Abstract
Dynamic nuclear polarization (DNP) enhanced solid-state NMR can provide orders of magnitude in signal enhancement. One of the most important aspects of obtaining efficient DNP enhancements is the optimization of the paramagnetic polarization agents used. To date, the most utilized polarization agents are nitroxide biradicals. However, the efficiency of these polarization agents is diminished when used with samples other than small molecule model compounds. We recently demonstrated the effectiveness of nitroxide labeled lipids as polarization agents for lipids and a membrane embedded peptide. Here, we systematically characterize, via electron paramagnetic (EPR), the dynamics of and the dipolar couplings between nitroxide labeled lipids under conditions relevant to DNP applications. Complemented by DNP enhanced solid-state NMR measurements at 600 MHz/395 GHz, a molecular rationale for the efficiency of nitroxide labeled lipids as DNP polarization agents is developed. Specifically, optimal DNP enhancements are obtained when the nitroxide moiety is attached to the lipid choline head group and local nitroxide concentrations yield an average e−-e− dipolar coupling of 47 MHz. Based on these measurements, we propose a framework for development of DNP polarization agents optimal for membrane protein structure determination.
TOC image
INTRODUCTION
Solid state NMR (ssNMR) is becoming a valuable tool for structural biology. In particular, it is well suited for the study of biomolecules and macromolecular complexes that are too large for standard solution state NMR techniques and/or too challenging to crystallize for diffraction studies, for instance membrane-associated proteins.1, 2 However, there remain many challenges when studying these complex systems by ssNMR. Primarily, the types of experiments required to obtain high-resolution structures, i.e., multidimensional 13C and 15N correlation experiments for resonance assignments or homonuclear/heteronuclear dipolar recoupling experiments for distance restraints can require days to weeks of instrument time for each experiment. The extensive time requirement is largely due to the inherent insensitivity of NMR and thus the requirement for substantial signal averaging. Herein we describe a molecular rationale for a sample preparation strategy utilizing nitroxide radical labeled lipids (spin labeled lipids, SL-lipids, Figure 1) that we recently demonstrated gave large dynamic nuclear polarization (DNP) NMR signal enhancements for membrane embedded peptides.3
Figure 1.
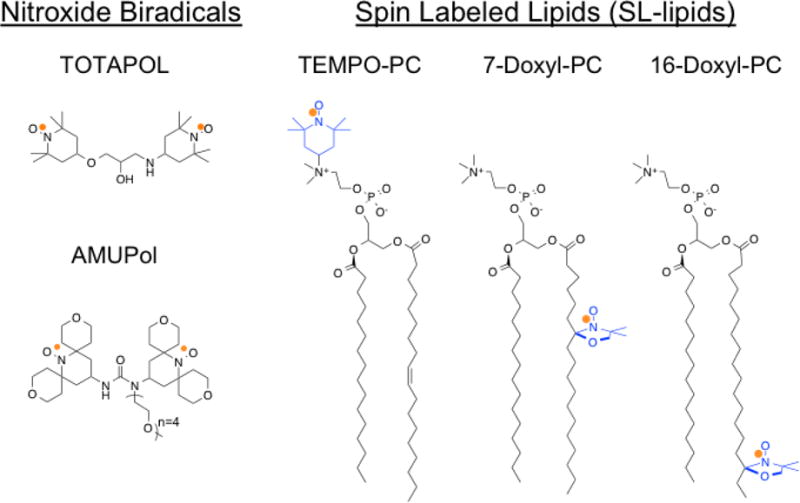
Molecular structures of (left) common water soluble nitroxide biradicals and (right) SL-lipids used in this study. The unpaired electron, indicated by an orange dot, is distributed over the N-O bond. The nitroxide moiety of the SL-lipids is highlighted in blue.
The inherent insensitivity of NMR stems from the low Boltzmann population difference between nuclear spin states in accessible magnetic fields. The limited sensitivity of NMR is exacerbated for membrane protein samples because physiologically relevant conditions oftentimes necessitate the dilution of the protein of interest within the lipid milieu. Compared to crystalline protein NMR samples, membrane protein concentrations are typically an order of magnitude more dilute. In recent years DNP has emerged as a powerful technique to enhance ssNMR signals by up to almost three orders of magnitude,4, 5 enabling up to ~ 400,000 fold savings in data collection time. DNP signal enhancements are procured by inducing mutual electron-nuclear spin transitions by irradiating a paramagnetic dopant with high power microwaves (MW) resulting in a larger nuclear spin state population difference.6, 7, 8
DNP signal enhancements have a theoretical maximum value that is fundamentally determined by the gyromagnetic ratios of the paramagnetic source (i.e. an unpaired electron) and the nucleus of interest; for a 1H nucleus, γe/γ1H ≈ 660. In practice, DNP enhancements are strongly influenced by factors such as the spectral characteristics of the paramagnetic species, temperature, magnetic field strength, and the sample type.4, 5, 8 To date, the most utilized high field (≥ 5 T) DNP MAS ssNMR polarization transfer mechanism is the cross effect which is mediated by a three spin, electron-electron-nuclear, process.9 The cross effect condition is met when the resonant frequency difference between the two dipolar-coupled electrons is approximately equal to the nuclear resonant frequency (Δωe ≈ ωn).10 In addition, recent computational analysis of the cross effect has given rise to a new theoretical framework for the cross effect DNP mechanism under the effects of MAS. 11, 12, 13 Specifically, it is postulated that sample rotation induces anti-crossings that are the predominant cause of polarization transfer from electrons to nuclei. Many recent advancements in DNP-enhanced MAS ssNMR have focused on the design of tailored paramagnetic reagents that satisfy the cross effect condition, namely the development of nitroxide biradicals.14, 15, 16, 17, 18 An enhancement value of 515, which is close to the theoretical maximum of 660, has been obtained for ssNMR DNP experiments on the model compound tetrachloroethane at lower magnetic fields (9.4 T/400 MHz 1H) using the designer biradical TEKPol.19 However, when samples of biological interest, such as membrane proteins, fibrils, or macromolecular assemblies, have been studied at higher magnetic fields (14.1–18.8 T) using DNP magic angle spinning (MAS) ssNMR, lesser enhancements have been observed.20, 21 Nuclear longitudinal relaxation time constants have been shown to impact the overall enhanced polarization in DNP experiments and the complex chemical environments of biological macromolecules can display a range of relaxation timescales.11, 12 Therefore, the diminutive enhancements on samples of biological interest can in part be attributed to the multitude of relaxation time constants of the macromolecules. In addition, the inhomogeneous broadening of the nitroxide EPR resonance with increasing magnetic field strength, due to its large g-anisotropy, may affect polarization transfer mechanisms.
