Abstract
α‐1,2 mannosidases, key enzymes in N‐glycosylation, are required for the formation of mature glycoproteins in eukaryotes. Aberrant regulation of α‐1,2 mannosidases can result in cancer, although the underlying mechanisms are unclear. Here, we report the distinct roles of α‐1,2 mannosidase subtypes (MAN1A, MAN1B, ERMAN1, MAN1C) in the formation of hepatocellular carcinoma (HCC). Clinicopathological analyses revealed that the clinical stage, tumor size, α‐fetoprotein level, and invasion status were positively correlated with the expression levels of MAN1A1, MAN1B1, and MAN1A2. In contrast, the expression of MAN1C1 was decreased as early as stage I of HCC. Survival analyses showed that high MAN1A1, MAN1A2, and MAN1B1 expression levels combined with low MAN1C1 expression levels were significantly correlated with shorter overall survival rates. Functionally, the overexpression of MAN1A1 promoted proliferation, migration, and transformation as well as in vivo migration in zebrafish. Conversely, overexpression of MAN1C1 reduced the migration ability both in vitro and in vivo, decreased the colony formation ability, and shortened the S phase of the cell cycle. Furthermore, the expression of genes involved in cell cycle/proliferation and migration was increased in MAN1A1‐overexpressing cells but decreased in MAN1C1‐overexpressing cells. MAN1A1 activated the expression of key regulators of the unfolded protein response (UPR), while treatment with endoplasmic reticulum stress inhibitors blocked the expression of MAN1A1‐activated genes. Using the MAN1A1 liver‐specific overexpression zebrafish model, we observed steatosis and inflammation at earlier stages and HCC formation at a later stage accompanied by the increased expression of the UPR modulator binding immunoglobulin protein (BiP). These data suggest that the up‐regulation of MAN1A1 activates the UPR and might initiate metastasis. Conclusion: MAN1A1 represents a novel oncogene while MAN1C1 plays a role in tumor suppression in hepatocarcinogenesis. (Hepatology Communications 2017;1:230‐247)
Abbreviations
- AFP
α‐fetoprotein
- ATF6
activating transcription factor 6
- BiP
binding immunoglobulin protein
- cDNA
complementary DNA
- CFSE
carboxyfluorescein succinimidyl ester
- DMJ
1‐deoxymannojirimycin
- EGFP
enhanced green fluorescent protein
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum‐associated protein degradation
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- IHC
immunohistochemistry
- IRE1
inositol‐requiring enzyme 1
- MAN1A1
Golgi α‐mannosidase IA
- MAN1A2
Golgi α‐mannosidase IB
- MAN1B1
endoplasmic reticulum α‐mannosidase I
- MAN1C1
Golgi α‐mannosidase IC
- MMP
matrix metalloproteinase
- mRNA
messenger RNA
- 4‐PBA
sodium 4‐phenylbutyrate
- PCR
polymerase chain reaction
- qPCR
quantitative polymerase chain reaction
- shRNA
short hairpin RNA
- SW
swainsonine
- TUDCA
tauroursodeoxycholic acid
- UPR
unfolded protein response
- XBP1
X‐box binding protein 1
Introduction
N‐glycosylation is important for the formation of mature glycoproteins in eukaryotes.1 Class I α‐1, 2‐mannosidases are important for Asn‐linked oligosaccharide maturation in the endoplasmic reticulum (ER) and Golgi complex.2 The process of N‐glycosylation consists of a covalent linkage of a specific oligosaccharide (Glc3Man9GlcNAc2) to a nascent protein. Once the oligosaccharide is transferred, several subsequent steps of maturation occur along the secretory pathway.3 The four class I α‐1,2 mannosidases in humans are ER α‐mannosidase I (MAN1B1) and three Golgi α1,2‐mannosidases (Golgi α‐mannosidase IA [MAN1A1], Golgi α‐mannosidase IB [MAN1A2], and Golgi α‐mannosidase IC [MAN1C1]). MAN1B1 trims the central branch of the mannose residue in Man9GlcNAc2 to produce Man8GlcNAc2, resulting in the transport of the protein to the Golgi apparatus for further processing. Glycoproteins can traffic to the Golgi with or without the removal of one mannose residue by ER mannosidase I. MAN1B1 can also trim more mannose residues to target a misfolded glycoprotein to the ER‐associated protein degradation (ERAD) pathway. The Golgi mannosidases MAN1A1, MAN1A2, and MAN1C1 trim the mannose residues of Man8GlcNAc2 to Man5GlcNAc2.4 Oligosaccharide complexes are formed after Man5GlcNAc2 is processed by specific enzymes in the medial and trans‐Golgi compartments. These modifications affect cell growth, cell–cell adhesion, cell motility, and protein phosphorylation. The activity of the enzymes involved in N‐glycosylation must therefore be tightly regulated because N‐glycan composition determines the fate of the protein, including whether or not the protein will be folded in the ER lumen or retrotranslocated into the cytosol and degraded.1, 5 Hence, α‐1,2‐mannosidases are not only involved in protein folding but also play a role in misfolded protein degradation.6
