Abstract
The liver is well known to possess high regenerative capacity in response to partial resection or tissue injury. However, liver regeneration is often impaired in the case of advanced liver fibrosis/cirrhosis when mature hepatocytes can hardly self‐proliferate. Hepatic progenitor cells have been implicated as a source of hepatocytes in regeneration of the fibrotic liver. Although alpha‐fetoprotein (AFP) is known as a clinical marker of progenitor cell induction in injured/fibrotic adult liver, the origin and features of such AFP‐producing cells are not fully understood. Here, we demonstrate a unique and distinct AFP‐expressing cell population that is induced by the Jagged1/Notch2 signal in murine fibrotic liver. Following repeated carbon tetrachloride injections, a significant number of AFP‐positive cells with high proliferative ability were observed along the fibrous septa depending on the extent of liver fibrosis. These AFP‐positive cells exhibited features of immature hepatocytes that were stained positively for hepatocyte‐lineage markers, such as albumin and hepatocyte nuclear factor 4 alpha, and a stem/progenitor cell marker Sox9. A combination of immunohistological examination of fibrotic liver tissues and coculture experiments with primary hepatocytes and hepatic stellate cells indicated that increased Jagged1 expression in activated hepatic stellate cells stimulated Notch2 signaling and up‐regulated AFP expression in adjacent hepatocytes. The mobilization and proliferation of AFP‐positive cells in fibrotic liver were further enhanced after partial hepatectomy, which was significantly suppressed in Jagged1‐conditional knockout mice. Finally, forced expression of the intracellular domain of Notch2 in normal liver induced a small number of AFP‐expressing hepatocytes in vivo. Conclusion: Insight is provided into a novel pathophysiological role of Jagged1/Notch2 signaling in the induction of AFP‐positive cells in fibrotic liver through the interaction between hepatocytes and activated hepatic stellate cells. (Hepatology Communications 2017;1:215‐229)
Abbreviations
- AFP
alpha‐fetoprotein
- BrdU
bromodeoxyuridine
- CCl4
carbon tetrachloride
- CK
cytokeratin
- cKO
conditional knockout
- Col1a1
type I α1 chain collagen
- DDC
3,5‐diethoxycarbonyl‐1,4‐dihydrocollidine
- EGFP
enhanced green fluorescent protein
- EpCAM
epithelial cell adhesion molecule
- GFAP
glial fibrillary acidic protein
- Hnf4α
hepatocyte nuclear factor 4 alpha
- HPC
hepatic progenitor cell
- HSC
hepatic stellate cells
- mRNA
messenger RNA
- NICD
intracellular domain of Notch
- PCR
polymerase chain reaction
- poly(I):poly(C)
polyinosinic‐polycytidylic acid
- RT‐PCR
reverse transcription‐PCR
- αSMA
α‐smooth muscle action
Introduction
The prevalence of liver cirrhosis, which represents the common consequence of chronic fibrotic liver diseases of various etiologies, is increasing and has become a serious medical issue worldwide.1 Liver cirrhosis not only leads to hepatic functional failure but also provides a pathological background for the development of hepatocellular carcinoma.2 In patients with cirrhosis‐associated hepatocellular carcinoma, it is sometimes difficult to remove the tumor surgically because of the expected high risk of postoperative regenerative failure of the remaining cirrhotic liver. Development of a novel therapeutic strategy for suppressing the progression of liver fibrosis and accelerating regeneration of fibrotic liver is urgently needed.
Normal liver is well known to exhibit remarkable regenerative capacity in response to partial resection or tissue injury. Following partial hepatectomy, mature hepatocytes vigorously self‐proliferate and restore the lost volume and function.3 In contrast, in the case of severe liver fibrosis/cirrhosis when mature hepatocytes show minimal division, it is generally considered that hepatic progenitor cells (HPCs) are mobilized to proliferate and differentiate into mature hepatocytes.4, 5 Two origins have been proposed as the cellular sources of HPCs. The first is the differentiation of bipotent oval cells located in the canal of Hering,6 while the other involves the dedifferentiation of mature hepatocytes.7
Alpha‐fetoprotein (AFP) is not only a representative tumor marker of hepatocellular carcinoma but is also known as a marker of HPC induction in both clinical4 and experimental8, 9 settings. AFP is produced by bipotent HPCs and yolk sac endoderm, and serum AFP concentrations are increased during embryonic development. On the other hand, serum AFP levels are elevated in the recovery phase of acute hepatic failure10 and active liver cirrhosis11 in adult patients, possibly indicating mobilization and proliferation of HPCs. However, the origin of such AFP‐producing cells induced in injured adult liver tissue is not fully understood.
In the present study, we show that the Jagged1/Notch2 signal accelerates the induction of AFP‐expressing cells in carbon tetrachloride (CCl4)‐induced murine fibrotic liver. A combination of immunohistological examination of fibrotic liver tissues and coculture experiments with primary hepatocytes and hepatic stellate cells (HSC) indicated that increased Jagged1 expression in activated HSC stimulated Notch2 signaling and up‐regulated AFP expression in adjacent hepatocytes. Experiments using Jagged1 conditional knockout (cKO) mice demonstrated that Jagged1 deletion suppressed the mobilization and proliferation of AFP‐positive cells after partial hepatectomy in CCl4‐treated fibrotic mice. Moreover, forced expression of the intracellular domain of Notch (NICD)2 in normal liver induced a small number of AFP‐expressing hepatocytes in vivo. These results provide insight into a novel important role of Jagged1/Notch2 signaling in liver pathophysiology.