Nitroxide radicals are extensively utilized for spin-labeling applications in the biological electron paramagnetic resonance (EPR) community and therefore are chemically and spectrally well characterized. Currently, nitroxide biradicals, such as TOTAPOL and AMUPol, that contain two nitroxides tethered together (Figure 1), are the standard in paramagnetic dopants for DNP MAS ssNMR experiments.4, 5, 8, 9 The strength of the electron-electron dipolar coupling is a parameter that influences the magnitude of DNP enhancements.16, 22 It has also been experimentally determined that the relative orientation between the g-tensors of the tethered nitroxide radicals impact the observed DNP enhancement, where roughly perpendicular orientations between the two g-tensors gives the highest probability of inducing DNP polarization transfer.14, 16 Thus far, the largest DNP MAS ssNMR enhancement values in an aqueous system have been obtained with the biradical AMUPol,17 where the relative orientation between the g-tensors is ~ 64° and the average inter-electron distance is ~12 Å corresponding to an electron-electron dipolar coupling of ~35 MHz.17 For heterogeneous biological macromolecular ssNMR samples such as membrane proteins, fibrils, and macromolecular assemblies, DNP enhancement gradients have been observed, with largest enhancement values observed for nuclei that are located at the interface between the aqueous phase and the molecule of interest with diminishing values obtained for nuclei located within the sample interior.3, 23, 24 For example, with the use of TOTAPOL or AMUPol in membrane protein DNP MAS ssNMR studies, the largest enhancement values are observed at the hydrophilic periphery of the membrane and the least enhancement is observed at the hydrophobic interior of the membrane, where the bulk of the protein is embedded thus demonstrating a distance dependence on polarization transfer efficiency.3 This observed enhancement gradient is attributed to the mechanism of polarization transfer in the DNP experiment. Specifically, polarization is first induced by the paramagnetic agent in local nuclei, which are NMR silent due to relaxation effects that originate from their close proximity to the paramagnetic species. Then, through nuclear spin-diffusion, polarization is transferred to other nuclei. 25, 26 A particular challenge when applying DNP enhanced ssNMR to the study of membrane proteins is that these water soluble biradicals do not partition into the hydrophobic region of the bilayer; this phase separation mitigates the overall DNP enhancements for nuclei in membrane proteins that reside within the lipid bilayer interior.
In an effort to improve DNP enhancements of integral membrane proteins, we explored the capabilities of SL-lipids as polarization agents in DNP MAS ssNMR investigations of lipid membrane embedded species.3 Several noteworthy advantages of the SL-lipid polarizing agents were demonstrated. (1) By locating the nitroxide within the lipid milieu, a more homogeneous distribution of DNP enhancements was observed throughout the various carbon nuclei of the phospholipids. (2) When compared to results obtained for TOTAPOL, the SL-lipids produced a nearly doubled DNP enhancement for a specific carbonyl of a membrane-embedded peptide. (3) The incorporation of the radical into the lipid bilayer removed the dependence on glycerol as a glassing agent upon freezing. Routinely, water soluble biradicals require a glassing agent to prevent ice crystal formation which creates concentrated radical micro-domains.27 Because lipids are self-glassing and the SL-lipids are spontaneously distributed throughout the membrane, phase separation of radicals during freezing is no longer a problem and additional glassing agents (i.e. glycerol) are not necessary. (4) The impact of removing excess glycerol is tri-fold: without bulk glycerol, the 13C resonances that arise from the phosphoglycerol moiety are readily discernable; any concerns about vesicle morphology changes due to osmotic effects of glycerol are removed;28, 29 and there is no further dilution of the protein beyond what is required by the requisite lipid bilayer. (5) Finally, the use of SL-lipids is compatible with common membrane protein sample preparation strategies.
Here, we provide a systematic spectroscopic evaluation of SL-lipid samples to develop a framework for understanding the molecular dynamics and organization underpinning the efficiency of SL-lipids as polarization agents for DNP enhanced MAS ssNMR experiments. Specifically, from analysis of X-band EPR line shapes of SL-lipids of varying molar concentrations and in different locations within the lipid, we determine the average distances and dipolar interactions between SL-lipids. Further analysis of nitroxide mobility gives estimates of order parameters, leading to insight into the orientational disorder of the nitroxide at different locations within the bilayer. From the strength of the dipolar interactions and relative orientational disorder, we propose a molecular level understanding of the concentration dependence and depth dependence of SL-lipids on DNP enhancements obtained at 600 MHz/395 GHz. These findings provide guidance for the development of next generation polarization agents for membrane proteins.
MATERIALS AND METHODS
Materials
1,2-dipalmitoyl-d62-sn-glycero-3-phosphocholine (DPPC-d62), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho(tempo)choline (TEMPO-PC), 1-palmitoyl-2-stearoyl-(7-doxyl)-sn-glycero-3-phosphocholine (7-Doxyl-PC), and 1-palmitoyl-2-stearoyl-(16-doxyl)-sn-glycero-3-phosphocholine (16-doxyl-PC) were purchased as chloroform solutions from Avanti Polar Lipids Inc. (Alabaster, AL) and used as received. Unless otherwise stated all other reagents were purchased from Sigma Aldrich (St. Louis, MO) and used as received.
Liposome Preparation
For each sample, 1:1 (molar ratio, ~10 mg total lipid) DPPC-d62:POPC liposomes were prepared by mixing DPPC-d62, POPC, and the stated amount of SL-lipid (0.4, 1.5, 3.0, 6.0, 8.0 mol%) as chloroform solutions. After mixing, the chloroform was evaporated under a N2 gas stream. The resulting lipid film was resuspended in an excess of cyclohexane, frozen in liquid N2, and lyophilized overnight. The dry lipid mixture was resuspended in excess 10 mM BisTris propane buffer, pH 7.4 made with 90:10 (v/v) D2O:H2O, and subjected to multiple freeze-thaw cycles to form multilamellar vesicles (MLVs). The suspension of MLVs was centrifuged (16,000 g), the excess buffer was removed, and the remaining liposomes were loaded into relevant sample containers. Details regarding optimal levels of lipid deuteration are given in the supporting information and Figure S1.
X-Band (9 GHz) Continuous Wave (CW) EPR
All X-band CW EPR experiments were performed on a Bruker E500 EPR spectrometer (University of Florida). Low temperature CW EPR experiments were performed using a Bruker high-Q resonator (4122 SHQE) that was placed in an Oxford continuous flow cryostat (ESR900) and cooled using liquid He. A Lakeshore temperature regulator was used to maintain the temperature at 100 K. Spectra were collected with 2 mW of MW power at a center field of 3358 G, a sweep width of 300 G, and a modulation frequency of 100 kHz at an amplitude of 1 G. CW EPR experiments at 282 K, above the phase transition temperature of the lipids, were collected on a Bruker E500 EPR spectrometer equipped with a Bruker dielectric resonator (4123D). The sample temperature was maintained by flowing N2 gas through a copper heat exchanger in a water bath and then through the sample space in the resonator. Spectra were collected with 2 mW of MW power at a center field of 3475 G, a sweep width of 300 G, and a modulation frequency of 100 kHz with 0.1 G modulation amplitude. The ratio d1/d0 was determined as described in the text and used to predict distances as previously described.30, 31, 32 All EPR spectra were baseline corrected and area normalized using software generously provided by Altenbach C. and Hubell W.L. (UCLA).