The unfolded protein response (UPR) is a cellular recovery mechanism that responds to the accumulation of misfolded proteins resulting from ER stress.7 The UPR is composed of three signaling cascades, consisting of the activating transcription factor 6 (ATF6) pathway, which acts through the regulation of proteolysis; the RNA‐activated protein kinase‐like ER kinase (PERK) pathway, which functions through translational control; and the type I transmembrane protein kinase and endoribonuclease inositol‐requiring enzyme 1 (IRE1) cascade, which acts through nonconventional messenger RNA (mRNA) splicing.8 The UPR pathway regulates the ER protein load and increases folding capacity to re‐establish homeostasis and also coordinates with the endoplasmic reticulim‐associated degradation (ERAD) pathway.8, 9 Folding‐deficient proteins are labeled with specific mannose residues for ERAD degradation. Increased ER stress causes many human diseases,10 including cancers.11 Recent studies have demonstrated that the overexpression of α‐1,2‐mannosidase accelerates ERAD.12, 13
Previous reports have indicated that the mannosidase inhibitors 1‐deoxymannojirimycin (DMJ) and swainsonine (SW) suppress fibronectin‐dependent adhesion and inhibit cancer metastasis. SW functions as an α‐mannosidase II inhibitor and can efficiently decrease tumor size in nude mice injected with leukemic cells.14, 15 DMJ functions as an α‐mannosidase I inhibitor, which induces apoptosis and decreases the migration ability16 of hepatocarcinoma cells.17 MAN1B1 has also been reported to promote hepatocellular carcinoma (HCC) formation.18 High expression levels of α‐1,2 mannosidases have been associated with specific cancers,19 and α‐1,2 mannosidase inhibitors may represent potential anticancer strategies.20 Therefore, it is important to understand how α‐1,2 mannosidases influence cancer development.
In this report, we demonstrate that the expression levels of different α‐1,2 mannosidase subtypes are correlated with the severity of different stages of liver cancer. Functional studies in vitro and in vivo indicate that two subtypes of α‐1,2 mannosidases, MAN1A1 and MAN1C1, may represent possible biomarkers for early stage HCC, with MAN1A1 possessing oncogenic qualities and MAN1C1 potentially acting as a tumor suppressor. Furthermore, transgenic zebrafish overexpressing MAN1A1 under the control of a liver‐specific promoter exhibit elevated mRNA levels of cell cycle/proliferation markers and an enhanced binding immunoglobulin protein (BiP) expression. Our work reveals a novel role for MAN1A1 and provides a molecular mechanism by which MAN1A1‐mediated activation of UPR promotes liver cancer formation.
Materials and Methods
HUMAN HCC SAMPLES
We used 49 human liver hepatitis B virus (HBV)‐positive cancer samples in this study. Specimens and related clinical data were obtained from the Taiwan Liver Cancer Network. The study protocol was approved by the Institutional Review Board of the National Health Research Institutes (Human Ethics Committee code EC0971102). RNA from these samples was reverse transcribed into complementary DNA (cDNA) for quantitative polymerase chain reaction (qPCR) analysis as described.21 The mRNA expression levels of MAN1A1, MAN1A2, MAN1B1, and MAN1C1 were quantified using qPCR.
CELL CULTURE, PLASMIDS, AND REAGENTS
Human liver cancer cell lines (PLC/PRF/5, Hep3B, and HepG2) and 293T were obtained from the Bioresource Collection and Research Center, Taiwan, as described.22 Plasmids and reagents are included in the Supporting Materials and Methods.
ZEBRAFISH HUSBANDRY AND XENOTRANSPLANTATION
Zebrafish husbandry and xenotransplantation were performed as previously described.23, 24, 25 Detailed protocols and reagents are included in the Supporting Materials and Methods.
