Abstract
The circadian clock machinery is responsible for biological timekeeping on a systemic level. The central clock system controls peripheral clocks through a number of output cues that synchronize the system as a whole. There is growing evidence that changing cellular metabolic states have important effects on circadian rhythms and can thereby influence neuronal function and disease. Epigenetic control has also been implicated in the modulation of biological timekeeping, and cellular metabolism and epigenetic state seem to be closely linked. We discuss the idea that cellular metabolic state and epigenetic mechanisms may work through the circadian clock to potentially regulate neuronal function and to influence disease states.
The circadian clock is an extensive molecular network of timing mechanisms that converge to maintain organismal physiological state. In mammals, biological rhythms are established and maintained by a central clock consisting of around 20,000 pacemaker neurons found in the suprachiasmatic nucleus (SCN)1. SCN neurons are entrained by light, the most powerful zeitgeber (time-giver), via the retinohypothalamic tract (RHT). The central SCN clock directs rhythms in a number of peripheral tissues using several output cues, including numerous secreted paracrine signals 2, 3, and thereby helps to synchronize the clock system. However, peripheral clocks are also entrained by extrinsic cues, specifically food intake, which operates as a powerful zeitgeber. Other findings have shown that various ‘nutrient sensors’ are linked to circadian rhythms2, 4, 5, reinforcing the idea that there is a tightly coupled relationship between metabolic state and the clock. A further layer of this complex timing mechanism lies in the emerging link between cellular metabolic state and epigenetics6.
Circadian clockwork relies on a highly regulated network of transcriptional and translational loops that drive clock-controlled gene (CCG) expression. It is believed that approximately 10% of the genome in a given cell is subject to circadian oscillation, although this is an estimate as most oscillatory genes are different from one tissue to another7, 8. The positive transcriptional limb of the clock machinery is composed of the core transcription factors CLOCK and BMAL1 (also known as ARNTL), which drive circadian gene expression. Among these transcribed clock genes are Period (Per) and Cryptochome (Cry) family members. Their products comprise the negative regulatory arm of the circadian clock system9, 10. The circadian clock machinery has been reviewed in detail elsewhere5, 11, 12. A variety of chromatin remodelers and epigenetic events have been associated with the oscillatory nature of circadian transcription (Table 1)3. These might be directly linked to cyclic changes in the levels of specific metabolites, potentially placing the clock at an interface between cellular metabolism and epigenetic control (Figure 1). As an example, SIRT1 is a NAD+-dependent histone deacetylase (HDAC) that is known to deacetylate the circadian transcription factors BMAL113 and PER214, as well as the histone H3 at Lys9/Lys14, thereby contributing to circadian gene expression3. SIRT1 is also a metabolic sensor, as it requires binding of its coenzyme NAD+ for enzymatic activity. As the levels of NAD+ oscillate over the circadian cycle15, 16, the enzymatic HDAC activity of SIRT1 oscillates, which links the metabolic state of the cell with an epigenetic mechanism dependent on the circadian clock.
Table 1.
Epigenetic Modifiers and the Circadian Clock
| Enzyme | Function | Target | Modification |
|---|---|---|---|
| Circadian Locomotor Output Cycles Kaput(CLOCK) | Transcription factor & histone acetyltransferase (HAT) | H3K9/1463 and BMAL190 | Acetylation |
| p300 or EP300 | HAT | H3K9/1491 | Acetylation |
| CREB-binding protein (CBP) | HAT | H3/H492 | Acetylation |
| p300/CBP-associated factor (PCAF) | HAT | H3/H492 | Acetylation |
| Sirtuin 1 (SIRT1) | Histone deacetylase (HDAC) | H3K9/1413, BMAL113, PER214 | Acetylation |
| Histone deacetylase 3 (HDAC3) and nuclear receptor corepressor 1 (Ncor1) | HDAC | H3K993, 94 | Acetylation |
| Mixed Lineage Leukemia (MLL1) | Histone methyltransferase (HMT) | H3K487 | Methylation |
| Enhancer of Zeste Homolog 2 (EZH2) | HMT | H3K2795 | Methylation |
| Jumonji/ARID domain-containing protein 1A (JARID1a) or KDM5A | Histone demethylase | H3K496 | Methylation |
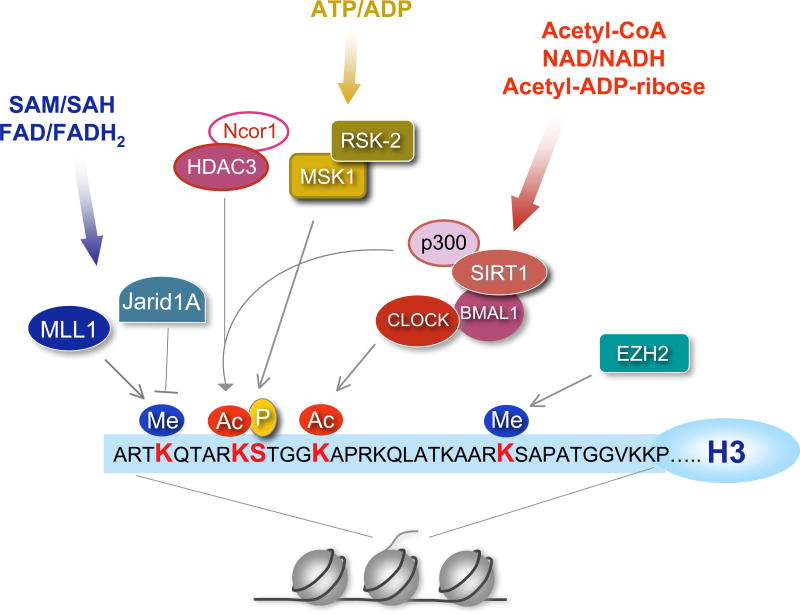
Figure 1. Histone modifications and the circadian clock.
