SUMMARY
Bacterial cell wall synthesis is the target for some of our most powerful antibiotics and has thus been the subject of intense research focus for more than 50 years. Surprisingly, we still lack a fundamental understanding of how bacteria build, maintain and expand their cell wall. Due to technical limitations, directly testing hypotheses about the coordination and biochemistry of cell wall synthesis enzymes or architecture has been challenging, and interpretation of data has therefore often relied on circumstantial evidence and implicit assumptions. A number of recent papers have exploited new technologies, like single molecule tracking and real-time, high resolution temporal mapping of cell wall synthesis processes, to address fundamental questions of bacterial cell wall biogenesis. The results have challenged established dogmas and it is therefore timely to integrate new data and old observations into a new model of cell wall biogenesis in rod-shaped bacteria.
Graphical abstract
The cell wall is an essential component of most bacterial cells, and has been a major focus of research for the last 50 years. Despite this intense research, we still have an incomplete understanding of how bacteria construct their cell wall. In this MicroReview, we will summarize important new data generated in the last few years, reexamine some established ideas in the field, and propose a new model of cell wall biogenesis in rod-shaped bacteria.
INTRODUCTION
Most bacteria surround themselves with a cell wall, a complex biopolymer with a crucial role in maintaining cellular integrity and cell shape. Due to its essentiality for bacterial growth and survival, the bacterial cell wall constitutes an ideal target for antibiotics, and there has been a longstanding scientific interest in the mechanisms of its synthesis and turnover. Pioneering work beginning over 50 years ago established the general composition of the cell wall (or sacculus) as a single large molecule made primarily of peptidoglycan (PG). The Gram-positive cell wall also contains a large amount of teichoic acid, including wall teichoic acids covalently linked to PG (Brown et al., 2013, Reichmann & Grundling, 2011)). PG is a mesh-like macromolecule comprising roughly parallel glycan strands of polymerized disaccharide (N-acetylglucosamine-N-acetylmuramic acid; or NAG-NAM), which are intermittently crosslinked with neighboring strands by peptide bonds between short oligopeptides (pentapeptides) attached to the NAM residues (Strominger et al., 1971, Strominger et al., 1959, Anderson et al., 1967, Anderson et al., 1966).
This basic PG structure is conserved amongst essentially all Bacteria, although there are variations in the details. These include differences in glycan strand chain length and a diversity of peptide crosslinks. The peptide crosslinks vary in amino acid composition and modifications (e.g. amidation), the addition in some species of interstrand bridging peptides, the precise site of interstrand linkage, and their overall density (Espaillat et al., 2016, Quintela et al., 1995). The PG sacculus can be further modified after synthesis by occasional additions to the glycan strands, for example of acetyl residues (O-acetylation, (Moynihan et al., 2014)). Despite these subtle variations, the overall PG structure is highly conserved when compared to other bacterial surface layers (including capsules, S-layers, and enterobacterial O-antigen) and therefore PG and its derivatives serve as effective pathogen-associated molecular patterns (PAMPs) for host recognition of bacterial infection (Mogensen, 2009).
Despite being the focus of intense research, conspicuous gaps in our knowledge of PG biogenesis have persisted over decades, and some long-entrenched ideas have been found to be either incorrect or incomplete. Recent studies of the enzymology, genetics and cell biology of PG synthesis have challenged many long-standing assumptions. Here, we review recent insights into PG synthesis, largely from studies of the rod-shaped model organisms Escherichia coli and Bacillus subtilis. The focus will be the enzymology and cell biology of proteins involved in cell elongation and division. We conclude by proposing a new model of cell wall biogenesis that incorporates these recent findings.
Enzymology of PG synthesis: variations, nuances, and the key points of controversy
Assembly of the PG layer requires three major stages: precursor synthesis in the cytoplasm to generate the key intermediate lipid II, the lipid II cycle (translocation, transglycosylation, and recycling of the carrier lipid), and glycan strand crosslinking and maturation. As would be expected for such a central process in bacterial cell biology, the key enzymes for PG synthesis are generally well-established. However, the overall process displays more plasticity than originally envisioned, some key enzymes have remained elusive or controversial, and several puzzling genetic observations have only recently been resolved.
PG synthesis starts in the cytoplasm, where the precursor molecule UDP-NAM-pentapeptide is produced by enzymes encoded by the mur genes as well as the D-Ala-D-Ala ligase Ddl as the last soluble precursor (Lovering et al., 2012). Ligation of this precursor to an undecaprenyl (C55) carrier lipid by the membrane-associated enzyme MraY generates lipid I, the first membrane-associated intermediate. MurG ligates a NAG residue to lipid I to generate the final, lipidated disaccharide-pentapeptide precursor referred to as lipid II (Scheffers & Tol, 2015). Once synthesis is complete on the cytoplasmic face of the inner membrane, the lipid II precursor must be translocated (flipped) to the outer face of the membrane by a flippase, where the final steps of PG assembly occur. Following assembly, PG may be further modified and serves as a scaffold for the anchoring of wall teichoic acids (in Gram-positive bacteria), proteins, and other surface structures and appendages (Siegel et al., 2016, Brown et al., 2013, Guest & Raivio, 2016).
Lipid II provides the subunits that are polymerized into glycan strands via a transglycosylation (TG) reaction. The lipid carrier is released as undecaprenyl-pyrophosphate and recycled into the cytoplasm by a putative, as yet unidentified C55-pyrophosphate flippase, which may or may not be the same as the lipid II flippase. The TG reaction has historically been thought to be mediated solely by bifunctional (class A) penicillin binding proteins, here designated as aPBPs (Goffin & Ghuysen, 1998, Sauvage et al., 2008). The polysaccharide strands resulting from the TG reaction are subsequently covalently linked via D, D transpeptidation (TP) reactions to form peptide bond crosslinks between the glycan strands (Sauvage et al., 2008). The acceptor amino group derives from the side chain of the third amino acid (typically diaminopimelic acid, DAP3 or lysine, Lys3, depending on the species) with D-Ala4 as the donor (generating a 4-3 crosslink); this results in the release of the terminal D-Ala5 from the donor strand (McDonough et al., 2002, Mainardi et al., 2008). The TP reaction can be mediated by either aPBPs (which possess both TG and TP activity) or by monofunctional D, D-transpeptidases (class B PBPs, designated here as bPBPs).
This basic, textbook version of PG synthesis provides a framework for a more detailed consideration of how this process may differ between organisms or be modified in response to stress. Moreover, some of the central steps in PG synthesis have retained an aura of mystery, with a lack of consensus about the identity of key enzymes and some confounding genetic observations. Recent excitement centers on three major advances. First, the proteins that translocate the lipid II precursor from the cytosolic to the external face of the membrane are now becoming clear. Second, a long predicted but elusive PBP-independent TG activity has been defined. Third, variations in the nature of the PG intra-strand crosslinking reactions, and in particular the presence and impact of 3-3 in place of 4-3 crosslinks, is an emerging area of focus.
MurJ and functionally redundant lipid II flippases
After its generation in the cytoplasm, the PG precursor lipid II must be translocated (“flipped”) across the cytoplasmic membrane to provide the substrate for cell wall synthesis enzymes. The identity of the lipid II flippase(s) has been the subject of a longstanding controversy. Using a reductionist bioinformatics approach, Ruiz first proposed the membrane-anchored protein MurJ as the lipid II flippase in E. coli and supported this notion by demonstrating that MurJ is essential and required for PG synthesis (both of which would be expected of a flippase) (Ruiz, 2008). This was later challenged by Mohammadi et al., who used an in vitro assay to demonstrate flippase activity of purified FtsW protein, and thus speculated that SEDS (shape, elongation, division, and sporulation) family proteins (including RodA, FtsW and SpoVE in B. subtilis), rather than MurJ, were flippases (Mohammadi et al., 2011). Another key point of their argument was that while a flippase is expected to be universally essential, all MurJ homologues could be deleted in B. subtilis (Fay & Dworkin, 2009).