IN VITRO MIGRATION ASSAY, COLONY FORMATION ASSAY, CELL PROLIFERATION ASSAY, AND FLOW CYTOMETRY
Detailed protocols and reagents are included in the Supporting Materials and Methods.
RNA EXTRACTION, REVERSE‐TRANSCRIPTION PCR, AND qPCR
RNA extraction and reverse‐transcription qPCR were performed as described.21 The specific primers used for PCR amplification are shown in Supporting Table S5.
SODIUM DODECYL SULFATE–POLYACRYLAMIDE GEL ELECTROPHORESIS AND WESTERN BLOTTING
Detailed protocols and reagents for polyacrylamide gel electrophoresis and western blotting are included in the Supporting Materials and Methods.
OIL RED O AND SIRIUS RED STAINING
Oil Red O and Sirius Red staining were performed as described.26 Detailed protocols and reagents are included in the Supporting Materials and Methods.
STATISTICAL ANALYSIS
All data were analyzed using SPSS 17.0 (SPSS, Inc.). The results are presented as the mean ± SD of multiple batches. The significance between two groups was analyzed using the Student t test. A P value less than 0.05 was considered to be statistically significant.
Results
DIFFERENTIAL EXPRESSION OF α‐1,2‐MANNOSIDASE I SUBTYPES IN HUMAN HCC TISSUES
Our previous study showed that reduced expression of α‐1,2 mannosidase I (mas1) extends the lifespan in both Drosophila melanogaster and Caenorhabditis elegans.27 As cancer is an aging‐related disease and the pathways related to longevity represent potential targets for anticancer therapies,28 we investigated the role of class I α‐1,2‐mannosidases in liver cancer.
In humans, there are four class I α‐1,2‐mannosidases, MAN1A1, MAN1A2, MAN1B1, and MAN1C1. To examine the roles of these molecules in HCC, the mRNA levels of MAN1A1, MAN1A2, MAN1B1, and MAN1C1 were analyzed by qPCR using cDNAs collected from paired tissues (tumor and adjacent normal) from patients with HCC. When compared to the adjacent normal tissues, different stages of HCC tumors revealed increased expression of MAN1A1, MAN1A2, and MAN1B1 and decreased MAN1C1 expression (Fig. 1A). The average increased expression values of MAN1A1, MAN1A2, and MAN1B1 were significantly higher in stages II and III than in stage I (Fig. 1A). However, the expression of MAN1C1 was down‐regulated as early as stage I in patients with HCC (Fig. 1A). The expression patterns were also analyzed in three different liver cancer cell lines, PLC/PRF/5, Hep3B, and HepG2. Similar to the expression patterns seen in patients with HCC, MAN1A1, MAN1A2, and MAN1B1 show tens of thousands of molecules while MAN1C1 exhibits only hundreds of molecules in those hepatoma cells (Fig. 1B). Because HCC progresses gradually from inflamed and cirrhotic hepatocytes, we used a liver disease spectrum tissue array containing all stages of liver specimens to verify the protein expression levels of two selected subtypes, MAN1A1 and MAN1C1, by using immunohistochemistry (IHC). Compared to normal liver, the expression levels of MAN1A1 increased proportionally with the grade of hepatocarcinogenesis, except for decreased expression in the inflamed and cirrhotic liver samples (Fig. 1C,D). Conversely, the MAN1C1 protein progressively decreased from inflammatory to cirrhotic tissue and was almost undetectable in the HCC samples (Fig. 1C,D). These data suggest that during HCC formation, MAN1A1 may act as oncogenes while MAN1C1 may function as a tumor suppressor.
Figure 1.