DNP MAS ssNMR
All 13C cross-polarization (CP) DNP MAS ssNMR experiments were carried out on a 600 MHz Bruker Avance spectrometer equipped with a 395 GHz Bruker gyrotron and a Bruker 3.2 mm 1H/13C/15N DNP MAS probe at the National High Magnetic Field Laboratory (NHMFL). Hydrated MLVs were packed into 3.2 mm sapphire rotors and sealed with silicone plugs. Rotors were inserted into a pre-cooled, ~100 K probe and spun at 5 kHz. For 13C CP MAS ssNMR experiments, a train of 100 kHz 1H 90° pulses were applied prior to each polarization to ensure an equilibrated bath of 1H spins; a ramped CP, centered at 50 kHz 1H B1 field, pulse program with a mixing time of 2 ms was used to transfer polarization to 13C spins; 100 kHz SPINAL-64 1H decoupling was used during 13C signal acquisition and a relaxation delay of 10 s was used. 1H T1 relaxation times and DNP buildup times were previously determined for SL-lipid and TOTAPOL liposome sample types respectively. 1H T1 relaxation times ranged from ~1 – 2 s and DNP buildup curves are reported in Figure S4 and Table S1. For DNP experiments, ~10 W of continuous MW irradiation (as measured at the gyrotron output) was applied to the sample.
RESULTS
DNP Enhancement as a Function of mol% SL-lipid
13C CP spectra of MLVs with increasing amounts of 7-Doxyl-PC were collected with either MW on (DNP) or off (control) to measure the DNP enhancement as a function of the amount of SL-lipid. Enhancement was calculated as ε = Ion/Ioff where Ion and Ioff are the signal intensities of the measured resonance for the lipid acyl chain with the MW on or off, respectively (Figure 2). The measured DNP enhancements for 1.5, 3.0, 6.0, and 8.0 mol% of 7-Doxyl-PC are shown in Figure 3A. The DNP enhancement increased from 3.6 to 6.1 when 7-Doxyl-PC increased from 1.5 to 3.0 mol% of the total lipid. In contrast, the enhancement decreased when the amount of 7-Doxyl-PC increased beyond 3 mol%. At 6 mol% 7-Doxyl-PC, ε = 4.9, and, at 8.0 mol% 7-Doxyl-PC, ε = 4.3.
Figure 2.
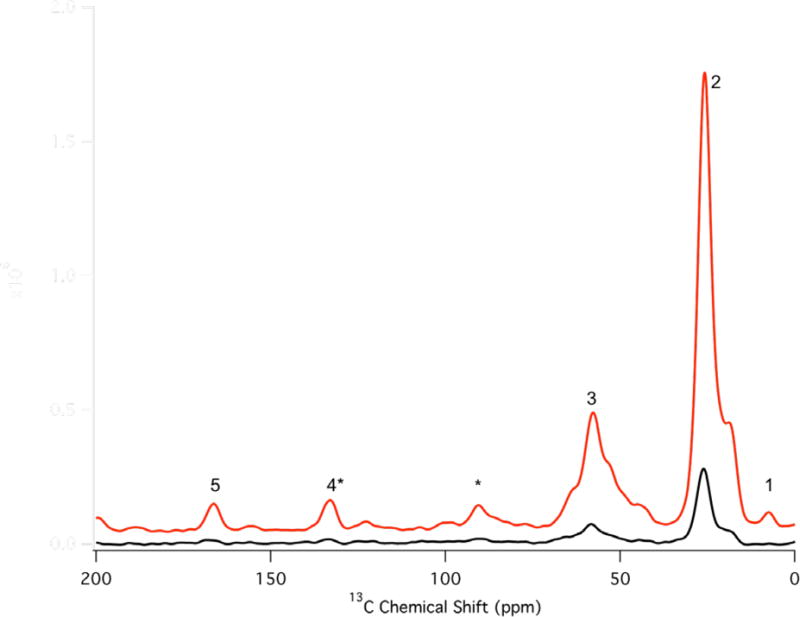
13C spectra for PC MLVs containing 3 mol% 7-Doxyl-PC with (red) and without (black) microwave irradiation. Resonances are labeled as follows: 1 terminal methyl of lipid acyl chain, 2 sp3 carbons of the lipid acyl chain, 3 lipid glycerol backbone, 4* sp2 carbons at the point of unsaturation in the lipid acyl chain which also includes the spinning sideband from 5, 5 lipid carbonyl groups, and * spinning sidebands resulting from 3 and 5. The lipid acyl chain (resonance 2) was used to measure all reported DNP enhancements.
Figure 3.
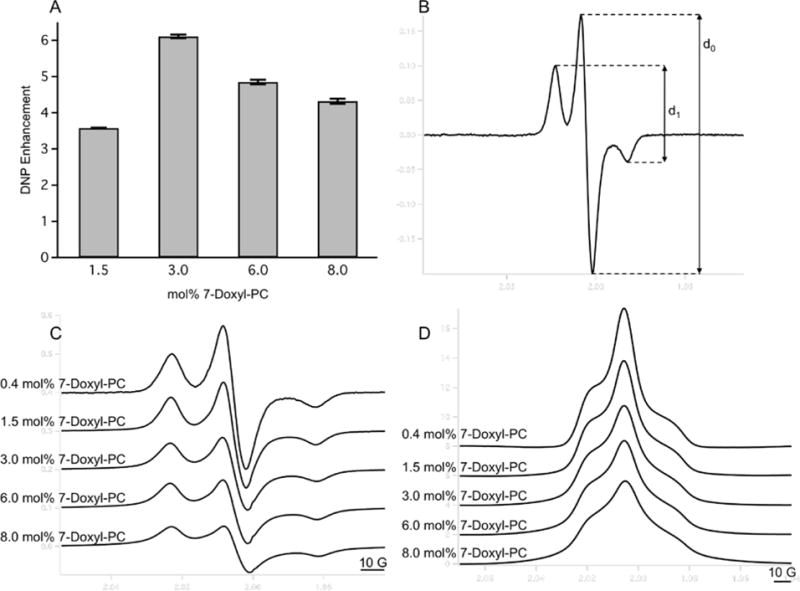
A) DNP enhancement as a function of mol% of 7-Doxyl PC as measured via 13C MAS ssNMR spectra of MLVs. Error bars were calculated by measuring the RMS of noise in the baseline of each spectrum and propagated through the DNP enhancement calculation. B) Shows how values of d1/d0 are measured from the EPR derivative spectra. C) Derivative and D) absorption area normalized 300 G X-band CW EPR spectra of PC MLVs containing the indicated amount of 7-Doxyl-PC. The electron-electron dipolar coupling between 7-Doxyl-PC nitroxide spins grows in strength as the concentration of 7-Doxyl-PC is increased, leading to spectral broadening. Spectra plotted vertically offset for clarity.