Two recent studies have shed some more light on this controversy. Using an in vivo biochemical assay, Sham et al. demonstrated that MurJ does have lipid II flippase activity (Sham et al., 2014). Importantly, in the same study, depleting FtsW in a ΔrodA background (essentiality of rodA was suppressed by overexpression of the ftsQAZ operon (Kruse et al., 2005)) did not affect precursor translocation, suggesting that RodA and FtsW are entirely dispensable for this process. Another recent study addressed the important question of why MurJ proteins were (collectively) non-essential in B. subtilis. Using a synthetic lethal screen (via transposon insertion sequencing), Meeske et al. searched for genes that become essential in the absence of all MurJ homologs, arguing that an alternative flippase must exist and should be synthetic lethal with MurJ (Meeske et al., 2015). The screen was answered by a locus that was renamed amj (“alternate to MurJ”); intriguingly, the predicted Amj protein bears no sequence or structural homology to MurJ. Using the in vivo biochemical assay mentioned above (Sham et al., 2014) it was demonstrated that both MurJ and Amj can mediate lipid II translocation across the inner membrane; in addition, Amj could functionally replace MurJ in E. coli. Interestingly, amj is induced in the absence of MurJ, and its expression depends on the cell wall stress responsive alternative sigma factor SigM (Helmann, 2016, Eiamphungporn & Helmann, 2008, Meeske et al., 2015). Thus, B. subtilis can respond to inhibition of one of its flippases, perhaps by currently unknown antibiotics, with the expression of an alternative, structurally unrelated enzyme. In summary, there are now strong data supporting the role of MurJ and Amj as lipid II flippases. The role of FtsW remains controversial; however, recent revelations about the similar SEDS family protein RodA provide us with some room to speculate on FtsW function (see next section).
Important open questions remain concerning the reverse side of the flippase reaction; after transglycosylation, the undecaprenyl pyrophosphate (UPP) portion of lipid II remains on the outer leaflet of the cytoplasmic membrane. UPP molecules in the cell membrane are limited and UPP must therefore be efficiently recycled. This is accomplished by known, membrane-associated enzymes (UPP phosphatases) that convert UPP to undecaprenyl phosphate (UP), which can be reintroduced into the lipid II cycle (El Ghachi et al., 2005, Zhao et al., 2016). Due to the size and charge of the lipid carrier, it is generally expected to be translocated back into the cytoplasm by an enzyme facilitator rather than via spontaneous flipping, but the identity of this putative facilitator remains unknown.
The SEDS protein RodA has TG activity
The transglycosylation (TG) reaction is a crucial step in periplasmic cell wall assembly. Until recently, two classes of enzymes were known or predicted to perform the TG reaction: monofunctional transglycosylases (MTGs) and the TG domains of aPBPs. While MTGs have a demonstrated role in cell wall synthesis in some coccoid Gram-positive bacteria like Staphylococcus aureus (Reed et al., 2011), they are not widely conserved (absent for example in B. subtilis) and their physiological role in rod-shaped bacteria is unclear as there are no strong phenotypes associated with deletion or overexpression mutants (Denome et al., 1999, Di Berardino et al., 1996). Hence, the aPBPs were generally considered as the principal TGases during cell wall biosynthesis.
This dogma was challenged over a decade ago, when David Popham’s group found that B. subtilis was able to grow (albeit poorly) in the absence of all aPBPs (McPherson & Popham, 2003). This striking finding strongly suggested that an unidentified TGase could compensate for the loss of aPBPs by collaborating with the TP function of a bPBP. Other groups have reported similar observations in Enterococcus spp. (Arbeloa et al., 2004, Rice et al., 2009). Intriguingly, a study from more than 30 years ago had already provided a candidate for Popham’s “missing” transglycosylase. Ishino et al., while conducting studies on PG synthesis processes mediated by the bPBP2 (for clarity, we will add the a/b class prefix to specific PBPs throughout the text), found that crude membrane extracts of E. coli produced cell wall material when they were isolated from a strain in which bPBP2 as well as RodA were overproduced (the aPBPs were at the same time inactivated using antibiotics) (Ishino et al., 1986). Using bPBP2-specific antibiotics and thermosensitive variants of both bPBP2 and RodA, these authors dissected the contribution of each protein to the PG synthesis process and found that while bPBP2 was, as expected, required for the crosslinking part of assembly, RodA was required for chain elongation. They then discussed the possibility that RodA itself possessed transglycosylase activity, but dismissed this as “unlikely” and rather concluded (in light of what was known about PBPs in 1986) that bPBP2 itself had TG activity that was somehow stimulated by RodA. These observations were thus not integrated into later models of cell wall synthesis. Later, the idea that RodA possessed TG activity was further obscured by the proposal (as noted above) that another SEDS protein, FtsW, functioned as a lipid II flippase based on an in vitro biochemical assay (Mohammadi et al., 2011), fueling the assumption that this was true for RodA as well. FtsW and RodA were thus tentatively assigned as flippases, as noted above.
Several recent papers from the Bernhardt, Ruiz, Rudner and Errington labs have provided new insights into the roles of SEDS proteins. First, the identification of MurJ (and Amj in B. subtilis, see previous section) as a lipid II flippase (Meeske et al., 2015), re-established the possibility that RodA and FtsW have activities other than (or in addition to) precursor translocation. Then, using independent approaches (homology search (Meeske et al., 2016) or candidate genes elimination (Emami et al., 2017)), it was discovered that RodA has TGase activity in vitro (Meeske et al., 2016), and that overexpression of RodA rescued the strong growth defect of the B. subtilis strain lacking all aPBPs (Meeske et al., 2016, Emami et al., 2017). Possible natural molecule inhibitors of RodA were also identified (Emami et al., 2017).
Interestingly, these data provided an explanation for another curious feature of B. subtilis: its resistance to moenomycin. Moenomycin is a potent aPBP transglycosylase inhibitor (Welzel, 2007, Gampe et al., 2013, Rebets et al., 2014) whereas RodA TG activity was found to be unaffected by moenomycin in vitro (Meeske et al., 2016, McPherson & Popham, 2003). In B. subtilis, resistance to moenomycin depends on the SigM dependent cell envelope damage response, and SigM induces expression of rodA (Eiamphungporn & Helmann, 2008, Meeske et al., 2016, Mascher et al., 2007). Thus, similar to Amj (see above), or PBP2a in Methicillin Resistant Staphylococcus aureus (Hao et al., 2012), B. subtilis enhances the expression of one cell wall synthesis enzyme (RodA) upon inhibition of another (aPBPs) (Meeske et al., 2015, Helmann, 2016).
RodA was also shown to contribute significant TG activity to cell wall synthesis mediated by the “elongasome” in E. coli (Cho et al., 2016). However, unlike its Gram-positive counterpart, this activity does not suffice to sustain growth in the absence of aPBPs. This might be a common feature in Gram-negative bacteria, as in these organisms depletion or inhibition of aPBPs typically leads to cessation of growth and/or lysis and death (Dorr et al., 2014, Yousif et al., 1985, Satta et al., 1995).
Whether FtsW possesses TG activity has not been completely resolved. Recent biochemical evidence suggests that in E. coli, FtsW forms a complex with bPBP3 and aPBP1B at the division site (Leclercq et al., 2017). FtsW was also shown to bind lipid II and to negatively regulate aPBP1b activity using in vitro assays, and this inhibition was alleviated by the presence of bPBP3 (Leclercq et al., 2017). Importantly, FtsW did not exhibit TGase activity under these experimental conditions.
L,D-transpeptidases and diversification of PG architecture
D-Ala4-D-DAP3 or D-Ala4-D-Lys3 (D,D) crosslinks (generally referred to as 4,3 crosslinks), whose formation is mediated by D,D transpeptidases (the PBPs), have been established as the major type of PG crosslink. However, many bacteria also harbor L,D transpeptidases (LDT) (Magnet et al., 2008, Lavollay et al., 2008, Hernandez et al., 2015, Mainardi et al., 2000, Lam et al., 2009, Cava et al., 2011, Bramkamp, 2010, Magnet et al., 2007). These enzymes also catalyze TP reactions between two amino acids, e.g. between two DAP molecules in neighboring PG strands (using the energy stored in the DAP3-D-Ala4 bond), which at least in principle could lead to fully crosslinked PG. Intriguingly, even mutants deleted in multiple or all L,D transpeptidases exhibit only minor phenotypes (Sanders & Pavelka, 2013) and the types of crosslinks these enzymes create (DAP-DAP or 3,3 crosslinks) are typically too rare to provide full structural integrity (Glauner et al., 1988, Desmarais et al., 2013). A notable exception is Agrobacterium tumefaciens, whose PG naturally consists of ~45% L,D crosslinks (Quintela et al., 1995). An increase in 3,3 crosslinks has been observed in multiple other species when cells enter stationary phase (where in Mycobacterium tuberculosis, up to 80% of PG can be crosslinked via DAP-DAP (Lavollay et al., 2008)), and under envelope stress conditions: activation of the Cpx response in E. coli for example led to a ~1.5-fold increase in DAP-DAP crosslinks (Lavollay et al., 2008, Bernal-Cabas et al., 2015). It is therefore possible that L,D transpeptidation serves a supporting role for D,D crosslinks to further strengthen the PG meshwork under certain conditions.
