Abstract
The natural history and treatment outcome of hepatitis B viruses (HBV) infection is largely dependent on genotype, subgenotype, and the presence or absence of virulence associated mutations. We have studied the prevalence of genotype and subgenotype as well as virulence and drug resistance associated mutations and prevalence of recombinant among HBV from Bangladesh. A prospective cross-sectional study was conducted among treatment naïve chronic HBV patients attending at Bangabandhu Sheikh Mujib Medical University, Dhaka, Bangladesh for HBV viral load assessment between June and August 2015. Systematical selected 50% of HBV DNA positive patients (every second patient) were enrolled. Biochemical and serological markers for HBV infection and whole genome sequencing (WGS) was performed on virus positive sample. Genotype, subgenotype, virulence, nucleos(t)ide analogue (NA) resistance (NAr) mutations, and the prevalence of recombinant isolates were determined. Among 114 HBV DNA positive patients, 57 were enrolled in the study and 53 HBV WGS were generated for downstream analysis. Overall, 38% (22/57) and 62% (35/57) of patients had acute and chronic HBV infections, respectively. The prevalence of genotypes A, C, and D was 18.9% (10/53), 45.3% (24/53), and 35.8% (19/53), respectively. Among genotype A, C and D isolates subgenotype A1 (90%; 9/10), C1 (87.5%; 21/24) and D2 (78.9%; 15/19) predominates. The acute infection, virulence associated mutations, and viral load was higher in the genotype D isolates. Evidence of recombination was identified in 22.6% (12/53) of the HBV isolates including 20.0% (2/10), and 16.7% (4/24) and 31.6% (6/19) of genotype A, C and D isolates, respectively. The prevalence of recombination was higher in chronic HVB patients (32.2%; 10/31 versus 9.1%; 2/22); p<0.05. NAr mutations were identified in 47.2% (25/53) of the isolates including 33.9% novel mutations (18/53). HBV genotype C and D predominated in this population in Bangladesh; a comparatively high prevalence of recombinant HBV are circulating in this setting.
Introduction
There are an estimated two billion people with serological markers of present or past Hepatitis B virus (HBV) infection globally; 257 million of these are chronically infected[1]. The outcomes of acute HBV infection range from complete recovery to fulminant liver disease. A failure to clear HBV after acute infection may lead to either inactive or active chronic infection, which can induce hepatic insufficiency, end-stage liver disease including liver cirrhosis (LC) and hepatocellular carcinoma (HCC) [2, 3].
Current classification segregates HBV into 10 different genotypes (A-J; segregated by <7.5% genomic sequence diversity); these are further classified into 40 different subgenotypes, which are separated by >4% genomic sequence diversity [4]. The dominant HBV genotypes and subgenotypes differ by geographical location, transmission dynamics, disease progression, and response to antiviral therapy [5]. The clinical progression of HBV infection is dependent on multiple factors, which includes age when infected, genetic factors, and the infecting genotypes and subgenotypes [6]. Notably, factors such as genotype and subgenotype as well as HBV e antigen (HBeAg) status, viral load, drug resistance mutations in the reverse transcriptase (RT) domain of polymerase gene, and mutations in the basal core promoter (BCP) precore (PC) and core gene have a major influence in determining disease progression and treatment outcome [7–9]. End-stage liver disease, and poor response to interferon therapy is commonly observed in chronic HBV infection associated with genotypes C and D [5]. It has also been observed that horizontal HBV transmission is more common with genotypes A and D [5]. Further, a number of genetic characteristics including mutations in preS1, preS2, and S genes have been shown to be associated with viral replication, progression of liver disease including HCC, occult HBV infections (OBI), immune escape, and therapy escape [10–13]. Mutations in the core promoter (CP) have been associated with severity of liver disease; a G1896A mutation in the precore and core gene creates a premature stop codon at position 28 and abolishes the synthesis of HBeAg [10, 14].
Bangladesh is a country with intermediate endemic HBV and a chronic HBV carriage rate of 2–6%. The prevalence of chronic HBV among the general population and various high-risks groups, including intravenous drug users, ranges from 0.8% to 6.2% [15] [16]. However, data regarding the prevalence of HBV genotypes and subgenotypes, the prevalence of recombinant viruses, virulence-associated characteristics such as drug resistance mutations are limited from Bangladesh. In order to address this paucity of data, we performed a prospective cross sectional study to determine the dominant HBV genotypes and subgenotypes. We additionally assessed the prevalence of recombination, resistance associated mutations, and the prevalence of virulence mutations in HBV in this setting.
Materials methods
Study population
A prospective cross-sectional study was conducted among treatment naïve patients attending the Bangabandhu Sheikh Mujib Medical University (BSMMU), a tertiary care hospital in Dhaka, for HBV viral load testing between June and August 2015. All patients attending at BSMMU for HBV viral load assay and providing written informed consent were eligible for enrollment in the study. Parental or guardian consent were collected for patient <18 years of age. Systematically selected 50% (every second) of the HBV DNA positive patients was enrolled in the present study. Venous blood samples were collected from enrolled patients for biochemical, virological, and molecular testing. Plasma samples were stored at -86°C before being shipped to Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam for further analysis. Clinical chemistry results of patients were obtained from the hospital database for analysis. The study was approved by Bangabandhu Sheikh Mujib Medical University ethical review committee (Approval No. BSMMU/2014/10612).
HBV, HCV, HIV serology
All plasma samples were tested for HBs Ag, HBe Ag, anti HBs, anti HBe, anti HBcTotal, and anti HBc IgM using serological tests as per the manufacturer’s recommendation (Beijing Wantai Biological Pharmacy Enterprise Co., Ltd., Beijing, China). Serum samples were classified as being from acute HBV infections (HBs Ag positive, anti HBcTotal positive, anti HBC IgM positive, anti HBs negative), or chronic HBV infections (HBs Ag positive, anti HBcTotal positive, and anti HBc IgM negative and anti HBs negative) according to the CDC guidelines for the interpretation of hepatitis B serological test results (http://www.cdc.gov/hepatitis/HBV/PDFs/SerologicChartv8.pdf). All samples are screened for anti hepatitis C (HCV) antibody by ELISA (Beijing Wantai Biological Pharmacy Enterprise Co., Ltd., Beijing, China) and for anti human immunodeficiency virus (HIV) antibody by ELISA (DIALAB ELISA, Biorad, France) as per manufacturer’s recommendation.
