Abstract
Cholera toxin (CT) is the principal virulence factor of Vibrio cholerae for fatal cholera diarrhoea. Serogroups O1 and O139 harbour CT and are known to be epidemic strains. The remaining serogroups (nonO1/nonO139) are non-toxigenic and may be associated with mild disease. O1 serogroup emerged with a variant of CT known as Haitian cholera toxin (HCT). The HCT strains are hypervirulent and have been associated with severe cholera outbreaks in India, Western Africa and Haiti. Here, we report the presence of HCT (ctxB7) in a nonO1/nonO139 isolate causing persistent diarrhoea.
Keywords: Haitian cholera toxin, hypervirulence, nonO1/nonO139, persistent diarrhoea, Vibrio cholerae
Introduction
Vibrio cholerae, the causative agent of fatal cholera diarrhoea, has more than 200 serogroups, out of this only O1 and O139 strains are toxigenic (i.e. harbour cholera toxin (CT)) and are known to be epidemic strains. Serogroups other than O1 and O139, are broadly grouped as non-O1, non-O139 strains, which may cause mild gastroenteritis [1]. CT is the principal virulence factor for the disease and is mainly produced by O1 and O139, whereas nonO1/nonO139 strains are non-toxigenic. These possess other virulence factors associated with mild gastroenteritis. Here, we report a nonO1/nonO139 strain harbouring an unusual Haitian cholera toxin (HCT) associated with persistent diarrhoea in a 5-month-old baby. Emergence of HCT has become an alarming transition after its first report in 2007 from Odisha, India, and subsequently from Western Africa and Haiti [2], [3], [4]. The HCT-carrying strains are hypervirulent strains and are associated with higher severity of the disease in these outbreaks [2], [3], [4].
Case report
A 5-month-old female child resident in Kachi colony, Ashok Vihar, Loni, Ghaziabad (UP) presented with acute gastroenteritis (10–15 episodes of loose stools per day and three or four episodes of vomiting per day) with mild dehydration (at admission) was first admitted at Aruna Asaf Ali hospital, Delhi on 10 May 2016. Next day, the child developed severe dehydration with anuria, resulting in a stuporous condition. The stool was negative for rotavirus antigen (latex agglutination test). Stool culture was negative for enteropathogens like V. cholerae, Salmonella and Shigella species. However, Escherichia coli (few colonies) in mixed bacteria were identified from the stool. Stool microscopy showed plenty of bacteria but no ova or cyst. There were no pus cells or red blood cells. Cefixime 20 mg twice daily was started on the second day of illness but the high purge rate continued for 3 days. Cefixime was continued for 4 days. By the 4th day, the child's purge rate had reduced to three or four times per day. She was discharged on the 5th day with apparent clinical improvement and advice was to continue cefixime for another 3 days.
The child was readmitted on 27th May 2016 with the same complaints of loose stools; 10–15 episodes per day and vomiting with mild to moderate dehydration to begin with. Intravenous injection of monocef (cefpodoxime) 200 mg twice daily was started after sending the stool sample for pathogen identification at the Diarrhoeal Disease Laboratory, National Centre for Disease Control, Delhi. The stool sample was enriched in alkaline peptone water for 6 h followed by sub-culture on selective media (thiosulphate–citrate–bile salts–sucrose agar). The bacterial colonies were phenotypically identified as V. cholerae. Vibrio cholerae was also identified on non-selective media in mixed culture. Stool was found negative for rotavirus, Salmonella and Shigella. The isolate (named as NCDC-A54) was Gram-negative, motile and positive for oxidase and string test. The isolates were further confirmed as V. cholerae by PCR analyses as described earlier [2], [5]. The isolates were broadly grouped as nonO1/nonO139 because it did not react with antisera specific for the O1 and O139 serogroups. Biotyping of the isolate showed it to be haemolytic on sheep blood agar and variably positive to a Voges–Proskauer test, indicating ElTor biotype. The child remained apparently stable for 3 days; however, after 3 days she started having diarrhoea again (seven to eight times per day), which continued for another 2 days (three or four times per day) with vomiting and reduced urine output, though there was no blood or mucus in the stools and she did not have any fever. Stool microscopy showed the presence of pus cells, three to four per high-power field, and absence of red blood cells, ova or cysts. Monocef was continued until the day of discharge, i.e. for 7 days. The patient was discharged on 2 June 2016. Syndromically the patient was diagnosed as a case of persistent diarrhoea; she had taken a course of septran before hospital admission.