At least 10% of the genes in any given cell are expressed in a cyclic manner under circadian control mediated by cyclic chromatin modifications at the promoters of clock-controlled genes. Here we show a schematic representation of the histone H3 tail, the relevant post-translational modifications and the chromatin remodelers involved in circadian control. Phosphorylation (serine 10 or S10), acetylation (lysine 9/14 or K9/14) and methylation (K4 and K27) are associated with circadian transcription. Some chromatin modifiers may be directly or indirectly modulated by the circadian system (Table 1). Methylation at K4 is consistently associated with gene activation and might be crucial for circadian gene transcription and recruitment of circadian chromatin remodeling complexes 87. Non-histone proteins can also undergo clock-dependent acetylation, as is the case for BMAL1 (Table 1). Several chromatin remodelers can be considered metabolic sensors, as they use metabolites for their enzymatic function; for example, the NAD+-dependent deacetylase SIRT1, whose role in circadian control and physical interaction with CLOCK revealed a link between the circadian clock and cellular metabolism. For enzymatic functions of these chromatin modifiers, references, and abbreviations, see Table 1. MSK1: mitogen- and stress-activated protein kinase (MSK1), RSK2: ribosomal S6 kinase 2 (see references 88, 89).
The importance of the circadian clock is highlighted by the fact that defects in timekeeping give rise to a variety of patho-physiological manifestations. Circadian dysfunction has been linked to sleep disorders, depression, bipolar disorder, and changes in cognitive function and memory formation. A detailed discussion of clock biology and neuronal function or disease state is presented elsewhere 17–19. Here, we discuss the connection between neuronal function and the circadian clock, especially as it relates to underlying metabolic states and an epigenetic framework, which might feedback and modulate the clock in ways not yet fully appreciated. This article will address the current understanding of metabolic feedback from the periphery to the central clock, the possible connection between metabolism and epigenetics in sleep regulation and mental disease state, including potential links with memory formation and synaptic plasticity in mammalian model systems of the clock.
Metabolic feedback to the SCN
It is known that the human brain uses about 20% of the body's energy even though it only comprises about 2% of total body mass. Thus, nutritional inputs and circadian metabolic changes are critical for brain functions. The control of peripheral functions by the hypothalamic central oscillator is well established1–3, but it has also been suggested that various hormones and metabolites that are generated by peripheral clocks could feed back to the SCN (Figure 2). Glucose is the dominant source of energy for the brain and modulation of hypothalamic adenosine monophosphate (AMP)-activated protein kinase (AMPK) activity is sufficient and necessary for hypothalamic nutrient-sensing mechanisms to alter glucose production in vivo20. Importantly, the brain contains structures and circuits able to sense changes in glucose concentration, so-called glucosensors21, 22. Projections from these ‘glucosensing elements’ into the hindbrain and hypothalamus transmit information about circulating glucose levels that can be integrated with inputs from other regions in the brain, such as circadian influences. Projections from the SCN via the sub-paraventricular zone and dorsomedial nucleus (DMH) can be integrated with glucosensing information from the hindbrain to affect ACTH and glucocorticoid responses to hypoglycemia across the day22.
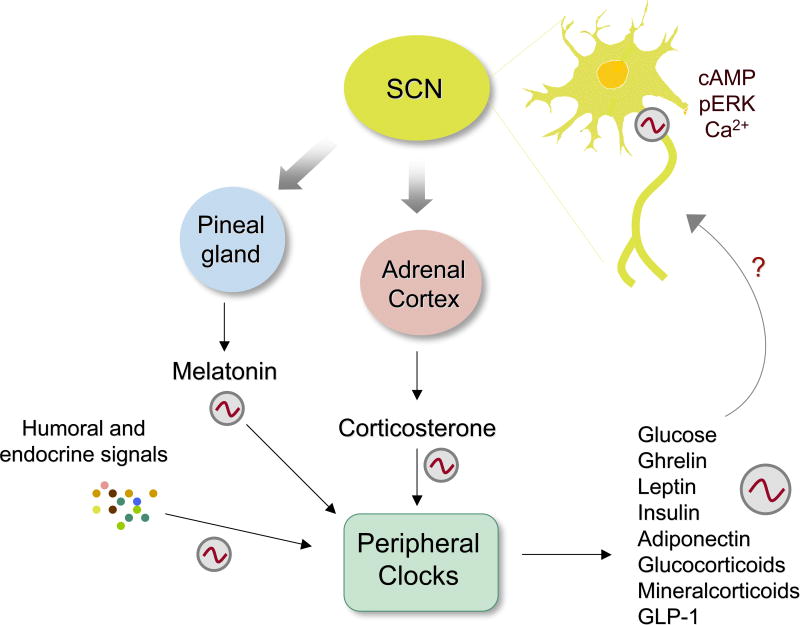
Figure 2. A network of clocks and their interplay.
The central clock in the SCN controls a variety of endocrine and metabolic functions. Neurons of the SCN undergo oscillations in depolarization, activation of the ERK/MAPK cascade and CREB activity, as well as oscillations in gene expression. In addition, several small molecules oscillate, including cAMP and calcium. While the SCN indirectly controls oscillations of humoral factors coming from other tissues such as the pineal and adrenal cortex, continuity within the SCN is mediated by loops within VIP and AVP-expressing neurons. Other tissues also maintain circadian output via positive and negative feedback loops within cells that make up different compartments of the tissue. Oscillations in humoral factors control the circadian release of factors from the periphery, such as ghrelin, leptin, insulin, and glucose and these in turn regulate CNS function. Melatonin, which is released in a circadian fashion from the pineal gland, is involved in feedback regulation of the SCN where melatonin receptors are abundantly expressed. Peripheral tissues provide positive and negative feedback to the brain by releasing soluble factors such as hormones and adipokines, including ghrelin (stomach), leptin (adipose tissue), insulin (pancreas), and glucose (liver and other tissues). Thus, the periphery may influence brain functions, and specifically SCN neurons, through a feedback mechanism.