Early evidence that L,D transpeptidases could assume a more fundamental role in cell wall biogenesis came from the work of the Gutmann and Arthur labs. In a series of papers (Mainardi et al., 2000, Mainardi et al., 2002, Mainardi et al., 2005), these authors described the selection for a β-lactam resistant mutant of Enterococcus faecalis, whose cell wall was found to be essentially devoid of the classical D,D crosslinks mediated by PBPs. This mutant could grow in the presence of β-lactam antibiotics by substituting the D,D crosslinks formed by the β-lactam sensitive PBPs with those formed by a β-lactam-insensitive L,D transpeptidase named Ldtfm, which catalyzed transpeptidation between D-asparagine and L-lysine residues situated in neighboring PG strands, resulting in L,D (3,3) bonds (Mainardi et al., 2005). Interestingly, the activity or abundance of the Ldtfm or PBPs was unaltered in the resistant mutant. Instead, this strain showed an increase in the activity of a carboxypeptidase that removes the terminal D-Ala5; the resulting tetrapeptide sidestem is recognized by L,D transpeptidases, but not PBPs as a substrate. Thus, E. faecalis provides intrinsic substrate cues to reprogram the activity of PG crosslinking enzymes and thus the nature of its PG crosslinks.
Another study recently reported that a similar mechanism of β-lactam resistance can evolve in E. coli (Hugonnet et al., 2016). Upon multistep selection on β-lactam antibiotics, a mutant emerged that had upregulated one of its L,D transpeptidases (YcbB) as well as the stringent response, a starvation response that leads to the accumulation of the alarmone ppGpp and subsequent reprogramming of transcription. While the connection between the stringent response and L,D-TP activity is unclear, this strain utilizes the TG activity of the aPBP1B in conjunction with TP activity of YcbB for cell wall synthesis and crosslinking. Like in E. faecium, the ability to form L,D crosslinks depended on the presence of a carboxypeptidase, in this case PBP5. Although these experiments involved mutants that were generated under severe and artificial selection conditions, these results clearly demonstrate that L,D-TPase activity can, at least in principle, be contribute significantly to the structural integrity of the cell wall. It remains to be seen whether the primary reliance on L,D transpeptidation for bacterial growth is an oddity resulting from stringent conditions of mutant selection or can be an adaptive response (for example as a stress response mechanism in the presence of β-lactam antibiotics) in nature as well.
Cell biology of PG synthesis: New insights into the roles of the cytoskeletal proteins MreB and FtsZ
The basic enzymology of PG synthesis was established in early studies following conventional approaches that integrated in vitro enzyme assays with chemical and structural characterization of reaction mechanisms and products. However, efforts to decipher the larger scale coordination of PG synthesis with cell growth and division did not make great strides until the advent of bacterial cell biology. The introduction of fluorescently labeled proteins, high resolution light microscopy methods, and, more recently, single-molecule tracking approaches has invigorated the field and enabled the development of new models of PG synthesis and its coordination. It is not enough to just be able to stitch together new PG; the newly synthesized glycan strands must be integrated into the existing sacculus in a manner that does not compromise the overall integrity and load-bearing properties of the wall, and old wall material must be simultaneously shed and recycled. How new areas of synthesis are defined in a manner appropriate for the maintenance of cell shape, as seen for example in rods, cocci, and helically shaped bacteria, has been a challenging problem. Here, we focus on the emerging view of the two primary biosynthetic, macromolecular complexes involved in synthesis of rod-shaped bacteria: the “elongasome” and the “divisome”.
MreB and the “elongasome”
The transmembrane and periplasmic proteins associated with cell wall synthesis processes have been shown, or at least implicitly assumed, to be part of a single multiprotein complex called the “elongasome” (Laddomada et al., 2016, Egan et al., 2017, Errington, 2015) that contains structural components, as well as aPBPs, bPBPs, cell wall lytic enzymes (“autolysins”), and presumably a flippase. The “elongasome” was assumed to be spatio-temporally directed by the cytoskeletal protein MreB, a homologue of eukaryotic actin (van den Ent et al., 2001) that localizes to the lateral wall of the bacterial cell (Jones et al., 2001). One model suggests that MreB mediates the formation of regions with increased fluidity (RIFs), which affect distribution and diffusion of membrane proteins and may contribute to the organization of the “elongasome” (Strahl et al., 2014). Until recently, a generally accepted model of cell wall synthesis proposed that MreB served to guide the aPBPs, which in turn produce peptidoglycan strands via their TG domains while the aPBPs and the bPBPs crosslink these strands into a tight PG mesh, fitting new material into cell wall gaps provided by the cleavage activity of autolysins (Figure 1A). However, the existence of an “elongasome” protein complex could never be demonstrated in vivo and recent single molecule tracking experiments revealed that MreB and aPBPs operate in distinct complexes (Cho et al., 2016). This, in addition to the recent revelation that RodA itself possesses TG activity, calls for a re-evaluation of MreB’s contribution to cell wall synthesis (Meeske et al., 2016, Emami et al., 2017).
Figure 1. Unified (A) and Interdependent (B) models of peptidoglycan (PG) synthesis complexes.
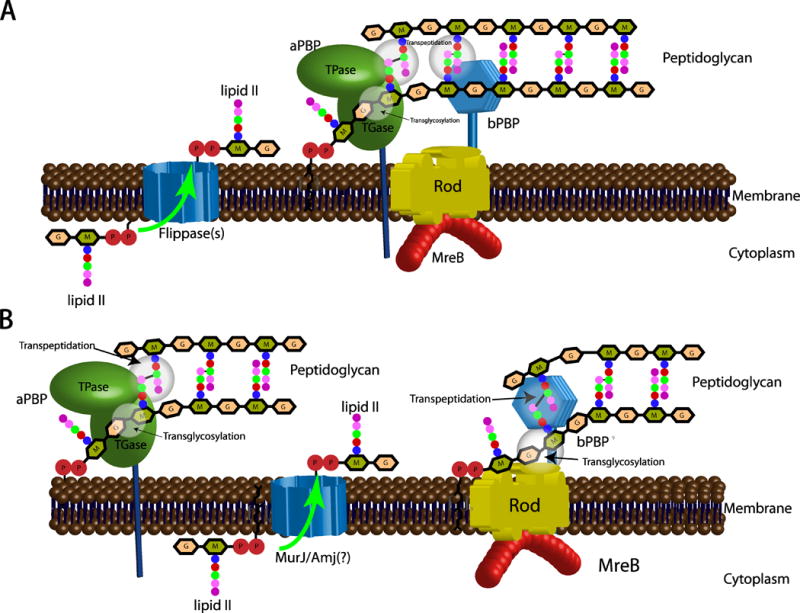
(A) In the unified model, RodAZ, MreB, aPBPs and bPBPs form one protein complex: guided by MreB, the aPBPs produce peptidoglycan strands via their TG domains while both aPBPs and bPBPs crosslink these strands into a tight PG mesh. (B) In the interdependent model, RodAZ, bPBP and MreB form one complex, while aPBP works in a different spatial and temporal frame. Glycan strands are produced by the transglycosylase RodA and are crosslinked by bPBP to existing PG. PG synthesis provides the force for pushing circumferential MreB movement. aPBPs exhibit a different movement pattern distinct from MreB, including two modes of movement: fast diffusion and slow movement (pause). These two systems are spatially distinct, but functionally interdependent for PG synthesis.
MreB is found in most rod-shaped bacteria and loss of MreB generally leads to the cessation of lateral cell wall synthesis and concomitant loss of rod-shape, establishing the cytoskeleton’s crucial role in directional PG insertion during cell elongation. MreB strongly interacts with RodA and RodZ in E. coli (Morgenstein et al., 2015) and the latter mediates the indirect interaction between MreB and a bPBP. This established complex (MreB-RodAZ-bPBP) will be referred to hereafter as the “Rod complex”.
Early localization studies using immunofluorescence and epifluorescence microscopy to visualize fluorescently tagged proteins in B. subtilis, E. coli and Caulobacter crescentus suggested that MreB localizes in helical filaments along the inner face of the cytoplasmic membrane spanning the lateral cell (Shih et al., 2003, Figge et al., 2004, Jones et al., 2001). However, using microscopy techniques that allowed for higher spatio-temporal resolution, it was later shown that instead of forming continuous filaments, MreB rotates around the cell in patches (arcs) whose motion depends on bPBP transpeptidation activity and the presence of RodA, but, at least in E. coli, not on the activity of aPBPs (Dominguez-Escobar et al., 2011, Garner et al., 2011, van Teeffelen et al., 2011). More recent data show that MreB locates to regions of negative curvature, and “corrects” this negative curvature by filling this region with newly synthesized PG, suggesting a self-correcting feedback mechanism for cells to maintain rod shape (Ursell et al., 2014). Overall these data strongly support a model in which cell wall synthesis during cell elongation is mediated primarily by the Rod complex.