The effect of increasing amounts of paramagnetic species on the observed NMR signal intensity must be acknowledged. Paramagnetic induced broadening, or paramagnetic relaxation enhancement (PRE), of NMR signals was analyzed by measuring the full width at half height (FWHH) of the observed resonances and these measurements are reported in Table S2. The FWHH of the acyl chain resonance increased from 670 to 696 Hz upon increasing the amount of 7-Doxyl-PC from 1.5 to 3.0 mol% and increased further to 708 Hz at 8 mol% 7-Doxyl-PC, indicating that there is some paramagnetic bleaching albeit to a minor extent. In addition, the FWHH of 3 mol% TEMPO-PC, 7-Doxyl-PC, and 16-Doxyl-PC are nearly identical, indicating that the position of the nitroxide radical on the lipid molecule does not have a large role in the observed PRE. This is in agreement with previously measured PRE of membrane proteins with the biradicals TOTAPOL and AMUPol, and the conclusion that line broadening of nuclear resonances appears to only affect nuclei within ~5 Å of the radical.21 Lastly, there has also been discussion of nuclear depolarization induced by electron-electron anti-crossings, which are mediated by the electron-electron dipolar interaction under MAS conditions.33 However, it has been observed that nuclear depolarization is attenuated at higher magnetic fields and moderate to slow MAS, and it has been hypothesized that stronger intramolecular electron-electron dipolar couplings could further mediate nuclear depolarization.33 Because our experiments were conducted at high field (600 MHz/395 GHz), at moderate MAS (5 kHz), and we have demonstrated stronger electron-electron dipolar couplings we postulate that there is minimal nuclear depolarization in this system. However, due to the nature of the sample preparation (see Liposome Preparation) it was not possible to directly compare signal intensities between the different samples, or between radical doped and undoped samples, due to uncertainty in the total amount of liposomes deposited into the rotor.
Characterization of the Dipolar Coupling between SL-lipids in MLVs
CW EPR is a standard technique used to measure the distance between two nitroxide labeled sites in biomolecules.30, 31, 34, 35, 36 Distances between nitroxide radicals below ~20 Å and above ~6 Å result in broadened EPR spectra; this broadening can almost entirely be attributed to the dipolar interaction between the radicals.37 It has been observed that strong electron-electron dipolar couplings (i.e. short distances between radicals) results in larger DNP enhancements due to more efficient polarization transfer through the cross effect. Here we employ an empirical method and measure the d1/d0 ratio, defined as the height between the high and low field transitions over the height of the central transition, to estimate electron-electron dipolar couplings (Figure 3B, 3C, and 3D). This particular method has been used to measure distances between two spin labeled sites in proteins and was initially calibrated by Kokorin et al. using frozen solutions of nitroxides with known radical distributions.32 A d1/d0 ratio < 0.40 is indicative of a very weak dipolar coupling at the upper limit of what is detectable by CW EPR, and an increasing d1/d0 value linearly correlates with a stronger dipolar coupling.30, 31 CW EPR spectra of frozen MLV solutions with increasing amounts of 7-Doxyl-PC (0.4, 1.5, 3.0, 6.0, and 8.0 mol% of total lipid) were recorded and the derivative and absorption area normalized spectra are shown in Figure 3C and 3D respectively. The area normalized absorption (Figure 3D) spectra are shown to better demonstrate the spectral broadening effects of the electron-electron dipolar coupling, and as expected increased spectral broadening is observed with increasing concentrations of 7-Doxyl-PC which is indicative of stronger electron dipolar couplings between the nitroxide radicals in the SL-lipids. The d1/d0 ratio was measured for each spectrum in Figure 3C and the results are tabulated in Table 1. For the lowest SL-lipid concentration (0.4 mol% 7-Doxyl-PC), the d1/d0 ratio is 0.37, indicating a very weak dipolar coupling and an average distance of ~20 Å between nitroxides. The d1/d0 ratio increases to 0.64 for 8.0 mol% 7-Doxyl-PC, which correlates to a distance of <6 Å between nitroxides. At 3.0 mol% 7-Doxyl-PC, a d1/d0 of 0.52, corresponding to an inter-nitroxide distance of 10 Å, was measured. The DNP enhancements in Figure 3A correlate well with what is expected based on the measured inter-nitroxide distances. When the distance decreases from 15 Å (1.5 mol% 7-Doxyl-PC) to 10 Å (3.0 mol% 7-Doxyl-PC), the DNP enhancement nearly doubles. However, when the distance further decreases from 10 Å to 7.3 Å (6.0 mol% 7-Doxyl-PC) the DNP enhancement decreases. This observation is consistent with similar studies performed with nitroxide biradicals. TOTAPOL, an efficient DNP biradical designed with a linker between the two nitroxide moieties, has an electron-electron distance of ~12 Å. However, when the distance between the radicals is decreased to ~5 Å by eliminating the linker and generating the biradical with a spiro connection to directly link the two nitroxides, the measured DNP enhancement decreases by a factor of 6.5.38
Table 1.
Values of d1/d0 and Corresponding Inter-Nitroxide Distances and Dipolar Couplings for Varying Concentrations of 7-Doxyl-PC in MLVs
| 7-Doxyl-PC concentration | d1/d0 (± 0.002)a | average distance (Å)b ± 2%c | dipolar coupling (MHz)d ± 8%c |
|---|---|---|---|
| 0.4 mol% | 0.371 | 19.9 | 6.5 |
| 1.5 mol% | 0.448 | 15.1 | 15 |
| 3.0 mol% | 0.525 | 10.3 | 47 |
| 6.0 mol% | 0.572 | 7.32 | 130 |
| 8.0 mol% | 0.640 | ≤ 6 Å | ≥ 240 |
Error was calculated by measuring the RMS of noise in the baseline of the 0.4 mol% 7-Doxyl-PC EPR spectrum at 100 K and propagated through the d1/d0 calculation.
Error was calculated as the percent residual of a fit to previously published data.31
Calculated using the point dipole approximation D = μ0g2μ2B/4πhr3.
We note that the exchange interaction, or spin-spin coupling, between electrons can also lead to alterations in the EPR line shape. This interaction occurs when either (1) there is a physical collision between paramagnetic species or (2) when an unpaired electron is delocalized over the molecule and interacts with another unpaired electron within the same molecule, i.e. a through-bond interaction in a tethered biradical.37 The latter has been observed in nitroxide-trityl biradicals, where the unpaired electron of trityl is highly delocalized over the triarylmethyl molecular scaffold and is able to interact with the localized unpaired electron of the N-O moiety of the nitroxide radical.39, 40 If the exchange interaction is strong enough such that J » A, where J is the magnitude of the exchange interaction and A is the hyperfine coupling constant, then additional multiplet structures would be observed in the EPR spectrum. Upon a further increase in J the multiplet structure would begin to exchange narrow until only a single transition is observed in the EPR spectrum.37 In addition to alterations in the fine structure of the EPR spectrum the observed transitions would resonate at different magnetic fields with a change in J.37 We do not observe these manifestations of the exchange interaction in most of our EPR spectra, as evidenced by Figure 3C, and we therefore conclude that the line broadening we observe with increased amount of 7-Doxyl-PC is due to increased electron-electron dipolar couplings and not the exchange interaction. The one exception is the spectrum of 8 mol% 7-Doxyl-PC, where we observe possible exchange narrowing, i.e. the collapse of a triplet derivative spectrum into a singlet, could be occurring. To recognize the early effects of the exchange interaction we only provide an upper limit for the average inter-nitroxide distance of 6 Å and a lower limit for the electron-electron dipolar coupling of 240 MHz.