The concepts outlined above lend credit to the hypothesis that cyclic nutritional inputs and peripheral clocks might influence specific brain functions3. Various neurotransmitters and hormones are regulated by and feed back to the SCN (Figure 2). Whereas peripheral tissues respond acutely to glucose demands, the central nervous system (CNS) anticipates glucose demands. It accomplishes this, in part, by communicating with the periphery to release glucose and insulin in a circadian manner. In humans, insulin release is highest during the early morning, when the body anticipates upcoming glucose metabolism23, 24. There is evidence that the SCN helps to maintain this balance, as lesions of the SCN eliminate plasma glucose and insulin rhythmicity25, 26. These oscillations seem to be independent of a loss of rhythmicity in food intake24. Circadian oscillations in glucose and insulin production are also controlled by inhibitory and excitatory inputs into the pre-autonomic neurons of the paraventricular nucleus (PVN) via the SCN. GABAergic stimulation of pre-autonomic neurons of the PVN produces hyperglycemia but only during the light period, whereas stimulation of PVN neurons by the glutamatergic agonist N-methyl-D-aspartic acid (NMDA) does not show this time-of-day effect 27. The SCN seems to use rhythmic GABAergic projections to control the activity of sympathetic pre-autonomic neurons of the PVN, as SCN-lesioned animals show no circadian variation in hyperglycemia after GABAergic antagonism. Although it is unclear how changes in glucose levels could contribute to the feedback mechanism characteristic of circadian regulation, glucose can down-regulate clock gene transcription in cultured fibroblasts28.
One issue that is of extreme interest, but yet virtually unexplored, relates to how peripheral signals are sensed by oscillators in the brain. Importantly, there is increasing evidence for the presence of a food-entrainable oscillator (FEO) in the hypothalamus. How this FEO senses metabolic variations from the periphery and potentially communicates these metabolic cues to the SCN are basically unknown (Figure 3). Despite the large gap in available evidence for how the SCN responds to metabolic cues, a recent study has shown that redox state of SCN neurons is dependent on a functional clock29. Furthermore, some drugs of abuse, such as methamphetamine, can circumvent and/or impinge on the dominance exerted by the SCN through molecular pathways that appear to be distinct from the canonical clock30, though this idea is still open to debate31. In this respect, the recent finding that ketamine might operate directly on the clock machinery could be relevant32 (also see below). Again, the circuitries and functional relationships between these seemingly separated neuronal structures within the hypothalamus and midbrain are unexplored (Figure 3).
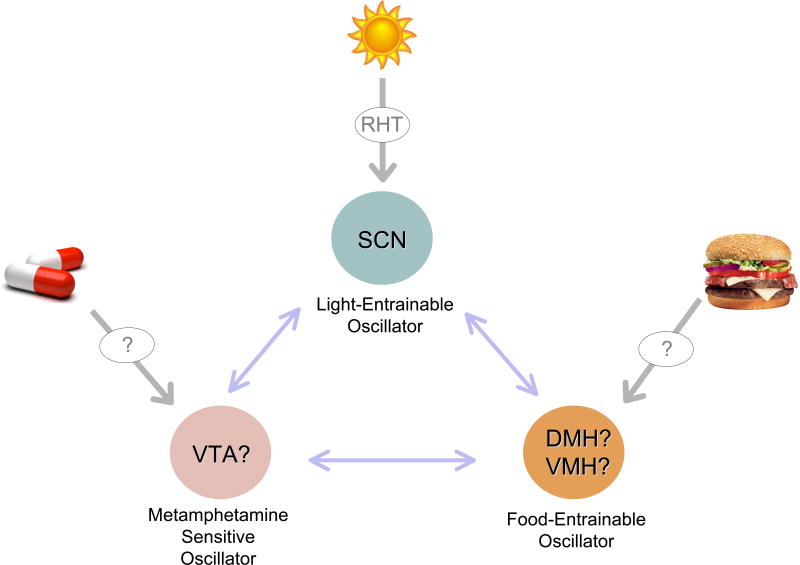
Figure 3. How many clock centers are in the brain?
The central role of the SCN as light-entrainable oscillator is well documented. The light signal, through the retinohypothalamic tract (RHT), exerts critical control over SCN neurons. Accumulating findings indicate the presence of a food-entrainable oscillator (FEO), probably in either the dorsomedial hypothalamus (DMH) or the ventromedial hypothalamus(VMH), as well as a methamphetamine-sensitive oscillator, which could be localized in the ventral tegmental area (VTA). The functional relationships and circuitry interplay among these centers remains unexplored.
Metabolism, sleep and the clock
Mutations in the circadian clock machinery have been linked with a number of sleep-related disorders. In humans, a point mutation in the Period gene Per2 causes familial advanced sleep phase syndrome (FASPS), in which the time of sleep is advanced33,34. Also, delayed sleep phase syndrome (DSPS), which is associated with delayed sleep schedule, has been associated with multiple polymorphisms in the Clock gene35. A number of mouse models with mutations in circadian genes have shown disruptions in sleep schedules, including the Bmal1 knockout, the Clock mutant, the Npas2 knockout and the Cry1/2 double knockout35. Alterations in sleep patterns have also been associated with a number of disorders, including depression, which will be discussed in a later section. The sleep/wake schedule is regulated through a number of neurotransmitters that function in an excitatory or sleep-promoting manner (for a detailed review of sleep, see references18, 36). Excitatory neurotransmitters include serotonin, histamine, acetylcholine, noradrenaline and orexins, which are released during wakefulness, whereas sleep-promoting signals such as melatonin, glycine, GABA and adenosine induce the switch from wake to sleep37. The circadian connection with sleep-wake cycles is intuitive and well documented, but the potential link whereby the central circadian clock is regulated by metabolic state that can feedback and modulate sleep-wake cycles is an intriguing idea.