Importantly, these single molecule studies have provided evidence for a spatial independence of the Rod complex and aPBPs (Figure 1B). In contrast, the Rod complex and bPBPs appear to move along the same trajectories, suggesting that they may be coupled (Cho et al., 2016) (though it has to be noted that another study had previously found that MreB and bPBP2 move at different velocities (Lee et al., 2014); this can be attributed to differences in imaging parameters and/or intrinsic differences between different fusion constructs, and may suggest that bPBP2’s circumferential motion is not essential for its function). In contrast, aPBPs showed a bimodal pattern of movement, with two distinct subpopulations: one exhibiting fast, diffusive motion, and another moving at a speed an order of magnitude slower (Cho et al., 2016, Lee et al., 2016). When considering the behavior of a single PBP molecule, these data can be interpreted as short periods of fast diffusion interspersed with temporary pauses. Although spatially independent, the partially redundant TG activities of the Rod complex and aPBPs are functionally coupled, as inactivation of one or the other leads to the same dramatic (~80%) decrease in incorporation of new cell wall material (Cho et al., 2016). This, together with the observation that in E. coli a bPBP interacts directly with aPBP1A (Banzhaf et al., 2012), suggests that the two seemingly independent activities of aPBPs and the Rod complex are somehow synergistic, and that they may at least transiently interface. A recent paper has exposed an additional layer of complexity about the relationship between the Rod complex and aPBPs. Using TIRF, Billaudeau et al. showed that in B. subtilis, MreB not only shows rotational movement, but has subpopulations that, similar to aPBPs, diffuse slowly or stop altogether (Billaudeau et al., 2017). This, coupled with indirect evidence of an interaction between MreB and aPBPs (Kawai et al., 2009) opens up the possibility that MreB and aPBPs associate. It is important to note that neither MreB patch rotational movement, nor aPBP activity, depend on each other (Cho et al., 2016, van Teeffelen et al., 2011); but whether diffusive MreB molecules functionally interact with aPBPs is not known. Further work is thus required to investigate the spatial and functional relationship between MreB (or at least a sub-population of it) and aPBPs.
How does Rod-mediated cell wall synthesis apparently drive its own motion, while aPBPs, in principle mediating the exact same reactions, are more diffusive? One possibility is that RodA’s TG activity drives directional movement, and that its interaction with short, dynamic MreB arcs essentially reinforces this movement, while the aPBPs move along a similar trajectory for a short time, but produce shorter PG chains and diffuse away when the TG reaction is terminated (e.g. through interaction with a putative chain termination factor, or due to an intrinsic capability to produce shorter PG chains). It is noteworthy that MreB was shown to interact with cytoplasmic cell wall precursor synthesis proteins and their localization changed during MreB depletion (Favini-Stabile et al., 2013, White et al., 2010, Rueff et al., 2014, Divakaruni et al., 2007), suggesting that MreB might coordinate the availability of precursors to generate a local pool of lipid II to support the activity of the Rod complex. This might be beneficial if RodA’s TG activity intrinsically generates longer PG strands than the aPBPs (generating shorter chains may enable the aPBPs to operate on a more limited local supply of lipid II).
In the light of the existence of two independent cell wall synthesis complexes, an important open question is whether flippase activity is associated with one of the complexes, both, or is completely independent. Current data favor the latter hypothesis: heterologous expression of the Helicobacter pylori O-antigen flippase Wzk (Elhenawy et al., 2016) or B. subtilis Amj (Meeske et al., 2015) can functionally replace MurJ in E. coli. Amj has no homologs in E. coli and is thus unlikely to specifically interact with this organism’s cell wall synthesis machinery, suggesting that unguided lipid II flipping may be sufficient to sustain bacterial growth. Furthermore, cell wall incorporation after inhibition of either the Rod complex or aPBPs is not zero, implying residual flippase activity. Thus, it is likely that lipid II flippase activity is not strictly dependent on either complex.
FtsZ and the divisome: FtsZ treadmilling drives PG synthesis at the septum
FtsZ, a homolog of tubulin, is essential for cell division in many bacteria. Cytoplasmic FtsZ molecules polymerize at the inner face of the cytoplasmic membrane as a dynamic ring of FtsZ filaments of varying lengths (Michie & Lowe, 2006). This so-called “Z-ring” and various accessory factors anchor the assembly of a dynamic, spatio-temporally ordered multiprotein complex called the divisome. The divisome contains what are generally assumed to be structural proteins, but also proteins involved in cell wall synthesis (PBPs) and turnover, like amidases and lytic transglycosylases (Egan & Vollmer, 2013). Ultimately, the FtsZ-guided divisome serves the function of facilitating cytokinesis, membrane constriction, synthesis of new cell wall material and finally daughter cell separation. Purified FtsZ is sufficient to initiate constriction of elongated liposomes (Osawa et al., 2009, Osawa et al., 2008), suggesting that FtsZ itself generates the forces for cell division, powered by GTP hydrolysis (RayChaudhuri & Park, 1992, de Boer et al., 1992). This was later challenged by results from experiments showing that constriction does not initiate in the absence of cell wall synthesis (Daley et al., 2016). Beyond these observations, the role of FtsZ outside of its anchor function remained largely mysterious.
Several recent studies have addressed this issue with newly available super-resolution techniques. In two parallel studies, Bisson-Filho et al. and Yang et al. used single molecule tracking and super-resolution microscopy combined with targeted perturbations of division processes to assess the role of FtsZ filaments in the division process in B. subtilis and E. coli (Bisson-Filho et al., 2017, Yang et al., 2017). Both groups found that short FtsZ filaments display a rotational, inward movement that coincides with the deposition of new cell wall material. Strikingly, and in contrast to MreB, FtsZ movement was independent of cell wall synthesis and driven by treadmilling, which depended on its GTPase activity. It thus appears that FtsZ generates its own motion, and induces cell wall synthesis during the constriction process. This observation could provide an explanation for previously inconsistent data: while FtsZ treadmilling by itself probably generates enough force to initiate membrane constriction, it is the reinforcement of these constrictions via guided traces of PG material that enables the completion of outer membrane constriction and cytokinesis. This more active, cytoskeleton-driven process of movement (as opposed to MreB’s passive motion) might thus be necessary to apply the forces needed for cell division.
An emerging model of PG synthesis for rod-shaped bacteria
One of the most influential unified models of cell wall growth coordination was put forth in a seminal review paper by Höltje (Holtje, 1998). Asking how PG lytic and synthetic processes might be coordinated without compromising cell wall structural integrity, the author proposed that bacteria synthesize a precursor of three parallel, crosslinked PG strands, which would substitute for a single strand concomitantly removed by the coordinated activity of PG hydrolases; he termed this the “3-for-1” model. Höltje assumed that the parallel strands were generated before breaking any bonds in the PG meshwork (to ensure structural integrity), or a “make before break” mode of sacculus expansion. This model has remained an important conceptual framework, but has not been re-evaluated in the light of subsequent new observations.
We will attempt here to integrate new data on the mechanisms of PG biogenesis with previous observations into an updated model of cell wall biogenesis (Figure 2). Perhaps one of the most striking recent realizations is that the Rod complex and aPBPs are spatially distinct, yet their activities are interdependent. A possible model is that one cell wall synthesis complex creates a template structure for the other, consistent with what has been suggested by Wientjes et al. (Wientjes & Nanninga, 1991) and Cho et al. (Cho et al., 2016). We speculate that template generation is accomplished by the circumferentially moving population of the Rod complex, as steady, circumferential motion would be conducive to providing a regular template structure. In contrast, the aPBPs exhibit a diffusive motion interspersed with prolonged local persistence (Cho et al., 2016, Lee et al., 2016), which could suggest that their role is to diffuse freely until they recognize a PG trace or gap (e.g. one generated by the Rod complex) and then add new material to the template. Inhibition of a bPBP (PBP2) in E. coli results in the generation of PG fragments that are not incorporated into the PG meshwork but rather are rapidly degraded and recycled (Cho et al., 2014, Uehara & Park, 2008), suggesting that in Rod-mediated PG synthesis, PBP2 provides the first point of attachment for nascent PG after (or while) it emerges from RodA-mediated transglycosylation. This first point of attachment would also be an important anchor providing a fulcrum for RodA-driven MreB movement; perhaps this is why inhibition of bPBPs stops MreB motion (van Teeffelen et al., 2011, Garner et al., 2011, Dominguez-Escobar et al., 2011) but allows futile synthesis of PG by RodA (Cho et al., 2016, Cho et al., 2014).
Figure 2. “Break before Make” model of peptidoglycan (PG) synthesis complexes.