Materials and Methods
MICE
All mice used in the present study received humane care and were maintained under specific pathogen‐free conditions. Animal experiments were approved by the Animal Experimentation Committee of Tokai University. Jagged1‐floxed knockin mice (Jagged1 lox/lox)12 were crossed with Mx‐Cre transgenic mice13 to generate polyinosinic‐polycytidylic acid [poly(I):poly(C)]‐inducible Jagged1 cKO mice. Notch2‐floxed knockin mice (Notch2 lox/lox)14 were mated with Albumin‐Cre transgenic mice15 [C57BL/6TgN(AlbCre)21Mgn strain from Jackson Laboratory, Bar Harbor, ME] to achieve hepatocyte‐specific Notch2 gene deletion. Both strains of mice were back‐crossed with C57BL/6 mice. Their genotypes were determined by polymerase chain reaction (PCR) using the primer sets listed in Supporting Table S1. At 8 weeks of age, Jagged1 cKO mice and control Mx‐Cre‐negative Jagged1 lox/lox littermates were given a total of four injections of 250 μg of poly(I):poly(C) (Sigma‐Aldrich, St. Louis, MO) every 3 days.16 Approximately 8 weeks after the poly(I):poly(C) injections, the mice started to receive subcutaneous injections of 1 mL/kg body weight of CCl4 (Wako Pure Chemical Industries, Ltd., Tokyo, Japan) mixed in olive oil every 3 days for a total of 28‐30 times.17 Acute liver injury was induced in wild‐type mice by an intraperitoneal injection of CCl4 or thioacetamide. In some experiments, mice were fed a 3,5‐diethoxycarbonyl‐1,4‐dihydrocollidine (DDC) diet (Oriental Yeast, Tokyo, Japan) or a methionine choline‐deficient ethionine‐supplemented diet (MP Biomedicals, Santa Ana, CA). Bile duct ligation and 70% partial hepatectomy were performed as described.17, 18
HISTOLOGICAL AND IMMUNOHISTOLOGICAL EXAMINATION
Histological and immunohistological studies were performed as described.17, 19, 20 Briefly, formalin‐fixed and paraffin‐embedded sections were subjected to hematoxylin/eosin or sirius red‐fast green FCF staining using standard protocols. For immunofluorescent/immunohistochemical staining, sections were incubated for 2 hours at room temperature with the specific primary antibodies listed in Supporting Table S2, followed by incubation with the fluorescent or horseradish peroxidase‐conjugated secondary antibodies shown in Supporting Table S3. They were examined under a fluorescence microscope (BZ9000; Keyence Corp., Osaka, Japan) or confocal laser‐scanning microscope (LSM510META; Carl Zeiss, Oberkochen, Germany). Nuclei were stained using 4′,6‐diamidino‐2‐phenylindole (Sigma‐Aldrich). The numbers of stained cells were counted, and the values per unit of area were calculated using AxioVision Rel 4.9.1 (Carl Zeiss) as described.19 The extent of fibrosis was calculated by the sirius red‐stained areas, which were measured using ImageJ software (National Institutes of Health, Bethesda, MD) as described.20
ISOLATION AND PRIMARY CULTURE OF PARENCHYMAL HEPATOCYTES AND HSC
HSC were prepared using the pronase/collagenase perfusion method17 and seeded on a type I collagen‐coated 24‐well plate or chamber slides (AGC Techno Glass, Shizuoka, Japan). They were cultured using Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum and nonessential amino acids. Hepatocytes isolated by the collagenase perfusion method20 were seeded on the HSC‐preseeded wells and cultured in the same medium as for HSC. Gamma secretase inhibitor IX (565770; Merck, Darmstadt, Germany) was diluted in dimethyl sulfoxide (Sigma‐Aldrich) before use.
IMMUNOFLUORESCENT STAINING OF CULTURED CELLS
Cultured cells were fixed with 4% paraformaldehyde for 20 minutes at 4 ºC and then incubated overnight at 4 ºC with the specific primary antibodies listed in Supporting Table S2. They were subsequently incubated with appropriate fluorescent secondary antibodies and examined under a fluorescence microscope (BZ9000; Keyence). The immunofluorescent intensity was measured using ImageJ software.
HYDRODYNAMIC GENE DELIVERY
Forced NICD2 expression in normal liver was achieved by the hydrodynamic gene delivery method as described.3 Briefly, an expression plasmid encoding Nicd2 complementary DNA or control empty vector21 was dissolved in TransIT‐EE Hydrodynamic Delivery Solution (Mirus Bio, Madison, WI) and injected through the tail vein. Efficient gene transfer into hepatocyte nuclear factor 4 alpha (Hnf4α)‐positive hepatocytes was confirmed by enhanced green fluorescent protein (EGFP) fluorescence expressed bicistronically through an internal ribosomal entry site located immediately upstream of the Egfp gene in the vector.
REAL‐TIME REVERSE TRANSCRIPTION PCR
Total RNA was isolated from liver tissues or culture cells using the RNeasy Plus Mini kit (Qiagen, Hilden, Germany). RNA was reverse transcribed using SuperScript Reverse Transcriptase (Invitrogen, Carlsbad, CA). Quantitative reverse transcription (RT)‐PCR was performed using the SYBR Green PCR master mix (Applied Biosystems, Foster City, CA) with the specific primers listed in Supporting Table S4.
STATISTICAL ANALYSIS
Values are indicated as the mean ± SD. Statistical analyses were performed using either the Student t test or the Mann‐Whitney U test to evaluate the differences between groups. Significant correlations were estimated by the Pearson product‐moment correlation coefficient. P values less than 0.05 were considered statistically significant.
Results
SIGNIFICANT NUMBER OF AFP‐EXPRESSING CELLS APPEARED IN CCl4‐INDUCED FIBROTIC LIVER TISSUE
We first compared the expression of α‐smooth muscle action (αSMA) and AFP in untreated normal liver and CCl4‐treated fibrotic liver tissues. While αSMA expression was observed only in the vessel walls in the untreated normal liver (Fig. 1A), a large number of αSMA‐positive cells were observed along the fibrous septa after 28 times of repeated CCl4 injections (Fig. 1B). Immunofluorescent staining using anti‐AFP antibodies revealed no AFP‐expressing cells in the untreated normal liver (Fig. 1A,C). In marked contrast to this, a significant number of AFP‐positive cells were detected adjacent to the αSMA‐positive myofibroblasts (Fig. 1B,C). AFP‐expressing cells were observed mainly along the fibrous septa connecting the blood vessels and preferentially located adjacent to the central veins rather than the portal areas (Fig. 1B). There was a significant correlation between the amount of type I α1 chain collagen (Col1a1) messenger RNA (mRNA) and that of Afp mRNA in the CCl4‐induced fibrotic liver tissues (Fig. 1D). These results suggested that AFP‐expressing cells are induced in parallel to the extent of liver fibrosis.
Figure 1.
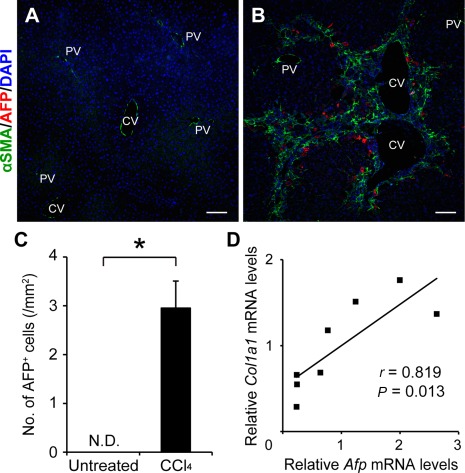
Appearance of AFP‐expressing cells in CCl4‐induced liver fibrosis. Representative pictures are shown for (A) immunofluorescent staining of untreated liver tissues and (B) CCl4‐treated liver tissues using antibodies recognizing αSMA and AFP together with DAPI nuclear staining. Scale bars, 200 μm. (C) Three sections with a mean area of 20 mm2 were prepared from each sample. Numbers of AFP‐expressing cells were counted in more than10 visual fields (4 mm2 each) that covered the whole area of the three sections without large vessels and were compared between untreated and CCl4‐treated liver tissues. The values are the means ± SD from eight mice in each group (four males, four females). An asterisk indicates that the difference between the groups was statistically significant (P < 0.01). (D) Correlation between the amounts of Afp and Col1a1 mRNAs was analyzed using CCl4‐induced fibrotic liver tissues obtained from eight mice (four males, four females). Abbreviations: CV, central vein; DAPI, 4´,6‐diamidino‐2‐phenylindole; N.D., not detected; PV, portal vein.