HBV DNA sequencing
Viral load was determined by real-time PCR using the Single Step HBV DNA assay kit (Genebio Inc. Ltd. USA) as per the manufacturer’s instructions. Viral DNA was extracted from 200μL of plasma using QIAamp viral DNA extraction kit (QIAgen GmbH, Hilden, Germany) before elution in 50μL tris borate EDTA (TBE) buffer. HBV nucleic acid was prepared for genome sequencing by PCR amplification of four overlapping fragments (800bp to 1.2 kb) using P1-P2, P3-P4, P5-P6, and P7-P8 primers (P1: 5'-TTTTTCACCTCTGCCTAATCA-3'; P2: 5'-TTGGGATTGAAGTCCCAA TCTGG-3'; P3: 5'-GGGTCACCTTATTCTTGG-3'; P4: 5'-ATAACTGAAAGCCAAACAGTG GG-3'; P5: 5'-GTCTTCTTGGTTGTTCTTCTAC-3'; P6: 5'-GCAGCACAGCCTAGCAGCCAT GG-3'; P7: 5'-CCATACTGCGGAACTCCTAGC-3'; P8: 5'-CAATGCTCAGGAGACTCTAAG GC-3') [17]. The PCR amplification reactions were performed in 40μL volumes containing 50mM Tris-HCl (pH 8.3), 50mM KCl, 1.5mM MgCl2, 200mM deoxynucleoside triphosphates (dNTPs), 1U of Taq DNA-Pwo Polymerase (Expand High Fidelity assay, Boehringer Mannheim) and 30pmol of primer. Amplification was performed for 35 cycles at 94°C for 1 minute, 58°C for 1 minute and 72°C for 1 minute in a thermal cycler (ABI 9800). PCR amplicons were visualized by 1% agarose electrophoresis and stained with Nancy 520 DNA gel stain.
PCR amplicons were purified using the QIAamp PCR product purification kit (QIAgen GmbH, Hilden, Germany). The eluted DNA was quantified by a fluorescence-based dsDNA quantification method using the Quant-iT dsDNA Assay Kit in a Qubit fluorometer (Invitrogen). For sequencing, genomic fragments were pooled into equal quantities of each individual PCR amplicon. One nanogram of pooled DNA from each individual sample was subjected to library preparation using the Nextera XT DNA sample preparation kit (Illumina, San Diego, CA, USA), in which each sample was assigned to a unique barcode sequence using the Nex-tera XT Index Kit (Illumina). Sequencing of the libraries was performed using MiSeq reagent kit v2 (300 cycles, Illumina) on an Illumina MiSeq platform. All samples were sequenced in a single run. The Illumina fastq sequence files were assembled using Genious 8.0.5 software package (Biomatters Ltd, Auckland, New Zealand) utilizing a reference-based mapping tool after primer sequences clipping (i.e. the consensus sequence was obtained by mapping individual reads of each sample to a reference sequence). A minimum variant frequency of 5% and 500-fold coverage were chosen as cut-off values and all analysis was done on dominant/consensus variants. The resulting sequences were deposited in GenBank under accession numbers MF925358 to MF925410.
HBV recombination and phylogenetic analysis
One hundred and three HBV whole genome sequences (WGS) representing all 10 genotypes and at least two sequences for each of the subgenotypes were downloaded from Genbank and combined with 53 HBV WGS from the current study. These 156 sequences were subjected to recombination and phylogenetic analysis (S1 File) [4, 18]. All sequences were analyzed for possible recombination by RDP4 v 4.55 software. Any recombination detected by at least 5 of the 7 programs (RDP, Geneconv, Bootscan, Maxchi, Chimaera, Siscan, and Topol) was considered as true recombination. RDP4 v4.55 standard default setting was used except for Bootscan and Siscan the window sizes 300bp, step size 30 were used. Data on the type of recombination, recombination breakpoint (start and end point), homology with major parent & minor recombinant parent, the size of the recombinant fragment, and the location of the recombination were determined.
For phylogenetic analysis, all 156 complete genome sequence were aligned using MUSCLE in the Genious software package (S1 File) [19]. Phylogenetic analysis was conducted in two stages. In the first stage, analysis was conducted using the full length genome sequences (data not shown). As recombination analysis revealed a highly recombinant population and the limits of the regions susceptible to recombination, we conducted second stage of analysis to determine the evolutionary relationships between the isolates. In this stage, two partial sequence alignments were created; the first alignment contained WGS without the recombination susceptible region (1–1,272bp and 2,028–3,215bp) and the second set of alignment contained the recombination susceptible region only (1,273bp—2,027 bp). The sequence alignments were subjected to Jmodel test to identify the best model for phylogenetic analysis. The suggested nucleotide substitution model (GTR+G+I) was subsequently used in the phylogenetic analysis using RAxML v7.2.8 (available in Genious package). To confirm the reliability of phylogenetic tree, bootstrap resampling and reconstruction were performed 1000 times [20].
HBV mutation analysis
The HBV polymerase gene (nt 2306..1623) consists of four domains (terminal protein (TP) domain; nt 2307–2843, spacer; 2844..3215, 1..135, reverse transcriptase (RT)domain; nt 136..1167, and RNaseH (RH) domain; nt 1168..1623. The RT domain (136bp to 1167bp) was analyzed for the presence of 42 potential nucleos(t)ide analogue (NA) resistance (NAr) mutations. This included the primary drug resistance mutation (rt169, rt180, rt181, rt184, rt194, rt202, rt204, rt236, and rt250), the secondary drug resistance mutations (rt80 and rt173), the putative NAr mutations (rt53, rt54, rt82, rt84,rt85, rt91, rt126, rt128, rt139, rt153,rt166, rt191,rt200, rt207,rt213, rt214,rt215,rt217, rt218, rt221, rt229, rt233, rt237, rt238, rt245, and rt256), and the pretreatment mutations (rt38, rt124, rt134, rt139, rt224, and rt242) as previously described [21]. The distribution of mutations in six functional regions (F, A, B, C, D, and E) of RT domain and the five the regions (F-A, A-B, B-C, C-D, and D-E) connecting them were analyzed. The RT domain was also analyzed for genotype dependent AA polymorphism at rt38, rt53, rt54, rt91, rt124, rt126, rt139, rt153, rt221, rt224, and rt256 positions [21].
The preS2/S1 regions were analyzed for preS1 deletion, preS1 mutations (A2962G, C3026A/T, C2964A, and C3116T), preS2 start codon deletion, and preS2 mutations (T53C). The S gene was analyzed for mutations in major hydrophilic region (MHR) (99aa-169aa) including the “a” determinant region (Y/L100C/I, Q101H/R, T113S, T115N, I/T126S/N, T127P, A128V, A143L, G145R, R160K, Y161F, E164D, and A168V), and other mutations associated with increased risk of HCC (N3S, R24K, P56Q, P62L, F85C, L126T/S, A168V, V184A, and S204R) [22–25] [26] [27]. Mutations in BCP (C1653T, T1753C, G1757A, A1762T, G1764A, C1766G, and T1768A), and the PC/core region associated with HCC (G1896A, G1899A, A2159G and A2189C/T, and G2203A/T) were also analyzed.
All data (socio demographic, biochemical, and virological) was recorded and analyzed using the Statistical Package for the Social Sciences (IBM SPSS version 23, NY, USA). Pearson’s Chi squared test was used for the comparison qualitative variables and Mann—Whitney U test for ordinal scale variables. The one-way ANOVA test was used for comparing the significance between three or more groups. A p value <0.05 was inferred to indicate statistical significance.