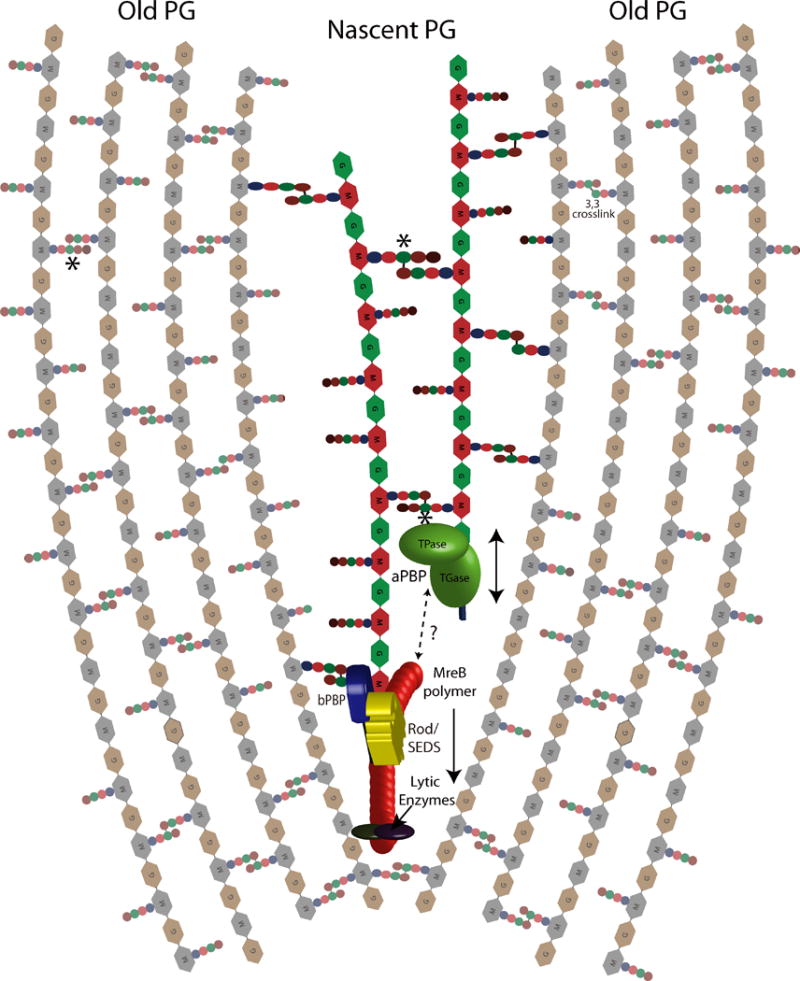
Rod/SEDS/MreB-associated endopeptidases locally cleave crosslinks in mature PG. RodA generates a PG template, which is attached to the sacculus via bPBPs (only one strand is shown here, note that in principle this could also be a raft structure of multiple parallel strands). The aPBPs then generate additional strands, which are crosslinked with nascent PG on one side and mature PG on the other, ensuring maintenance of structural integrity. Whether the Rod/SEDS/MreB complex interacts with aPBPs remains an open question. Crosslinked pentapeptide (asterisk) is formed when a nascent PG strand containing pentapeptide is crosslinked with another one. PBP-independent 3,3 crosslinks also exist albeit at low abundance under normal growth conditions.
Interestingly, at least in E. coli, a bPBP (PBP2) was shown to activate an aPBP (PBP1A) (Banzhaf et al., 2012). The fact that the Rod-associated bPBP stimulates the aPBP’s TG activity may suggest that rod-driven PG synthesis starts prior to aPBP-driven PG synthesis. We propose that the aPBPs likely use the Rod-mediated PG template to attach parallel (or possibly antiparallel) PG strands (Figure 2). In addition to providing lateral directionality of sacculus expansion, this is consistent with the observed existence of crosslinked PG strands containing pentapeptide: enzymatically, the only way to generate PG containing pentapeptides is when nascent PG is crosslinked with another strand of nascent PG (since the terminal D-Ala of the donor strand is lost in the crosslinking reaction). Moreover, pentapeptides are likely rapidly processed by carboxypeptidases associated with cell wall synthesis (Potluri et al., 2010, Santos et al., 2002, Atrih et al., 1999, Moll et al., 2015)), but nascent-nascent crosslinks are indeed observed during pulse-chase PG labeling experiments (Burman & Park, 1984). Alternatively, these crosslinked PG strands containing pentapeptide may come from region with presumably less carboxypeptidase activity, for example the septum (Morales Angeles et al., 2017), or from PG generated by several Rod complexes working in parallel, as recently suggested for B. subtilis (Billaudeau et al., 2017). It is therefore possible that the Rod template actually consists of a PG “raft” structure of several strands that are then woven tightly into the cell wall by aPBPs. Why are the aPBPs partially dispensable in B. subtilis (dependent on stress-response mediated upregulation of RodA) and not in E. coli? Perhaps the Rod complex template itself is enough to mediate sacculus expansion, as long as it is made in sufficient quantity (B. subtilis upregulates RodA upon aPBP deletion via the Sigma M cell wall stress sensing pathway) and as long as there is a sufficient stress-bearing “buffer”, i.e. a thick cell wall that can partially compensate for localized, inefficient crosslinking.
Our model suggests that the Rod complex might coordinate its cell wall synthesis activity with PG cleavage by endopeptidases, which are required for the insertion of new PG material during cell elongation (Vollmer, 2012, Singh et al., 2012, Dorr et al., 2013) and should therefore immediately precede template attachment. Consistent with this hypothesis, MreB homologs in B. subtilis have been shown to direct the activities of elongation-specific endopeptidases LytE and (indirectly) CwlO (Dominguez-Cuevas et al., 2013, Meisner et al., 2013). Further, at least in Gram-negative bacteria, inhibition or depletion of aPBPs (leaving the putative RodA-autolysin complex active) typically leads to a catastrophic loss of the cell wall (Dorr et al., 2014, Yousif et al., 1985, Satta et al., 1995), while defects associated with the Rod complex (which might cause concurrent lack of major autolysin activation) are generally milder and simply result in an abrogation of cell elongation with loss of cellular integrity only after prolonged exposure (Tybring & Melchior, 1975, Iwai et al., 2002). Measurements of the incorporation of new cell wall material in the presence of antibiotics previously showed that inhibition of aPBPs resulted in a delayed inhibition of cell wall incorporation, and cessation of cell wall synthesis coincided with the onset of lysis (Wientjes & Nanninga, 1991). These observations are also consistent with Rod-associated cell wall cleavage and template generation, which would proceed even in the absence of aPBPs until a “tipping point” is reached where accumulated damage caused by the lack of subsequent aPBP-mediated crosslinking results in catastrophic failure of structural integrity. Recent results demonstrate that E. coli endopeptidase activity increases aPBP-mediated cell wall attachment during inhibition of bPBP2 (Lai et al., 2017), suggesting that endopeptidases might, in addition to priming the Rod system, provide Rod-independent starting points (gaps) for PG synthesis by aPBPs.
It remains to be seen what the roles of lytic transglycosylases (LTGs) are in the cell wall biosynthesis process. The typical PG breakdown products of these enzymes are detected during growth and at increased levels upon exposure to cell wall synthesis inhibitors; however, it is currently unclear whether removal of a strand (or strands) of mature PG is actually necessary for the insertion of new material. Alternatively, LTG breakdown products could be the result of the removal of the outer cell layer in Gram-positive bacteria, a proofreading capacity (removing erroneously crosslinked and thus potentially unstable cell wall material), as has been suggested previously (Cho et al., 2014), or simply the fact that nascent PG is produced in longer chains at first and then trimmed down to the length most appropriate for the current growth condition (Yunck et al., 2016, Vollmer & Holtje, 2004).
In summary, we propose a “break before make” model. Endopeptidases locally cleave crosslinks in mature PG. RodA generates a PG template, which is attached to the sacculus via bPBPs. Since the Rod complex is not expected to perform an entire rotation around the cell (based on short PG chain lengths measured in various bacteria), these PG degradation events are initially localized and overall structural integrity is thus not immediately compromised. The aPBPs then generate additional strands, which are crosslinked with nascent PG on one side and mature PG on the other, ensuring that the local degradation events initiated by the Rod complex do not accumulate with harmful consequences. Previous simulations and observations regarding bPBP2 inactivation have indeed shown that the cell wall synthesis machinery, even in Gram-negative bacteria, is surprisingly well-buffered and can sustain a fairly high amount of degradation before experiencing catastrophic failure (Lee et al., 2014, Huang et al., 2008).
Important questions remain unanswered in the context of this model; most importantly, how the Rod complex defines start sites for PG synthesis, how aPBPs recognize the putative Rod template and what role the modulators of PBP activity play (such as the outer membrane localized activators in Gram-negative bacteria, (Typas et al., 2010, Paradis-Bleau et al., 2010)). After all, more than 50 years after its emergence, bacterial cell wall research still holds surprises, and is expected to continue doing so.
