Abstract
The majority of bat rabies cases in Europe are attributed to European bat 1 lyssavirus (EBLV-1), circulating mainly in serotine bats (Eptesicus serotinus). Two subtypes have been defined (EBLV-1a and EBLV-1b), each associated with a different geographical distribution. In this study, we undertake a comprehensive sequence analysis based on 80 newly obtained EBLV-1 nearly complete genome sequences from nine European countries over a 45-year period to infer selection pressures, rates of nucleotide substitution, and evolutionary time scale of these two subtypes in Europe. Our results suggest that the current lineage of EBLV-1 arose in Europe ∼600 years ago and the virus has evolved at an estimated average substitution rate of ∼4.19×10−5 subs/site/year, which is among the lowest recorded for RNA viruses. In parallel, we investigate the genetic structure of French serotine bats at both the nuclear and mitochondrial level and find that they constitute a single genetic cluster. Furthermore, Mantel tests based on interindividual distances reveal the absence of correlation between genetic distances estimated between viruses and between host individuals. Taken together, this indicates that the genetic diversity observed in our E. serotinus samples does not account for EBLV-1a and -1b segregation and dispersal in Europe.
Keywords: European bat 1 lyssavirus (EBLV-1), RNA virus evolution, serotine bat, next generation sequencing, genome sequence
Introduction
Bats are recognized as reservoir hosts for many viruses and some of them are associated with zoonotic viral diseases such as SARS, Hendra, Nipah, rabies, and Ebola (Hayman et al. 2013; Smith and Wang 2013; Wynne and Wang 2013; Han et al. 2015). Rabies, caused by lyssaviruses, is the oldest known viral zoonotic disease and was the first recognized bat-associated viral infection in humans.
Lyssaviruses (genus Lyssavirus, family Rhabdoviridae) are RNA viruses with a single-stranded negative-sense genome that infect a variety of mammals, causing rabies-like diseases. Their genome size of ∼12 kb in length encodes five proteins: the nucleoprotein (N), the phosphoprotein (P), the matrix protein (M), the glycoprotein (G), and the polymerase (L) (Tordo et al. 1986, 1988; Delmas et al. 2008). Currently, lyssaviruses are classified into 14 recognized species defined on the basis of their genetic similarity: rabies lyssavirus (RABV), Lagos bat lyssavirus (LBV), Mokola lyssavirus (MOKV), Duvenhage lyssavirus (DUVV), European bat 1 lyssavirus (EBLV-1), European bat 2 lyssavirus (EBLV-2), Australian bat lyssavirus (ABLV), Irkut lyssavirus (IRKV), Aravan lyssavirus (ARAV), Khujand lyssavirus (KHUV), West Caucasian bat lyssavirus (WCBV), Shimoni bat lyssavirus (SHIBV), Ikoma lyssavirus (IKOV), and Bokeloh bat lyssavirus (BBLV) (https://talk.ictvonline.org/taxonomy/). Two other putative lyssaviruses do not yet have a taxonomic status; one is Lleida bat lyssavirus (LLEBV), identified in Miniopterus schreibersii in Spain (Ceballos et al. 2013; Marston et al. 2017), whereas the other lyssavirus is the most recently described Gannoruwa bat lyssavirus (GBLV) that has been isolated from brains of Indian flying foxes (Pteropus medius) in Sri Lanka (Gunawardena et al. 2016). Bats are natural reservoirs for all lyssaviruses except for IKOV and MOKV, for which a reservoir has not yet been identified.
In Europe, the first record of a rabid bat dates back to 1954 in Germany (Mohr 1957). To date, five different lyssavirus species have been isolated from European insectivorous bats: EBLV-1 and EBLV-2, which are responsible of the majority of rabid bat cases; and WCBV, BBLV, and LLEBV, which were isolated mainly from sporadic cases. These lyssavirus species were almost exclusively found in a few natural hosts: serotine bats (Eptesicus serotinus in Europe and Eptesicus isabellinus in south-east Spain) for EBLV-1, Daubenton’s bats (Myotis daubentonii) and pond bats (Myotis dasyceme) for EBLV-2, Natterer’s bats (Myotis nattereri) for BBLV, and Schreiber’s long-fingered bats (Miniopterus schreibersii) for WCBV and LLEBV (Bourhy et al. 1992; Botvinkin et al. 2003; Freuling et al. 2011; Ceballos et al. 2013; McElhinney et al. 2013; Picard-Meyer et al. 2013) although some serological and molecular data suggest a larger number of insectivorous bat species being exposed and infected (Serra-Cobo et al. 2013; López-Roig et al. 2014; Schatz et al. 2014).
To date, natural spill-over of EBLV-1 has only been reported in a limited number of terrestrial mammals, including five sheep on two separate occasions in Denmark in 1998 and 2002 (Tjørnehøj et al. 2006), one stone marten in Germany in 2001 (Müller et al. 2004), and two cats in France in 2003 and in 2007 (Dacheux et al. 2009). EBLV-2 spillover into terrestrial animals has not yet been documented. Albeit constituting a restricted number of cases, the fact that EBLVs have caused human deaths underscores the relevance of bat rabies for public health in Europe (Johnson et al. 2010).
Between 1977 and 2015, 1,126 rabid bat cases have been reported in Europe (http://www.who-rabies-bulletin.org/). The majority was characterized as EBLV-1, which can be subdivided into two distinct lineages or subtypes designated EBLV-1a and EBLV-1b, showing distinct geographic distributions (Amengual et al. 1997; Davis et al. 2005). EBLV-1a has an east-west distribution from Russia to France and most of the cases have been reported in Germany, France, Poland, and the Netherlands. In contrast, EBLV-1b has a south-north distribution from Spain to the Netherlands with at least four lineages that are genetically diverse and have a complex history (Amengual et al. 1997; Davis et al. 2005). Germany, France, Poland, and the Netherlands are the only countries in which both EBLV-1a and EBLV-1b have been isolated (Davis et al. 2005; Smreczak et al. 2009; McElhinney et al. 2013; Schatz et al. 2013, 2014; Picard-Meyer et al. 2014).
In this study, we undertake a comprehensive sequence analysis of 82 EBLV-1 complete genome sequences from nine European countries over a 45-year period to infer selection pressures, rates of nucleotide substitution, and the evolutionary time scale of EBLV-1a and -1b in Europe. In parallel, we analyze eight microsatellite loci and two variable mitochondrial genome sequences from about 80 Eptesicus serotinus bats originating from France to investigate the bat population genetic structure and the dispersal of this bat species in relation to the spatio-temporal dynamic of EBLV-1. This study provides new insights into the propagation of EBLV-1 in serotine bat populations and the evolutionary interaction between the virus and its host.
Materials and Methods
Viruses Analyses
Virus Samples
We analyzed a total of 82 nearly complete genome sequences from EBLV-1 isolates, collected in nine countries between 1968 and 2015. Details of these isolates are provided in supplementary table S1, Supplementary Material online. Except for two, all genomes were generated in this study based on 35 EBLV-1 samples from the archives of the World Health Organization Collaborative Centre for Reference and Research on Rabies, or from the National Reference Centre for Rabies, both located at Institut Pasteur, Paris, France and 45 EBLV-1 samples from ANSES’s Nancy Laboratory for Rabies and Wildlife, EU and OIE reference laboratory for rabies located in Malzeville, France. These data were combined with two full-length genome sequences extracted from GenBank (KF155003 and KP241939) selected to be representative of the overall phylogenetic diversity of EBLV-1 in bats. We did not consider sequences for viral isolates that were also sequenced as part of this study or sequences that were obtained from other hosts than bats.
RNA Extraction and Next-Generation Sequencing
Total RNA was extracted using Trizol (Ambion) according to the manufacturer’s instructions from primary brain samples or after an amplification passage on suckling mouse brain (supplementary table S1, Supplementary Material online). RNA was then reverse transcribed using Superscript III reverse transcriptase with random hexamers (Invitrogen) according to the manufacturer’s instructions.