Results
From June to August 2015, a total of 274 treatment naive patients attending BSMMU for HBV viral load measurement were invited to participate the study. Among these, 159 patients provide informed consent to join the study. Of these 159 patients, 114 were HBV DNA positive and 57 patients (50%; every second patient) were enrolled in this study. The sex, demographics, liver enzyme profiles, HBV serology, and hepatitis status of all 57 patients are presented in Table 1. Approximately 80% (47/57) of the patients were male and the median age was 32.2 years (12 to 65 years; IQR 20); 38.5% (22/57) of patients had an acute HBV infection and 61.5% (35/57) had a chronic HBV infection. AST, ALT, and serum bilirubin was significantly higher in those with an acute HBV infection (p<0.05). HBe Ag seropositivity was higher amongst the acute HBV patients than the chronic HBV patients, although this was not statistically significant. The median viral loads of the acute HBV infection cases were significantly higher than chronic HBV infection (2.9x106 versus 3.2x 103; p<0.001) (Table 1). All patients were HCV and HIV negative (data not shown).
Table 1. Socio demographic, biochemical, serological and virological profile of 57 treatment naïve acute and chronic HBV DNA positive patients enrolled in the study during June to August 2014.
| Variable | All | Acute infection | Chronic infection | p value | |
|---|---|---|---|---|---|
| % (n) | % (n) | % (n) | |||
| N = 57 | 38.5 (22) | 61.5 (35) | |||
| Age a | 32.25 (12–65) | 29.5 (19–65) | 28.0 (12–60) | 0.104 c | |
| Sex | |||||
| Male | 82.5 (47) | 90.9 (20) | 77.1 (27) | ||
| Female | 17.5 (10) | 9.1 (2) | 22.9 (8) | ||
| Biochemical (mean; min–max) | |||||
| AST(SGOT) a | 74.82 (18.0–482.0) | 101.95 (22.0–482.0) | 57.2 (18.0–319.0) | 0.01b | |
| ALT (GSPT) a | 87.69 (22.0–405.0) | 116.27 (28.0–405.0) | 69.2 (22.0–313.0) | 0.002 b | |
| Creatinin a | 73.2 (34.0–134.0) | 75.8 (48.0–134.0) | 71.4 (34.0–119.0) | 0.486 b | |
| Bilirubin a | 21.4 (2.5–189.1) | 36.2 (2.9–189.1) | 5.7 (2.5–21.0) | 0.006 b * | |
| HBV Serology | |||||
| Anti HBs | 5.3 (3) | 4.5 (1) | 5.7 (2) | 0.847 c | |
| HBe Ag | 33.3 (19) | 50.0 (11) | 22.9 (8) | 0.034 c * | |
| Anti HBe | 47.4 (27) | 36.4 (8) | 54.3 (19) | 0.187 c | |
| Anti HBc Total | 96.5 (55) | 95.5 (21) | 97.1 (34) | 0.736 c | |
| Anti HBc IgM | 38.6 (22) | 100 (22) | 0.0 (0) | 0.001 c ** | |
| Genotype (n = 53) | |||||
| A | 18.9 (10) | 13.6 (3) | 22.6 (7) | 0.422 c | |
| C | 45.3 (24) | 45.5 (10) | 45.2 (14) | 0.984 c | |
| D | 35.8 (19) | 40.9 (9) | 32.3 (10) | 0.527 c | |
| HBV viral loadc | 7.3x103 (1.1x102–4.9x108) | 2.9x106 (3.9x102–4.9x108) | 3.2x 103 (1.1x102–5.7x107) | 0.001 b** | |
a = mean (minimum–maximum)
b = Mann-Whitney U test
c = Person’s chi square test
* = <0.05
** = <0.01
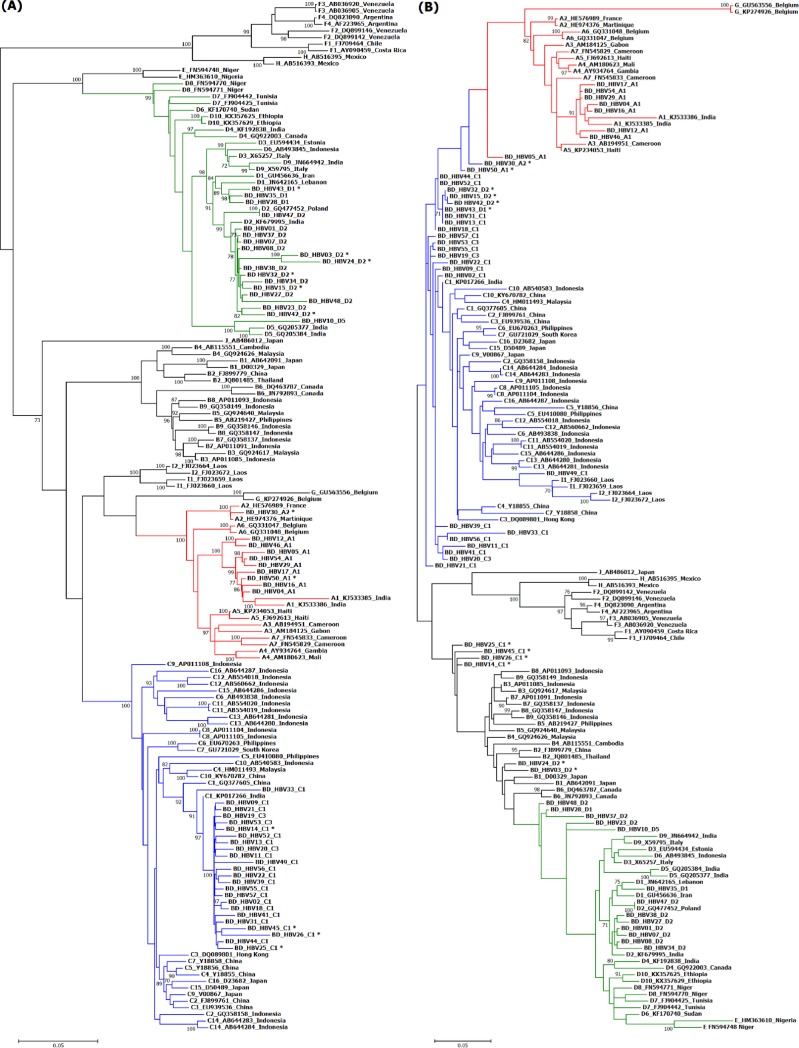
HBV DNA positive samples (n = 57) were subjected to WGS, resulting in 53 HBV whole genome sequences. The mean length of the genome sequences was 3,202 bp (range: 3,125–3,227bp; Genotype A; 3,221–3,221bp, Genotype C; 3,125–3,227bp, and genotype D; 3,131–3,182bp). The genotypes and subgenotypes of all isolates as determined by phylogenetic analysis are shown in Fig 1A. Among the 53 HBV whole genome sequences, 18.6% (10/53) were genotype A, 42.1% (24/53) were genotype C, and 33.3% (19/53) were genotype D. Of the 10 genotype A HBV, 90% (9/10) were subgenotype A1, and 10% (1/10) were subgenotype A2. Of the 24 genotype C viruses, 87.5% (21/24) were subgenotype C1 and 12.5% (3/24) were subgenotype C3. Of the 19 genotype D viruses, 15.7% (3/19) were subgenotype D1, 78.9% (15/19) were subgenotype D2, and 5.2% (1/19) were genotype D5 (Fig 1A). Besides this, HBV isolates from Bangladesh clustered with isolates from neighboring India.