PROLIFERATING AFP‐EXPRESSING CELLS IN FIBROTIC LIVER TISSUE EXHIBITED FEATURES OF IMMATURE HEPATOCYTES
We next characterized the AFP‐expressing cells detected in CCl4‐induced fibrotic liver tissue. First, we examined the expression of several stem/progenitor cell markers in AFP‐expressing cells. While Sox9 was co‐expressed in more than half (57.3% ± 2.6%) of the AFP‐positive cells (Fig. 2A), expression of either CD44 (Fig. 2B) or epithelial cell adhesion molecule (EpCAM) (Fig. 2C) was not detected in AFP‐expressing cells. In addition, all of the AFP‐positive cells expressed hepatocyte‐lineage markers, such as albumin (Fig. 2D) and Hnf4α (Fig. 2F). However, the expression levels of albumin were lower than those in the neighboring AFP‐negative mature hepatocytes (Fig. 2E). In contrast, ductal lineage markers, such as cytokeratin‐19 (CK19; Fig. 2G) and Hnf1β (Fig. 2H), were not expressed in AFP‐positive cells. Furthermore, nearly all AFP‐expressing cells (99.5% ± 0.8%) were stained positively for proliferating cell nuclear antigen, a marker of cell proliferation (Fig. 2I). This finding was contrary to the relatively low proliferating cell nuclear antigen‐positive ratio (9.0% ± 2.2%) in the AFP‐negative mature hepatocytes present in the fibrotic liver tissue (Fig. 2I). Collectively, these results indicated that the AFP‐expressing cells exhibit features of immature hepatocytes22 and possess high proliferative ability.
Figure 2.
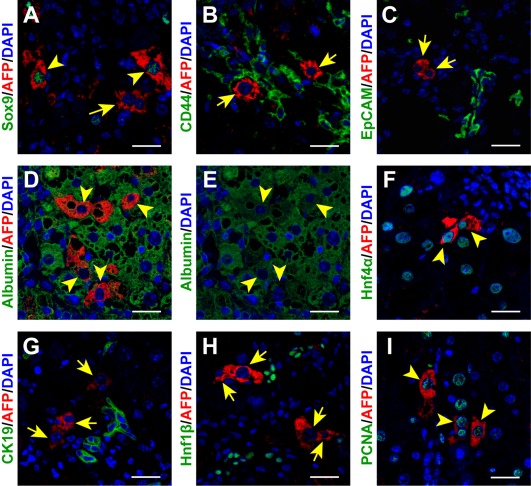
Characterization of AFP‐expressing cells present in the CCl4‐treated fibrotic liver tissue. AFP‐expressing cells were costained with the indicated antibodies recognizing HPC markers, such as (A) Sox9, (B) CD44, and (C) EpCAM; hepatocyte‐lineage markers, such as (D,E) albumin and (F) Hnf4α; or ductal cell markers, such as (G) CK19 and (H) Hnf1β, together with DAPI nuclear staining. (I) Cells were also tested for proliferating ability by staining PCNA. Arrowheads and arrows indicate AFP‐positive cells with and without co‐expression of another cell marker, respectively. Note that the arrowheads in panels D and E show identical cells. Scale bars, 50 μm. Abbreviations: DAPI, 4´,6‐diamidino‐2‐phenylindole; PCNA, proliferating cell nuclear antigen.
INCREASED JAGGED1 EXPRESSION IN MYOFIBROBLASTS STIMULATED NOTCH SIGNALING IN ADJACENT AFP‐EXPRESSING CELLS
Because Sox9' a downstream target molecule of Notch signaling19, 23, 24, was frequently co‐expressed in AFP‐positive cells (Fig. 2A), we next examined the relative gene expression levels of canonical Notch ligands in normal and fibrotic liver tissues by real‐time RT‐PCR.25 The results indicated that the gene expression of Jagged1 and Jagged2 was significantly increased after repeated CCl4 injections, and Jagged1 gene expression was predominantly up‐regulated in fibrotic liver tissue (Fig. 3A). Immunofluorescent staining of Jagged1 further confirmed that, compared with the normal liver tissue exhibiting few Jagged1‐positive cells (Fig. 3B), the protein was expressed predominantly in αSMA‐positive myofibroblasts present along the fibrous septa (Fig. 3C). Interestingly, most AFP‐expressing cells were located adjacent to the Jagged1‐expressing myofibroblasts (Fig. 3C). Consistent with these findings, the expression of Hes1, the major effector of Notch signaling, was detected in 66.9% ± 2.2 % of AFP‐positive cells present in fibrotic liver tissue (Fig. 3E), while its expression was restricted to ductal epithelial cells and endothelial cells in the normal liver (Fig. 3D). These results suggested that in the fibrotic liver Notch signaling is activated in most AFP‐expressing cells by Jagged1, which is expressed in adjacent myofibroblasts (Fig. 3F). To further assess the role of myofibroblast‐derived Jagged1 in the induction of AFP‐expressing cells, we also performed immunofluorescent staining of Jagged1, AFP, and αSMA in acute liver injury models. While αSMA‐expressing myofibroblasts detected in the CCl4‐induced centrilobular necrotic areas co‐expressed Jagged1, similarly activated αSMA‐positive cells did not express significant levels of Jagged1 after a single thioacetamide injection (Supporting Fig. S1). A small number of AFP‐positive cells were observed around the necrotic areas only in the CCl4 model, indicating the close association between the Jagged1 signal derived from activated myofibroblasts and the induction of AFP‐expressing cells (Supporting Fig. S1).
Figure 3.
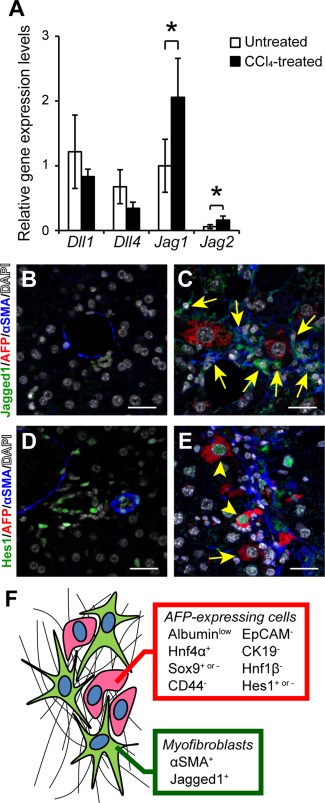
Activation of Notch signaling in AFP‐positive cells by the adjacent Jagged1‐expressing myofibroblasts. (A) Relative gene expression levels of Delta‐like 1 (Dll1), Delta‐like 4 (Dll4), Jagged1 (Jag1), and Jagged2 (Jag2) were compared between untreated and CCl4‐treated fibrotic liver tissues. The values are the means ± SD from four mice (two males, two females) in each group. An asterisk indicates that the difference between groups was statistically significant (P < 0.05). Expression of Jagged1, AFP, and αSMA was examined by immunofluorescent staining in (B) untreated and (C) CCl4‐treated liver tissues. The arrows indicate myofibroblasts co‐expressing Jagged1 and αSMA. Scale bars, 50 μm. Expression of Hes1, AFP, and αSMA was also examined in (D) untreated and (E) CCl4‐treated liver tissues. The arrowheads and arrow indicate AFP‐positive cells with and without co‐expression of Hes1, respectively. Scale bars, 50 μm. (F) Schematic representation of AFP‐positive cells located adjacent to Jagged1‐expressing activated myofibroblasts in fibrotic liver. Abbreviation: DAPI, 4´,6‐diamidino‐2‐phenylindole.