ATP follows a circadian oscillation in the rat SCN38, and its breakdown product adenosine is crucial for the transition from wake to sleep; accumulation of adenosine in the basal forebrain ultimately signals the release of GABA and initiates the sleep phase18. Sleep deprivation leads to elevated levels of adenosine in the basal forebrain37. On a molecular level, adenosine is phosphorylated by adenosine kinase to produce AMP, which activates AMPK, a sensor of AMP/ATP intracellular levels39. AMPK is a master regulator of intracellular metabolism and is involved in glucose uptake, lipogenesis and fatty acid oxidation40; sleep deprivation has been reported to lead to elevated levels of phosphorylated AMPK in the hypothalamus41. AMPK is both directly and indirectly linked with the circadian clock, demonstrating how interconnected these networks are. Activated AMPK destabilizes and degrades protein levels of the circadian repressors CRY142 and indirectly, PER243, and thereby to adversely affect circadian rhythms in the periphery. Also, AMPK is further connected to peripheral circadian clocks, as AMPK enhances SIRT1 activity by inducing levels of NAD+, both in a nicotinamide phosphoribosyltransferase (NAMPT)-dependent44 and independent manner45. As previously discussed, the reported connection between the deacetylase SIRT1 and the circadian clock is established13, 14, and this connection further links cellular metabolic state governed by AMPK with the circadian epigenome (Figure 4). It could be speculated that oscillating ATP levels in the SCN modulate levels of adenosine – also involved in sleep regulation18 – revealing metabolic control via AMPK and a fascinating link with the circadian clock circuitry. Indeed, it has been shown that the changing ATP levels during sleep lead to modulation of AMPK activity, setting the stage for increased anabolic processes during sleep46. Thus, the clock system, through an enzymatic and epigenetic regulatory network, could contribute to the molecular events leading to the restorative biosynthetic processes that occur during sleep. The question that remains open relates to the contribution of the central SCN clock to peripheral clocks: does the AMPK/clock connection exclusively modulate metabolic tissues in the periphery or does it also operate in the SCN, or even other brain regions as well? These questions require further exploration, but it is possible that a detailed network might exist that connects metabolism and epigenetic control with sleep.
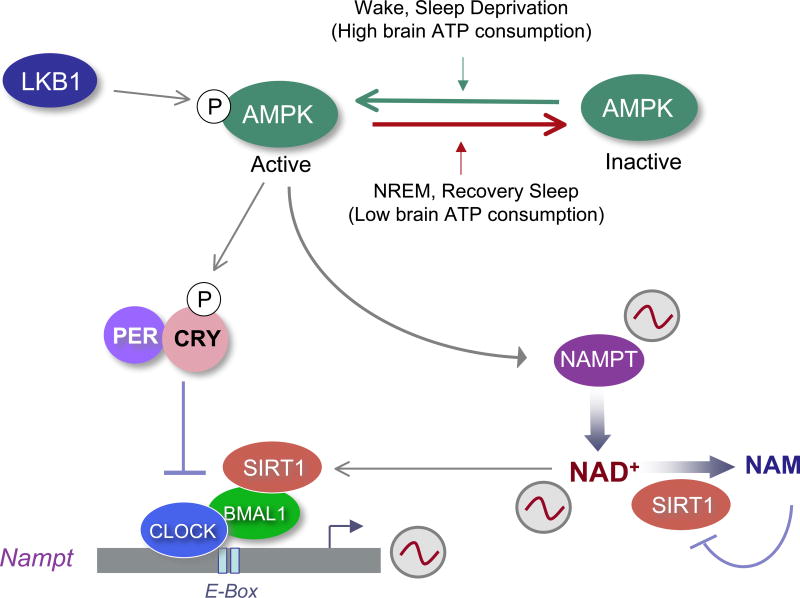
Figure 4. Linking NAD+ metabolism to circadian clock and sleep.
The circadian clock controls the expression of nicotinamidephosphoribosyltransferase (Nampt), the rate limiting enzyme in mammalian NAD+ biosynthesis from nicotinamide (NAM), via the NAD+ salvage pathway. Oscillating levels of NAMPT result in cyclic variations in NAD+, which determine the circadian activity of SIRT1 and possibly other NAD+-dependent enzymes. Consequently, SIRT1 determines the oscillatory levels of its own co-enzyme, NAD+. In addition, Liver kinase B1 (LKB1)-activated AMPK has been shown to control the phosphorylation of CRY proteins as well as the activity of NAMPT. Thus, an enzymatic network that depends on cellular metabolism is intimately associated with the circadian clock machinery. This may constitute a critical link between sleep control and the circadian clock, as AMPK activity is coupled to sleep state.
An additional feature of sleep and metabolic control may be found in the melatonin pathway. The hormone melatonin is produced by the pineal gland during the dark period, and melatonin levels oscillate, as the rate-limiting enzyme in melatonin synthesis arylalkylamine-N-acetyltransferase (Aanat) depends in large part on the cyclic AMP pathway and the yin-yang effect of CREB and CREM-mediated promoter activity47. Melatonin is released in a circadian fashion by the pineal gland to control physiological events as disparate as heart rate and sleep homeostasis. Indeed, melatonin receptors have been identified in many peripheral tissues 48. Melatonin is metabolized quickly in the liver and excreted as 6-sulphatoxymelatonin (aMT6)49. Patients with liver cirrhosis have disturbances in normal sleep patterns as a result of disrupted oscillation of melatonin50 and impaired melatonin metabolism51. This clinical finding suggests that impairments in the underlying metabolic state can also lead to disturbances in the biological timekeeping system. In addition, the peripheral liver clock might feedback to the central clock in the SCN to regulate sleep-wake cycles. As mentioned above, the concept that the peripheral circadian clocks can provide feedback cues to the central clock is an emerging idea in the field3. What is not understood is the role of metabolites, for example aMT6, as feedback cues to the central and peripheral clock systems. An even deeper layer of complexity is suggested by the idea that metabolic sensors such as AMPK or SIRT1 could provide an epigenetic link to feedback to the clock and regulate sleep state.