The complete viral genome was amplified using the whole-transcription amplification (WTA) protocol (QuantiTect Whole Transcriptome kit; Qiagen) as previously described (Dacheux et al. 2010).
Briefly, two different protocols were used for the preparation of libraries and next-generation sequencing on Illumina platforms: 1) dsDNA was fragmented by ultrasound; libraries were prepared using the TruSeq protocol (Illumina) and sequenced using a 100 nucleotides single-end strategy on the HiSeq2000 platform, 2) dsDNA libraries were constructed using the Nextera XT kit (Illumina) and sequenced using a 2×150 nucleotides paired-end strategy on the NextSeq500 platform.
Genome Sequence Analyses
All reads were preprocessed to remove low-quality or artifactual bases. Library adapters at 5′ and 3′ ends and base pairs with a Phred quality score <25 were trimmed using AlienTrimmer as implemented in Galaxy (Giardine et al. 2005; Blankenberg et al. 2010; Goecks et al. 2010; Criscuolo and Brisse 2013) (https://research.pasteur.fr/en/tool/pasteur-galaxy-platform/). Reads with length <50 bp after these preprocessing steps or those containing >20% of bp with a Phred score of <25 were discarded. The filtered reads were mapped to specific complete genome sequences: EU293109 (isolate 03002FRA) and EU293112 (isolate 8918FRA) for the EBLV-1a and -1b viruses, respectively, using the CLC Genomics Assembly Cell (http://www.clcbio.com/products/clc-assembly-cell/) implemented in Galaxy. The majority nucleotide (>50%) at each position with generally a minimum coverage of 200 was used to generate the consensus sequence.
All consensus sequences generated were manually inspected for accuracy, such as the presence of intact open reading frames, using BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html). A multiple sequence alignment of the 80 newly sequenced genomes combined with the two complete genome sequences from GenBank (KF155003 and KP241939) was constructed using ClustalW2 with default parameters (Larkin et al. 2007) (http://www.ebi.ac.uk/Tools/msa/clustalw2) implemented in Galaxy and manually adjusted when necessary. All the nearly full-length genome sequences generated in the present study have been submitted to GenBank (supplementary table S1, Supplementary Material online).
Maximum-Likelihood Phylogenetic Analysis
jModelTest2 (Darriba et al. 2012) was used to determine the best-fit model of nucleotide substitution. This revealed that the general time reversible model with a proportion of invariable sites and gamma-distributed rate heterogeneity (GTR + I + Γ4) was optimal. We reconstructed a maximum-likelihood tree from the concatenated genes sequences using an SPR branch-swapping heuristic search in PhyML 3.0 (Guindon et al. 2003, 2010). The robustness of individual nodes in the phylogeny was estimated using 1,000 bootstrap replicates.
Estimates of RABV Evolutionary Dynamics and Time Scale
To ensure that the data set contained sufficient temporal signal, we performed linear regression of root-to-tip divergences as a function of sampling time using TempEst (Rambaut et al. 2016). We subsequently estimated a posterior distribution of time-measured trees using Bayesian inference through Markov chain Monte Carlo (MCMC), as implemented in BEAST (Drummond and Rambaut 2007). In this analysis, we specified separate partitions for each gene. The substitution process was modeled using the GTR + Γ4 model of nucleotide substitution for each gene, but with hierarchical priors over the parameters in order to pool information across genes (Suchard et al. 2003). We specified a relaxed (uncorrelated lognormal) molecular clock and a Bayesian skygrid model as a coalescent prior (Minin et al. 2008). In our partitioning scheme, we allowed for a different relative substitution rate for each gene; the product of the relative rate and the overall substitution rate provides evolutionary rate estimates for each gene in units time per site. We ran three independent MCMC analyses for 50 million steps sampling every 10,000 states. We used the BEAGLE high-performance computational library (Suchard and Rambaut 2009) in conjunction with BEAST to speed up the calculations.
The log and tree files of each MCMC chain were combined using LogCombiner v1.8.2 (http://tree.bio.ed.ac.uk/software/beast/), with a burn-in of 10%. We assessed convergence of each parameter in this combined file using TRACER v1.6 (http://tree.bio.ed.ac.uk/software/tracer/) based on effective sample size (ESS) >200. The degree of statistical uncertainty in each parameter estimate was given by the 95% highest posterior density (HPD) values. A Maximum Clade Credibility (MCC) tree was summarized using TreeAnnotator v1.8.2 (http://tree.bio.ed.ac.uk/software/beast/) and visualized in FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Analysis of Selection Pressures
To reveal the selection pressures acting on the EBLV-1 genome, including the possible occurrence of positive selection, we compared the numbers of nonsynonymous (dN) and synonymous (dS) substitutions per site for the different EBLV-1 genes in the two subtypes. Because different methods may arrive at different estimates, we compared the Single Likelihood Ancestor Counting (SLAC), Fixed Effect Likelihood (FEL), the Mixed Effects of Model Evolution (MEME), and the Fast Unbiased Bayesian Approximation (FUBAR) models (Kosakovsky Pond and Frost 2005; Murrell et al. 2012, 2013) implemented on the DATAMONKEY server (Pond and Frost 2005; Delport et al. 2010). Only codon positions with a P value <0.05 for the SLAC, FEL, and MEME models and with a posterior of probability >0.95 for the FUBAR method were considered as showing evidence for positive selection.
Host Species Analyses
Sample Collection
A total of 80 French dead bats from 2000 to 2015 were used to perform host genetic analysis. 31 samples were provided by the archives of the World Health Organization Collaborative Centre for Reference and Research on Rabies, or by the National Reference Centre for Rabies, both located at Institut Pasteur, Paris, France, and 49 samples were provided by ANSES’s Nancy Laboratory for Rabies and Wildlife, EU and OIE reference laboratory for rabies located in Malzeville, France. Dead bats from Anses Nancy were morphologically determined either by the French national bat conservation network (SFEPM) or by the laboratory. Out of these bat samples, 10 were identified to be infected by EBLV1-a subtype, 29 by the EBLV1-b subtype, whereas 41 have been identified as noninfected bats. Details of these bat samples are described in supplementary table S2, Supplementary Material online.
Microsatellites Genotyping
A subset of samples (n = 68) were genotyped using a panel of eight microsatellite markers (Smith et al. 2011; Moussy et al. 2015) as described in supplementary table S3, Supplementary Material online. PCRs were performed in 50 µl total volume containing 50–100 ng of template DNA, 1× amplification buffer, 0.8 mM of each dNTP, 0.2 µM of each primer, and 1,25 unit of ExTaq polymerase (Takara, Japan). PCR reactions consisted of denaturation at 94 °C for 3 min, followed by 35–45 cycles with denaturation at 94 °C for 30 s, annealing between 40 °C and 60 °C according to locus, and elongation at 72 °C for 30 s. The final elongation occurred at 72 °C for 7 min. Details of annealing temperature for each microsatellite marker are provided in supplementary table S3, Supplementary Material online. All PCR products were visualized on a 1.5% agarose gel containing ethidium bromide. Further, PCR products were diluted and mixed into two sets according to their size and dyes (supplementary table S3, Supplementary Material online) and run on an ABI Prism 3730 genetic analyzer (Applied Biosystems) with Genescan Rox 500 size standard (Applied Biosystems). Microsatellite alleles were sized using GeneMapper 4.0 software (Applied Biosystems).