Acknowledgments
This work was supported by a grant to JDH from the National Institutes of Health (R35GM122461). We thank Thomas Bernhardt, Felipe Cava, KC Huang, Kevin Young and two anonymous reviewers for helpful comments.
References
- Anderson JS, Matsuhashi M, Haskin MA, Strominger JL. Biosythesis of the peptidoglycan of bacterial cell walls. II. Phospholipid carriers in the reaction sequence. J Biol Chem. 1967;242:3180–3190. [PubMed] [Google Scholar]
- Anderson JS, Meadow PM, Haskin MA, Strominger JL. Biosynthesis of the peptidoglycan of bacterial cell walls. I. Utilization of uridine diphosphate acetylmuramyl pentapeptide and uridine diphosphate acetylglucosamine for peptidoglycan synthesis by particulate enzymes from Staphylococcus aureus and Micrococcus lysodeikticus. Arch Biochem Biophys. 1966;116:487–515. doi: 10.1016/0003-9861(66)90056-7. [DOI] [PubMed] [Google Scholar]
- Arbeloa A, Segal H, Hugonnet JE, Josseaume N, Dubost L, Brouard JP, Gutmann L, Mengin-Lecreulx D, Arthur M. Role of class A penicillin-binding proteins in PBP5-mediated beta-lactam resistance in Enterococcus faecalis. J Bacteriol. 2004;186:1221–1228. doi: 10.1128/JB.186.5.1221-1228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atrih A, Bacher G, Allmaier G, Williamson MP, Foster SJ. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J Bacteriol. 1999;181:3956–3966. doi: 10.1128/jb.181.13.3956-3966.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banzhaf M, van den Berg van Saparoea B, Terrak M, Fraipont C, Egan A, Philippe J, Zapun A, Breukink E, Nguyen-Disteche M, den Blaauwen T, Vollmer W. Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol Microbiol. 2012;85:179–194. doi: 10.1111/j.1365-2958.2012.08103.x. [DOI] [PubMed] [Google Scholar]
- Bernal-Cabas M, Ayala JA, Raivio TL. The Cpx envelope stress response modifies peptidoglycan cross-linking via the L,D-transpeptidase LdtD and the novel protein YgaU. J Bacteriol. 2015;197:603–614. doi: 10.1128/JB.02449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billaudeau C, Chastanet A, Yao Z, Cornilleau C, Mirouze N, Fromion V, Carballido-Lopez R. Contrasting mechanisms of growth in two model rod-shaped bacteria. Nat Commun. 2017;8:15370. doi: 10.1038/ncomms15370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson-Filho AW, Hsu YP, Squyres GR, Kuru E, Wu F, Jukes C, Sun Y, Dekker C, Holden S, VanNieuwenhze MS, Brun YV, Garner EC. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science. 2017;355:739–743. doi: 10.1126/science.aak9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramkamp M. The putative Bacillus subtilis L,D-transpeptidase YciB is a lipoprotein that localizes to the cell poles in a divisome-dependent manner. Arch Microbiol. 2010;192:57–68. doi: 10.1007/s00203-009-0532-5. [DOI] [PubMed] [Google Scholar]
- Brown S, Santa Maria JP, Jr, Walker S. Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol. 2013;67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burman LG, Park JT. Molecular model for elongation of the murein sacculus of Escherichia coli. Proc Natl Acad Sci U S A. 1984;81:1844–1848. doi: 10.1073/pnas.81.6.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cava F, de Pedro MA, Lam H, Davis BM, Waldor MK. Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J. 2011;30:3442–3453. doi: 10.1038/emboj.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Uehara T, Bernhardt TG. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell. 2014;159:1300–1311. doi: 10.1016/j.cell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Wivagg CN, Kapoor M, Barry Z, Rohs PD, Suh H, Marto JA, Garner EC, Bernhardt TG. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol. 2016:16172. doi: 10.1038/nmicrobiol.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley DO, Skoglund U, Soderstrom B. FtsZ does not initiate membrane constriction at the onset of division. Sci Rep. 2016;6:33138. doi: 10.1038/srep33138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- Denome SA, Elf PK, Henderson TA, Nelson DE, Young KD. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol. 1999;181:3981–3993. doi: 10.1128/jb.181.13.3981-3993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmarais SM, DePedro MA, Cava F, Huang KC. Peptidoglycan at its peaks: how chromatographic analyses can reveal bacterial cell wall structure and assembly. Mol Microbiol. 2013;89:1–13. doi: 10.1111/mmi.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Berardino M, Dijkstra A, Stuber D, Keck W, Gubler M. The monofunctional glycosyltransferase of Escherichia coli is a member of a new class of peptidoglycan-synthesising enzymes. FEBS Lett. 1996;392:184–188. doi: 10.1016/0014-5793(96)00809-5. [DOI] [PubMed] [Google Scholar]
- Divakaruni AV, Baida C, White CL, Gober JW. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol Microbiol. 2007;66:174–188. doi: 10.1111/j.1365-2958.2007.05910.x. [DOI] [PubMed] [Google Scholar]
- Dominguez-Cuevas P, Porcelli I, Daniel RA, Errington J. Differentiated roles for MreB-actin isologues and autolytic enzymes in Bacillus subtilis morphogenesis. Mol Microbiol. 2013;89:1084–1098. doi: 10.1111/mmi.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, Carballido-Lopez R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- Dorr T, Cava F, Lam H, Davis BM, Waldor MK. Substrate specificity of an elongation-specific peptidoglycan endopeptidase and its implications for cell wall architecture and growth of Vibrio cholerae. Mol Microbiol. 2013;89:949–962. doi: 10.1111/mmi.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr T, Lam H, Alvarez L, Cava F, Davis BM, Waldor MK. A novel peptidoglycan binding protein crucial for PBP1A-mediated cell wall biogenesis in Vibrio cholerae. PLoS Genet. 2014;10:e1004433. doi: 10.1371/journal.pgen.1004433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan AJ, Cleverley RM, Peters K, Lewis RJ, Vollmer W. Regulation of bacterial cell wall growth. FEBS J. 2017;284:851–867. doi: 10.1111/febs.13959. [DOI] [PubMed] [Google Scholar]
- Egan AJ, Vollmer W. The physiology of bacterial cell division. Ann N Y Acad Sci. 2013;1277:8–28. doi: 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- Eiamphungporn W, Helmann JD. The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses. Mol Microbiol. 2008;67:830–848. doi: 10.1111/j.1365-2958.2007.06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Ghachi M, Derbise A, Bouhss A, Mengin-Lecreulx D. Identification of multiple genes encoding membrane proteins with undecaprenyl pyrophosphate phosphatase (UppP) activity in Escherichia coli. J Biol Chem. 2005;280:18689–18695. doi: 10.1074/jbc.M412277200. [DOI] [PubMed] [Google Scholar]
- Elhenawy W, Davis RM, Fero J, Salama NR, Felman MF, Ruiz N. The O-Antigen Flippase Wzk Can Substitute for MurJ in Peptidoglycan Synthesis in Helicobacter pylori and Escherichia coli. PLoS One. 2016;11:e0161587. doi: 10.1371/journal.pone.0161587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami K, Guyet A, Kawai Y, Devi J, Wu LJ, Allenby N, Daniel RA, Errington J. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat Microbiol. 2017;2:16253. doi: 10.1038/nmicrobiol.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J. Bacterial morphogenesis and the enigmatic MreB helix. Nat Rev Microbiol. 2015;13:241–248. doi: 10.1038/nrmicro3398. [DOI] [PubMed] [Google Scholar]
- Espaillat A, Forsmo O, El Biari K, Bjork R, Lemaitre B, Trygg J, Canada FJ, de Pedro MA, Cava F. Chemometric Analysis of Bacterial Peptidoglycan Reveals Atypical Modifications That Empower the Cell Wall against Predatory Enzymes and Fly Innate Immunity. J Am Chem Soc. 2016;138:9193–9204. doi: 10.1021/jacs.6b04430. [DOI] [PubMed] [Google Scholar]
- Favini-Stabile S, Contreras-Martel C, Thielens N, Dessen A. MreB and MurG as scaffolds for the cytoplasmic steps of peptidoglycan biosynthesis. Environ Microbiol. 2013;15:3218–3228. doi: 10.1111/1462-2920.12171. [DOI] [PubMed] [Google Scholar]
- Fay A, Dworkin J. Bacillus subtilis homologs of MviN (MurJ), the putative Escherichia coli lipid II flippase, are not essential for growth. J Bacteriol. 2009;191:6020–6028. doi: 10.1128/JB.00605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge RM, Divakaruni AV, Gober JW. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol. 2004;51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- Gampe CM, Tsukamoto H, Doud EH, Walker S, Kahne D. Tuning the moenomycin pharmacophore to enable discovery of bacterial cell wall synthesis inhibitors. J Am Chem Soc. 2013;135:3776–3779. doi: 10.1021/ja4000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauner B, Holtje JV, Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263:10088–10095. [PubMed] [Google Scholar]