LOSS OF JAGGED1 DECREASED THE NUMBER OF AFP‐EXPRESSING CELLS WITHOUT INFLUENCING THE EXTENT OF FIBROSIS OR DUCTULAR HYPERPLASIA
Because the results obtained so far indicated the possible role of Notch signaling in the induction of AFP‐positive cells in the fibrotic liver, we used poly(I):poly(C)‐inducible Jagged1 cKO mice for subsequent experiments. Following intraperitoneal poly(I):poly(C) injections, both Jagged1 cKO mice and control Mx‐Cre‐negative littermates received repeated CCl4 administration to induce liver fibrosis. PCR detection of the wild‐type and deleted alleles (Supporting Fig. S2A) and immunofluorescent staining of Jagged1 (Supporting Fig. S2B,C) confirmed the efficient ablation of Jagged1 in poly(I):poly(C)‐treated Mx‐Cre‐positive Jagged1 lox/lox mice. There was no difference between the control and Jagged1 cKO mice in the extent of liver fibrosis estimated based on sirius red staining (Fig. 4A‐C). Real‐time RT‐PCR analyses quantifying the amounts of Col1a1 mRNA also revealed no difference between the two groups of mice (Fig. 4D). In contrast, both the number of AFP‐expressing cells (Fig. 4E‐G) and the amount of Afp mRNA (Fig. 4H) were significantly decreased in the Jagged1 cKO mice compared with those in the control mice. On the other hand, CK19 expression remained unchanged by Jagged1 deletion when estimated by either the number of CK19‐positive cells, including oval cells (Fig. 4I‐K), or Krt19 mRNA levels (Fig. 4L). These results indicated that the lack of Jagged1 primarily affects the induction of AFP‐expressing cells rather than influencing the ductular hyperplasia or oval cell recruitment in the fibrotic liver. Therefore, we also examined the correlation between Jagged1 expression and AFP‐expressing cell induction or ductular hyperplasia using several other models. Despite the remarkable proliferation of CK19‐positive cells and αSMA‐positive myofibroblasts, there were few AFP‐positive cells detected following feeding of a DDC diet or bile duct ligation (Supporting Fig. S3). Importantly, few Jagged1‐expressing myofibroblasts were observed in these models (Supporting Fig. S3). These results suggested that the levels of Jagged1 expression in activated myofibroblasts may vary depending on the experimental models and that the induction of AFP‐expressing cells is quantitatively related with the Jagged1 expression levels in adjacent myofibroblasts independent of the extent of ductular hyperplasia.
Figure 4.
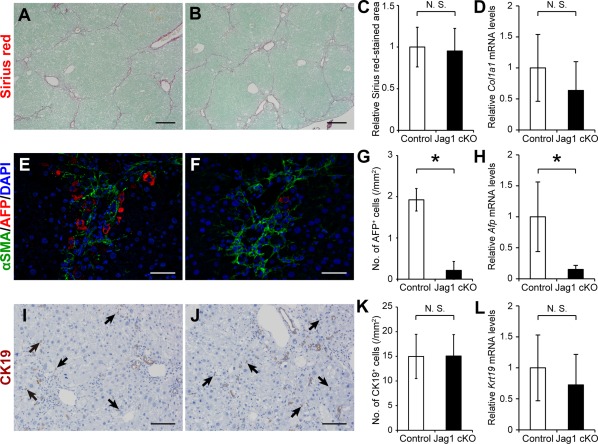
Suppression of AFP‐expressing cell mobilization in CCl4‐treated Jagged1 cKO mice. (A,B) Sirius red staining estimating the extent of fibrosis; immunofluorescent staining of (E) αSMA and (F) AFP; and (I,J) immunohistochemical detection of CK19 were performed using liver tissues obtained from (A,E,I) control and (B,F,J) Jagged1 cKO (Jag1 cKO) mice. Statistical analyses were performed to compare the differences between the two groups of mice in the (C) sirius red‐stained areas, the numbers of (G) AFP‐positive and (K) CK19‐positive cells, or the relative expression levels of (D) Col1a1, (H) Afp, and (L) Krt19 mRNAs. The values are the means ± SD from (C,G,K) five mice (three males, two females) or (D,H,L) seven mice (three males, four females) in each group. An asterisk indicates that the difference between groups was statistically significant (P < 0.01). Scale bars, 200 μm in A and B, 50 μm in E and F, and 100 μm in I and J. Abbreviations: DAPI, 4´,6‐diamidino‐2‐phenylindole; N.S., not significant.
JAGGED1/NOTCH SIGNALING‐DEPENDENT MOBILIZATION AND PROLIFERATION OF AFP‐EXPRESSING CELLS WERE ASSOCIATED WITH PROLONGED SURVIVAL AFTER PARTIAL RESECTION OF FIBROTIC LIVER
In the next set of experiments, we evaluated the impact of Jagged1 deletion on the mobilization and proliferation of AFP‐positive cells after 70% partial hepatectomy of fibrotic liver. Immunofluorescent studies indicated that the number of AFP‐positive cells was significantly decreased in the fibrotic liver tissue of Jagged1 cKO mice compared with that in control littermates (Fig. 5A‐C) on day 2 after partial hepatectomy. Furthermore, while a significant number of AFP‐expressing cells showed positive staining for bromodeoxyuridine (BrdU) in the regenerating fibrotic liver of control mice (Fig. 5A,C,E), few cells were co‐stained with AFP and BrdU antibodies in Jagged1 cKO mice (Fig. 5B,C,E). There was no difference in the number of CK19‐positive cells between the control and Jagged1 cKO mice (Fig. 5D), and only a limited number of CK19‐positive cells were stained positively for BrdU in both groups of mice on day 2 after partial hepatectomy (Fig. 5D,E). These findings suggested that Jagged1/Notch signaling stimulates the mobilization and proliferation of AFP‐positive cells but not CK19‐positive cells after partial resection of fibrotic liver. In contrast to these results obtained with fibrotic mice, no AFP‐positive cells were detected in the regenerating nonfibrotic liver tissue from either control or Jagged1 cKO mice without CCl4 intoxication (Supporting Fig. S4). To further assess the functional relevance of AFP‐expressing cells in fibrotic liver regeneration, we compared the survival rates for 7 days after 70% partial hepatectomy between Jagged1 cKO mice and the control littermates. In the absence of AFP‐positive proliferating cells, the survival rate of CCl4‐treated Jagged1 cKO mice was significantly lower than that of the control animals that had been treated similarly with repeated CCl4 injections (Fig. 6). In contrast, all Jagged1 cKO mice without CCl4 intoxication survived up to day 7 after 70% partial hepatectomy, as did the control untreated littermates (Fig. 6).
Figure 5.