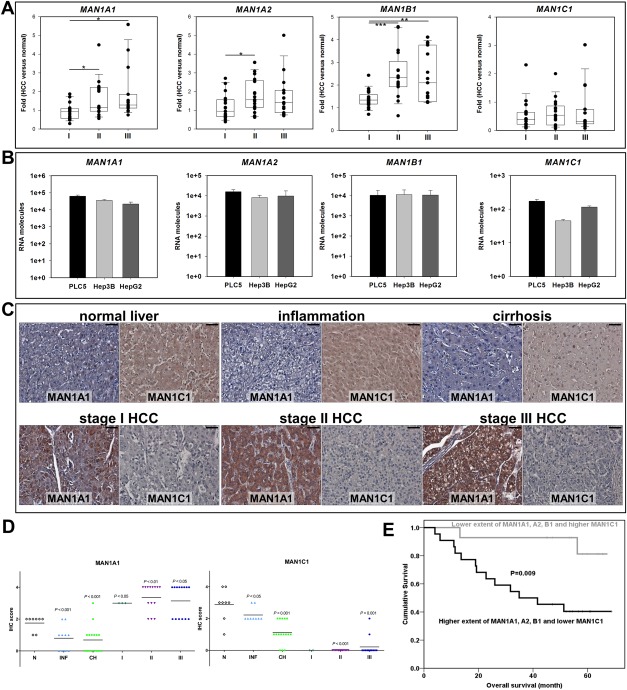
Expression patterns of the four human MAN1 genes in different stages of hepatocellular carcinoma. (A) The mRNA levels of MAN1A1, MAN1A2, MAN1B1, and MAN1C1 were correlated with the progression of HCC. I, II, and III represent liver tissues from stages I, II, and III of patients with HBV(+) HCC. The mRNA expression levels were calculated after normalizing to 18s and were compared to normal counterparts. The data are presented as dot plots with a horizontal line for the median from 17 samples from each stage and repeated in triplicate. (B) The mRNA molecules of four MAN1 subtypes were measured in triplicate using absolute qPCR analysis and known molecules (serial dilution of EGFP PCR fragment) as standards. In the PLC5, Hep3B, and HepG2 hepatoma cell lines, the RNA molecules encoding MAN1A1, MAN1A2, and MAN1B1 were from 10,000 to 70,000 per cell line, indicating high expression levels, while the three hepatoma cell lines only expressed hundreds of MAN1C1 RNA molecules, a low level of expression. (C) Representative IHC staining of MAN1A1 and MAN1C1 at different stages are shown. Magnification ×400; scale shown is for 30 μm. (D) Statistical analysis of the protein levels of MAN1A1 and MAN1C1 showed an up‐regulation of MAN1A1 and down‐regulation of MAN1C1 during hepatocarcinogenesis. The specimens were from a tissue array where N represents normal liver tissue (n = 8 each group); INF indicates inflammation (n = 10 each group); CH stands for cirrhosis (n = 16 each group); and I, II, and III represent liver tissues from stages I (n = 3 in MAN1A1 group; n = 2 in MAN1C group), II (n = 14 each group), and III (n = 14 each group) of HCC. The data are presented as a dot plot with a horizontal line for the mean, and P values were calculated using the Student t test. (E) Survival curve analysis comparing high levels of expression of MAN1A1, MAN1A2, and MAN1B1 and low expression of MAN1C1 versus low expression of MAN1A1, MAN1A2, and MAN1B1 and higher expression of MAN1C1. The statistical significance was calculated using the log rank test to determine the χ2 value.
Furthermore, clinicopathological analysis indicated that the expression levels of MAN1A1, MAN1A2, and MAN1B1 were significantly correlated with the levels of α‐fetoprotein (AFP), a biomarker for HCC diagnosis, suggesting that they may be applied as HCC biomarkers (Supporting Tables S1‐S3). In addition, higher MAN1A1 levels were significantly associated with tumors of a higher grade and larger size as well as multiple tumor types and invasion (Supporting Table S1). Higher MAN1B1 levels were correlated with invasion and relapse (Supporting Table S3). Importantly, the patients with HCC with higher levels of MAN1A1, MAN1A2, and MAN1B1 and lower levels of MAN1C1 showed poorer overall survival rates (Fig. 1E), providing clinical evidence for the possible oncogenic roles of MAN1A1, MAN1A2, and MAN1B1 and a tumor suppressive role for MAN1C1.
IN IN VITRO AND IN VIVO ASSAYS, OVEREXPRESSION OF MAN1A1 INCREASES CELL MIGRATION ABILITY WHILE MAN1C1 OVEREXPRESSION DECREASES CELL MIGRATION
To study the roles of class I α‐1,2‐mannosidases in liver cancer formation, MAN1A1 and MAN1C1 were overexpressed in cell lines to examine their effects on cell migration, colony formation, proliferation, and cell cycle. Initially, the endogenous migration ability of various cell lines, including the noncancerous human embryonic kidney cell line (293T) and hepatoma cell lines (PLC/PRF/5 and Hep3B), were examined by in vivo xenotransplantation. Only 5.5% and 6% of the embryos injected with the 293T or PLC5 cells exhibited migratory ability, whereas 81.5% of the embryos injected with Hep3B cells displayed cell migration ability at 3 days postinjection (Supporting Fig. S1A). We performed in vitro transwell assays, which revealed that Hep3B cells had a higher migration ability than PLC5 cells (Supporting Fig. S1B). Based on these results, PLC5 or 293T lines were used to examine whether the migration ability of cells can be enhanced upon overexpression of MAN1A1. Hep3B cells were used to study whether down‐regulation of MAN1C1 has any effect on decreasing cellular migration ability. Hep3B cells were used for the knockdown of MAN1A1, MAN1A2, and MAN1B1 to examine whether migration ability was affected. The strategy and selection of cell lines are shown in Supporting Fig. S1C.