- Goffin C, Ghuysen JM. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest RL, Raivio TL. Role of the Gram-Negative Envelope Stress Response in the Presence of Antimicrobial Agents. Trends Microbiol. 2016;24:377–390. doi: 10.1016/j.tim.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Hao H, Dai M, Wang Y, Huang L, Yuan Z. Key genetic elements and regulation systems in methicillin-resistant Staphylococcus aureus. Future Microbiol. 2012;7:1315–1329. doi: 10.2217/fmb.12.107. [DOI] [PubMed] [Google Scholar]
- Helmann JD. Bacillus subtilis extracytoplasmic function (ECF) sigma factors and defense of the cell envelope. Curr Opin Microbiol. 2016;30:122–132. doi: 10.1016/j.mib.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez SB, Cava F, Pucciarelli MG, Garcia-Del Portillo F, de Pedro MA, Casadesus J. Bile-induced peptidoglycan remodelling in Salmonella enterica. Environ Microbiol. 2015;17:1081–1089. doi: 10.1111/1462-2920.12491. [DOI] [PubMed] [Google Scholar]
- Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS. Cell shape and cell-wall organization in Gram-negative bacteria. Proc Natl Acad Sci U S A. 2008;105:19282–19287. doi: 10.1073/pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugonnet JE, Mengin-Lecreulx D, Monton A, den Blaauwen T, Carbonnelle E, Veckerle C, Brun YV, van Nieuwenhze M, Bouchier C, Tu K, Rice LB, Arthur M. Factors essential for L, D-transpeptidase-mediated peptidoglycan cross-linking and beta-lactam resistance in Escherichia coli. Elife. 2016;5 doi: 10.7554/eLife.19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino F, Park W, Tomioka S, Tamaki S, Takase I, Kunugita K, Matsuzawa H, Asoh S, Ohta T, Spratt BG, et al. Peptidoglycan synthetic activities in membranes of Escherichia coli caused by overproduction of penicillin-binding protein 2 and rodA protein. J Biol Chem. 1986;261:7024–7031. [PubMed] [Google Scholar]
- Iwai N, Nagai K, Wachi M. Novel S-benzylisothiourea compound that induces spherical cells in Escherichia coli probably by acting on a rod-shape-determining protein(s) other than penicillin-binding protein 2. Biosci Biotechnol Biochem. 2002;66:2658–2662. doi: 10.1271/bbb.66.2658. [DOI] [PubMed] [Google Scholar]
- Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Daniel RA, Errington J. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol Microbiol. 2009;71:1131–1144. doi: 10.1111/j.1365-2958.2009.06601.x. [DOI] [PubMed] [Google Scholar]
- Kruse T, Bork-Jensen J, Gerdes K. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol. 2005;55:78–89. doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- Laddomada F, Miyachiro MM, Dessen A. Structural Insights into Protein-Protein Interactions Involved in Bacterial Cell Wall Biogenesis. Antibiotics (Basel) 2016;5 doi: 10.3390/antibiotics5020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai GC, Cho H, Bernhardt TG. The mecillinam resistome reveals a role for peptidoglycan endopeptidases in stimulating cell wall synthesis in Escherichia coli. PLoS Genet. 2017;13:e1006934. doi: 10.1371/journal.pgen.1006934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, Mainardi JL. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L,D-transpeptidation. J Bacteriol. 2008;190:4360–4366. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Derouaux A, Olatunji S, Fraipont C, Egan AJ, Vollmer W, Breukink E, Terrak M. Interplay between Penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis. Sci Rep. 2017;7:43306. doi: 10.1038/srep43306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TK, Meng K, Shi H, Huang KC. Single-molecule imaging reveals modulation of cell wall synthesis dynamics in live bacterial cells. Nat Commun. 2016;7:13170. doi: 10.1038/ncomms13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TK, Tropini C, Hsin J, Desmarais SM, Ursell TS, Gong E, Gitai Z, Monds RD, Huang KC. A dynamically assembled cell wall synthesis machinery buffers cell growth. Proc Natl Acad Sci U S A. 2014;111:4554–4559. doi: 10.1073/pnas.1313826111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovering AL, Safadi SS, Strynadka NC. Structural perspective of peptidoglycan biosynthesis and assembly. Annu Rev Biochem. 2012;81:451–478. doi: 10.1146/annurev-biochem-061809-112742. [DOI] [PubMed] [Google Scholar]
- Magnet S, Arbeloa A, Mainardi JL, Hugonnet JE, Fourgeaud M, Dubost L, Marie A, Delfosse V, Mayer C, Rice LB, Arthur M. Specificity of L,D-transpeptidases from gram-positive bacteria producing different peptidoglycan chemotypes. J Biol Chem. 2007;282:13151–13159. doi: 10.1074/jbc.M610911200. [DOI] [PubMed] [Google Scholar]
- Magnet S, Dubost L, Marie A, Arthur M, Gutmann L. Identification of the L,D-transpeptidases for peptidoglycan cross-linking in Escherichia coli. J Bacteriol. 2008;190:4782–4785. doi: 10.1128/JB.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainardi JL, Fourgeaud M, Hugonnet JE, Dubost L, Brouard JP, Ouazzani J, Rice LB, Gutmann L, Arthur M. A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant transpeptidation pathway. J Biol Chem. 2005;280:38146–38152. doi: 10.1074/jbc.M507384200. [DOI] [PubMed] [Google Scholar]
- Mainardi JL, Legrand R, Arthur M, Schoot B, van Heijenoort J, Gutmann L. Novel mechanism of beta-lactam resistance due to bypass of DD-transpeptidation in Enterococcus faecium. J Biol Chem. 2000;275:16490–16496. doi: 10.1074/jbc.M909877199. [DOI] [PubMed] [Google Scholar]
- Mainardi JL, Morel V, Fourgeaud M, Cremniter J, Blanot D, Legrand R, Frehel C, Arthur M, Van Heijenoort J, Gutmann L. Balance between two transpeptidation mechanisms determines the expression of beta-lactam resistance in Enterococcus faecium. J Biol Chem. 2002;277:35801–35807. doi: 10.1074/jbc.M204319200. [DOI] [PubMed] [Google Scholar]
- Mainardi JL, Villet R, Bugg TD, Mayer C, Arthur M. Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol Rev. 2008;32:386–408. doi: 10.1111/j.1574-6976.2007.00097.x. [DOI] [PubMed] [Google Scholar]
- Mascher T, Hachmann AB, Helmann JD. Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function sigma factors. J Bacteriol. 2007;189:6919–6927. doi: 10.1128/JB.00904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough MA, Anderson JW, Silvaggi NR, Pratt RF, Knox JR, Kelly JA. Structures of two kinetic intermediates reveal species specificity of penicillin-binding proteins. J Mol Biol. 2002;322:111–122. doi: 10.1016/s0022-2836(02)00742-8. [DOI] [PubMed] [Google Scholar]
- McPherson DC, Popham DL. Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J Bacteriol. 2003;185:1423–1431. doi: 10.1128/JB.185.4.1423-1431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, Riley EP, Robins WP, Uehara T, Mekalanos JJ, Kahne D, Walker S, Kruse AC, Bernhardt TG, Rudner DZ. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature. 2016;537:634–638. doi: 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, Sham LT, Kimsey H, Koo BM, Gross CA, Bernhardt TG, Rudner DZ. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc Natl Acad Sci U S A. 2015;112:6437–6442. doi: 10.1073/pnas.1504967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J, Montero Llopis P, Sham LT, Garner E, Bernhardt TG, Rudner DZ. FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol Microbiol. 2013;89:1069–1083. doi: 10.1111/mmi.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michie KA, Lowe J. Dynamic filaments of the bacterial cytoskeleton. Annu Rev Biochem. 2006;75:467–492. doi: 10.1146/annurev.biochem.75.103004.142452. [DOI] [PubMed] [Google Scholar]
- Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240–273. doi: 10.1128/CMR.00046-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi T, van Dam V, Sijbrandi R, Vernet T, Zapun A, Bouhss A, Diepeveen-de Bruin M, Nguyen-Disteche M, de Kruijff B, Breukink E. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011;30:1425–1432. doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll A, Dorr T, Alvarez L, Davis BM, Cava F, Waldor MK. A D, D-carboxypeptidase is required for Vibrio cholerae halotolerance. Environ Microbiol. 2015;17:527–540. doi: 10.1111/1462-2920.12779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales Angeles D, Liu Y, Hartman AM, Borisova M, de Sousa Borges A, de Kok N, Beilharz K, Veening JW, Mayer C, Hirsch AK, Scheffers DJ. Pentapeptide-rich peptidoglycan at the Bacillus subtilis cell-division site. Mol Microbiol. 2017;104:319–333. doi: 10.1111/mmi.13629. [DOI] [PubMed] [Google Scholar]