Mitochondrial DNA Sequencing
DNA was extracted from patagium biopsies using the DNeasy Blood and Tissue kit (Qiagen), according to the manufacturer’s protocol. We performed a first PCR to amplify a 1,130-bp region in the cytochrome B (CytB) gene using primers LGL-765-F (5′-GAAAAACCAYCGTTGTWATTCAACT-3′) and LGL-766-R (5′-GTTTAATTAGAATYTYAG CTT TGGG-3′) (Bickham et al. 2004) and a second PCR on a 460-bp portion of the hypervariable domain II of the mtDNA control (D-loop) region using primers L-strand (5′-CTACCTCCGTGAAACCAGCAAC-3′) and H-strand (5′-CGTACACGTATTCGTATGTATGTCCT-3′) (Moussy et al. 2015). PCRs were performed in 50 µl total volume containing 100 ng of template DNA, 1× amplification buffer, 0.8 mM of each dNTP, 0.2 µM of each primer, and 1,25 unit of ExTaq polymerase (Takara). PCR reactions consisted of denaturation at 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 50 °C for 30 s, 72 °C for 1 or 1.5 min and final elongation at 72 °C for 7 min. All PCR products were visualized on a 1% agarose gel containing ethidium bromide. Sequencing was carried out in a ABI Prism 3100 sequencer (Applied Biosystems). Using the Basic Local Alignment Search Tool (BLAST, http://blast.ncbi.nlm.nih.gov/Blast.cgi), most closely related CytB sequences were identified and only bats identified as E. serotinus were kept for the further analysis.
The resulting sequences were assembled with Sequencher (GeneCodes) aligned with ClustalX (http://www.clustal.org) and trimmed with BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html) to create a 1,064- and 424-bp alignment for CytB and D-loop regions, respectively. (Accession numbers are available in supplementary table S2, Supplementary Material online).
Microsatellite Data Analyses
To estimate the genetic variability in the microsatellite data set, we first assessed the size range of PCR products and the number of alleles. The observed (Ho) and unbiased expected heterozygosity (He) for each locus were calculated using GENETIX 4.05.2 (Belkhir et al. 2004).
We then used microsatellite data to analyze the population genetic structure of Eptesicus serotinus with two different clustering algorithms: the methods implemented in STRUCTURE 2.3 (Pritchard et al. 2000) and GENELAND 4.5.0 (Guillot et al. 2005). STRUCTURE proposes a Bayesian algorithm to identify clusters of individuals respecting the Hardy–Weinberg/linkage equilibrium. With this first method, we estimated the number of clusters (K) with ten independent runs of K = 1–10 carried out with 106 MCMC iterations after a burn-in period of 105 iterations, using the model with correlated allele frequencies and assuming admixture. The most probable number of clusters was identified based on the log-likelihood values (and their convergence) associated with each K and visualized with STRUCTURE HARVESTER (Earl and VonHoldt 2012). GENELAND also implements a Bayesian clustering model, but also considers the sampling coordinates of each individual when inferring population genetic structure. With GENELAND, the number of genetic clusters was determined by running the algorithm 10 times, allowing K to vary from 1 to 10, with the following parameters: 106 MCMC iterations with a thinning of 1,000, maximum rate of the Poisson process fixed to 100, and maximum number of nuclei in the Poisson–Voronoi tessellation fixed to 300.
Finally, we estimated Bray–Curtis dissimilarities (BCD) (Bray and Curtis 1957) as interindividual distances based on microsatellite data, as well as FST (Weir and Cockerham 1984) distances estimated with the R package “hierfstat” (Goudet 2005) between defined groups of individuals. Two group partitions were investigated: 1) the group of E. serotinus individuals tested negative for EBLV-1 (and for the presence of the other lyssaviruses) (n = 34) versus the group of individuals tested positive for EBLV-1 (n = 36), and 2) the group of individuals tested positive for EBLV subtype 1a (n = 10) versus the group of individuals tested positive for EBLV subtype 1b (n = 26). Statistical significances of FST statistics were assessed by recalculating them with 10,000 random permutations of the original data sets.
Mitochondrial DNA Sequence Analyses
Alignments obtained for the two mitochondrial gene fragments CytB and D-loop were concatenated in a single alignment (1,488 bp) for subsequent analyses. Median-joining networks (Bandelt et al. 1999) were inferred for the concatenated alignment using the software NETWORK 4.6.6 (available at http://www.fluxus-engineering.com). We used the program SPADS 1.0 (Dellicour and Mardulyn 2014) to compute interindividual distances (IID, number of mismatches divided by the sequences length), and the pairwise distance ΦST (Excoffier et al. 1992) among the same groups of individuals defined above: 1) the group of E. serotinus individuals tested negative for EBLV-1 versus the group of individuals tested positive for EBLV-1, and 2) the group of individuals tested positive for EBLV-1 subtype a versus the group of individuals tested positive for EBLV-1 subtype b. Statistical significance for the ΦST statistics was assessed by recalculating them with 10,000 random permutations of the original data sets.
Correlation of Bat and Virus Genetic Distances
We performed Mantel tests 1) to assess the isolation-by-distance pattern for E. serotinus based on both microsatellite and mtDNA sequence data, and 2) to test for a potential correlation between viral and host genetic differentiation. In the first case, matrices of pairwise interindividual genetic distances (BCD for host microsatellite data and IID for host mtDNA sequence data) were directly compared with a matrix of pairwise geographic distances between individuals. As developed in (Lecocq et al. 2017), Mantel tests based on interindividual rather than on interpopulation metrics avoid having to arbitrarly define “groups” (or “populations”) a priori among sampled sequences. Having to define such groups is indeed associated with several limitations: The partition is often completely arbitrary, hides intrapopulation differentiation, and decreases the number of available pairwise values available for Mantel tests. Geographic distances were computed as great circle geographic distances (i.e., distances on the surface of the earth, measured in kilometers) with the R package “fields” (https://cran.r-project.org/web/packages/fields/index.html). In the second case, “mirror” data sets were first generated to only contain genetic data corresponding to host individuals for which viral sequence, host mtDNA sequences and microsatellite data are available. This resulted in matrices of viral/host sequences and viral sequences/host microsatellite data with 25 entries. The different Mantel tests (Mantel 1967) were performed with the R package “vegan” (https://cran.r-project.org/web/packages/vegan/index.html) and based on 10,000 permutations.
Results
Phylogenetic Relationships of EBLV-1 Isolates
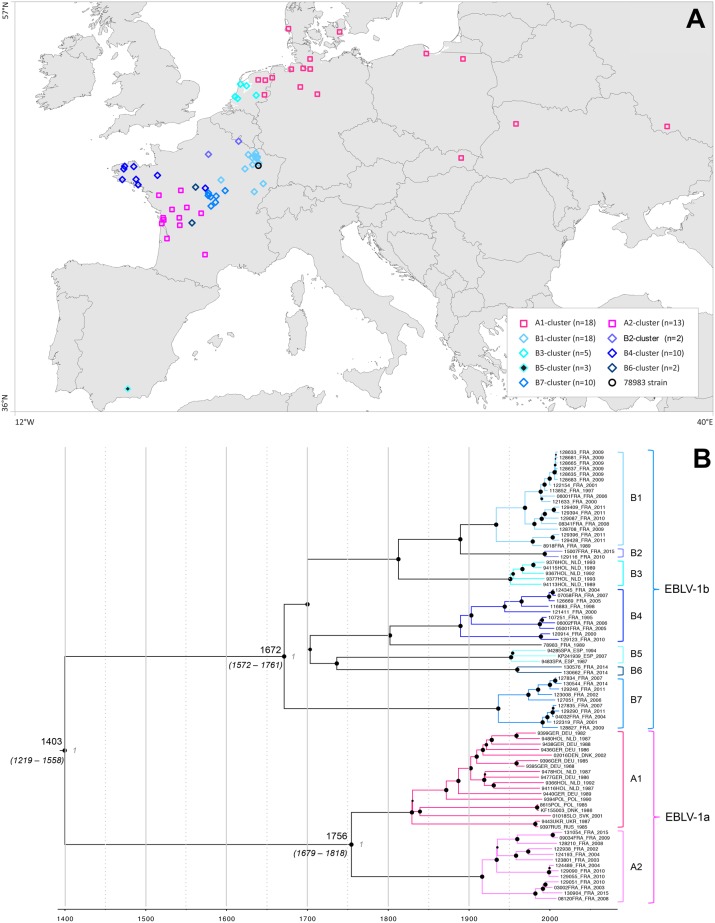
We performed a phylogenetic analysis on the five concatenated genes sequences, corresponding to 90% of the full-length genome, of 82 EBLV-1 sequences sampled from nine European countries over a 45-year period (fig. 1A and supplementary table S1, Supplementary Material online). As previously shown, two major phylogenetic groups were apparent, corresponding to the EBLV-1a and -1b subtypes, each of which can be further subdivided into several clusters (fig. 1B and supplementary fig. S1, Supplementary Material online). This is consistent with previous analyses performed on smaller data sets and on individual complete or partial RABV genes such as N and/or G genes (Amengual et al. 1997; Davis et al. 2005; McElhinney et al. 2013; Picard-Meyer et al. 2014; Schatz et al. 2014).