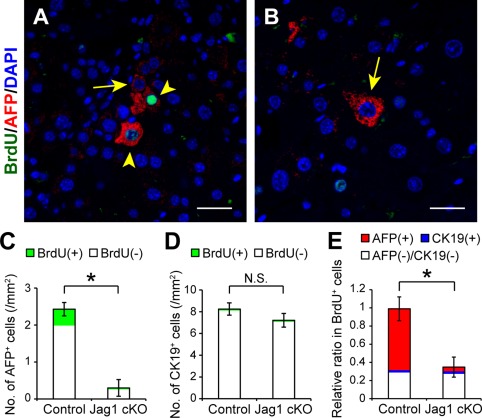
Suppression of mobilization and proliferation of AFP‐positive cells after partial hepatectomy in CCl4‐treated Jagged1 cKO mice. Cell proliferation was evaluated by BrdU staining in regenerating fibrotic liver 48 hours after partial hepatectomy. BrdU (100 μmol/kg body weight) was injected intraperitoneally 2 hours before excising the liver. Costaining of AFP and BrdU together with DAPI nuclear staining was performed using liver specimens obtained from (A) CCl4‐treated control and (B) Jag1 cKO mice. The arrowheads and arrows indicate AFP‐expressing cells with and without BrdU costaining, respectively. Scale bars, 100 μm. The total numbers of (C) AFP‐positive or (D) CK19‐positive cells with or without BrdU co‐expression and (E) the relative ratio of each cell population in BrdU‐positive cells were compared between control and Jag1 cKO mice as described in Fig. 1. The values indicate the means ± SD from three mice in each group (two males, one female). An asterisk indicates that the difference between groups was statistically significant (P < 0.01). Abbreviations: DAPI, 4´,6‐diamidino‐2‐phenylindole; N.S., not significant.
Figure 6.
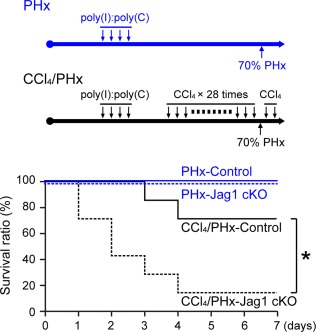
Reduced survival after partial hepatectomy in CCl4‐treated Jagged1 cKO mice. Control (solid lines) and Jagged1 cKO mice (dotted lines) either untreated (blue) or treated with repeated CCl4 injections (black) were subjected to 70% PHx. Survival rates were compared between control and Jagged1 cKO mice (five mice: three males, two females) in each untreated group and seven animals (three males, four females) in each CCl4‐treated group. An asterisk indicates that the survival rate of CCl4‐treated Jag1 cKO mice was significantly lower than that in the CCl4‐treated control animals (P < 0.05). Abbreviation: PHx, partial hepatectomy.
JAGGED1/NOTCH2 SIGNALING STIMULATED AFP AND SOX9 EXPRESSION IN PRIMARY CULTURED HEPATOCYTES
Although AFP‐positive cells expressed hepatocyte‐specific markers, such as albumin and Hnf4α (Fig. 2D,F), their location was distant from the periportal areas where oval cells exist (Fig. 1B). These findings suggested that these cells might be different from the oval cell‐derived hepatocytes26 and possibly represent a unique and distinct cell population converted from mature hepatocytes by Jagged1 stimulation from adjacent myofibroblasts. To test this hypothesis in vitro, primary hepatocytes were cocultured with quiescent or activated HSC. HSC underwent activation during culture on a collagen type I‐coated dish for 7 days estimated by the relative expression levels of glial fibrillary acidic protein (GFAP) and αSMA (Fig. 7A). Consistent with the results of in vivo immunostaining shown in Fig. 3C, Jagged1 expression was up‐regulated in activated HSC cultured for 7 days compared to that in quiescent cells cultured for only 1 day (Fig. 7A). Immunofluorescent staining and real‐time RT‐PCR analysis failed to detect AFP production or Afp mRNA expression, respectively, in these cultured HSC (data not shown). The amounts of AFP and Sox9 proteins were significantly increased in primary hepatocytes cocultured with activated HSC compared to those with quiescent HSC (Fig. 7B). In parallel to the increased AFP production, Afp gene expression levels were significantly higher in hepatocytes cocultured with activated HSC than those with quiescent HSC (Fig. 8A). This elevation of Afp gene expression levels was completely suppressed by adding gamma secretase inhibitor IX, an inhibitor of Notch signaling27 into the culture media (Fig. 8A). Coculture experiments were also performed using HSC obtained from either control or Jagged1 cKO mice. Efficient Jagged1 deletion was confirmed by PCR analysis (Supporting Fig. S5A) and the lack of Jagged1expression in activated HSC derived from Jagged1 cKO mice (Supporting Fig. S5E). Immunofluorescent staining of GFAP and αSMA (Supporting Fig. S5B‐E) as well as quantitative RT‐PCR analysis of Acta2, Col1a1, and Gfap gene expression (Supporting Fig. S5F‐H) indicated the same extent of in vitro activation of HSC derived from Jagged1 cKO mice and the control animals. The induction of Afp gene expressions in primary hepatocytes cocultured with activated control HSC was not observed when the same hepatocytes were cocultured with activated Jagged1 cKO HSC (Fig. 8B). Similarly, the induction of Sox9 gene expression in hepatocytes was significantly reduced by adding gamma secretase inhibitor IX (Supporting Fig. S6A) or coculture with Jagged1 cKO HSC (Supporting Fig. S6B). Conversely, when primary hepatocytes obtained from Notch2 cKO mice were cocultured with wild‐type activated HSC, the induction of Afp (Fig. 8C) and Sox9 (Supporting Fig. S6C) gene expression was completely suppressed compared with hepatocytes derived from the control littermates. On the other hand, gene expression of ductal lineage markers, such as CK19 and Hnf1β, was not induced by coculture with activated HSC (Supporting Fig. S6A‐C). Collectively, the results of these in vitro studies suggested that the AFP‐expressing cells observed in the fibrotic liver tissue in vivo might be derived from mature hepatocytes via Jagged1/Notch2 signaling.
Figure 7.
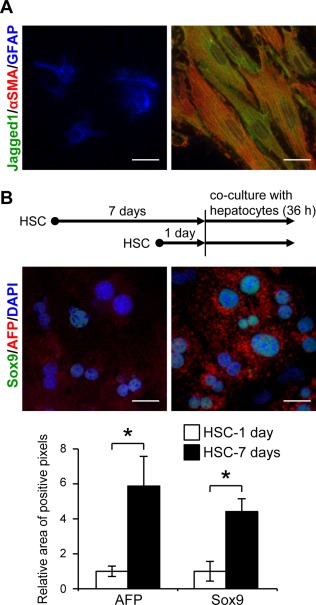
Induction of AFP and Sox9 expression in primary hepatocytes cocultured with activated HSC. (A) Expression of Jagged1, αSMA, and GFAP was analyzed by immunofluorescent staining in quiescent HSC cultured for 1 day (left) or activated cells cultured for 7 days (right) on collagen type I‐coated dishes. Scale bars, 25 μm. (B) Immunofluorescent analysis for AFP and Sox9 in primary hepatocytes cocultured with either quiescent HSC (left) or activated HSC (right). Expression levels of AFP and Sox9 proteins were semiquantified by readout of positive pixels using ImageJ software. An asterisk indicates that the difference between groups was statistically significant (P < 0.01). Scale bars, 25 μm. Abbreviation: DAPI, 4´,6‐diamidino‐2‐phenylindole.
Figure 8.