Fig 1. A midpoint rooted tree showing the relationship between the Bangladeshi HBV genome sequences with 103 reference sequences.
The tree was constructed using RAxML v7.2.8 available in Geneious software using GTR+G+I nucleotide substitution model with 1000 bootstrapping replicates. Bootstrap values greater than 70% are shown at the branch nodes. The Bangladeshi HBV strains are presented as BD HBV followed by isolate number, genotype and subgenotype and reference genomes are presented as genotype, subgenotype followed by Gene Bank accession number and country of origin. The scale bar indicates the number of nucleotide substitution. (A) presents the full-length genome excluding the recombination susceptible region (1–1272 and 2028–3215 bp). Isolates with recombination are marked with a asterics and are clustered with major parent genotype. (B) present the recombination susceptible region (1273 to 2027 bp). Isolates with recombination are marked with a asterics and are clustered with minor parent genotype.
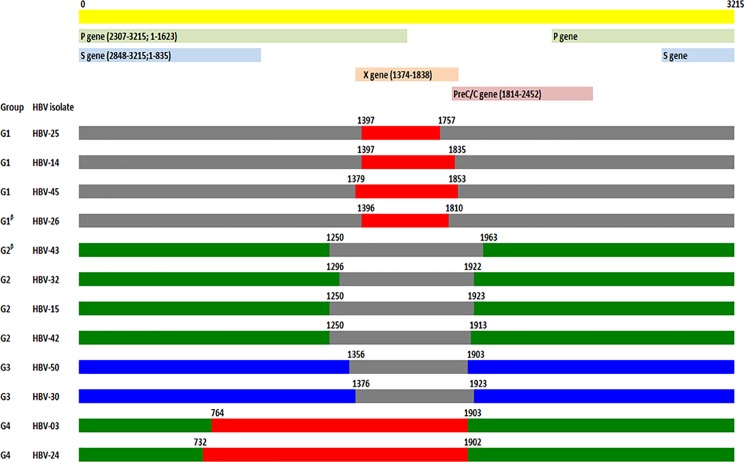
Recombination analysis using RDP identified evidence of recombination in 22.6% (12/53) of the HBV isolates including 20.0% (2/10), and 16.7% (4/24) and 31.6% (6/19) of genotype A, C and D isolates, respectively. The recombination events were classified into four groups based on the size of the recombinant fragment and the major parent genotypes; in group 1 (4 isolates), group 2 (4 isolates), group 3 (2 isolates), and group 4 (2 isolates) recombination was evident between genotype C/B, D/C, A/C and D/B, respectively. The recombinant groups, types, fragment lengths, breakpoints, major and minor parents, and sequence homologies with these parents are shown in Fig 2, Fig 1B and S1 Table. The majority of the recombination events were identified in the X gene and in the early part of the PC/C gene. The number of recombination events was significantly greater in the HBV associated with chronic infections (32.2% (10/31) versus 9.1% (2/22); p<0.05).
Fig 2. Schematic representation of the mosaic structure of the HBV recombinant sequences of the present study.
The recombinant sequences are divided into four groups (G1 to G4); based on the major and minor parent of recombination fragments. The recombinant between C/B, D/C, A/C, D/B genotypes were presented as Group1, 2, 3, and 4 respectively. Genotype A sequence is presented in blue, genotype B in red, genotype C in gray and genotype D in green. A linear physical map of the HBV genome including the position of different gene(s) is depicted. Numbering starts from the hypothetical EcoRI restriction site.
We identified an 18bp (incorporating the start codon) preS1 deletion in 39.6% (21/53) of HBV samples, which included all the genotype D isolates (p<0.001). A preS2 M1V/T start codon mutation was identified in two isolates. The presence of an F22L mutation in preS2 was significantly more common in genotype A viruses than other viruses (50% (5/10); p<0.002) (Table 2). Mutations in S gene can be found within or outside the MHR region. Mutations in N3S, L126T/S, A168V were significantly more common in genotype C HBV (p<0.001) and genotype D HBV (p<0.001) than other genotypes (Table 2). MHR mutations were identified in 17.5% (10/53) isolates including 10.5% (6/53) associated with HBsAg detection failure, 12.7% (7/53) associated with escape mutants, 7% (4/53) associated with therapy failure, and 17.5% (10/53) associated with OBI. However, there was no significant difference in the distribution of MHR mutations between genotypes (Table 3).
Table 2. Mutations in the preS2/S1/S gene reported to be associated with HCC.
| Variable | All | Genotype A | Genotype C | Genotype D | p value d | Reference | ||
|---|---|---|---|---|---|---|---|---|
| % (no) | % (no) | % (no) | % (no) | |||||
| N = 53 | n = 10 | n = 24 | n = 19 | |||||
| Gene | Nucleotide | Amino acid | ||||||
| Genome length | 3125–3227 | 3221–3221 | 3125–3227 | 3131–3182 | ||||
| PreS1 gene (nts 2848–3204) | ||||||||
| Pre S1 deletion | 39.6 (21) | 0 (0) | 8.3 (2) | 100 (19) | 0.000** | [13] [12] | ||
| 6 bp insertion in 2374 | 20.8 (11) | 90 (9) | 8.3 (2) | 0 (0) | 0.000** | |||
| PreS2 gene (nts 3205–154) | ||||||||
| Start codon | M1V/T/I | 5.7 (3) | 20 (2) | 4.2 (1) | 0 (0) | 0.010* | [13] [12] | |
| PreS1/S2 continuous | 3.8 (2) | 20 (2) | 0.0 (0) | 0 (0) | 0.010* | [13] | ||
| T53C | F22L | 15.1 (8) | 50 (5) | 12.5 (3) | 0 (0) | 0.002* | [13] [12] | |
| S gene mutation associated with HCC (nts 155–835) | ||||||||
| A162G | N3S | 35.8 (19) | 0 (0) | 75.0 (18) | 5.3 (1) | 0.000** | [12] | |
| G225A | R24K | 5.7 (3) | 0 (0) | 0 (0) | 15.7 (3) | 0.058 | [28] | |
| C321A | P56Q | 1.9 (1) | 0 (0) | 0 (0) | 5.3 (1) | 0.417 | [28] | |
| C339T | P62L | 3.8 (2) | 0 (0) | 8.3 (2) | 0 (0) | 0.297 | [29] | |
| T408G | F85C | 1.9 (1) | 0 (0) | 0 (0) | 5.3 (1) | 0.417 | [30] | |
| T531C/G | L126T/S | 66.0 (35) | 100 (10) | 29.1 (7) | 94.7 (18) | 0.000** | [12] | |
| T657C | A168V | 33.9 (18) | 0 (0) | 0 (0) | 94.7 (18) | 0.000** | [27] | |
| A706C | V184A | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0.116 | [12] | |
| T766A | S204R | 1.9 (1) | 0 (0) | 0 (0) | 5.3 (1) | 0.162 | [12] | |
| Stop codon on S | 3.8 (2) | 10 (1) | 4.2 (1) | 0 (0) | 0.417 | |||
d = One way ANOVA test
* = <0.05
** = <0.001
Table 3. Mutations in MHR (aa 99–169) including “a” determinant region (aa 124–147).