- Morgenstein RM, Bratton BP, Nguyen JP, Ouzounov N, Shaevitz JW, Gitai Z. RodZ links MreB to cell wall synthesis to mediate MreB rotation and robust morphogenesis. Proc Natl Acad Sci U S A. 2015;112:12510–12515. doi: 10.1073/pnas.1509610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan PJ, Sychantha D, Clarke AJ. Chemical biology of peptidoglycan acetylation and deacetylation. Bioorg Chem. 2014;54:44–50. doi: 10.1016/j.bioorg.2014.03.010. [DOI] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP. Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 2009;28:3476–3484. doi: 10.1038/emboj.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell. 2010;143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potluri L, Karczmarek A, Verheul J, Piette A, Wilkin JM, Werth N, Banzhaf M, Vollmer W, Young KD, Nguyen-Disteche M, den Blaauwen T. Septal and lateral wall localization of PBP5, the major D,D-carboxypeptidase of Escherichia coli, requires substrate recognition and membrane attachment. Mol Microbiol. 2010;77:300–323. doi: 10.1111/j.1365-2958.2010.07205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintela JC, Caparros M, de Pedro MA. Variability of peptidoglycan structural parameters in gram-negative bacteria. FEMS Microbiol Lett. 1995;125:95–100. doi: 10.1111/j.1574-6968.1995.tb07341.x. [DOI] [PubMed] [Google Scholar]
- RayChaudhuri D, Park JT. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992;359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- Rebets Y, Lupoli T, Qiao Y, Schirner K, Villet R, Hooper D, Kahne D, Walker S. Moenomycin resistance mutations in Staphylococcus aureus reduce peptidoglycan chain length and cause aberrant cell division. ACS Chem Biol. 2014;9:459–467. doi: 10.1021/cb4006744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed P, Veiga H, Jorge AM, Terrak M, Pinho MG. Monofunctional transglycosylases are not essential for Staphylococcus aureus cell wall synthesis. J Bacteriol. 2011;193:2549–2556. doi: 10.1128/JB.01474-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann NT, Grundling A. Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol Lett. 2011;319:97–105. doi: 10.1111/j.1574-6968.2011.02260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LB, Carias LL, Rudin S, Hutton R, Marshall S, Hassan M, Josseaume N, Dubost L, Marie A, Arthur M. Role of class A penicillin-binding proteins in the expression of beta-lactam resistance in Enterococcus faecium. J Bacteriol. 2009;191:3649–3656. doi: 10.1128/JB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueff AS, Chastanet A, Dominguez-Escobar J, Yao Z, Yates J, Prejean MV, Delumeau O, Noirot P, Wedlich-Soldner R, Filipe SR, Carballido-Lopez R. An early cytoplasmic step of peptidoglycan synthesis is associated to MreB in Bacillus subtilis. Mol Microbiol. 2014;91:348–362. doi: 10.1111/mmi.12467. [DOI] [PubMed] [Google Scholar]
- Ruiz N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci U S A. 2008;105:15553–15557. doi: 10.1073/pnas.0808352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AN, Pavelka MS. Phenotypic analysis of Eschericia coli mutants lacking L,D-transpeptidases. Microbiology. 2013;159:1842–1852. doi: 10.1099/mic.0.069211-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Lobo M, Matos AP, De Pedro MA, Arraiano CM. The gene bolA regulates dacA (PBP5), dacC (PBP6) and ampC (AmpC), promoting normal morphology in Escherichia coli. Mol Microbiol. 2002;45:1729–1740. doi: 10.1046/j.1365-2958.2002.03131.x. [DOI] [PubMed] [Google Scholar]
- Satta G, Cornaglia G, Mazzariol A, Golini G, Valisena S, Fontana R. Target for bacteriostatic and bactericidal activities of beta-lactam antibiotics against Escherichia coli resides in different penicillin-binding proteins. Antimicrob Agents Chemother. 1995;39:812–818. doi: 10.1128/aac.39.4.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- Scheffers DJ, Tol MB. LipidII: Just Another Brick in the Wall? PLoS Pathog. 2015;11:e1005213. doi: 10.1371/journal.ppat.1005213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham LT, Butler EK, Lebar MD, Kahne D, Bernhardt TG, Ruiz N. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science. 2014;345:220–222. doi: 10.1126/science.1254522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih YL, Le T, Rothfield L. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci U S A. 2003;100:7865–7870. doi: 10.1073/pnas.1232225100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel SD, Liu J, Ton-That H. Biogenesis of the Gram-positive bacterial cell envelope. Curr Opin Microbiol. 2016;34:31–37. doi: 10.1016/j.mib.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, SaiSree L, Amrutha RN, Reddy M. Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol Microbiol. 2012;86:1036–1051. doi: 10.1111/mmi.12058. [DOI] [PubMed] [Google Scholar]
- Strahl H, Burmann F, Hamoen LW. The actin homologue MreB organizes the bacterial cell membrane. Nat Commun. 2014;5:3442. doi: 10.1038/ncomms4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strominger JL, Blumberg PM, Suginaka H, Umbreit J, Wickus GG. How penicillin kills bacteria: progress and problems. Proc R Soc Lond B Biol Sci. 1971;179:369–383. doi: 10.1098/rspb.1971.0103. [DOI] [PubMed] [Google Scholar]
- Strominger JL, Park JT, Thompson RE. Composition of the cell wall of Staphylococcus aureus: its relation to the mechanism of action of penicillin. J Biol Chem. 1959;234:3263–3268. [PubMed] [Google Scholar]
- Tybring L, Melchior NH. Mecillinam (FL 1060), a 6beta-amidinopenicillanic acid derivative: bactericidal action and synergy in vitro. Antimicrob Agents Chemother. 1975;8:271–276. doi: 10.1128/aac.8.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, von Rechenberg M, Breukink E, den Blaauwen T, Gross CA, Vollmer W. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Park JT. Growth of Escherichia coli: significance of peptidoglycan degradation during elongation and septation. J Bacteriol. 2008;190:3914–3922. doi: 10.1128/JB.00207-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursell TS, Nguyen J, Monds RD, Colavin A, Billings G, Ouzounov N, Gitai Z, Shaevitz JW, Huang KC. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc Natl Acad Sci U S A. 2014;111:E1025–1034. doi: 10.1073/pnas.1317174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F, Amos LA, Lowe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci U S A. 2011;108:15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W. Bacterial growth does require peptidoglycan hydrolases. Mol Microbiol. 2012;86:1031–1035. doi: 10.1111/mmi.12059. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Holtje JV. The architecture of the murein (peptidoglycan) in gram-negative bacteria: vertical scaffold or horizontal layer(s)? J Bacteriol. 2004;186:5978–5987. doi: 10.1128/JB.186.18.5978-5987.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welzel P. A long research story culminates in the first total synthesis of moenomycin A. Angew Chem Int Ed Engl. 2007;46:4825–4829. doi: 10.1002/anie.200700765. [DOI] [PubMed] [Google Scholar]
- White CL, Kitich A, Gober JW. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol Microbiol. 2010;76:616–633. doi: 10.1111/j.1365-2958.2010.07108.x. [DOI] [PubMed] [Google Scholar]
- Wientjes FB, Nanninga N. On the role of the high molecular weight penicillin-binding proteins in the cell cycle of Escherichia coli. Res Microbiol. 1991;142:333–344. doi: 10.1016/0923-2508(91)90049-g. [DOI] [PubMed] [Google Scholar]
- Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, Xiao J. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science. 2017;355:744–747. doi: 10.1126/science.aak9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousif SY, Broome-Smith JK, Spratt BG. Lysis of Escherichia coli by beta-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J Gen Microbiol. 1985;131:2839–2845. doi: 10.1099/00221287-131-10-2839. [DOI] [PubMed] [Google Scholar]
- Yunck R, Cho H, Bernhardt TG. Identification of MltG as a potential terminase for peptidoglycan polymerization in bacteria. Mol Microbiol. 2016;99:700–718. doi: 10.1111/mmi.13258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sun Y, Peters JM, Gross CA, Garner EC, Helmann JD. Depletion of Undecaprenyl Pyrophosphate Phosphatases Disrupts Cell Envelope Biogenesis in Bacillus subtilis. J Bacteriol. 2016;198:2925–2935. doi: 10.1128/JB.00507-16. [DOI] [PMC free article] [PubMed] [Google Scholar]