Memory and the clock connection
Synaptic plasticity and long-term potentiation (LTP) are believed to follow circadian trends52, 53 and are considered to constitute underlying mechanisms of learning and memory 17. The circadian clock has been implicated in learning and memory54, though the molecular pathways remain elusive. The role of CREB in synaptic plasticity and memory formation has been long appreciated55. An interesting addition to the role of CREB in memory formation was recently reported, whereby the mammalian sirtuin, SIRT1, was demonstrated to play a role in synaptic plasticity. SIRT1 regulates the expression of a brain-specific microRNA, miR-134, which is critical in maintaining levels of CREB and brain-derived neurotrophic factor (BDNF) that are necessary for proper cognitive function and memory formation56. This study establishes a direct link between cognition/memory formation and epigenetic mechanisms, which can also be connected to the circadian clock. Based on the known oscillation of NAD+ and SIRT1 activity, it would be intriguing to determine how this would affect miR-134 levels in the brain, and the consequent outcome on synaptic plasticity and memory. This possibility is not so far-fetched since another micro-RNA, miR-132, modulates the circadian clock57, 58. miR-132 is induced by photic entrainment cues via the MAPK/CREB signaling pathway, modulates clock-gene expression and attenuates the entraining effect of light58. Interestingly, some miR-132 target genes encode proteins involved in chromatin remodeling (MeCP2, p300, Jarid1a) and are connected to the circadian epigenome (Figure 1; Table 1), revealing a possible coordinated regulation of chromatin remodeling by this miRNA within SCN neurons57. Finally, SIRT1 has been linked with the modulation of genes involved in lipid metabolism59, 60 as well as glucose metabolism60 in the hippocampus, a regulatory pathway that has been linked with cognition and synaptic plasticity. It would be intriguing to determine if the circadian clock plays a role in this pathway.
Epigenetic changes or chromatin modifications have also been implicated in the early stages of long-term memory (LTM) formation. Histone H3 becomes acetylated in neurons within the CA1 area of the hippocampus in a fear conditioning model, specifically in the early stage of LTM61. Histone acetylation depends on activation of NMDA receptors and ERK/MAPK, with a role for PKA and/or PKC61. Interestingly, many of these molecules have been reported to modulate circadian rhythms. NMDA specific receptors are involved in entrainment of the circadian clock in SCN neurons62. The possibility that NMDA-dependent pathways could activate histone acetyltransferase (HAT) activity of the CLOCK63 or a CLOCK-dependent complex that facilitates oscillations in histone modifications64, would link the circadian clock and memory with an epigenetic pathway. Also, phosphorylation of histone H3 at serine 10 (H3S10) has been linked with light-induction in the SCN through an NMDA receptor-dependent mechanism, which is responsible for immediate-early gene expression and circadian gene expression65. This suggests another possible involvement of kinase signaling events (most likely MAPK- or PKC-directed) that could subsequently lead to H3 acetylation, a pathway that has been described previously whereby H3-S10 phosphorylation generates a privileged substrate for efficient H3 acetylation at Lys 14 (H3K14)66, 67.
Mental disorders and the clock
A number of mental disorders have been associated with dysfunctions in the circadian clock system. We discuss two of these conditions in the context of a potential underlying network that might link the clock to metabolic state and epigenetic changes that could be altered in disease.
Depression and SAD
Alterations in the expression of circadian genes — specifically Clock, Bmal1 and Per1— have been observed in patients with a history of depression68. Animal models with mutations or disruptions in the circadian machinery show changes in mood and behavior. Knockdown of Clock in the ventral tegmental area (VTA) of mice using a short hairpin RNA (shRNA) resulted in increased depression-like behavior, as measured by immobility in a forced swim test (FST) and delayed escape time in a learned helplessness (LH) model69. Clock knockdown also resulted in increased firing of dopaminergic neurons in the VTA, which was attributed to altered expression of genes involved in dopamine synthesis and metabolism in the VTA69. The enzyme tyrosine hydroxylase (which is essential for dopamine synthesis), dopamine transporters and dopamine receptors have all been reported to follow rhythmic oscillation70. Also, mice deficient in the D2 receptor show aberrant light-masking (light-induced suppression of locomotion)71 and dopamine can regulate circadian gene expression72, 73. These data suggest that defects in the clock system of the VTA can alter dopamine synthesis and metabolism and might thereby contribute to depression-like symptoms. These defects in dopamine metabolism could potentially feedback and functionally disrupt the transcriptional potential of the circadian machinery. As mice lacking D2 receptors show reduced induction of Per1 expression by light73, and light-induced Per1 expression has been linked with chromatin modifications65, we can envision a far more complex regulatory network that could connect metabolism, epigenetic regulation and depression.
In seasonal affective disorder (SAD; also known as “winter depression”) patients experience depression symptoms during the winter season as a result of delayed dawn and subsequent alterations in circadian rhythms. Phase delays in circadian secretion of cortisol and melatonin at specific times of the day have been observed in patients with either SAD or depression, which further links the clock on a molecular level with these diseases74. Long periods of phototherapy are an important treatment for patients with SAD and have been shown to be effective in mimicking dawn simulation, or gradual light illumination75.Light is known to influence chromatin remodeling in clock neurons65, so it is possible that light-induced chromatin modifications could ‘jump-start’ immediate-early gene transcription and circadian gene expression – a cascade of events which could contribute to long-term effects in the treatment of SAD and depression. In this context, the long-lasting effects of light on the circadian clock are relevant. Mice were exposed to seasonal variations in light exposure (short versus long days) from birth until weaning and subsequently subjected to behavioral studies or ex vivo fluorescence imaging of circadian gene expression in the SCN. Varying the length of light exposure caused alterations in the period length of circadian behavior and gene expression that persisted for weeks, suggesting that the circadian clock can be developmentally imprinted by light76. This also raises the possibility that neuronal cells in the SCN might have an epigenetic memory that regulates circadian rhythms. Furthermore, shortened light periods consistent with the winter season are correlated with depression symptoms and SAD76. This notion establishes a conceptual framework for light imprinting and epigenetic state early during development that can directly affect the circadian clock and depression symptoms later in life.