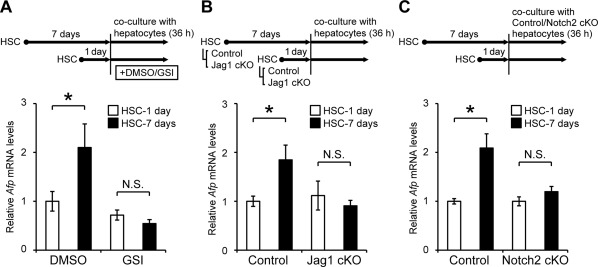
Regulation of Afp gene induction in primary hepatocytes by Jagged1/Notch2 signaling. (A) Primary hepatocytes isolated from wild‐type mice were cocultured with either quiescent or activated HSC in a mixed condition in the presence of DMSO or 10 μM GSI for 36 hours. (B) Primary hepatocytes isolated from wild‐type mice were cocultured with HSC derived from either control or Jag1 cKO mice for 36 hours. (C) Primary hepatocytes isolated from either wild‐type or Notch2 cKO mice were cocultured with the wild‐type HSC for 36 hours. Afp gene expression levels were quantified by real‐time RT‐PCR. The values indicate the means ± SD from four samples in each group and represent two or three independent experiments. An asterisk indicates that the difference between groups was statistically significant (P < 0.01). Abbreviations: DMSO, dimethyl sulfoxide; GSI, gamma secretase inhibitor IX; N.S., not significant.
FORCED NICD2 EXPRESSION INDUCED AFP EXPRESSION IN HEPATOCYTES
In the last set of experiments, we examined the effects of forced NICD2 expression in hepatocytes on induction of HPC markers by injecting control vector or Nicd2 expression plasmid through the tail vein. Preferential gene transfer into Hnf4α‐positive hepatocytes was confirmed by bicistronically expressed EGFP fluorescence (Fig. 9A, D). Expression of the major Notch effector Hes1 was increased in Hnf4α‐positive hepatocytes, and a small number of AFP‐expressing hepatocytes were induced only in mice that had been injected with an Nicd2 expression plasmid (Fig. 9C,F) but not in the control animals (Fig. 9B,E). Consistently, forced expression of NICD2 significantly increased gene expression levels of Hes1 and AFP as well as another HPC marker Sox9 compared with the injection of the control vector (Fig. 9G). Interestingly, NICD2 overexpression also stimulated Krt19 gene expression (Fig. 9G).
Figure 9.
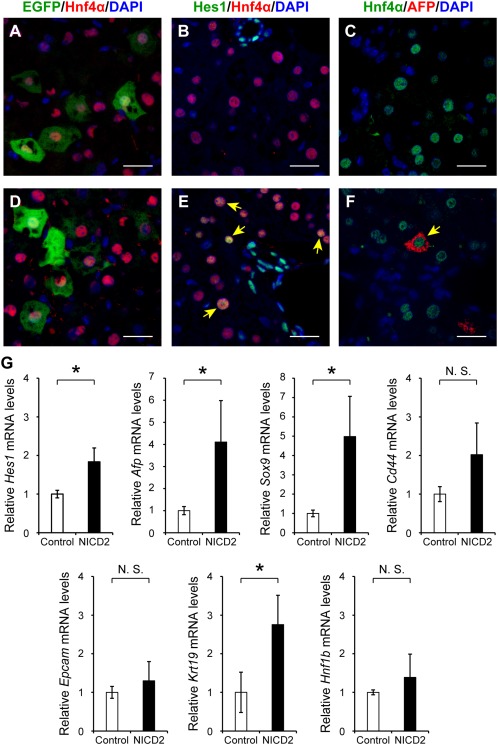
Induction of AFP‐expressing hepatocytes by forced expression of NICD2. Ten milligrams of a control empty vector or expression plasmid encoding Nicd2 complementary DNA was injected through the tail vein into 5‐week‐old wild‐type female mice (approximately 20 g body weight). (A,D) After 24 hours, excised liver tissues were subjected to immunofluorescence microscopic examination to confirm preferential EGFP expression in Hnf4α‐positive hepatocytes. Expression of (B,E) Hes1 and (C,F) AFP in Hnf4α‐positive hepatocytes was also examined using liver tissues injected with either (A‐C) control vector or (D‐F) an Nicd2 expression plasmid. The arrows indicate Hes1‐positive or AFP‐positive cells with co‐expression of Hnf4α. Scale bars, 50 μm. (G) Gene expression levels of AFP, Sox9, CD44, EpCAM, CK19, and Hnf1β were quantified by real‐time RT‐PCR. The values indicate the means ± SD from three female mice in each group. An asterisk indicates that the difference between groups was statistically significant (P < 0.05). Abbreviation: N.S., not significant.
Discussion
In this study, we demonstrate the mobilization of AFP‐expressing cells in CCl4‐induced liver fibrosis. A series of immunohistological examinations of liver tissues indicate that the AFP‐expressing cells exhibit features of immature hepatocytes and possess a high proliferative ability. The induction of AFP‐expressing cells is regulated by Notch signaling, and adjacent myofibroblasts enhance this process by expressing one of the Notch ligands, Jagged1. Experiments using Jagged1 cKO mice indicate that the Jagged1/Notch signal further enhances the mobilization and proliferation of AFP‐expressing cells after partial resection of fibrotic liver. Moreover, in vitro coculture experiments with primary hepatocytes and HSC suggest that these AFP‐expressing cells might be derived from mature hepatocytes. Consistent with this hypothesis, forced NICD2 expression in normal liver induced a small number of AFP‐expressing hepatocytes in vivo.
Notch signaling is essential for maintaining the stem cell niche in various organs.28, 29, 30, 31 Regarding its functions in the liver, it was originally implicated in the regulation of the terminal differentiation of HPCs into biliary epithelial cells23 and/or ductal structure formation in liver development.32, 33, 34 Consistently, mutations in Jagged1 or its major receptor gene Notch2 lead to Alagille syndrome in humans, which is characterized by abnormal formation of the ductal network, causing jaundice.35 In addition to such a fundamental role in fetal liver development, several recent studies have indicated another functional role of Notch signaling in ductular hyperplasia (so‐called pseudoductal reaction) induced by feeding of a DDC diet and the biliary type of liver injury.36, 37 In contrast to those studies, the results of the present study showed that the extent of ductular cell proliferation was not altered by Jagged1 deletion in CCl4‐induced fibrotic liver tissue (Fig. 4I‐K). In addition, our preliminary experiment using Jagged1 cKO mice fed a DDC diet indicated that the extent of ductular hyperplasia was not affected by the lack of Jagged1 (data not shown).
In the previous studies, the contribution of Notch signaling to the pseudoductal reaction was suggested by using gamma secretase inhibitors and mice either overexpressing NICD1 or lacking the Notch target molecule Hes1.36, 37, 38 Therefore, it is unknown which Notch ligand(s) plays a central role in DDC diet‐induced ductular hyperplasia. We therefore examined Jagged1 expression in the DDC diet and bile duct ligation models and failed to detect Jagged1 expression in αSMA‐positive myofibroblasts despite remarkable ductular proliferation and periportal fibrosis (Supporting Fig. S3). Although we need to rule out the possibility that Jagged1 is expressed temporally in an earlier or later stage of fibrosis in these models, we speculate that Jagged1 expression is not essential for inducing ductular hyperplasia in adult liver. Alternatively, another Notch ligand, such as Jagged2, may compensate for the Jagged1 deletion in the pseudoductal reaction in Jagged1 cKO mice.