Fig. 1.
—EBLV-1 isolates sampling and Maximum Clade Credibility tree. The map of geographical distribution in Europe of these isolates (A) is labeled according to the clusters revealed by the MCC tree (B) and the ML tree (supplementary fig. S1, Supplementary Material online), EBLV-1a strains are indicated by the magenta gradation squares and EBLV-1b strains are indicated by blue gradation losanges (except the isolate 78983 that is indicated by a black circle). The MCC tree shows the division between the two subtypes. The size of the black circle is proportional to the posterior probability values of each nodes. Tip times represent the time (year) of sampling. Bayesian estimates of divergence time, upper and lower limits of the 95% highest posterior density (HPD) estimates and the posterior probability values are shown for major nodes. The name of each cluster is also indicated.
The EBLV-1a group (n = 31) is subdivided in two clusters, the A1 cluster that includes isolates from Denmark, Germany, Netherlands, Poland, Russia, Slovakia, and Ukraine and the A2 cluster that appears to be specific to French isolates (fig. 1B and supplementary fig. S1, Supplementary Material online). The EBLV-1b group (n = 51) is exhibiting more diversity and for the purpose of description has been arbitrarily subdivided into seven geographical clusters (named B1 to B7) supported by high bootstrap values. Each of these clusters presents a specific geographical distribution, with B1: 18 French strains from the north-east of France; B2: 2 isolates from Picardie and Champagne–Ardenne regions in France; B3: 5 isolates from the Netherlands; B4: 7 isolates from the north-west of France and 3 from the center of France near Bourges city; B5: 3 isolates from the south-east of Spain; B6: 2 French isolates from Centre and Limousin regions; and B7: 10 isolates from the center of France. The 78983 isolate which originated from Bainville-sur-Mandon isolated in the north-east of France remained isolated as a divergent strain from the other clusters.
This EBLV-1b group is characterized by a strong phylogenetic and geographic structure at the European level with the Dutch, Spanish, and French isolates forming separate groups. For the isolates from France, the geographic structure is less clear, but still present. Interestingly, the branching structure of the EBLV-1a is different. Specifically, the A1 cluster is characterized by a weak geographical structure, with Danish, Dutch, German, Russian, Ukraininan, Polish, and Slovakian isolates intermixed suggesting a free and rapid movement of EBLV-1a among these countries while isolates from Eastern Europe occupied more basal positions. In contrast, all French EBLV-1a isolates are grouped together in the A2 cluster.
Temporal Dynamics and Spread of EBLV-1 Isolates
To estimate the evolutionary dynamics of EBLV-1, we first determined that the data set contained sufficient temporal structure to undertake detailed molecular clock analysis by performing a regression of root-to-tip genetic distance against the year of sampling (not shown), allowing us to estimate substitution rates, and times of most common ancestors (TMRCA), using a Bayesian approach.
The mean rate of evolutionary change in the EBLV-1 was 4.19×10−5 subs/site/year (95% HPD of 2.95–5.83×10−5 subs/site/year) for the whole genome. The evolutionary rate of each gene is slightly different and varies between 3.51×10−5 subs/site/year (95% HPD of 2.47–4.52×10−5 and 2.41–4.65×10−5 subs/site/year) for the slowest (nucleoprotein and matrix proteins, respectively) and 4.18×10−5 subs/site/year (95% HPD of 2.92–5.44×10−5 subs/site/year) for the fastest (phosphoprotein). As expected, the evolutionary rate in the noncoding regions is slightly higher than those of the coding regions, indicative of overall weaker selective constraints exerted on these genomic regions (table 1). A separate analysis of each subtype shows that the mean evolutionary rate is slightly slower for EBLV-1a subtype compared with the EBLV-1b subtype, which is consistent with previous studies (Davis et al. 2005; Hughes 2008).
Table 1.
Evolutionary Characteristics of EBLV-1
| EBLV-1 (n= 82) | EBLV-1a (n = 31) | EBLV-1b (n = 51) | |
|---|---|---|---|
| Evolution rate | |||
| All genomea (11,976 bp) | 4.19 ×10−5 (2.95–5.83 ×10−5) | 3.01×10−5 (1.30–5.60×10−5) | 4.07×10−5 (2.87–5.77×10−5) |
| Nucleoprotein (1,353 bp) | 3.51×10−5 (2.47–4.52×10−5) | 2.58×10−5 (0.91–4.13×10−5) | 3.48×10−5 (2.37–4.67×10−5) |
| Phosphoprotein (936 bp) | 4.18×10−5 (2.92–5.44×10−5) | 3.33×10−5 (1.11–5.30×10−5) | 3.91×10−5 (2.65–5.33×10−5) |
| Matrix (609 bp) | 3.51×10−5 (2.41–4.65×10−5) | 2.72×10−5 (0.92–4.49×10−5) | 3.47×10−5 (2.27–4.85×10−5) |
| Glycoprotein (1,575 bp) | 3.83×10−5 (2.72–4.90×10−5) | 2.88×10−5 (1.11–5.30×10−5) | 3.80×10−5 (2.60–5.07×10−5) |
| Polymerase (6,384 bp) | 3.84×10−5 (2.74–4.81×10−5) | 2.77×10−5 (0.98–4.31×10−5) | 3.83×10−5 (2.76–5.01×10−5) |
| Noncodingb (1,119 bp) | 5.74×10−5 (4.06–7.34×10−5) | 4.20×10−5 (1.43–6.65×10−5) | 5.71×10−5 (4.02–7.67×10−5) |
| Inter noncodingc (918 bp) | 5.72×10−5 (4.08–7.40×10−5) | 4.55×10−5 (1.52–7.21×10−5) | 5.37×10−5 (3.69–7.23×10−5) |
| TRMCAd | 1,403 (1,219–1,558) | 1,756 (1,679–1,818) | 1,672 (1,572–1,761) |
Mostly complete for all sequences.
All intergenic regions + leader/trailer sequences (nearly complete for all of them).
Only intergenic regions sequences.
95% HPD values are given in the brackets.
This estimation of a reliable substitution rate allowed us to estimate the TMRCA for each EBLV-1 subtypes (fig. 1B); thanks to the nearly complete genome sequences, these estimates exhibited less uncertainty compared with previous estimates obtained from only N or G genes (Davis et al. 2005). We estimated the emergence of EBLV-1 in Europe was approximately around the year 1400 (95% HPD 1219–1558). The TMRCAs of the EBLV-1a and -1b subtypes were estimated to date back to 1756 (95% HPD 1679–1818) and to 1672 (95% HPD 1572–1761), respectively.
Within the EBLV-1a subtype, the A1 cluster emerged between 1780 and 1873, whereas the A2 cluster grouping only French isolates appeared between 1888 and 1942.