| Amino acid position | All | Genotype A | Genotype C | Genotype D | Reference |
|---|---|---|---|---|---|
| % (no) | % (no) | % (no) | % (no) | ||
| N = 53 | n = 10 | n = 24 | n = 19 | ||
| Y100CIV | 3.8 (2) | 0 (0) | 8.3 (2) | 0 (0) | [22] |
| Q101K | 3.8 (2) | 0 (0) | 8.3 (2) | 0 (0) | [22] |
| Q101R IV | 1.9 (1) | 0 (0) | 4.2 (1) | 0 (0) | [22] |
| L100I IV | 1.9 (1) | 0 (0) | 4.2 (1) | 0 (0) | [22] |
| T113S IV | 1.9 (1) | 0 (0) | 4.2 (1) | 0 (0) | [22] |
| T115N II,IV | 1.9 (1) | 0 (0) | 0 (0) | 5.3 (1) | [22] |
| I/T126N I, II,III,IV | 3.8 (2) | 0 (0) | 4.2 (1) | 5.3 (1) | [22][23] |
| I/T126S I, II, IV | 1.9 (1) | 0 (0) | 4.2 (1) | 0 (0) | [22][23] |
| T127P | 5.7 (3) | 0 (0) | 0 (0) | 15.8 (3) | [22] |
| A128V | 7.6 (4) | 0 (0) | 0 (0) | 21.0 (4) | [22] |
| S143L I, II, IV | 1.9 (1) | 0 (0) | 0 (0) | 5.3 (1) | [22][24][26] |
| G145R I, II, III, IV | 3.8 (2) | 0 (0) | 8.3 (2) | 0 (0) | [22][24][26] |
| R160N | 1.9 (1) | 0 (0) | 4.2 (1) | 0 (0) | [22] |
| Y161F | 1.9 (1) | 10 (1) | 0 (0) | 0 (0) | [26] |
| E164D | 1.9 (1) | 0 (0) | 4.2 (1) | 0 (0) | [26] |
| A168VIV | 1.9 (1) | 0 (0) | 0 (0) | 5.3 (1) | [27] |
I; Mutations associated with HBsAg detection failure
II; mutations associated with escape mutant
III; mutations associated with therapy escape
IV; mutations associated OBI. Mutation in “a” determinant region is marked in bold. Amino acid positions are relative to HBV reference sequence
GenBank accession number AB014381
BCP mutations at C1653T T1753C, G1757A, A1762T, G1764T and C1766G were investigated and 70.2% (40/53) of all isolates, which included 100% (10/10), 66.6% (16/24), and 73.6% (14/19) of genotypes A, C, and D viruses, respectively. There was no significant difference in the combination of BCP mutations among different genotypes (Table 4). A G1896A mutation in the PC gene was present in 26.4% (14/53) of viruses, which included 47.4% (9/19) of the genotype D viruses and none of the genotype A viruses and 20.8% (5/24) of genotype C viruses.
Table 4. Mutations in the BCP, PC/core gene associated with HCC in 53 HBV isolates from Bangladesh.
| Variable | All | Genotype A | Genotype C | Genotype D | p value d | |
|---|---|---|---|---|---|---|
| % (no) | % (no) | % (no) | % (no) | |||
| N = 53 | n = 10 | n = 24 | n = 19 | |||
| Nucleotide | ||||||
| Mutation in BCP region | ||||||
| C1653T | 5.7 (3) | 10 (1) | 8.3 92) | 0 (0) | 0.297 | |
| T1753C | 13.2 (7) | 20 (2) | 16.7 (4) | 5.3 (1) | 0.443 | |
| G1757A | 37.7 (20) | 30 (3) | 29.2 (7) | 52.6 (10) | 0.258 | |
| A1762T | 32.1 (17) | 60 (6) | 29.2 (7) | 21.1 (4) | 0.097 | |
| G1764A | 37.7 (20) | 70 (7) | 33.3 (8) | 26.3 (5) | 0.059 | |
| A1762T/G1764A | 32.1 (17) | 60 (6) | 29.2 (7) | 21.1 (4) | 0.097 | |
| C1766G | 1.9 (1) | 0 (0) | 00.0 (0) | 5.3 (1) | 0.417 | |
| T1768A | ||||||
| Mutation in PC/Core | ||||||
| G1896A | 26.4 (14) | 0 (0) | 20.8 (5) | 47.4 (9) | 0.014* | |
| G1899A | 8.8 (5) | 0 (0) | 8.3 (2) | 15.8 (3) | 0.387 | |
| A2159G | 5.7 (3) | 0 (0) | 12.5 (3) | 0 (0) | 0.152 | |
| A2189C/T | 50.1(27) | 0 (0) | 33.3 (8) | 100 (19) | 0.000** | |
d = One way ANOVA test
* = <0.05
** = <0.01
Reverse transcriptase sequence analysis identified genotype dependant AA polymorphisms, previously reported NAr mutations, and additional novel NAr mutations. The genotype dependant AA polymorphism was identified in 11 sites (Table 5). The presence of threonine at rt38, serine at rt 53, tyrosine at rt 124, isoleucine at rt 224 were significantly associated with genotype C (p<0.0001). Similarly, the presence of isoleucine at rt 91, glutamine at rt 139, tryptophan at rt 153, tyrosine at rt 221 and aspartic acid at rt 53, histidine at rt 54, arginine at rt 126, and cysteine at rt 256 were all significantly associated with genotype A and genotype D viruses respectively (all p< 0.0001) (Table 5).