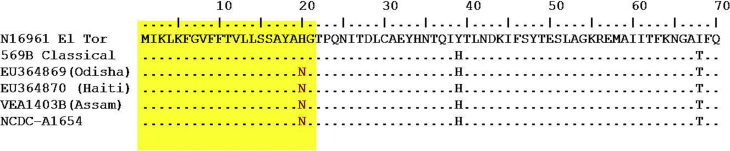
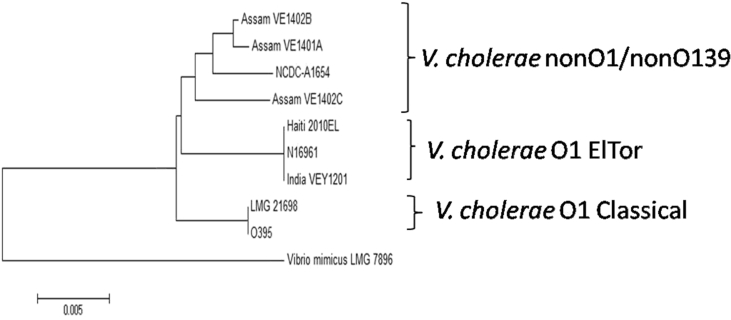
The presence of the ctxAB gene in the isolate was confirmed by PCR followed by DNA sequencing of the amplicon [6]. DNA sequencing also revealed the ctxB7 allele or HCT (Fig. 1). The isolate was positive for other virulence-associated genes: ompW, zot, rtxC and tcpA. A partial sequence of housekeeping genes recA and rpoA was used to establish the genetic relationship with other strains of classical, ElTor and Haiti types. The isolate was grouped in a clade of nonO1/nonO139 strains with environmental strains from Assam, India (Fig. 2).
Fig. 1.
Multiple sequence alignment of translated ctxB gene of Vibrio cholerae nonO1/nonO139 (NCDC-A1654) with ctxB sequence for reference strains.
Fig. 2.
Genetic relatedness of the NCDC-A1654 Vibrio cholerae isolate with reference O1 ElTor, classical and nonO1/nonO139 strains based on partial sequence of recA and rpoA. Neighbour-joining tree was constructed by MEGA6 using Kimura-2 method. VEA1401A, VEA1402B and VEA1403C are environmental nonO1/non139 strains from Assam, India [5].
We were unable to isolate V. cholerae from the stool sample of the patient after first admission. This could be due to the early institution of antibiotics. Syndromically, as the patient had gone into acute dehydration with anuria, a clinical picture of V. cholerae infection was suggested in the first instance. The second time, it was confirmed as cholera based on clinical presentation, culture characteristics, and biochemical, serological and PCR assays.
Discussion
The HCT strains originated from India and were disseminated to Haiti through Nepal by United Nations peacekeepers from Nepal who were deployed in Haiti after an earthquake [3], [7], [8]. All the epidemic outbreaks mediated by HCT strains were associated with severe diarrhoea and higher rate of mortality [2], [3], [4]. Now that the prevalence of the HCT strain is evident, retrospective consideration of recent epidemics of Odisha (2007), Western Africa (2009) and Haiti (2010) indicates that outbreaks of this strain are associated with mortality rates far greater than the WHO international target of 1% or less [9], [10]. A very recent study demonstrated that these strains are hyper-virulent [11]. Recently, we found that HCT genetic background was present in V. cholerae nonO1/nonO139 strains isolated from environmental water in Assam, India [5].
Harbouring of the CT (including HCT) gene by nonO1/nonO139 strains is an indication of the emergence of a new toxigenic serogroup with epidemic potential. The two biotypes of V. cholerae O1 (classically causing six earlier pandemics) and El Tor (responsible for a recent seventh pandemic) have been the culprits in all cholera pandemics. The biotype alteration has resulted in the appearance of hybrid or variant El Tor strains causing severe purging diarrhoea that have spread worldwide [12]. The hybrid V. cholerae El Tor strains possessing CT of classical biotype were associated with more severity and have now become a new threat worldwide because their hybrid property enhances the survival and virulence of the strains. Persistent diarrhoea in children has been associated with enteric pathogens including V. cholerae in Bangladesh [13]. Vibrio cholerae nonO1/nonO139 strains lacking CT have also been associated with cholera-like diarrhoea (mild to severe) in Kolkata [14]. This is probably the first report of HCT genes harboured in V. cholerae nonO1/nonO139 strains being associated with persistent diarrhoea. The study showed successful management of persistent diarrhoea associated with new toxigenic V. cholerae. There is a need for continuous tracking if this could emerge as an outbreak strain or a different genotype causing persistent diarrhoea.
Transparency declaration
None declared.