Selection Pressure in EBLV-1
We compared the ratio of nonsynonymous (dN) to synonymous (dS) substitutions per site of each EBLV-1 gene using the SLAC method. For each gene, the overall dN/dS ratios of the two subtypes were very low (between 0.019 and 0.215) revealing strong selective constraints, and followed the same ascending order between genes: N, L, G, P, and M genes (table 2). The dN/dS ratios of the two subtypes are very close to one another, even if the dN/dS ratios are slightly higher for EBLV-1a compared with EBLV-1b. We also explored the number of positively selected sites using several different approaches (SLAC, FUBAR, FEL, and MEME; Kosakovsky Pond and Frost 2005; Murrell et al. 2012, 2013). Only the position 244 in the G protein was identified as positively selected by at least two of these methods when the two subtypes were analyzed together, whereas no position was identified using the same criteria when the two subtypes were analyzed independently.
Table 2.
Selection Pressures in the Five Genes from EBLV-1
| Data set | Gene | dN/dS | SLACa | FELa | MEMEa | FUBARb |
|---|---|---|---|---|---|---|
| EBLV-1 (n=82) | N | 0.025 | – | – | – | – |
| P | 0.131 | – | – | – | – | |
| M | 0.142 | – | – | – | – | |
| G | 0.077 | – | 244 | 244 | – | |
| L | 0.038 | – | – | 1,597 | – | |
| EBLV-1a (n=31) | N | 0.021 | – | – | – | – |
| P | 0.157 | – | – | – | – | |
| M | 0.215 | – | – | – | – | |
| G | 0.087 | – | 244 | – | – | |
| L | 0.043 | – | – | 1,597 | – | |
| EBLV-1b (n=51) | N | 0.019 | – | – | – | – |
| P | 0.118 | – | – | – | – | |
| M | 0.108 | – | – | – | – | |
| G | 0.069 | – | – | – | – | |
| L | 0.038 | – | – | – | – |
dN/dSratios are calculated using SLAC.
Putatively positively selected codons identified by more than one method are underlined.
Codons with P value <0.05.
Codons with posterior of probability >0.95.
Microsatellite and Mitochondrial DNA Sequences Data Analyses
To further explore the origin of the differences observed in terms of temporal dynamics and spread of the different subtypes and clusters of EBLV-1, we investigated the genetic structure of the EBLV-1 major hosts in France, that is, E. serotinus, by analyzing eight nuclear microsatellite and two mitochondrial DNA (mtDNA) sequence loci.
All analyzed microsatellite loci were polymorphic with a number of alleles ranging from 4 (EF1) to 13 (EF6). The allele sizes are consistent with previous studies performed on E. serotinus in Europe (Smith et al. 2011; Bogdanowicz et al. 2013; Moussy et al. 2015). The highest observed heterozygosity (Ho) was found in Paur05 and the lowest in NN8. One of the eight microsatellites loci, AF141650, demonstrated a high level of null alleles (supplementary table S4, Supplementary Material online).
Both STRUCTURE and GENELAND analyses identified only a single genetic cluster. We also estimated the Bray–Curtis dissimilarities (BCD) as interindividual distances based on microsatellite data (Bray and Curtis 1957) and FST distances (Weir and Cockerham 1984). Two group partitions were investigated: 1) the group of E. serotinus individuals tested negative for EBLV-1 versus the group of individuals tested positive for EBLV-1, and 2) the group of individuals tested positive for EBLV-1a subtype versus the group of individuals tested positive for EBLV-1b. Pairwise FST values were not significant (P > 0.05) for both partitions tested, thus arguing against host genetic differentiation associated with EBLV-1 infection at the nuclear genetic level.
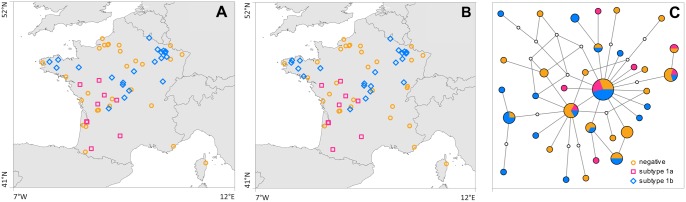
In order to analyze the host genetic variability at the mitochondrial level, we selected two independent mitochondrial DNA regions, the CytB and the D-loop regions and we concatenated them to perform analysis at a higher resolution. The resulting 1,488-bp alignment revealed the presence of 34 haplotypes. The most likely relationships among haplotypes were estimated by constructing an haplotype network based on the concatened alignment of the two mitochondrial loci (CytB + D-loop). The haplotype network showed no apparent clustering between host sequences sampled in host individuals tested negative for EBLV-1, positive for EBLV-1a subtype, and positive for EBLV-1b subtype (fig. 2C).
Fig. 2.
—Host microsatellites and mtDNA samplings, as well as corresponding mtDNA haplotype network. Sampling maps of host microsatellites (A) and host mtDNA sequences (B) data and median-joining network for the concatenated alignment of two mtDNA gene fragments (Cyt-B and D-loop) sequenced for the host species (Eptesicus serotinus) (C). In the haplotype network, each sequenced haplotype is represented by a circle, the size of which is proportional to its overall frequency. Each line segment in the network represents a single mutational change. Haplotype colours indicate if the host individual was tested negative (orange) or positive to EBLV-1 and, in the later case, if the subtype 1a (magenta) or 1b (blue) has been identified.
To investigate this in further detail, we computed interindividual distances (IID), and the pairwise distance ΦST (Excoffier et al. 1992) among the previous defined groups of individuals. Pairwise ΦST values were not significant (P >0.05) for both tested partitions. Hence, analogous to the microsatellite data, these results suggested that the genetic differentiation among host individuals was not related to EBLV-1 infection at the mitochondrial genetic level.
Correlation of Bat and Virus Genetic Distances
We performed Mantel tests 1) to preliminarily assess the isolation-by-distance pattern for E. serotinus based on both microsatellite data and mtDNA sequence data, and 2) to test for a potential correlation between viral and host genetic differentiation. Mantel tests between geographic distance and pairwise interindividual genetic distances (BCD for microsatellite data and IID for mtDNA sequence data) were not significant. Furthermore, the EBLV-1 genetic distances neither correlated with those of the host nuclear DNA nor with those of the host mitochondrial DNA.
Discussion
The central aim of this study was to explore the evolutionary history of EBLV-1 by undertaking a comprehensive sequence analysis of 82 EBLV-1 nearly complete genome sequences from nine European countries over a 45-year period, almost all of which were newly generated by this study. The results we present here reveal important aspects of EBLV-1 evolutionary history in Europe. First, our molecular clock analyses allowed us to hypothesize that the current lineage of EBLV-1 arose in Europe between 1219 and 1558 (95% HPD; mean of 1403), consistent with a previous study performed by our team (Davis et al. 2005). However, the estimates provided by our complete genome sequence analysis reduce the uncertainty of estimates previously obtained for N or G genes (Davis et al. 2005). Thus, the current genetic diversity in EBLV-1 has a relatively recent origin, as observed in some other lyssaviruses (Guyatt et al. 2003; Streicker, Lemey, et al. 2012; Troupin et al. 2016). The emergence of the EBLV-1a and -1b subtypes was estimated to be between 1679–1818 (mean of 1756) and 1572–1761 (mean of 1672), respectively, which raised the possibility of two independent introductions. However, the estimated date of the TMRCA could also be biased by the sampling of the EBLV-1 sequences used in this study. In addition, the topology of the tree and the different geographical distribution of the two subtypes (fig. 1B and supplementary fig. S1, Supplementary Material online) may be compatible with different introductions of the two subtypes in Europe. Further, the branching structure of the cluster A1 of the EBLV-1a subtype suggests that there is more population mixing in this cluster than in the cluster A2 and in the seven clusters of EBLV-1b subtype which are, all consistently characterized by a strong phylogenetic and geographic structure. However, additional sampling and statistical analyses are needed to confirm these mixing patterns. In the A1 cluster, the phylogenetic mixing of isolates from different geographic regions suggests extensive viral movement among serotine bat populations in northern Europe confirming previous hypothesis (Davis et al. 2005). However, this does not seem to be the case for the southern part of Europe. Finally, this study using 82 EBLV-1 nearly complete genome sequences does not support the existence of a third lineage named EBLV-1c as recently proposed by Eggerbauer et al. (2017). The sequences designed as EBLV-1c in this study correspond to lineage B5 in our study (fig. 1B and supplementary fig. S1, Supplementary Material online).