DNP Enhancement as a Function of Radical Position in a Lipid Bilayer
In addition, the position of attachment for the nitroxide to the lipid molecule affects the observed DNP enhancement. 13C CP NMR spectra of MLVs with 3 mol% of either TEMPO-PC, 7-Doxyl-PC or16-Doxyl-PC were compared to a control sample containing 20 mM TOTAPOL; DNP enhancements are presented in Figure 4A. It is evident that TEMPO-PC gives the most enhancement and that the enhancement decreases as the radical is positioned more deeply into the membrane. Also, as we previously observed,3 the water soluble nitroxide biradical TOTAPOL gives the least enhancement when compared to all of the SL-lipids used in this study. Lipid moieties demonstrate a range of motional anisotropies depending on their position in the bilayer. Therefore, a distribution of distances and orientations between nitroxide radicals of the SL-lipids depends upon where in the phospholipid molecule the nitroxide is attached. It is well known that, when in a bilayer, the acyl chain of a phospholipid has the smallest degree of motional freedom near the glycerol backbone and the largest degree of motional freedom at the terminus of the acyl chain.41 The head group region of the phospholipid can have varying degrees of motional freedom that are dependent upon the type of head group and the neighboring molecules.41 Solution (282 K) X-band CW EPR spectra were acquired for PC MLVs containing 3 mol% of either TEMPO-PC, 7-Doxyl-PC, or 16-Doxyl-PC, and are shown in Figure 4B. Upon inspection of the spectra it is evident that the 7-Doxyl-PC nitroxide moiety has the smallest range of motion, the 16-Doxyl-PC nitroxide moiety exhibits the largest range of motion, and the TEMPO-PC nitroxide moiety has an intermediate range of motion compared to the other nitroxide positions. To quantify this observation, the breadth of the hyperfine extrema, 2Amax, was measured for each spectrum (Figure 4B) and values are presented in Table 2. The 2Amax parameter has been used extensively to measure the relative degree of motional freedom at nitroxide labeled sites on a variety of biomolecules, including lipids.42, 43 Our results corroborate those of previous measurements and demonstrate that 7-Doxyl-PC has the smallest degree of motional freedom and 16-Doxyl-PC the largest degree of motional freedom and the measured DNP enhancements reflect this. The large distribution of orientations adopted by 16-Doxyl-PC approaches that of a frozen solution of nitroxide monoradicals and it is the least efficient at polarization transfer. 7-Doxyl-PC has a more efficient polarization transfer due to the narrow distribution of nitroxide conformations; however, TEMPO-PC, which demonstrates some degree of motional freedom, is the most efficient SL-lipid at polarization transfer. The distribution of conformations adopted by the various SL-lipids relative to the bilayer normal are depicted in Figure 4C. We hypothesize that the large orientational distribution adopted by 16-Doxyl-PC decreases the average electron-electron dipolar coupling, while the intermediate orientational distributions adopted by TEMPO-PC allows for the most efficient electron-electron dipolar couplings. In fact, at 3 mol% TEMPO-PC the d1/d0 = 0.603 which is larger than 3 mol% of 7-Doxyl-PC where d1/d0 = 0.525 and is indicative of a stronger electron-electron dipolar coupling.
Figure 4.
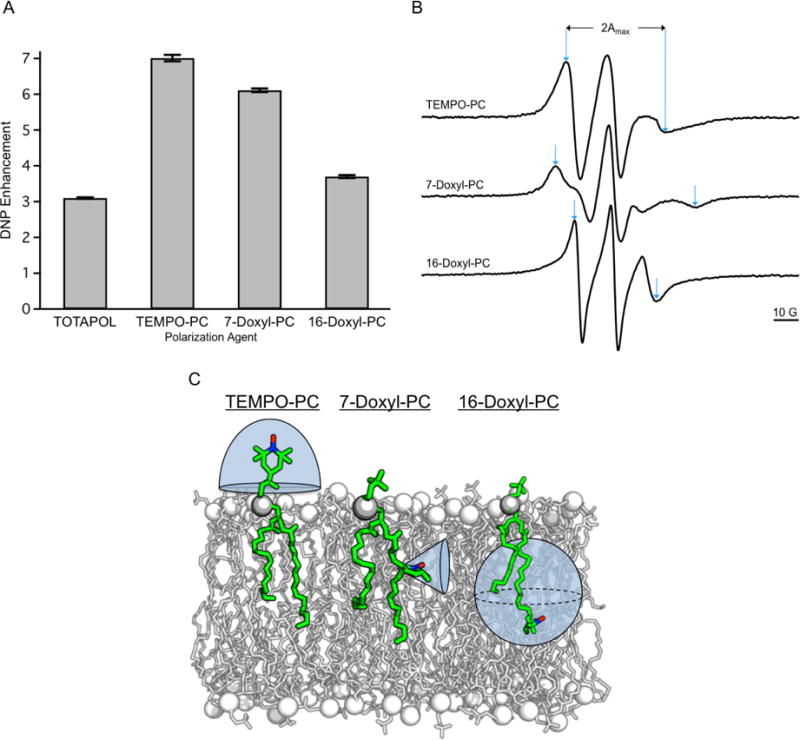
A) DNP enhancements for 3 mol% of indicated SL-lipids in MLVs vs. 20 mM TOTAPOL3 suspended with MLVs, measured via 13C CP NMR spectra. Error bars were calculated by measuring the RMS noise in the baseline of each 13C CP spectrum and propagating through the DNP enhancement calculation. B) Area normalized X-band CW EPR spectra of 3 mol% of the indicated SL-lipids reconstituted in MLVs at 282 K. Measurement of 2Amax is indicated with blue arrows for each spectrum. C) The distribution of available conformations for nitroxides in TEMPO-PC, 7-Doxyl-PC, and 16-Doxyl-PC relative to the lipid bilayer normal are depicted by the shaded blue region around the respective nitroxide moiety.
Table 2.
Measured 2Amax Values for the Various SL-lipid nitroxide Positions
| SL-lipid | 2Amax (G) |
|---|---|
| TEMPO-PC | 40 |
| 7-Doxyl-PC | 56 |
| 16-Doxyl-PC | 33 |
Increasing values of 2Amax indicate fewer degrees of motional freedom.
In addition to the strength of the electron dipolar coupling it has been demonstrated that the relative orientation between electron g-tensors may play a role in the DNP efficiency. Nitroxide g-tensors are very anisotropic and when orthogonal relative to one another have a high probability of meeting the cross effect matching condition (Δωe ≈ ωn).14, 38 This type of observation has been made with various nitroxide biradicals; for example, TOTAPOL, where the linker connecting the nitroxide moieties allows for multiple orientations between the g-tensors to be adopted, gives a high DNP efficiency relative to either a more constrained biradical or a random distribution of radical orientations.14, 16, 38
DISCUSSION
To date, many studies have focused on developing the most efficient exogenous DNP MAS ssNMR polarization agents, with the most successful being nitroxide biradicals. The efficiency of nitroxide biradicals as DNP polarization agents is attributed to their strong intramolecular electron-electron dipolar coupling. However, with the growth in biological applications of MAS DNP, there is a renewed focus on using DNP polarization agents that are endogenous to the system of study. For instance, we previously demonstrated that using SL-lipids yielded larger DNP enhancements of a membrane embedded peptide when compared to TOTAPOL.3 In addition, Wylie B.J., et al. used site-specific spin labeling to position nitroxide radicals at the interface of the dimeric transmembrane peptide Gramicidin,44 Voinov M.A., et al. developed an analog of TOTAPOL that can be used to site-specifically label cysteine residues with the biradical and van der Cruijsen E.A.W. et al. developed a similar approach using AMUPol,45, 46 Fernández-de-Alba C., et al. palmitylated TOTAPOL to generate a lipid anchored biradical (N-Propyl Palmipol),47 and Wenk P., et al. used a paramagnetic Mn2+ atom endogenous to a ribozyme as a polarization agent.48 Utilizing an endogenous DNP polarization agent results in a matrix free sample preparation strategy that enables better control of the radical location and does not require co-solvents to maintain a homogeneous distribution of radical, as is needed when an exogenous polarization agent is used. Our approach of using SL-lipids as the DNP polarization agent is a general strategy with the added advantage that it requires no manipulation of the membrane proteins, the SL-lipids are added at low enough concentrations to minimize perturbation of the membrane environment, sample preparation is straightforward, and the membrane protein is not further diluted by more than what is required to ensure it is in a native-like lipid bilayer environment.