Acknowledgements
Pramod Kumar acknowledges the support of Council of Scientific and Industrial Research (CSIR) Senior Research Associateship No. 8757A/2015, India for providing a Senior Research Associateship under the scientist's pool scheme. The study is part of routine identification of enteric pathogens associated with diarrhoea in children admitted to/visiting Aruna Asaf Ali government hospital, Delhi.
Contributor Information
P. Kumar, Email: pramodjnu1@gmail.com.
S. Karmakar, Email: somenathk@gmail.com.
References
- 1.Kaper J.B., Morris J.G., Jr., Levine M.M. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar P., Jain M., Goel A.K., Bhadauria S., Sharma S.K., Kamboj D.V. A large cholera outbreak due to a new cholera toxin variant of Vibrio cholerae O1 El Tor biotype in Orissa, Eastern India. J Med Microbiol. 2009;58:234–238. doi: 10.1099/jmm.0.002089-0. [DOI] [PubMed] [Google Scholar]
- 3.Chin C.S., Sorenson J., Harris J.B., Robins W.P., Charles R.C., Jean-Charles R.R. The origin of the Haitian cholera outbreak strain. N Engl J Med. 2011;364:33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quilici M.L., Massenet D., Gake B., Bwalki B., Olson D.M. Vibrio cholerae O1 variant with reduced susceptibility to ciprofloxacin, Western Africa. Emerg Infect Dis. 2010;16:1804–1805. doi: 10.3201/eid1611.100568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhuyan S.K., Vairale M.G., Arya N., Yadav P., Veer V., Singh L. Molecular epidemiology of Vibrio cholerae associated with flood in brahamputra river valley, Assam, India. Infect Genet Evol. 2016;40:352–356. doi: 10.1016/j.meegid.2015.11.029. [DOI] [PubMed] [Google Scholar]
- 6.Jain M., Goel A.K., Bhattacharya P., Ghatole M., Kamboj D.V. Multidrug resistant Vibrio cholerae O1 El Tor carrying classical ctxB allele involved in a cholera outbreak in South Western India. Acta Tropica. 2011;117:152–156. doi: 10.1016/j.actatropica.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Hendriksen R.S., Price L.B., Schupp J.M., Gillece J.D., Kaas R.S., Engelthaler D.M. Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak. mBio. 2011;2:e00157–11. doi: 10.1128/mBio.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar P., Mishra D.K., Deshmukh D.G., Jain M., Zade A.M., Ingole K.V. Vibrio cholerae O1 Ogawa El Tor strains with the ctxB7 allele driving cholera outbreaks in south-western India in 2012. Infect Genet Evol. 2014;25:93–96. doi: 10.1016/j.meegid.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 9.Enserink M. Infectious diseases: Haiti’s outbreak is latest in cholera’s new global assault. Science. 2010;330:738–739. doi: 10.1126/science.330.6005.738. [DOI] [PubMed] [Google Scholar]
- 10.Kumar P., Mishra D.K., Deshmukh D.G., Jain M., Zade A.M., Ingole K.V. Haitian variant ctxB producing Vibrio cholerae O1 with reduced susceptibility to ciprofloxacin is persistent in Yavatmal, Maharashtra, India after causing a cholera outbreak. Clin Microbiol Infect. 2014;20:292–293. doi: 10.1111/1469-0691.12393. [DOI] [PubMed] [Google Scholar]
- 11.Satchell K.J.F., Jones C.J., Wong J., Queen J., Agarwal S., Yildiz F.H. Phenotypic analysis reveals that the 2010 Haiti cholera epidemic is linked to a hypervirulent strain. Infect Immun. 2016;84:2473–2481. doi: 10.1128/IAI.00189-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh-Banerjee J., Senoh M., Takahashi T., Hamabata T., Barman S., Koley H. Cholera toxin production by the El Tor variant of Vibrio cholerae O1 compared to prototype El Tor and classical biotypes. J Clin Microbiol. 2010;48:4283–4286. doi: 10.1128/JCM.00799-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahfuz M., Alam M.A., Islam S.B., Naila N.N., Chisti M.J., Alam N.H. Treatment outcome of children with persistent diarrhoea admitted to an urban hospital, Dhaka during 2012–2013. BMC Pediatrics. 2017;17:142. doi: 10.1186/s12887-017-0896-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutta D., Chowdhury G., Pazhani G.P., Guin S., Dutta S., Ghosh S. Vibrio cholerae non-O1, non-O139 serogroups and cholera-like diarrhea, Kolkata, India. Emerg Infect Dis. 2013;19:464–467. doi: 10.3201/eid1903.121156. [DOI] [PMC free article] [PubMed] [Google Scholar]