Bipolar Disorder
Bipolar disorder is defined by a spectrum of mood fluctuations, ranging from minor to major depression and extending to mania. A number of polymorphisms in human circadian genes have been reported to be correlated with the incidence of bipolar disorder70. The clock mutant mouse model (ClockΔ19) that carries a deletion of exon 19 of the Clock gene, demonstrates hyperactivity and other behavioral phenotypes that are similar to manic-state bipolar disorder77. Moreover, anti-depressants and mood stabilizers have been reported to entrain the circadian clock, which may affect drug efficacy and therefore treatment viability. For example, the mood stabilizer lithium can phase delay the circadian clock70 and ClockΔ19 mice, when given lithium, revert towards normal behavior77. The molecular target of lithium could be glycogen synthase kinase 3 beta (GSK-3β), which has been implicated in modulating the clock machinery11 and has been suggested as a therapeutic target for bipolar disorder78. GSK-3β phosphorylates CRY and PER proteins11, and regulates protein stability of BMAL179 and REV-ERBα80, a negative regulator of Bmal1 expression, in cultured cells. Interestingly, GSK-3β signaling and dopamine pathways have been suggested to modulate each other78, 81. In addition, lithium has effects on localized glucose levels in the brain82, suggesting that it could have other side effects on the clock system.
The anti-depressant ketamine also affects the expression of Bmal1,Cry1 and Per2, possibly through decreased recruitment of the CLOCK:BMAL1 transcriptional complex on circadian gene promoters32. This study defines a mode of action of ketamine that could provide an epigenetic connection: repeated treatments with the drug could engender the consolidation of epigenetic information by progressive enzymatic modifications, leading to long-lasting changes in the circadian transcriptional program83. Other agents such as melatonin receptor agonists have been described as mood stabilizers70, which may also feedback and modulate the clock machinery, especially considering that the metabolic state of melatonin is highly regulated at the intracellular level. A better understanding of how these anti-depressants and mood stabilizers target the clock machinery is needed, in order to achieve optimum treatment regimens.
Future directions
Emerging evidence connects metabolic state with epigenetic mechanisms that are coupled to the circadian clock machinery6. These complex networks of regulation by the clock dictate physiological functions such as sleep, cognition and memory, and relate to a number of mental disorders. We have highlighted a number of potential links between the clock and an epigenetic framework coupled to cellular metabolic state. The extent to which the clock regulates metabolic and epigenetic states in the brain is an area that requires further understanding. In this context, our knowledge to date is limited to metabolic tissues. Also, future studies are needed to elucidate the communication between the central clock and peripheral clocks, as a number of secreted factors could be responsible for circadian entrainment via a FEO. These concepts should then be applied to a number of disease states to determine what metabolic and epigenetic factors are altered in the brain. Considering these ideas, a valuable tool is the use of tissue-specific metabolomics analysis that can be performed to compare metabolite levels in peripheral tissues and in the brain. This analysis would provide information regarding changes in tissue-specific metabolites, especially those occurring in a rhythmic or circadian manner. The recent, comprehensive analysis of the circadian metabolome has revealed the remarkable extent and specificity of its clock regulation84–86. It would be of great interest to further understand the cellular epigenome and how it intersects with metabolomic profiles. A deeper understanding of these interactions will contribute to the development of successful pharmacological and therapeutic strategies for a variety of pathological conditions.
References
- 1.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–76. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 2.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–54. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masri S, Sassone-Corsi P. Plasticity and specificity of the circadian epigenome. Nat Neurosci. 2010;13:1324–9. doi: 10.1038/nn.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckel-Mahan K, Sassone-Corsi P. Metabolism control by the circadian clock and vice versa. Nat Struct Mol Biol. 2009;16:462–7. doi: 10.1038/nsmb.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–42. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katada S, Imhof A, Sassone-Corsi P. Connecting threads: epigenetics and metabolism. Cell. 2012;148:24–8. doi: 10.1016/j.cell.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Akhtar RA, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–50. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 8.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 9.Thresher RJ, et al. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282:1490–4. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 10.Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–77. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–48. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 12.Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–96. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 13.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–28. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 15.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–7. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–4. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerstner JR, Yin JC. Circadian rhythms and memory formation. Nat Rev Neurosci. 2010;11:577–88. doi: 10.1038/nrn2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wulff K, Gatti S, Wettstein JG, Foster RG. Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nat Rev Neurosci. 2010;11:589–99. doi: 10.1038/nrn2868. [DOI] [PubMed] [Google Scholar]
- 19.Huang W, Ramsey KM, Marcheva B, Bass J. Circadian rhythms, sleep, and metabolism. J Clin Invest. 2011;121:2133–41. doi: 10.1172/JCI46043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang CS, et al. Hypothalamic AMP-activated protein kinase regulates glucose production. Diabetes. 2010;59:2435–43. doi: 10.2337/db10-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oomura Y, Ono T, Ooyama H, Wayner MJ. Glucose and osmosensitive neurones of the rat hypothalamus. Nature. 1969;222:282–4. doi: 10.1038/222282a0. [DOI] [PubMed] [Google Scholar]
