24458. doi: 10.5812/ircmj.24458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samet JM. Epidemiology and policy:the pump handle meets the new millennium. Epidemiol Rev. 2000;22:145–54. doi: 10.1093/oxfordjournals.epirev.a018013. [DOI] [PubMed] [Google Scholar]
- Shafaghi B, Jarollahi A, Yousefzadeh B, et al. Human papilloma virus prevalence and types among Iranian women attending regular gynecological visits. Rep Pract Oncol Radiother. 2013;1:73–9. [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- Syrjänen K, Syrjänen S, Lamberg M, Pyrhönen S, Nuutinen J. Morphological and immunohistochemical evidence suggesting human papillomavirus (HPV) involvement in oral squamous cell carcinogenesis. Int J oral surg. 1983;12:418–24. doi: 10.1016/s0300-9785(83)80033-7. [DOI] [PubMed] [Google Scholar]
- Tabatabaie F, Tafreshi ZA, Shahmohammad N, Pirestani M. Molecular detection of microsporidiosis in various samples of Iranian immunocompromised patients. J parasit dis. 2015;39:634–8. doi: 10.1007/s12639-014-0432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaudeau E, Fortin B, Coutlee F, et al. HPV prevalence and prognostic value in a prospective cohort of 255 patients with locally advanced HNSCC:a single-centre experience. Int J Otolaryngol. 2013;2013:1–9. doi: 10.1155/2013/437815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wang C, Xu S, et al. Detection of HPV-2 and identification of novel mutations by whole genome sequencing from biopsies of two patients with multiple cutaneous horns. J Clin Virol. 2007;39:34–42. doi: 10.1016/j.jcv.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Yahyapour Y, Shamsi-Shahrabadi M, Mahmoudi M, et al. High-risk and low-risk human papillomavirus in esophageal squamous cell carcinoma at Mazandaran, Northern Iran. Pathol Oncol Res. 2013;19:385–91. doi: 10.1007/s12253-012-9590-0. [DOI] [PubMed] [Google Scholar]
- Yan-Jun L, Chen G, Chen W, et al. Molecular epidemiological study on prevalence of human papillomaviruses in patients with common warts in Beijing area. Biomed Environ Sci. 2009;22:55–61. doi: 10.1016/S0895-3988(09)60023-4. [DOI] [PubMed] [Google Scholar]