Table 5. Genotype–dependent AA polymorphic sites in RT domain of HBV polymerase gene identified in this study.
| rt38 | rt53 | rt54 | rt91 | rt124 | rt126 | rt139 | rt153 | rt221 | rt224 | rt256 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ab | Ea | T | D | I | Ka | Na | S | Va | H | T | Ya | I | Lc | Hb | N | Y | H | R | Ya | Da | Ha | N | Q | Qa | R | W | F | Yc | I | Vb | C | S | ||
| Genotype A | N = 10 | 10 | 0 | 0 | 0 | 5 | 0 | 0 | 1 | 4 | 0 | 10 | 0 | 10 | 0 | 1 | 9 | 0 | 7 | 0 | 3 | 0 | 0 | 0 | 10 | 0 | 0 | 10 | 0 | 10 | 0 | 10 | 0 | 10 |
| Genotype C | N = 24 | 1 | 0 | 23 | 0 | 0 | 0 | 0 | 24 | 0 | 0 | 24 | 0 | 2 | 22 | 2 | 0 | 22 | 24 | 0 | 0 | 1 | 0 | 23 | 0 | 2 | 21 | 1 | 24 | 0 | 22 | 2 | 0 | 24 |
| Genotype D | N = 19 | 16 | 1 | 2 | 14 | 0 | 1 | 3 | 1 | 0 | 14 | 2 | 3 | 1 | 18 | 18 | 1 | 0 | 4 | 15 | 0 | 0 | 1 | 18 | 0 | 0 | 19 | 0 | 17 | 2 | 0 | 19 | 17 | 2 |
| P value | p < 0.0001 | p < 0.0001 | p < 0.0001 | p < 0.0001 | p < 0.0001 | p < 0.0001 | p < 0.0001 | p < 0.0001 | p < 0.0001 | p < 0.0001 | ||||||||||||||||||||||||
A: alanine; E: glutamic acid; T: threonine; D: aspartic acid; I: isoleucine; K: Lysine; N: asparagine; S: serine, H: histidine; Y: tyrosine; R: Arginine; Q: Glutamine; W: Tryptophan; F: Phenylalanine; G: glycine; F: phenylalanine; V: valine; C: cystine.
a Described as naturally occurring polymorphic mutations in this study
b Pretreatment mutation found to be Genotype–dependent AA
c Putative NAr mutation found to be Genotype–dependent AA
The NAr mutation was identified in 47.2% (25/53) of isolates, including previously reported mutations in 22.6% (12/53) of viruses and novel NAr mutations in 33.9% (18/53) of the isolates. 28.3% (15/53) of the viruses had at least one NAr mutations and 17.5% (10/53) had two or more mutations. The prevalence of previously reported primary, secondary, putative and pretreatment mutations, were 1.9% (1/35), 3.8% (2/53), 15.1% (8/53), and 7.6% (4/53), respectively (S2 File). The prevalence of any previously reported mutations were significantly higher in the genotype D viruses than the other genotypes (Genotype A; 10% (1/10), Genotype C; 20.8% (5/24), and Genotype D; 31.5% (6/19) (p< = 0.05).
NAr mutations were further characterized for their presence in RT domains or interdomian regions. Interdomian mutations were more common than domain mutations for previously reported (74.6%; (11/15) versus 26.4%; (4/15) and novel mutations (82.7% (19/23) versus 17.3% (4/23) (S1 Fig).
Discussion
A description of genotypes and subgenotypes is important for a better understanding of the epidemiology, transmission, virulence potential, and clinical outcome of HBV infections [5]. One of the key criteria for assigning a virus to a subgenotype is generating a whole genome sequence. Here, we analyzed a collection of HBV genome sequences collected in a single healthcare facility in Bangladesh. To our knowledge, this is the first study reporting genotypes and subgenotype of HBV in Bangladesh through WGS. Considering the prevalence of chronic HBV and the limited availability facilities for characterizing HBV in Bangladesh, this genotyping and subgenotype data from the Bangladeshi population is important for clinical management decision making, disease modeling, and health resource allocation for the management of chronic HBV [31].
The majority of the HBV in our study belonged to genotype C and D, which have a higher risk of HCC and chronic infection then genotype A and B. The progression to chronic HBV infection has been shown to be commonly associated with genotypes A and D then with other genotypes [31]. Our data is in agreement with recently published HBV genotyping data from Bangladesh, and data from neighboring countries including India [32] [33]. Genotypes A and D are known to be horizontally transmitted, more than half of the HBV identified here were genotype A or D, indicating possible horizontal transmission through blood or blood products [31]. Additionally, we observed a high degree of recombination among these HBV isolates; it is not apparent if this recombination occurred a result of co-infection/super infection with two genotypes within the patient or if the patient was infected with the recombinant strain. In half of the recombinant sequences, the recombination fragment (minor parent) was a genotype C virus. As the prevalence of genotype C HBV is high in Bangladesh, it is possible that patients were co-infected/super infected with two genotypes. The majority of the recombination breakpoints identified here, were in agreement with previous studies where recombination breakpoint hotspots have been observed in the X gene and the preCore/core gene [34]. Although the prevalence of recombination (HBV B/C) in HBV genotype B/Ba (B2-B5) from Vietnam, China, Hong Kong, Indonesia, and Thailand is high [35], the prevalence of genotype D/C or D/B recombination are relatively infrequent [34].
We chose to select patients who were treatment naïve to identify preexisting drug resistance mutations not likely influenced by treatment selective pressure. It is not unexpected that the majority of the viruses did not harbor primary or secondary drug resistance associated mutations, as the majority of the patients were treatment naive. Approximately half of the HBV had putative and pretreatment NAr mutations, including a third of the viruses exhibiting a novel mutation, and the prevalence of novel mutations was higher in the genotype D viruses. Eleven genotype-dependent AA polymorphic positions were identified for A-, C- and D- genotypes; similar observations have been reported previously. The cause of novel amino acid substitution associated with NAr and the aa dependant polymorphism is not known, however, it has been suggested that such mutations may be associated with the evolution and adaption of HBV in a defined population [21]. We identified an isoleucine at rt91 and tyrosine at rt221 is genotype A dependant; however, these positions (isoleucine at rt91 and tyrosine at rt22) have been reported as putative NAr mutations. Therefore, potential NAr mutations need further investigation regarding nucleos(t)ide resistance in vitro and in vivo. Moreover, the AA sites in interdomians displayed the highest mutation frequency (S1 Fig). It is likely that the interdomians are less crucial for RT function and antiviral resistance, rather the mutations within which might be driven by host immune responses [36].
All genotype D isolates and 8% of the genotype C isolates had an 18bp preS1 deletion. Longitudinal studies have shown that the preS deletion mutations occur during the long course of liver disease, but not at the beginning of HBV infection [12]. It is thought that such deletions evolve during the course of long lasting infections and are associated with higher risk of HCC. We found that HCC associated mutations in at A168V and V184A were significantly higher in genotype D isolates [12]. Analysis of MRH region showed that 17% of the isolates had mutations in the “a” determinant region. These mutations can affect the antigenicity of HBsAg, and have shown to be responsible for false-negative results by commercial assays for HBsAg, evasion of anti-HBV immunoglobulin therapy, and evasion of vaccine induced immunity. These “vaccine-escape” mutants are more common in countries with high rates of endemic infections and universal immunization programs [23].
Mutations in BCP region, specifically the G1762A/G1764A double mutation, have suggested to be closely associated with HCC [13]. One-third of the isolates in the present study across all genotypes harbored these mutations, indicating the presence of HBV with increased risk for HCC in Bangladesh. The G1896A mutation in the precore/core gene results in a stop codon in 28 aa of core gene, and has been shown to be associated with fulminant hepatitis [10]. Approximately half of the genotype D isolates in this study harbored this mutation, indicating a potential for hepatic flare in these patients.