- 22.Watts AG, Donovan CM. Sweet talk in the brain: glucosensing, neural networks, and hypoglycemic counterregulation. Front Neuroendocrinol. 2010;31:32–43. doi: 10.1016/j.yfrne.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trumper BG, Reschke K, Molling J. Circadian variation of insulin requirement in insulin dependent diabetes mellitus the relationship between circadian change in insulin demand and diurnal patterns of growth hormone, cortisol and glucagon during euglycemia. Horm Metab Res. 1995;27:141–7. doi: 10.1055/s-2007-979926. [DOI] [PubMed] [Google Scholar]
- 24.la Fleur SE, Kalsbeek A, Wortel J, Fekkes ML, Buijs RM. A daily rhythm in glucose tolerance: a role for the suprachiasmatic nucleus. Diabetes. 2001;50:1237–43. doi: 10.2337/diabetes.50.6.1237. [DOI] [PubMed] [Google Scholar]
- 25.Nagai K, et al. SCN output drives the autonomic nervous system: with special reference to the autonomic function related to the regulation of glucose metabolism. Prog Brain Res. 1996;111:253–72. doi: 10.1016/s0079-6123(08)60413-6. [DOI] [PubMed] [Google Scholar]
- 26.Nagai K, Nagai N, Sugahara K, Niijima A, Nakagawa H. Circadian rhythms and energy metabolism with special reference to the suprachiasmatic nucleus. Neurosci Biobehav Rev. 1994;18:579–84. doi: 10.1016/0149-7634(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 27.Kalsbeek A, et al. Circadian control of the daily plasma glucose rhythm: an interplay of GABA and glutamate. PLoS One. 2008;3:e3194. doi: 10.1371/journal.pone.0003194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirota T, et al. Glucose down-regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured Rat-1 fibroblasts. J Biol Chem. 2002;277:44244–51. doi: 10.1074/jbc.M206233200. [DOI] [PubMed] [Google Scholar]
- 29.Wang TA, et al. Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science. 2012;337:839–42. doi: 10.1126/science.1222826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohawk JA, Baer ML, Menaker M. The methamphetamine-sensitive circadian oscillator does not employ canonical clock genes. Proc Natl Acad Sci U S A. 2009;106:3519–24. doi: 10.1073/pnas.0813366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pendergast JS, Oda GA, Niswender KD, Yamazaki S. Period determination in the food-entrainable and methamphetamine-sensitive circadian oscillatoRs) Proc Natl Acad Sci U S A. 2012;109:14218–23. doi: 10.1073/pnas.1206213109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bellet MM, Vawter MP, Bunney BG, Bunney WE, Sassone-Corsi P. Ketamine influences CLOCK:BMAL1 function leading to altered circadian gene expression. PLoS One. 2011;6:e23982. doi: 10.1371/journal.pone.0023982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones CR, et al. Familial advanced sleep-phase syndrome: A short-period circadian rhythm variant in humans. Nat Med. 1999;5:1062–5. doi: 10.1038/12502. [DOI] [PubMed] [Google Scholar]
- 34.Toh KL, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–3. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 35.Kyriacou CP, Hastings MH. Circadian clocks: genes, sleep, and cognition. Trends Cogn Sci. 2010;14:259–67. doi: 10.1016/j.tics.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Mignot E, Taheri S, Nishino S. Sleeping with the hypothalamus: emerging therapeutic targets for sleep disorders. Nat Neurosci. 2002;5(Suppl):1071–5. doi: 10.1038/nn944. [DOI] [PubMed] [Google Scholar]
- 37.Albrecht U. Circadian rhythms and sleep-the metabolic connection. Pflugers Arch. 2011 doi: 10.1007/s00424-011-0986-6. [DOI] [PubMed] [Google Scholar]
- 38.Womac AD, Burkeen JF, Neuendorff N, Earnest DJ, Zoran MJ. Circadian rhythms of extracellular ATP accumulation in suprachiasmatic nucleus cells and cultured astrocytes. Eur J Neurosci. 2009;30:869–76. doi: 10.1111/j.1460-9568.2009.06874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–37. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 40.Kahn BB, Alquier T, Carling D, Hardie DG. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Chikahisa S, Fujiki N, Kitaoka K, Shimizu N, Sei H. Central AMPK contributes to sleep homeostasis in mice. Neuropharmacology. 2009;57:369–74. doi: 10.1016/j.neuropharm.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 42.Lamia KA, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–40. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Um JH, et al. Activation of 5'-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J Biol Chem. 2007;282:20794–8. doi: 10.1074/jbc.C700070200. [DOI] [PubMed] [Google Scholar]
- 44.Fulco M, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–73. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Canto C, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–60. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dworak M, McCarley RW, Kim T, Kalinchuk AV, Basheer R. Sleep and brain energy levels: ATP changes during sleep. J Neurosci. 2010;30:9007–16. doi: 10.1523/JNEUROSCI.1423-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zawilska JB, Skene DJ, Arendt J. Physiology and pharmacology of melatonin in relation to biological rhythms. Pharmacol Rep. 2009;61:383–410. doi: 10.1016/s1734-1140(09)70081-7. [DOI] [PubMed] [Google Scholar]
- 48.Slominski RM, Reiter RJ, Schlabritz-Loutsevitch N, Ostrom RS, Slominski AT. Melatonin membrane receptors in peripheral tissues: distribution and functions. Mol Cell Endocrinol. 2012;351:152–66. doi: 10.1016/j.mce.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arendt J, Bojkowski C, Franey C, Wright J, Marks V. Immunoassay of 6-hydroxymelatonin sulfate in human plasma and urine: abolition of the urinary 24-hour rhythm with atenolol. J Clin Endocrinol Metab. 1985;60:1166–73. doi: 10.1210/jcem-60-6-1166. [DOI] [PubMed] [Google Scholar]
- 50.Steindl PE, et al. Disruption of the diurnal rhythm of plasma melatonin in cirrhosis. Ann Intern Med. 1995;123:274–7. doi: 10.7326/0003-4819-123-4-199508150-00005. [DOI] [PubMed] [Google Scholar]
- 51.Steindl PE, Ferenci P, Marktl W. Impaired hepatic catabolism of melatonin in cirrhosis. Ann Intern Med. 1997;127:494. doi: 10.7326/0003-4819-127-6-199709150-00025. [DOI] [PubMed] [Google Scholar]
- 52.Nishikawa Y, Shibata S, Watanabe S. Circadian changes in long-term potentiation of rat suprachiasmatic field potentials elicited by optic nerve stimulation in vitro. Brain Res. 1995;695:158–62. doi: 10.1016/0006-8993(95)00717-5. [DOI] [PubMed] [Google Scholar]
- 53.Harris KM, Teyler TJ. Age differences in a circadian influence on hippocampal LTP. Brain Res. 1983;261:69–73. doi: 10.1016/0006-8993(83)91284-2. [DOI] [PubMed] [Google Scholar]