As a key aspect of our work, we highlight the low substitution rate of EBLV-1 as recently suggested by Hayman et al. (2016). Indeed, the substitution rate estimated in this study, that is, 4.19×10−5 subs/site/year (95% HPDs of 2.95–5.83×10−5 subs/site/year), is among the lowest observed in RNA viruses (Jenkins et al. 2002; Hanada et al. 2004; Duchêne et al. 2014) and lower than those described for the other lyssaviruses (Streicker, Lemey, et al. 2012; Troupin et al. 2016). There are several plausible explanations for the low evolutionary rates in EBLV-1. First, it is possible that EBLV-1 may experience a lower number of replication cycles per time unit compared with other lyssavirus species adapted to nonflying mammals but there is no data supporting this hypothesis. A second, but unlikely, explanation could be that the EBLV-1 polymerase produces a lower error rate per replication cycle than that of other lyssaviruses leading to a lower evolutionary rate per time unit. Further experiments will be needed, to compare the replication rate of polymerase of EBLV-1 with that of other lyssaviruses by in vitro RNA replication essay, before attributing any importance to this hypothesis. Third, this low rate of evolution could also be explained by a peculiarity of the biological cycle of bats. Bats protect themselves during the cold season when there are fewer insects available to eat and when the balance between the cost in terms of energy required for hunting and the benefit in terms of food is disadvantageous. This period known as the hibernation period in bats has a duration which is highly variable in Europe and depends of the type of climate as, for example, the temperature in the Northern part of Europe drops during winter. Further, the type of landscape may also influence the duration of hibernation in the way that the presence of mountains and altitude can be associated with a very cold winter. In these conditions bats may hibernate for several months. This would indirectly lower the number of replication cycles of EBLV-1 genome per time unit. On the other hand, in more austral regions of Europe and those under the influence of the Mediterranean and oceanic climate, this hibernation may be of little importance. However the complete genome EBLV-1 evolutionary rate is lower than the one estimated for the RABV nucleoprotein gene in Northern American bats, despite the very cold climate during winter that they have to face (Streicker, Lemey, et al. 2012).
Our analysis also showed that the nucleotide substitution rate does not vary substantially among genes and among subtypes. As expected, the evolutionary rate in the noncoding regions is slightly higher than those of the coding regions, indicative of weaker selective constraints. These results are slightly different to those obtained by our team in a similar study performed on dog-related rabies virus which showed that the nucleotide substitution rate varied markedly according to the gene analyzed in the ascending order: N, L, G, M, and P (Troupin et al. 2016). As previously described for EBLV-1 and for some other lyssaviruses, the dN/dS ratios of each gene were very low, revealing strong selective constraints (Davis et al. 2005; Streicker, Altizer, et al. 2012; Troupin et al. 2016). The same ascending order of the dN/dS ratios between genes was observed in the two subtypes: N, L, G, P, and M genes. However, the dN/dS ratios of the EBLV-1a are slightly higher than those of EBLV-1b, with that of the EBLV-1a M-gene being 2 times higher than the same gene for EBLV-1b. This may suggest a slighty difference in selective pressure between the two subtypes. Only one position was identified as positively selected by our analyses. It is position 244 of the G protein which is located in one of the four structural domain of the G protein ectodomain: the plectkstrin homology (PH) domain (domain III) (Bastida-González et al. 2016). To date, the role of this aa in the G protein is unknown.
Serotine bats infected by EBLV-1a were geographically separated from the regions displaying EBLV-1b cases, indicating a different geographical distribution of the two subtypes. Thus, several hypothesis can be put forward to explain these different geographical distributions between these two subtypes in Europe. First, according to the Bayesian analyses performed, the TMRCAs in Europe of the two subtypes around 1679–1818 (mean of 1756) for EBLV-1a subtype and 1572–1761 (mean of 1672) for the EBLV-1b subtype could be associated with two distinct introductions of each subtype in Europe. However, these results as well as those obtained using more sophisticated phylogeographic methods (Faria et al. 2011; Bielejec et al. 2014, 2016), are not sufficient to reveal the precise geographic origin of EBLV-1 in Europe or outside Europe. Indeed these complex analyses may be impaired by the lack of isolates originating from the southern part of Europe (excepted Spain). This absence of isolates could either reflect the absence of circulation of EBLV-1 in these countries or a weaker surveillance system. Finally, it has been proposed that EBLV-1 had its origins in Africa, being closely related to DUVV (Davis et al. 2005; Hayman et al. 2016). This hypothesis was not tested in our analysis. It would imply that subtypes EBLV-1a and -1b would represent different routes of spreading (and potentially introductions) of DUVV from Africa to Europe.
The second hypothesis is that serotines might be a paraphyletic group composed of differentiated lineages at species level, like the Natterer’s bat (Salicini et al. 2011; Halczok et al. 2017), one lineage being specific to subtype 1a and the other lineage specific to subtype 1b. To explore this hypothesis, we analyzed eight microsatellite loci and two distinct mtDNA regions of 80 French serotine bats. However, all the analyses performed in this study at the host genetic level showed no difference between the group of French E. serotinus tested positive for subtype 1a versus the group of individuals tested positive for subtype 1b. Moreover, we found no differences when comparing the group of E. serotinus individuals tested negative for EBLV-1 to the group of individuals tested positive for EBLV-1. Thus, it seems that the E. serotinus that circulated in France are monophyletic which is in accordance with previous few studies performed on the population genetic structure of E. serotinus in Europe (Bogdanowicz et al. 2013; Moussy et al. 2015).
The last hypothesis is that serotines bats are the sentinel species for EBLV-1 and other bat species are also implicated in the diffusion of EBLV-1 that are underdiagnosed and/or remain asymptomatic. Indeed, severals studies have previously shown that different indicators (neutralizing antibodies, viral RNAs) of ongoing or previous infection with EBLV-1 can be detected in other bat species like in Rhinolophus ferrumequinum, Myotis myotis, Myotis blythii, Miniopterus schreibersii, Barbastella barbastellus in France (Picard-Meyer et al. 2011) in Rhinolophus ferrumequinum, Myotis myotis, Pipistrellus pipistrellus, Miniopterus schreibersii, Tadarida teniotis, and Myotis capaccinii in Spain (Serra-Cobo et al. 2013) and in P. nathusii, P. pipistrellus, and Plecotus auritus in Germany (Schatz et al. 2014). However, only few studies have been undertaken in order to assess the seroprevalence of EBLV-1 infection in European bats (Nokireki et al. 2013; Serra-Cobo et al. 2013; López-Roig et al. 2014). In a seroprevalence study performed in Spain between 2001 and 2011 that tested 2,393 blood samples collected from 25 localities and 20 bat species, samples positive for EBLV-1-neutralizing antibodies were detected in 68% of the localities sampled and in 13 bat species, seven of which were found for the first time (even in Myotis daubentonii, a species to date always linked to EBLV-2). EBLV-1 seroprevalence (20.7%) ranged between 11.1% and 40.2% among bats .and was significantly associated with colony size and species richness. Indeed, higher percentages of seroprevalence were found in large multispecific colonies, suggesting that intra- and interspecific contacts are major risk factors for EBLV-1 transmission in bat colonies (Serra-Cobo et al. 2013) Thus, it is possible that EBLV-1 circulates in one or several other nonsynanthropic bat species that are not monophyletic and responsible for the two distinct geographical distribution of the two EBLV-1 subtypes.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Authors’ Contributions
C.T., E.P.M., S.D., L.D., P.L., and H.B. conceived and designed the experiments. C.T., E.P.M., S.D., I.C., L.K,. A.L., G.B., and P..L performed experiments. C.T., S.D., L.D., G.B., P.L., and H.B. analyzed the data. C.T., E.P.M., S.D., L.D., G.B., E.M.L., P.L., and H.B. wrote the paper. All the authors gave final approval for publications.