This study has limitations. First, specimens was collected over a short period of time and from a single tertiary care hospital and may not be representative of the population in Dhaka or Bangladesh as a whole. Second, a lack of data on the clinical presentation from the patients whom the sample was collected limits the clinical relevance of the viral subgenotype. Longitudinal studies on patients with specific subgenotype infection are essential to fill this knowledge gap.
Supporting information
Genotype, subgenotype, GenBank accession number and country of origin of the reference sequences used for phylogenetic analysis. BD_HBV followed by isolate number, GenBank accession number and subgenotype of the HBV isolates from this study.
(DOCX)
Potential NAr mutations including reported NAr mutations and novel NAr mutations identified in the present study among different genotypes are presented.
(DOCX)
HBV isolates were considered recombinant if detected by 5 out of 6 program (RDP, BootScan, Max Chi, Chimaera, SisScan and Topol). Recombinant isolate, recombination group major and minor parents and identity, recombinant break points, size of the recombinant fragment and location of the recombination are presented.
(DOCX)
Schematic presentation of six HVB RT domain functional regions and (F, A, B, C, D, and E) five regions (F-A, A-B, B-C, C-D, and D-E) connecting the functional regions. Functional regions are presented as box and regions between functional regions as lines. The start and end amino acid positions of each functional region are presented at the top of each box. Novel mutations are presented as brown bars and previously reported mutations as blue bars.
(DOCX)
Acknowledgments
We thank Wellcome Trust for supporting next generation sequencing facility at OUCRU.
Data Availability
ALL sequence data has been submitted to GenBank. Gene Bank accession number is MF925358 to MF925410.
Funding Statement
This study was supported by Oxford University Clinical Research Unit Core funding to Motiur Rahman. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Global Hepatitis Report 2017. World Health Organization, 2017.
- 2.Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386(10003):1546–55. doi: 10.1016/S0140-6736(15)61412-X . [DOI] [PubMed] [Google Scholar]
- 3.Te HS, Jensen DM. Epidemiology of hepatitis B and C viruses: a global overview. Clinics in liver disease. 2010;14(1):1–21, vii. doi: 10.1016/j.cld.2009.11.009 . [DOI] [PubMed] [Google Scholar]
- 4.Pourkarim MR, Amini-Bavil-Olyaee S, Kurbanov F, Van Ranst M, Tacke F. Molecular identification of hepatitis B virus genotypes/subgenotypes: revised classification hurdles and updated resolutions. World journal of gastroenterology: WJG. 2014;20(23):7152–68. Epub 2014/06/27. doi: 10.3748/wjg.v20.i23.7152 ; PubMed Central PMCID: PMC4064061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanwar S, Dusheiko G. Is there any value to hepatitis B virus genotype analysis? Current gastroenterology reports. 2012;14(1):37–46. doi: 10.1007/s11894-011-0233-5 . [DOI] [PubMed] [Google Scholar]
- 6.Chen YC, Chu CM, Liaw YF. Age-specific prognosis following spontaneous hepatitis B e antigen seroconversion in chronic hepatitis B. Hepatology. 2010;51(2):435–44. Epub 2009/11/18. doi: 10.1002/hep.23348 . [DOI] [PubMed] [Google Scholar]
- 7.Chandra PK, Biswas A, Datta S, Banerjee A, Panigrahi R, Chakrabarti S, et al. Subgenotypes of hepatitis B virus genotype D (D1, D2, D3 and D5) in India: differential pattern of mutations, liver injury and occult HBV infection. Journal of viral hepatitis. 2009;16(10):749–56. doi: 10.1111/j.1365-2893.2009.01129.x . [DOI] [PubMed] [Google Scholar]
- 8.McMahon BJ. The influence of hepatitis B virus genotype and subgenotype on the natural history of chronic hepatitis B. Hepatology international. 2009;3(2):334–42. doi: 10.1007/s12072-008-9112-z ; PubMed Central PMCID: PMC2716762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin J, Zhang H, Li C, Gao C, He Y, Zhai Y, et al. Role of hepatitis B virus genotype mixture, subgenotypes C2 and B2 on hepatocellular carcinoma: compared with chronic hepatitis B and asymptomatic carrier state in the same area. Carcinogenesis. 2008;29(9):1685–91. doi: 10.1093/carcin/bgm301 . [DOI] [PubMed] [Google Scholar]
- 10.Huy TT, Ushijima H, Quang VX, Ngoc TT, Hayashi S, Sata T, et al. Characteristics of core promoter and precore stop codon mutants of hepatitis B virus in Vietnam. Journal of medical virology. 2004;74(2):228–36. Epub 2004/08/28. doi: 10.1002/jmv.20175 . [DOI] [PubMed] [Google Scholar]
- 11.Kim H, Kim BJ. Association of preS/S Mutations with Occult Hepatitis B Virus (HBV) Infection in South Korea: Transmission Potential of Distinct Occult HBV Variants. International journal of molecular sciences. 2015;16(6):13595–609. doi: 10.3390/ijms160613595 ; PubMed Central PMCID: PMC4490511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qu LS, Liu JX, Liu TT, Shen XZ, Chen TY, Ni ZP, et al. Association of hepatitis B virus pre-S deletions with the development of hepatocellular carcinoma in Qidong, China. PloS one. 2014;9(5):e98257 doi: 10.1371/journal.pone.0098257 ; PubMed Central PMCID: PMC4029943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Ding HG. Key role of hepatitis B virus mutation in chronic hepatitis B development to hepatocellular carcinoma. World journal of hepatology. 2015;7(9):1282–6. doi: 10.4254/wjh.v7.i9.1282 ; PubMed Central PMCID: PMC4438503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman PF. Detecting hepatitis B surface antigen mutants. Emerging infectious diseases. 2006;12(2):198–203. doi: 10.3201/eid1202.050038 ; PubMed Central PMCID: PMC3293431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashraf H, Alam NH, Rothermundt C, Brooks A, Bardhan P, Hossain L, et al. Prevalence and risk factors of hepatitis B and C virus infections in an impoverished urban community in Dhaka, Bangladesh. BMC infectious diseases. 2010;10:208 doi: 10.1186/1471-2334-10-208 ; PubMed Central PMCID: PMC2918606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akbar SMF, Hossain M, Hossain MF, Sarker S, Hossain SAS, Tanimoto K, et al. Seroepidemiolgy of hepatitis viruses of chronic liver diseases in Bangladesh: high prevalence of HCV among blood donors and healthy person. Hepatology Research. 1997;7(2):113–20. http://dx.doi.org/10.1016/S0928-4346(97)00027-3. [Google Scholar]