In this work we have demonstrated that nitroxide monoradicals, when used in the appropriate environment, can yield the same optimal parameters that are exploited in nitroxide biradicals for effecting efficient DNP polarization. Because MLVs containing SL-lipids are hydrated in excess buffer and then pelleted via centrifugation to pack into a MAS rotor we are unable to calculate an accurate electron spin concentration, however, we estimate that at 3.0 mol% 7-Doxyl-PC the upper limit of the electron spin concentration is ~ 12 mM in the MAS rotor. In contrast to the measured distances between SL-lipid nitroxide moieties, a 10 mM solution of nitroxide monoradicals has an average distance of 30 Å between radicals, nearly 3× longer than the measured distance (10 Å) for 3.0 mol% 7-Doxyl-PC.49 This dramatic decrease in distance between similar spin concentrations of SL-lipids compared to monoradicals in solution is not unexpected as the monoradicals are homogenously dispersed throughout the sample while SL-lipids are effectively constrained to a two-dimensional lattice that is the lipid bilayer. This constraint to a planar lipid bilayer results in a stronger intermolecular electron-electron dipolar coupling that is manifested in the broadened EPR spectra. When the appropriate amount of SL-lipid is used (3 mol% in this case), a dipolar coupling of ~ 47 MHz is observed which is comparable to what is observed for the most efficient nitroxide biradicals, TOTAPOL and AMUPol. 38, 17 We acknowledge that Fernández-de-Alba C., et al. observed a DNP enhancement at 9.4 T of 8.1 on liposomes with N-Propyl Palmipol, which was achieved with ~ 1 mol% radical. However, N-Propyl Palmipol is a biradical; therefore the effective electron spin concentration was ~ 2 mol%, which is near the 3 mol% of monoradical that we utilized. In addition, it is thought that DNP enhancements should scale as 1/B0, when mediated by the bis-nitroxide radical based cross effect. Therefore, we contend that our approach is comparable to that of Fernández-de-Alba C., et al.
If the SL-lipid concentration is increased beyond 3 mol%, the electron-electron dipolar interaction starts to interfere with DNP via the cross effect. Mathies G., et al. have shown that the trityl-nitroxide biradicals can be used as efficient polarization agents at high magnetic fields, i.e. 18.8 T. The success of this class of biradical polarization agents was attributed to the relatively strong exchange interaction between the two electrons. In a similar fashion to the electron-electron dipolar coupling the exchange interaction can lead to the state mixing required for cross effect DNP. 40 However, Mathies G., et al. note that the interaction between electrons should not equal or exceed the larmor frequency of the target nuclei for hyperpolarization.40 Therefore, we contend that as the SL-lipid concentration is increased beyond 3 mol% the electron-electron dipolar coupling starts to approach the 1H larmor frequency and begins to interfere with the cross effect DNP mechanism.
Despite the success of using SL-lipids as DNP polarization agents there is a need to further improve their polarization transfer efficiency. One successful strategy, which might be employed with SL-lipids, is to substitute the protonated methyl nitroxide protecting groups with more rigid or deuterated moieties, as was demonstrated in the development of the water-soluble biradical AMUPol and deuteration of TOTAPOL.17, 50 At the working temperature of ~100 K common to most DNP MAS ssNMR experiments, methyls can still freely rotate. This rotation generates fluctuating dipolar fields that lead to the shortening of T1e and T2e relaxation times. Removing or deuterating the methyl groups blocks this non-productive cross relaxation mechanism. Second, other types of lipophilic biradicals could be synthesized that build upon the successful traits of the water-soluble nitroxide biradicals. Third, the use of lipids with a metal chelating head group provides another avenue to explore for efficient DNP of membrane proteins.51 In addition, high field electron spin relaxation measurements would inevitably add clarity in efforts to ascertain more efficient membrane protein DNP polarization agents. Specifically, they would elucidate the interplay between polarization agent orientation with respect to the membrane and the efficiency of the polarization agent as well as the effects that high electron spin concentrations have on electron spectral diffusion and cross effect efficiency.
CONCLUSION
We have developed a molecular rationale for the efficiency of SL-lipids as DNP polarization agents. Specifically, we have shown that monoradical SL-lipids at concentrations effective for maximal DNP exhibit electron-electron dipolar couplings similar to that of nitroxide biradicals. In addition, the distribution of nitroxide conformations differs based upon the location of the radical within the lipid molecule, which affects the relative orientations between the nitroxides’ g-tensors and subsequent DNP efficiency. Overall, a SL-lipid concentration of 3 mol% gave an average electron-electron dipolar coupling of 47 MHz (~10 Å) and the intermediate mobility of lipid head group bound nitroxide radicals yielded the most efficient DNP enhancement.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. James Collins for help with calculations and the National High Magnetic Field Laboratory (NHFML) for instrument time on their Bruker 600 MHz/395 GHz DNP spectrometer. The NHMFL is supported by the National Science Foundation (NSF) cooperative agreement number DMR-1157490 (JRL) and the State of Florida. This work was also supported by NSF MCB-1329467 (GEF) and MCB-1412700 (GEF), NSF CHE-1229170 (JRL), NIH S10 OD018519 and NIH S10 RR031603 (JRL).
Footnotes
Author Contributions
Research was designed by ANS, GEF, and JRL and carried out by ANS. The manuscript was written through contributions of ANS, GEF, and JRL. UTT provided technical assistance and hardware setup for low temperature EPR experiments. TD assisted in gyrotron operations.