Supplementary Material
Acknowledgments
The author want to acknowledge Karin Eiglmeier from Unit of Insect Vector Genetics and Genomics for access and help on ABI Prism 3730 for microsatellite genotyping. We would like to thank the SFEPM group (Societe Francaise pour l’Etude et la Protection des Mammiferes, groupe Chiropteres), and especially the bat naturalists, for their collaboration in the surveillance of bat rabies in France. This work was supported by European Union Seventh Framework Programme (FP7/2007-2013) PREDEMICS (under grant agreement number 278433), Santé Publique France and the Institut Pasteur. SD is funded by the Fonds Wetenschappelijk Onderzoek (FWO, Flanders, Belgium). PL acknowledges the Research Foundation - Flanders (G0D5117N & G066215) and the Research Fund KU Leuven (OT/14/115). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Literature Cited
- Amengual B, Whitby JE, King A, Cobo JS, Bourhy H.. 1997. Evolution of European bat lyssaviruses. J Gen Virol. 789:2319–2328.http://dx.doi.org/10.1099/0022-1317-78-9-2319 [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A.. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 161:37–48. [DOI] [PubMed] [Google Scholar]
- Bastida-González F, Celaya-Trejo Y, Correa-Basurto J, Zárate-Segura P.. 2016. Predicted 3D model of the rabies virus glycoprotein trimer. BioMed Res Int. 2016:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhir KP, Borsa P, Chikhi L, Raufaste N, Bonhomme F.. 2004. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions [cited 2017 May 20]. Available from: https://www.scienceopen.com/document?vid=9a3e2cf3-2971-405c-8297-258227c3cb30.
- Bickham JW, Patton JC, Schlitter DA, Rautenbach IL, Honeycutt RL.. 2004. Molecular phylogenetics, karyotypic diversity, and partition of the genus Myotis (Chiroptera: Vespertilionidae). Mol Phylogenet Evol. 332:333–338. [DOI] [PubMed] [Google Scholar]
- Bielejec F, et al. 2016. SpreaD3: interactive visualization of spatiotemporal history and trait evolutionary processes. Mol Biol Evol. 338:2167–2169.http://dx.doi.org/10.1093/molbev/msw082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielejec F, Lemey P, Baele G, Rambaut A, Suchard MA.. 2014. Inferring heterogeneous evolutionary processes through time: from sequence substitution to phylogeography. Syst Biol. 634:493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, et al. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. Chapter 19:Unit 19.10.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanowicz W, Lesiński G, Sadkowska-Todys M, Gajewska M, Rutkowski R.. 2013. Population genetics and bat rabies: a case study of Eptesicus serotinus in Poland. Acta Chiropterol. 151:35–56. [Google Scholar]
- Botvinkin AD, et al. 2003. Novel lyssaviruses isolated from bats in Russia. Emerg Infect Dis. 912:1623–1625.http://dx.doi.org/10.3201/eid0912.030374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhy H, Kissi B, Lafon M, Sacramento D, Tordo N.. 1992. Antigenic and molecular characterization of bat rabies virus in Europe. J Clin Microbiol. 309:2419–2426.1401009 [Google Scholar]
- Bray JR, Curtis JT.. 1957. An ordination of the upland forest communities of Southern Wisconsin. Ecol Monogr. 274:326–349. [Google Scholar]
- Ceballos NA, et al. 2013. Novel Lyssavirus in bat, Spain. Emerg Infect Dis. 195:793–795.http://dx.doi.org/10.3201/eid1905.121071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo A, Brisse S.. 2013. AlienTrimmer: a tool to quickly and accurately trim off multiple short contaminant sequences from high-throughput sequencing reads. Genomics 102(5–6):500–506.http://dx.doi.org/10.1016/j.ygeno.2013.07.011 [DOI] [PubMed] [Google Scholar]
- Dacheux L, et al. 2009. European bat Lyssavirus transmission among cats, Europe. Emerg Infect Dis. 152:280–284.http://dx.doi.org/10.3201/eid1502.080637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacheux L, et al. 2010. Application of broad-spectrum resequencing microarray for genotyping rhabdoviruses. J Virol. 8418:9557–9574.http://dx.doi.org/10.1128/JVI.00771-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 98:772..http://dx.doi.org/10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis PL, et al. 2005. Phylogeography, population dynamics, and molecular evolution of European bat lyssaviruses. J Virol. 7916:10487–10497.http://dx.doi.org/10.1128/JVI.79.16.10487-10497.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellicour S, Mardulyn P.. 2014. spads 1.0: a toolbox to perform spatial analyses on DNA sequence data sets. Mol Ecol Resour. 143:647–651.http://dx.doi.org/10.1111/1755-0998.12200 [DOI] [PubMed] [Google Scholar]
- Delmas O, et al. 2008. Genomic diversity and evolution of the Lyssaviruses. PLoS One 34:e2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delport W, Poon AFY, Frost SDW, Kosakovsky Pond SL.. 2010. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 2619:2455–2457.http://dx.doi.org/10.1093/bioinformatics/btq429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A.. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 7:214.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchêne S, Holmes EC, Ho SYW.. 2014. Analyses of evolutionary dynamics in viruses are hindered by a time-dependent bias in rate estimates. Proc Biol Sci. 2811786:20140732.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA, VonHoldt BM.. 2012. Structure Harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Cons Genet Res 4:359–361. [Google Scholar]
- Eggerbauer E, et al. 2017. Comparative analysis of European bat lyssavirus 1 pathogenicity in the mouse model. PLoS Negl Trop Dis. 116:e0005668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM.. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 1312:479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria NR, Suchard MA, Rambaut A, Lemey P.. 2011. Toward a quantitative understanding of viral phylogeography. Curr Opin Virol. 15:423–429.http://dx.doi.org/10.1016/j.coviro.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freuling C, et al. 2011. Novel Lyssavirus in Natterer’s bat, Germany. Emerg Infect Dis. 178:1519–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, et al. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 1510:1451–1455.http://dx.doi.org/10.1101/gr.4086505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J, Galaxy Team. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 118:R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. 2005. Hierfstat, a package for r to compute and test hierarchical F-statistics. Mol Ecol Notes 51:184–186.http://dx.doi.org/10.1111/j.1471-8286.2004.00828.x [Google Scholar]
- Guillot G, Estoup A, Mortier F, Cosson JF.. 2005. A spatial statistical model for landscape genetics. Genetics 1703:1261–1280.http://dx.doi.org/10.1534/genetics.104.033803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 593:307–321.http://dx.doi.org/10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O, Rannala B.. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 525:696–704.http://dx.doi.org/10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Gunawardena PS, et al. 2016. Lyssavirus in Indian flying foxes, Sri Lanka. Emerg Infect Dis. 228:1456–1459.http://dx.doi.org/10.3201/eid2208.151986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyatt KJ, et al. 2003. A molecular epidemiological study of Australian bat lyssavirus. J Gen Virol. 84(Pt 2):485–496. [DOI] [PubMed] [Google Scholar]