- 17.Friedt M, Gerner P, Wintermeyer P, Wirth S. Complete hepatitis B virus genome analysis in HBsAg positive mothers and their infants with fulminant hepatitis B. BMC gastroenterology. 2004;4:11 Epub 2004/06/10. doi: 10.1186/1471-230X-4-11 ; PubMed Central PMCID: PMC425580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi W, Zhu C, Zheng W, Carr MJ, Higgins DG, Zhang Z. Subgenotype reclassification of genotype B hepatitis B virus. BMC gastroenterology. 2012;12:116 doi: 10.1186/1471-230X-12-116 ; PubMed Central PMCID: PMC3523008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32(5):1792–7. doi: 10.1093/nar/gkh340 ; PubMed Central PMCID: PMC390337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic biology. 2003;52(5):696–704. . [DOI] [PubMed] [Google Scholar]
- 21.Liu BM, Li T, Xu J, Li XG, Dong JP, Yan P, et al. Characterization of potential antiviral resistance mutations in hepatitis B virus reverse transcriptase sequences in treatment-naive Chinese patients. Antiviral research. 2010;85(3):512–9. doi: 10.1016/j.antiviral.2009.12.006 . [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Liu Y, Zhao J, Xu Z, Chen R, Si L, et al. Characterization of Novel Hepatitis B Virus PreS/S-Gene Mutations in a Patient with Occult Hepatitis B Virus Infection. PloS one. 2016;11(5):e0155654 doi: 10.1371/journal.pone.0155654 ; PubMed Central PMCID: PMCPMC4868315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazarevic I. Clinical implications of hepatitis B virus mutations: recent advances. World journal of gastroenterology: WJG. 2014;20(24):7653–64. doi: 10.3748/wjg.v20.i24.7653 ; PubMed Central PMCID: PMCPMC4069294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Q, Wang Y. Comprehensive analysis of the prevalence of hepatitis B virus escape mutations in the major hydrophilic region of surface antigen. Journal of medical virology. 2012;84(2):198–206. doi: 10.1002/jmv.23183 . [DOI] [PubMed] [Google Scholar]
- 25.Avellon A, Echevarria JM. Frequency of hepatitis B virus 'a' determinant variants in unselected Spanish chronic carriers. Journal of medical virology. 2006;78(1):24–36. doi: 10.1002/jmv.20516 . [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez Lay LA, Corredor MB, Villalba MC, Frometa SS, Wong MS, Valdes L, et al. Genetic Diversity of the Hepatitis B Virus Strains in Cuba: Absence of West-African Genotypes despite the Transatlantic Slave Trade. PloS one. 2015;10(5):e0125052 doi: 10.1371/journal.pone.0125052 ; PubMed Central PMCID: PMCPMC4433336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziaee M, Javanmard D, Sharifzadeh G, Hasan Namaei M, Azarkar G. Genotyping and Mutation Pattern in the Overlapping MHR Region of HBV Isolates in Southern Khorasan, Eastern Iran. Hepat Mon. 2016;16(10):e37806 doi: 10.5812/hepatmon.37806 ; PubMed Central PMCID: PMCPMC5111392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramezani F, Norouzi M, Sarizade GR, Poortahmasebi V, Kalantar E, Magnius L, et al. Mutation hot spots in hepatitis B surface antigen in chronic carriers from Khoozestan province, southern of Iran. Iran J Allergy Asthma Immunol. 2013;12(3):269–75. . [PubMed] [Google Scholar]
- 29.Shih HH, Jeng KS, Syu WJ, Huang YH, Su CW, Peng WL, et al. Hepatitis B surface antigen levels and sequences of natural hepatitis B virus variants influence the assembly and secretion of hepatitis d virus. J Virol. 2008;82(5):2250–64. doi: 10.1128/JVI.02155-07 ; PubMed Central PMCID: PMCPMC2258943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moradi A, Zhand S, Ghaemi A, Javid N, Tabarraei A. Mutations in the S gene region of hepatitis B virus genotype D in Golestan Province-Iran. Virus Genes. 2012;44(3):382–7. doi: 10.1007/s11262-012-0715-z . [DOI] [PubMed] [Google Scholar]
- 31.Sunbul M. Hepatitis B virus genotypes: global distribution and clinical importance. World journal of gastroenterology: WJG. 2014;20(18):5427–34. doi: 10.3748/wjg.v20.i18.5427 ; PubMed Central PMCID: PMC4017058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahman MA, Hakim F, Ahmed M, Ahsan CR, Nessa J, Yasmin M. Prevalence of genotypes and subtypes of hepatitis B viruses in Bangladeshi population. SpringerPlus. 2016;5:278 doi: 10.1186/s40064-016-1840-2 ; PubMed Central PMCID: PMC4779089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saikia A, Bose M, Barman NN, Deka M, Thangkhiew RS, Bose S. Molecular epidemiology of HBV infection in chronic hepatitis B virus infected patients in northeast India. Journal of medical virology. 2015;87(9):1539–48. doi: 10.1002/jmv.24207 . [DOI] [PubMed] [Google Scholar]
- 34.Shi W, Carr MJ, Dunford L, Zhu C, Hall WW, Higgins DG. Identification of novel inter-genotypic recombinants of human hepatitis B viruses by large-scale phylogenetic analysis. Virology. 2012;427(1):51–9. doi: 10.1016/j.virol.2012.01.030 . [DOI] [PubMed] [Google Scholar]
- 35.Sugauchi F, Orito E, Ichida T, Kato H, Sakugawa H, Kakumu S, et al. Hepatitis B virus of genotype B with or without recombination with genotype C over the precore region plus the core gene. Journal of virology. 2002;76(12):5985–92. doi: 10.1128/JVI.76.12.5985-5992.2002 ; PubMed Central PMCID: PMC136227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheldon J, Soriano V. Hepatitis B virus escape mutants induced by antiviral therapy. The Journal of antimicrobial chemotherapy. 2008;61(4):766–8. doi: 10.1093/jac/dkn014 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotype, subgenotype, GenBank accession number and country of origin of the reference sequences used for phylogenetic analysis. BD_HBV followed by isolate number, GenBank accession number and subgenotype of the HBV isolates from this study.
(DOCX)
Potential NAr mutations including reported NAr mutations and novel NAr mutations identified in the present study among different genotypes are presented.
(DOCX)
HBV isolates were considered recombinant if detected by 5 out of 6 program (RDP, BootScan, Max Chi, Chimaera, SisScan and Topol). Recombinant isolate, recombination group major and minor parents and identity, recombinant break points, size of the recombinant fragment and location of the recombination are presented.
(DOCX)
Schematic presentation of six HVB RT domain functional regions and (F, A, B, C, D, and E) five regions (F-A, A-B, B-C, C-D, and D-E) connecting the functional regions. Functional regions are presented as box and regions between functional regions as lines. The start and end amino acid positions of each functional region are presented at the top of each box. Novel mutations are presented as brown bars and previously reported mutations as blue bars.
(DOCX)
Data Availability Statement
ALL sequence data has been submitted to GenBank. Gene Bank accession number is MF925358 to MF925410.