References
- 1.Renault M, Cukkemane A, Baldus M. Solid-State NMR Spectroscopy on Complex Biomolecules. Angew Chem, Int Ed. 2010;49:8346–8357. doi: 10.1002/anie.201002823. [DOI] [PubMed] [Google Scholar]
- 2.Weingarth M, Baldus M. Solid-State NMR-Based Approaches for Supramolecular Structure Elucidation. Acc Chem Res. 2013;46:2037–2046. doi: 10.1021/ar300316e. [DOI] [PubMed] [Google Scholar]
- 3.Smith AN, Caporini MA, Fanucci GE, Long JR. A Method for Dynamic Nuclear Polarization Enhancement of Membrane Proteins. Angew Chem Int Ed. 2015;54:1542–1546. doi: 10.1002/anie.201410249. [DOI] [PubMed] [Google Scholar]
- 4.Maly T, Debelouchina G, Bajaj V, Hu K, Joo C, Mak-Jurkauskas M, Sirigiri J, van der Wel P, Herzfeld J, Temkin R, Griffin R. Dynamic nuclear polarization at high magnetic fields. J Chem Phys. 2008;128 doi: 10.1063/1.2833582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ni Q, Daviso E, Can T, Markhasin E, Jawla S, Swager T, Temkin R, Herzfeld J, Griffin R. High Frequency Dynamic Nuclear Polarization. Acc Chem Res. 2013;46:1933–1941. doi: 10.1021/ar300348n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carver TR, Slichter CP. Polarization of Nuclear Spins in Metals. Phys Rev. 1953;92:212–213. [Google Scholar]
- 7.Carver TR, Slichter CP. Experimental Verification of the Overhauser Nuclear Polarization Effect. Phys Rev. 1956;102:975–980. [Google Scholar]
- 8.Smith AN, Long JR. Dynamic Nuclear Polarization as an Enabling Technology for Solid State Nuclear Magnetic Resonance Spectroscopy. Anal Chem. 2016;88:122–132. doi: 10.1021/acs.analchem.5b04376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Can TV, Ni QZ, Griffin RG. Mechanisms of dynamic nuclear polarization in insulating solids. J Magn Reson. 2015;253:23–35. doi: 10.1016/j.jmr.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu K, Debelouchina G, Smith A, Griffin R. Quantum mechanical theory of dynamic nuclear polarization in solid dielectrics. J Chem Phys. 2011;134 doi: 10.1063/1.3564920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mentink-Vigier F, Akbey U, Hovav Y, Vega S, Oschkinat H, Feintuch A. Fast passage dynamic nuclear polarization on rotating solids. J Magn Reson. 2012;224:13–21. doi: 10.1016/j.jmr.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Mentink-Vigier F, Akbey U, Oschkinat H, Vega S, Feintuch A. Theoretical aspects of Magic Angle Spinning - Dynamic Nuclear Polarization. J Magn Reson. 2015;258:102–120. doi: 10.1016/j.jmr.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Thurber KR, Tycko R. Theory for cross effect dynamic nuclear polarization under magic-angle spinning in solid state nuclear magnetic resonance: The importance of level crossings. J Chem Phys. 2012;137 doi: 10.1063/1.4747449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu K, Song C, Yu H, Swager T, Griffin R. High-frequency dynamic nuclear polarization using biradicals: A multifrequency EPR lineshape analysis. J Chem Phys. 2008;128 doi: 10.1063/1.2816783. [DOI] [PubMed] [Google Scholar]
- 15.Song C, Hu K, Joo C, Swager T, Griffin R. TOTAPOL: A biradical polarizing agent for dynamic nuclear polarization experiments in aqueous media. J Am Chem Soc. 2006;128:11385–11390. doi: 10.1021/ja061284b. [DOI] [PubMed] [Google Scholar]
- 16.Ysacco C, Karoui H, Casano G, Le Moigne F, Combes S, Rockenbauer A, Rosay M, Maas W, Ouari O, Tordo P. Dinitroxides for Solid State Dynamic Nuclear Polarization. Appl Magn Reson. 2012;43:251–261. [Google Scholar]
- 17.Sauvee C, Rosay M, Casano G, Aussenac F, Weber R, Ouari O, Tordo P. Highly Efficient, Water-Soluble Polarizing Agents for Dynamic Nuclear Polarization at High Frequency. Angew Chem Int Ed. 2013;52:10858–10861. doi: 10.1002/anie.201304657. [DOI] [PubMed] [Google Scholar]
- 18.Zagdoun A, Casano G, Ouari O, Schwarzwalder M, Rossini A, Aussenac F, Yulikov M, Jeschke G, Coperet C, Lesage A, Tordo P, Emsley L. Large Molecular Weight Nitroxide Biradicals Providing Efficient Dynamic Nuclear Polarization at Temperatures up to 200 K. J Am Chem Soc. 2013;135:12790–12797. doi: 10.1021/ja405813t. [DOI] [PubMed] [Google Scholar]
- 19.Kubicki D, Rossini A, Purea A, Zagdoun A, Ouari O, Tordo P, Engelke F, Lesage A, Emsley L. Amplifying Dynamic Nuclear Polarization of Frozen Solutions by Incorporating Dielectric Particles. J Am Chem Soc. 2014;136:15711–15718. doi: 10.1021/ja5088453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreas L, Barnes A, Corzilius B, Chou J, Miller E, Caporini M, Rosay M, Griffin R. Dynamic Nuclear Polarization Study of Inhibitor Binding to the M2(18–60) Proton Transporter from Influenza A. Biochemistry. 2013;52:2774–2782. doi: 10.1021/bi400150x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koers E, van der Cruijsen E, Rosay M, Weingarth M, Prokofyev A, Sauvee C, Ouari O, van der Zwan J, Pongs O, Tordo P, Maas W, Baldus M. NMR-based structural biology enhanced by dynamic nuclear polarization at high magnetic field. J Biomol NMR. 2014;60:157–168. doi: 10.1007/s10858-014-9865-8. [DOI] [PubMed] [Google Scholar]
- 22.Hu K-N. Polarizing agents and mechanisms for high-field dynamic nuclear polarization of frozen dielectric solids. Solid State Nucl Magn Reson. 2011;40:31–41. doi: 10.1016/j.ssnmr.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Wel P, Hu K, Lewandowski J, Griffin R. Dynamic nuclear polarization of amyloidogenic peptide nanocrystals: GNNQQNY, a core segment of the yeast prion protein Sup35p. J Am Chem Soc. 2006;128:10840–10846. doi: 10.1021/ja0626685. [DOI] [PubMed] [Google Scholar]
- 24.Lafon O, Thankamony A. S. L.; Kobayashi, T.; Carnevale, D.; Vitzthum, V.; Slowing, I. I.; Kandel, K.; Vezin, H.; Amoureux, J.-P.; Bodenhausen, G.; Pruski, M. Mesoporous Silica Nanoparticles Loaded with Surfactant: Low Temperature Magic Angle Spinning C-13 and Si-29 NMR Enhanced by Dynamic Nuclear Polarization. J Phys Chem C. 2013;117:1375–1382. [Google Scholar]
- 25.Smith A, Corzilius B, Barnes A, Maly T, Griffin R. Solid effect dynamic nuclear polarization and polarization pathways. J Chem Phys. 2012;136 doi: 10.1063/1.3670019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hovav Y, Feintuch A, Vega S. Dynamic nuclear polarization assisted spin diffusion for the solid effect case. J Chem Phys. 2011;134 doi: 10.1063/1.3526486. [DOI] [PubMed] [Google Scholar]
- 27.Lama B, Collins JHP, Downes D, Smith AN, Long JR. Expeditious dissolution dynamic nuclear polarization without glassing agents. NMR Biomed. 2016;29:226–231. doi: 10.1002/nbm.3473. [DOI] [PubMed] [Google Scholar]