Jagged1/Notch signal‐induced mobilization and proliferation of AFP‐expressing cells were associated with prolonged survival after partial hepatectomy in CCl4‐treated fibrotic mice (Fig. 6). In contrast, the lack of Jagged1 did not affect the survival rate after partial resection of nonfibrotic liver, when HPCs are not recruited for liver regeneration. These results therefore suggest that AFP‐positive proliferating cells may contribute, at least in part, to fibrotic liver regeneration. Meanwhile, the limitation of use of an Mx‐Cre recombination that induces Jagged1 deletion in a non‐cell type‐specific manner has to be carefully considered. A recent study has implicated Jagged1 expression in macrophages,39 and the lack of Jagged1 expression in many types of cells, including immune cells, may have influenced the results obtained in the present study. Work is in progress in our laboratory to establish a myofibroblast‐specific model of Jagged1 deletion.
A previous study reported that AFP‐expressing cells were induced by feeding of a methionine‐deficient and choline‐deficient ethionine‐supplemented diet.40 However, in contrast to the results of the present study, the AFP‐positive cells reported previously did not express Sox9, a downstream target molecule of Notch signaling. In addition, our preliminary experiment indicated that the AFP‐positive cells induced by the same methionine‐deficient and choline‐deficient ethionine‐supplemented diet were not in contact with the αSMA‐positive myofibroblasts (data not shown), which differs from the findings obtained with the CCl4‐induced liver fibrosis model (Fig. 3). Taken together, we conclude that AFP‐expressing cells observed in fibrotic liver tissue exhibit heterogeneous phenotypes and differential Notch signal dependency.
It should be noted that the AFP‐expressing cells observed in the fibrotic liver tissue did not express either HPC markers (CD44 and EpCAM) or ductal lineage markers (CK19 and Hnf1β). Furthermore, Krt19 or Hnf1b gene expression was not up‐regulated in primary hepatocytes following coculture with activated HSC (Supporting Fig. S6); nor did the AFP‐expressing hepatocytes differentiate into ductal cells using our established differentiation system41 (data not shown). These results suggest that the AFP‐expressing cells induced by repeated CCl4 injections do not have the full potential as bipotent HPCs. On the other hand, forced NICD2 expression in normal liver significantly increased gene expression of not only the HPC markers, such as AFP and Sox9, but also a ductal lineage marker CK19 (Fig. 9G). A recent study has shown the conversion of terminally differentiated hepatocytes to bipotent AFP‐positive HPCs with regenerative capacity by adding a chemical cocktail to the culture media.42 It is therefore possible that the AFP‐expressing immature hepatocytes observed in the present study acquire the ability to differentiate into both hepatocytes and cholangiocytes in certain experimental conditions. From this point of view, further studies, such as cell‐fate tracing experiments, are needed to determine both the origin and functional relevance of the AFP‐expressing cells in fibrotic liver regeneration in vivo. It is also important to examine whether the same type of cells are present in human fibrotic liver tissue. These studies will lead to a better understanding of the molecular and cellular mechanisms responsible for regeneration of the fibrotic liver and may eventually contribute to the establishment of a novel therapeutic strategy for intractable liver cirrhosis.
Author names in bold designate shared co‐first authorship.
Supporting information
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1002/hep4.1026/suppinfo.
Supporting Information Figure 1.
Supporting Information Figure 2.
Supporting Information Figure 3.
Supporting Information Figure 4.
Supporting Information Figure 5.
Supporting Information Figure 6.
Supporting Information Table S1.
Supporting Information Table S2.
Supporting Information Table S3.
Supporting Information Table S4.
Supporting Information Figure Legends.
Acknowledgment
We are grateful to A. Miyajima (University of Tokyo, Tokyo, Japan), T. Sudo (Toray Industries Inc., Tokyo, Japan), K. Rajewsky (Harvard University, Boston, MA), S. Chiba (University of Tsukuba, Tsukuba, Japan), and M. A. Magnuson (Vanderbilt University, Nashville, TN) for the generous gifts of anti‐CK19 antibodies, anti‐Hes1 antibodies, Mx‐Cre transgenic mice, Notch2‐floxed mice and an Nicd2 expression plasmid, and albumin‐Cre transgenic mice, respectively. We also thank Y. Kawaguchi (Kyoto University, Kyoto, Japan) for valuable discussions and N. Kawabe, Y. Kameyama, C. Okada, and T. Sato (Support Center for Medical Research and Education, Tokai University, Isehara, Japan) for supporting the experiments.
Potential conflict of interest: Nothing to report.
Supported in part by Grants‐in‐Aid for Scientific Research (Nos. 22390152 and 25293179 to Y.I. and 16K19369 to Y.N.) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan, 2016 Tokai University School of Medicine Research Aid (to Y.N.), and a MEXT‐Supported Program for the Strategic Research Foundation at Private Universities (2015‐2019) to Tokai University.
REFERENCES
- 1. Popov Y, Schuppan D. Targeting liver fibrosis: strategies for development and validation of antifibrotic therapies. Hepatology 2009;50:1294‐1306. [DOI] [PubMed] [Google Scholar]
- 2. Zhang DY, Friedman SL. Fibrosis‐dependent mechanisms of hepatocarcinogenesis. Hepatology 2012;56:769‐775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, Miyajima A. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol 2012;22:1166‐1175. [DOI] [PubMed] [Google Scholar]
- 4. Alison MR, Islam S, Lim S. Stem cells in liver regeneration, fibrosis and cancer: the good, the bad and the ugly. J Pathol 2009;217:282‐298. [DOI] [PubMed] [Google Scholar]
- 5. Kuramitsu K, Sverdlov DY, Liu SB, Csizmadia E, Burkly L, Schuppan D, et al. Failure of fibrotic liver regeneration in mice is linked to a severe fibrogenic response driven by hepatic progenitor cell activation. Am J Pathol 2013;183:182‐194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Williams MJ, Clouston AD, Forbes SJ. Links between hepatic fibrosis, ductular reaction, and progenitor cell expansion. Gastroenterology 2014;146:349‐356. [DOI] [PubMed] [Google Scholar]
- 7. Kopp JL, Grompe M, Sander M. Stem cells versus plasticity in liver and pancreas regeneration. Nat Cell Biol 2016;18:238‐245. [DOI] [PubMed] [Google Scholar]
- 8. Kuhlmann WD, Peschke P. Hepatic progenitor cells, stem cells, and AFP expression in models of liver injury. Int J Exp Pathol 2006;87:343‐359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karaca G, Swiderska‐Syn M, Xie G, Syn WK, Krüger L, Machado MV, et al. TWEAK/Fn14 signaling is required for liver regeneration after partial hepatectomy in mice. PLoS One 2014;9:e83987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kakisaka K, Kataoka K, Onodera M, Suzuki A, Endo K, Tatemichi Y, et al. Alpha‐fetoprotein: A biomarker for the recruitment of progenitor cells in the liver in patients with acute liver injury or failure. Hepatol Res 2015;45:E12‐20. [DOI] [PubMed] [Google Scholar]