- Halczok TK, et al. 2017. Evidence for genetic variation in Natterer’s bats (Myotis nattereri) across three regions in Germany but no evidence for co-variation with their associated astroviruses. BMC Evol Biol. 171:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H-J, et al. 2015. Bats as reservoirs of severe emerging infectious diseases. Virus Res. 205:1–6.http://dx.doi.org/10.1016/j.virusres.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Suzuki Y, Gojobori T.. 2004. A large variation in the rates of synonymous substitution for RNA viruses and its relationship to a diversity of viral infection and transmission modes. Mol Biol Evol. 216:1074–1080.http://dx.doi.org/10.1093/molbev/msh109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman DTS, et al. 2013. Ecology of zoonotic infectious diseases in bats: current knowledge and future directions. Zoonoses Public Health 601:2–21.http://dx.doi.org/10.1111/zph.12000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman DTS, Fooks AR, Marston DA, Garcia-R JC.. 2016. The global phylogeography of Lyssaviruses – challenging the ‘Out of Africa’ hypothesis. PLoS Negl Trop Dis. 1012:e0005266.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes GJ. 2008. A reassessment of the emergence time of European bat lyssavirus type 1. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 86:820–824. [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Rambaut A, Pybus OG, Holmes EC.. 2002. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J Mol Evol. 542:156–165.http://dx.doi.org/10.1007/s00239-001-0064-3 [DOI] [PubMed] [Google Scholar]
- Johnson N, et al. 2010. Human rabies due to lyssavirus infection of bat origin. Vet Microbiol. 142(3–4):151–159. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW.. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 225:1208–1222.http://dx.doi.org/10.1093/molbev/msi105 [DOI] [PubMed] [Google Scholar]
- Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 2321:2947–2948. Available from: http://www.clustal.org/. [DOI] [PubMed] [Google Scholar]
- Lecocq T, Gérard M, Michez D, Dellicour S.. 2017. Conservation genetics of European bees: new insights from the continental scale. Conserv Genet. 183:585–596. [Google Scholar]
- López-Roig M, Bourhy H, Lavenir R, Serra-Cobo J.. 2014. Seroprevalence dynamics of European bat lyssavirus type 1 in a multispecies bat colony. Viruses 69:3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 272:209–220. [PubMed] [Google Scholar]
- Marston DA, et al. 2017. Complete genome sequence of Lleida bat Lyssavirus. Genome Announc. 52:e01427–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhinney LM, et al. 2013. Molecular epidemiology of bat lyssaviruses in Europe: molecular epidemiology of EBLV. Zoonoses Public Health 601:35–45.http://dx.doi.org/10.1111/zph.12003 [DOI] [PubMed] [Google Scholar]
- Minin VN, Bloomquist EW, Suchard MA.. 2008. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol Biol Evol. 257:1459–1471.http://dx.doi.org/10.1093/molbev/msn090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr W. 1957. [Rabies]. Med Klin. 5224:1057–1060. [PubMed] [Google Scholar]
- Moussy C, et al. 2015. Population genetic structure of serotine bats (Eptesicus serotinus) across Europe and implications for the potential spread of bat rabies (European bat lyssavirus EBLV-1). Heredity 1151:83–92.http://dx.doi.org/10.1038/hdy.2015.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller T, et al. 2004. Spill-over of European bat lyssavirus type 1 into a stone marten (Martes foina) in Germany. J Vet Med B Infect Dis Vet Public Health 512:49–54. [DOI] [PubMed] [Google Scholar]
- Murrell B, et al. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 87:e1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, et al. 2013. FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol. 305:1196–1205.http://dx.doi.org/10.1093/molbev/mst030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nokireki T, et al. 2013. Bat rabies surveillance in Finland. BMC Vet Res. 9:174.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard-Meyer E, et al. 2011. Active surveillance of bat rabies in France: a 5-year study (2004-2009). Vet Microbiol. 151(3–4):390–395. [DOI] [PubMed] [Google Scholar]
- Picard-Meyer E, et al. 2013. Isolation of Bokeloh bat lyssavirus in Myotis nattereri in France. Arch Virol. 15811:2333–2340. [DOI] [PubMed] [Google Scholar]
- Picard-Meyer E, et al. 2014. Bat rabies in France: a 24-year retrospective epidemiological study. PLoS One 96:e98622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW.. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 2110:2531–2533.http://dx.doi.org/10.1093/bioinformatics/bti320 [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P.. 2000. Inference of population structure using multilocus genotype data. Genetics 1552:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Lam TT, Max Carvalho L, Pybus OG.. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 21:vew007.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salicini I, Ibáñez C, Juste J.. 2011. Multilocus phylogeny and species delimitation within the Natterer’s bat species complex in the Western Palearctic. Mol Phylogenet Evol. 613:888–898. [DOI] [PubMed] [Google Scholar]
- Schatz J, et al. 2013. Bat rabies surveillance in Europe. Zoonoses Public Health 601:22–34.http://dx.doi.org/10.1111/zph.12002 [DOI] [PubMed] [Google Scholar]
- Schatz J, et al. 2014. Enhanced passive bat rabies surveillance in indigenous bat species from Germany–a retrospective study. PLoS Negl Trop Dis. 85:e2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Cobo J, et al. 2013. Ecological factors associated with European bat lyssavirus seroprevalence in spanish bats. PLoS One 85:e64467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GC, et al. 2011. Bat population genetics and Lyssavirus presence in Great Britain. Epidemiol Infect. 13910:1463–1469. [DOI] [PubMed] [Google Scholar]
- Smith I, Wang L-F.. 2013. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr Opin Virol. 31:84–91.http://dx.doi.org/10.1016/j.coviro.2012.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smreczak M, Orlowska A, Zmudzinski F.. 2009. First case of the European bat lyssavirus type 1b in bats (Eptesicus serotinus) in Poland in retrospective study. Bull Vet Inst Puławy 53 [cited 2017 May 20]. Available from: http://agro.icm.edu.pl/agro/element/bwmeta1.element.agro-41766c9d-ed5b-4c7c-b0e7-78e93f093d2c. [Google Scholar]
- Streicker DG, Altizer SM, Velasco-Villa A, Rupprecht CE.. 2012. Variable evolutionary routes to host establishment across repeated rabies virus host shifts among bats. Proc Natl Acad Sci U S A. 10948:19715–19720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streicker DG, Lemey P, Velasco-Villa A, Rupprecht CE.. 2012. Rates of viral evolution are linked to host geography in bat rabies. PLoS Pathog. 85:e1002720.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard MA, Kitchen CMR, Sinsheimer JS, Weiss RE, Yang Z.. 2003. Hierarchical phylogenetic models for analyzing multipartite sequence data. Syst Biol. 525:649–664. [DOI] [PubMed] [Google Scholar]
- Suchard MA, Rambaut A.. 2009. Many-core algorithms for statistical phylogenetics. Bioinformatics 2511:1370–1376.http://dx.doi.org/10.1093/bioinformatics/btp244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjørnehøj K, Fooks AR, Agerholm JS, Rønsholt L.. 2006.. Natural and experimental infection of sheep with European bat lyssavirus type-1 of Danish bat origin. J Comp Pathol. 134(2–3):190–201. [DOI] [PubMed] [Google Scholar]
- Tordo N, Poch O, Ermine A, Keith G, Rougeon F.. 1986. Walking along the rabies genome: is the large G-L intergenic region a remnant gene? Proc Natl Acad Sci U S A. 8311:3914–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordo N, Poch O, Ermine A, Keith G, Rougeon F.. 1988. Completion of the rabies virus genome sequence determination: highly conserved domains among the L (polymerase) proteins of unsegmented negative-strand RNA viruses. Virology 1652:565–576.http://dx.doi.org/10.1016/0042-6822(88)90600-9 [DOI] [PubMed] [Google Scholar]
- Troupin C, et al. 2016. Large-scale phylogenomic analysis reveals the complex evolutionary history of rabies virus in multiple carnivore hosts. PLoS Pathog. 1212:e1006041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC.. 1984. Estimating F-statistics for the analysis of population structure. Evolution 386:1358–1370. [DOI] [PubMed] [Google Scholar]
- Wynne JW, Wang L-F.. 2013. Bats and viruses: friend or foe? PLoS Pathog. 910:e1003651.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.