- 28.Oleary TJ, Levin IW. Raman-Spectroscopic Study of an Interdigitated Lipid Bilayer Dipalmitoylphosphatidylcholine Dispersed in Glycerol. Biochim Biophys Acta. 1984;776:185–189. doi: 10.1016/0005-2736(84)90207-4. [DOI] [PubMed] [Google Scholar]
- 29.Fanucci GE, Lee JY, Cafiso DS. Spectroscopic evidence that osmolytes used in crystallization buffers inhibit a conformation change in a membrane protein. Biochemistry. 2003;42:13106–13112. doi: 10.1021/bi035439t. [DOI] [PubMed] [Google Scholar]
- 30.Gross A, Columbus L, Hideg K, Altenbach C, Hubbell WL. Structure of the KcsA potassium channel from Streptomyces lividans: A site-directed spin labeling study of the second transmembrane segment. Biochemistry. 1999;38:10324–10335. doi: 10.1021/bi990856k. [DOI] [PubMed] [Google Scholar]
- 31.Fanucci GE, Coggshall KA, Cadieux N, Kim M, Kadner RJ, Cafiso DS. Substrate-induced conformational changes of the perplasmic N-terminus of an outer-membrane transporter by site-directed spin labeling. Biochemistry. 2003;42:1391–1400. doi: 10.1021/bi027120z. [DOI] [PubMed] [Google Scholar]
- 32.Kokorin AI, Zamaraev KI, Rozantse Eg, Grigorya Gl, Ivanov VP. Measuring of distance between paramagnetic groups in solid-solutions of nitroxide radicals, biradicals and spin-labeled proteins. Biofizika. 1972;17:34. [PubMed] [Google Scholar]
- 33.Mentink-Vigier F, Paul S, Lee D, Feintuch A, Hediger S, Vega S, De Paepe G. Nuclear depolarization and absolute sensitivity in magic-angle spinning cross effect dynamic nuclear polarization. Phys Chem Chem Phys. 2015;17:21824–21836. doi: 10.1039/c5cp03457d. [DOI] [PubMed] [Google Scholar]
- 34.Rabenstein MD, Shin YK. Determination of the distance between 2 spin labels attached to a macromolecule. Proc Natl Acad Sci U S A. 1995;92:8239–8243. doi: 10.1073/pnas.92.18.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinhoff HJ, Radzwill N, Thevis W, Lenz V, Brandenburg D, Antson A, Dodson G, Wollmer A. Determination of interspin distances between spin labels attached to insulin: Comparison of electron paramagnetic resonance data with the x-ray structure. Biophys J. 1997;73:3287–3298. doi: 10.1016/S0006-3495(97)78353-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinhoff HJ. Inter- and intra-molecular distances determined by EPR spectroscopy and site-directed spin labeling reveal protein-protein and protein-oligonucleotide interaction. Biol Chem. 2004;385:913–920. doi: 10.1515/BC.2004.119. [DOI] [PubMed] [Google Scholar]
- 37.Salikhov KM. Contributions of Exchange and Dipole-Dipole Interactions to the Shape of EPR Spectra of Free Radicals in Diluted Solutions. Appl Magn Reson. 2010;38:237–256. [Google Scholar]
- 38.Ysacco C, Rizzato E, Virolleaud M, Karoui H, Rockenbauer A, Le Moigne F, Siri D, Ouari O, Griffin R, Tordo P. Properties of dinitroxides for use in dynamic nuclear polarization (DNP) Phys Chem Chem Phys. 2010;12:5841–5845. doi: 10.1039/c002591g. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Villamena FA, Rockenbauer A, Song Y, Zweier JL. Structural Factors Controlling the Spin-Spin Exchange Coupling: EPR Spectroscopic Studies of Highly Asymmetric Trityl-Nitroxide Biradicals. J Am Chem Soc. 2013;135:2350–2356. doi: 10.1021/ja311571v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathies G, Caporini MA, Michaelis VK, Liu Y, Hu K-N, Mance D, Zweier JL, Rosay M, Baldus M, Griffin RG. Efficient Dynamic Nuclear Polarization at 800 MHz/527 GHz with Trityl-Nitroxide Biradicals. Angew Chem Int Ed. 2015;54:11770–11774. doi: 10.1002/anie.201504292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gawrisch K. The Dynamics of Membrane Lipids. CRC Press; 2005. [Google Scholar]
- 42.Hubbell WL, McConnel HM. Orientation and motion of amphiphilic spin labels in membranes. Proc Natl Acad Sci U S A. 1969;64:20. doi: 10.1073/pnas.64.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rottem S, Hubbell WL, Hayflick L, McConnel HM. Motion of fatty acid spin labels in plasma membrane of mycoplasma. Biochim Biophys Acta. 1970;219:104. doi: 10.1016/0005-2736(70)90065-9. [DOI] [PubMed] [Google Scholar]
- 44.Wylie BJ, Dzikovski BG, Pawsey S, Caporini M, Rosay M, Freed JH, McDermott AE. Dynamic nuclear polarization of membrane proteins: covalently bound spin-labels at protein-protein interfaces. J Biomol NMR. 2015;61:361–367. doi: 10.1007/s10858-015-9919-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voinov MA, Good DB, Ward ME, Milikisiyants S, Marek A, Caporini MA, Rosay M, Munro RA, Ljumovic M, Brown LS, Ladizhansky V, Smirnov AI. Cysteine-Specific Labeling of Proteins with a Nitroxide Biradical for Dynamic Nuclear Polarization NMR. J Phys Chem B. 2015;119:10180–10190. doi: 10.1021/acs.jpcb.5b05230. [DOI] [PubMed] [Google Scholar]
- 46.van der Cruijsen EAW, Koers EJ, Sauvee C, Hulse RE, Weingarth M, Ouari O, Perozo E, Tordo P, Baldus M. Biomolecular DNP-Supported NMR Spectroscopy using Site-Directed Spin Labeling. Chem - Eur J. 2015;21:12971–12977. doi: 10.1002/chem.201501376. [DOI] [PubMed] [Google Scholar]
- 47.Fernandez-de-Alba C, Takahashi H, Richard A, Chenavier Y, Dubois L, Maurel V, Lee D, Hediger S, De Paepe G. Matrix-Free DNP-Enhanced NMR Spectroscopy of Liposomes Using a Lipid-Anchored Biradical. Chem - Eur J. 2015;21:4512–4517. doi: 10.1002/chem.201404588. [DOI] [PubMed] [Google Scholar]
- 48.Wenk P, Kaushik M, Richter D, Vogel M, Suess B, Corzilius B. Dynamic nuclear polarization of nucleic acid with endogenously bound manganese. J Biomol NMR. 2015;63:97–109. doi: 10.1007/s10858-015-9972-1. [DOI] [PubMed] [Google Scholar]
- 49.Siaw TA, Fehr M, Lund A, Latimer A, Walker SA, Edwards DT, Han S-I. Effect of electron spin dynamics on solid-state dynamic nuclear polarization performance. Phys Chem Chem Phys. 2014;16:18694–18706. doi: 10.1039/c4cp02013h. [DOI] [PubMed] [Google Scholar]
- 50.Perras FA, Reinig RR, Slowing II, Sadow AD, Pruski M. Effects of biradical deuteration on the performance of DNP: towards better performing polarizing agents. Phys Chem Chem Phys. 2016;18:65–69. doi: 10.1039/c5cp06505d. [DOI] [PubMed] [Google Scholar]
- 51.Corzilius B, Smith AA, Barnes AB, Luchinat C, Bertini I, Griffin RG. High-Field Dynamic Nuclear Polarization with High-Spin Transition Metal Ions. J Am Chem Soc. 2011;133:5648–5651. doi: 10.1021/ja1109002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