- 54.Kondratova AA, Dubrovsky YV, Antoch MP, Kondratov RV. Circadian clock proteins control adaptation to novel environment and memory formation. Aging (Albany NY) 2010;2:285–97. doi: 10.18632/aging.100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silva AJ, Kogan JH, Frankland PW, Kida S. CREB and memory. Annu Rev Neurosci. 1998;21:127–48. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- 56.Gao J, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–9. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alvarez-Saavedra M, et al. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum Mol Genet. 2011;20:731–51. doi: 10.1093/hmg/ddq519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng HY, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–29. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Michan S, et al. SIRT1 is essential for normal cognitive function and synaptic plasticity. J Neurosci. 2010;30:9695–707. doi: 10.1523/JNEUROSCI.0027-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu D, Qiu Y, Gao X, Yuan XB, Zhai Q. Overexpression of SIRT1 in mouse forebrain impairs lipid/glucose metabolism and motor function. PLoS One. 2011;6:e21759. doi: 10.1371/journal.pone.0021759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levenson JM, et al. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–59. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 62.Ding JM, et al. Resetting the biological clock: mediation of nocturnal circadian shifts by glutamate and NO. Science. 1994;266:1713–7. doi: 10.1126/science.7527589. [DOI] [PubMed] [Google Scholar]
- 63.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 64.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–74. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 65.Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat Neurosci. 2000;3:1241–7. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- 66.Cheung P, et al. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–15. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 67.Lo WS, et al. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell. 2000;5:917–26. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- 68.Gouin JP, et al. Altered expression of circadian rhythm genes among individuals with a history of depression. J Affect Disord. 2010;126:161–6. doi: 10.1016/j.jad.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mukherjee S, et al. Knockdown of Clock in the ventral tegmental area through RNA interference results in a mixed state of mania and depression-like behavior. Biol Psychiatry. 2010;68:503–11. doi: 10.1016/j.biopsych.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McClung CA. Circadian rhythms and mood regulation: insights from pre-clinical models. Eur Neuropsychopharmacol. 2011;21(Suppl 4):S683–93. doi: 10.1016/j.euroneuro.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Doi M, et al. Impaired light masking in dopamine D2 receptor-null mice. Nat Neurosci. 2006;9:732–4. doi: 10.1038/nn1711. [DOI] [PubMed] [Google Scholar]
- 72.Hood S, et al. Endogenous dopamine regulates the rhythm of expression of the clock protein PER2 in the rat dorsal striatum via daily activation of D2 dopamine receptors. J Neurosci. 2010;30:14046–58. doi: 10.1523/JNEUROSCI.2128-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yujnovsky I, Hirayama J, Doi M, Borrelli E, Sassone-Corsi P. Signaling mediated by the dopamine D2 receptor potentiates circadian regulation by CLOCK:BMAL1. Proc Natl Acad Sci U S A. 2006;103:6386–91. doi: 10.1073/pnas.0510691103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Monteleone P, Martiadis V, Maj M. Circadian rhythms and treatment implications in depression. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:1569–74. doi: 10.1016/j.pnpbp.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 75.Golden RN, et al. The efficacy of light therapy in the treatment of mood disorders: a review and meta-analysis of the evidence. Am J Psychiatry. 2005;162:656–62. doi: 10.1176/appi.ajp.162.4.656. [DOI] [PubMed] [Google Scholar]
- 76.Ciarleglio CM, Axley JC, Strauss BR, Gamble KL, McMahon DG. Perinatal photoperiod imprints the circadian clock. Nat Neurosci. 2011;14:25–7. doi: 10.1038/nn.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roybal K, et al. Mania-like behavior induced by disruption of CLOCK. Proc Natl Acad Sci U S A. 2007;104:6406–11. doi: 10.1073/pnas.0609625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rowe MK, Wiest C, Chuang DM. GSK-3 is a viable potential target for therapeutic intervention in bipolar disorder. Neurosci Biobehav Rev. 2007;31:920–31. doi: 10.1016/j.neubiorev.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sahar S, Zocchi L, Kinoshita C, Borrelli E, Sassone-Corsi P. Regulation of BMAL1 protein stability and circadian function by GSK3beta-mediated phosphorylation. PLoS One. 2010;5:e8561. doi: 10.1371/journal.pone.0008561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yin L, Wang J, Klein PS, Lazar MA. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science. 2006;311:1002–5. doi: 10.1126/science.1121613. [DOI] [PubMed] [Google Scholar]
- 81.Beaulieu JM, et al. Regulation of Akt signaling by D2 and D3 dopamine receptors in vivo. J Neurosci. 2007;27:881–5. doi: 10.1523/JNEUROSCI.5074-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kohno T, et al. Effects of lithium on brain glucose metabolism in healthy men. J Clin Psychopharmacol. 2007;27:698–702. doi: 10.1097/jcp.0b013e31815a23c2. [DOI] [PubMed] [Google Scholar]
- 83.Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60:961–74. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eckel-Mahan KL, et al. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci U S A. 2012;109:5541–6. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hatori M, et al. Time-Restricted Feeding without Reducing Caloric Intake Prevents Metabolic Diseases in Mice Fed a High-Fat Diet. Cell Metab. 2012;15:848–60. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci U S A. 2012;109:2625–9. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol. 2010;17:1414–21. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Crosio C, Heitz E, Allis CD, Borrelli E, Sassone-Corsi P. Chromatin remodeling and neuronal response: multiple signaling pathways induce specific histone H3 modifications and early gene expression in hippocampal neurons. J Cell Sci. 2003;116:4905–14. doi: 10.1242/jcs.00804. [DOI] [PubMed] [Google Scholar]
- 89.Butcher GQ, Lee B, Cheng HY, Obrietan K. Light stimulates MSK1 activation in the suprachiasmatic nucleus via a PACAP-ERK/MAP kinase-dependent mechanism. J Neurosci. 2005;25:5305–13. doi: 10.1523/JNEUROSCI.4361-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hirayama J, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–90. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 91.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–82. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 92.Curtis AM, et al. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J Biol Chem. 2004;279:7091–7. doi: 10.1074/jbc.M311973200. [DOI] [PubMed] [Google Scholar]
- 93.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–9. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alenghat T, et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Etchegaray JP, et al. The polycomb group protein EZH2 is required for mammalian circadian clock function. J Biol Chem. 2006;281:21209–15. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- 96.DiTacchio L, et al. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science. 2011;333:1881–5. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]