- 11. Liu YR, Lin BB, Zeng DW, Zhu YY, Chen J, Zheng Q, et al. Alpha‐fetoprotein level as a biomarker of liver fibrosis status: a cross‐sectional study of 619 consecutive patients with chronic hepatitis B. BMC Gastroenterol 2014;14:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development 2006;133:1277‐1286. [DOI] [PubMed] [Google Scholar]
- 13. Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science 1995;269:1427‐1429. [DOI] [PubMed] [Google Scholar]
- 14. Saito T, Chiba S, Ichikawa M, Kunisato A, Asai T, Shimizu K, et al. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity 2003;18:675‐685. [DOI] [PubMed] [Google Scholar]
- 15. Postic C, Magnuson MA. DNA excision in liver by an albumin‐Cre transgene occurs progressively with age. Genesis 2000;26:149‐150. [DOI] [PubMed] [Google Scholar]
- 16. Hozumi K, Negishi N, Suzuki D, Abe N, Sotomaru Y, Tamaoki N, et al. Delta‐like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat Immunol 2004;5:638‐644. [DOI] [PubMed] [Google Scholar]
- 17. Higashiyama R, Moro T, Nakao S, Mikami K, Fukumitsu H, Ueda Y, et al. Negligible contribution of bone marrow‐derived cells to collagen production during hepatic fibrogenesis in mice. Gastroenterology 2009;137:1459‐1466.e1. [DOI] [PubMed] [Google Scholar]
- 18. Inagaki Y, Truter S, Bou‐Gharios G, Garrett LA, de Crombrugghe B, Nemoto T, et al. Activation of Proalpha2(I) collagen promoter during hepatic fibrogenesis in transgenic mice. Biochem Biophys Res Commun 1998;250:606‐611. [DOI] [PubMed] [Google Scholar]
- 19. Nakano Y, Negishi N, Gocho S, Mine T, Sakurai Y, Yazawa M, et al. Disappearance of centroacinar cells in the Notch ligand‐deficient pancreas. Genes Cells 2015;20:500‐511. [DOI] [PubMed] [Google Scholar]
- 20. Moro T, Nakao S, Sumiyoshi H, Ishii T, Miyazawa M, Ishii N, et al. A combination of mitochondrial oxidative stress and excess fat/calorie intake accelerates steatohepatitis by enhancing hepatic CC chemokine production in mice. PLoS One 2016;11:e0146592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hozumi K, Abe N, Chiba S, Hirai H, Habu S. Active form of Notch members can enforce T lymphopoiesis on lymphoid progenitors in the monolayer culture specific for B cell development. J Immunol 2003;170:4973‐4979. [DOI] [PubMed] [Google Scholar]
- 22. Miyajima A, Tanaka M, Itoh T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell 2014;14:561‐574. [DOI] [PubMed] [Google Scholar]
- 23. Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, et al. Notch signaling controls liver development by regulating biliary differentiation. Development 2009;136:1727‐1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hosokawa S, Furuyama K, Horiguchi M, Aoyama Y, Tsuboi K, Sakikubo M, et al. Impact of Sox9 Ddosage and Hes1‐mediated Notch signaling in controlling the plasticity of adult pancreatic duct cells in mice. Sci Rep 2015;5:8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abe N, Hozumi K, Hirano K, Yagita H, Habu S. Notch ligands transduce different magnitudes of signaling critical for determination of T‐cell fate. Eur J Immunol 2010;40:2608‐2617. [DOI] [PubMed] [Google Scholar]
- 26. Tarlow BD, Pelz C, Naugler WE, Wakefield L, Wilson EM, Finegold MJ, et al. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell 2014;15:605‐618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shih HP, Kopp JL, Sandhu M, Dubois CL, Seymour PA, Grapin‐Botton A, et al. A Notch‐dependent molecular circuitry initiates pancreatic endocrine and ductal cell differentiation. Development 2012;139:2488‐2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pellegrinet L, Rodilla V, Liu Z, Chen S, Koch U, Espinosa L, et al. Dll1‐ and dll4‐mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology 2011;140:1230‐1240.e1231‐1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, et al. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med 2008;14:306‐314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Engin F, Yao Z, Yang T, Zhou G, Bertin T, Jiang MM, et al. Dimorphic effects of Notch signaling in bone homeostasis. Nat Med 2008;14:299‐305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, et al. Notch/gamma‐secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 2005;435:959‐963. [DOI] [PubMed] [Google Scholar]
- 32. Hofmann JJ, Zovein AC, Koh H, Radtke F, Weinmaster G, Iruela‐Arispe ML. Jagged1 in the portal vein mesenchyme regulates intrahepatic bile duct development: insights into Alagille syndrome. Development 2010;137:4061‐4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sparks EE, Huppert KA, Brown MA, Washington MK, Huppert SS. Notch signaling regulates formation of the three‐dimensional architecture of intrahepatic bile ducts in mice. Hepatology 2010;51:1391‐1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tanimizu N, Kaneko K, Itoh T, Ichinohe N, Ishii M, Mizuguchi T, et al. Intrahepatic bile ducts are developed through formation of homogeneous continuous luminal network and its dynamic rearrangement in mice. Hepatology 2016;64:175‐188. [DOI] [PubMed] [Google Scholar]
- 35. Kamath BM, Spinner NB, Rosenblum ND. Renal involvement and the role of Notch signalling in Alagille syndrome. Nat Rev Nephrol 2013;9:409‐418. [DOI] [PubMed] [Google Scholar]
- 36. Sekiya S, Suzuki A. Hepatocytes, rather than cholangiocytes, can be the major source of primitive ductules in the chronically injured mouse liver. Am J Pathol 2014;184:1468‐1478. [DOI] [PubMed] [Google Scholar]
- 37. Boulter L, Govaere O, Bird TG, Radulescu S, Ramachandran P, Pellicoro A, et al. Macrophage‐derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med 2012;18:572‐579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM, et al. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev 2013;27:719‐724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Terada M, Horisawa K, Miura S, Takashima Y, Ohkawa Y, Sekiya S, et al. Kupffer cells induce Notch‐mediated hepatocyte conversion in a common mouse model of intrahepatic cholangiocarcinoma. Sci Rep 2016;6:34691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, et al. Continuous cell supply from a Sox9‐expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet 2011;43:34‐41. [DOI] [PubMed] [Google Scholar]
- 41. Anzai K, Chikada H, Tsuruya K, Ida K, Kagawa T, Inagaki Y, et al. Foetal hepatic progenitor cells assume a cholangiocytic cell phenotype during two‐dimensional pre‐culture. Sci Rep 2016;6:28283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Katsuda T, Kawamata M, Hagiwara K, Takahashi RU, Yamamoto Y, Camargo FD, et al. Conversion of Terminally Committed Hepatocytes to Culturable Bipotent Progenitor Cells with Regenerative Capacity. Cell Stem Cell 2017;20:41‐55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1002/hep4.1026/suppinfo.
Supporting Information Figure 1.
Supporting Information Figure 2.
Supporting Information Figure 3.
Supporting Information Figure 4.
Supporting Information Figure 5.
Supporting Information Figure 6.
Supporting Information Table S1.
Supporting Information Table S2.
Supporting Information Table S3.
Supporting Information Table S4.
Supporting Information Figure Legends.