Upon transient expression, the expression levels of MAN1A1 and MAN1C1 were greatly increased relative to the mock control (Fig. 2A, left). Overexpression of MAN1A1 in PLC5 cells significantly enhanced cell migration relative to the mock control. In contrast, overexpression of MAN1C1 in Hep3B cells reduced cell migration (Fig. 2A, right). Specific short hairpin RNAs (shRNAs) were used to knock down the high endogenous levels of MAN1A1, MAN1A2, and MAN1B1 in Hep3B cells, and the subsequent effects on cell migration ability were examined. The expression levels of MAN1A1, MAN1A2, and MAN1B1 were significantly decreased by about 40% using shRNA knockdown compared to the mock control as measured by qPCR analysis (Fig. 2B, left). The resultant knockdown significantly reduced migration ability by about 65% (Fig. 2B, right). The migration ability with knockdown of MAN1C1 was examined by using shRNA against MAN1C1. Because the hepatoma cells contained only hundreds of MAN1C1 molecules, MAN1C1 was stably overexpressed in Hep3B cells and then treated with shRNA knockdown. The expression levels of MAN1C1 were significantly decreased by about 40% using shRNA knockdown compared to the stable MAN1C1‐overexpressing cells as measured by qPCR analysis (Fig. 2C, left), and the resultant knockdown significantly increased migration ability 3‐fold (Fig. 2C, right).
Figure 2.
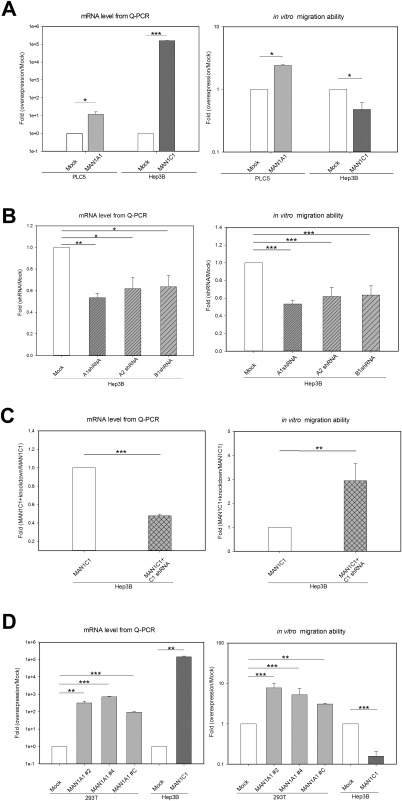
MAN1A1 enhances and MAN1C1 decreases the migration ability of cells in vitro. (A) Left: qPCR result of the mRNA level for transient overexpression of MAN1A1 and MAN1C1. Right: In vitro migration ability of MAN1A1‐ and MAN1C1‐overexpressing cells. The overexpression of MAN1A1 in PLC5 cells enhanced their migration ability compared to the DsRed vector control (Mock). In contrast, the overexpression of MAN1C1 in Hep3B cells decreased their migration ability compared to the mock control. (B) Left: qPCR result of mRNA levels following shRNA knockdown of MAN1A1, MAN1A2, and MAN1B1 individually. Right: In vitro migration ability as decreased on knockdown of MAN1A1, MAN1A2, and MAN1B1 in Hep3B cells. (C) Left: qPCR result of mRNA levels following overexpression of MAN1C1 and then shRNA knockdown of MAN1C1. Right: In vitro migration ability as increased on knockdown of MAN1C1 in Hep3B cells with overexpressing MAN1C1. (D) Left: Stable overexpression of MAN1A1 and MAN1C1 increased the mRNA level as measured by qPCR. Right: In vitro migration ability of the stable cell line showed that overexpression of MAN1A1 in 293T cells significantly enhanced their migration ability compared to the DsRed vector control (Mock). In contrast, the stable overexpression of MAN1C1 in Hep3B cells dramatically decreased their migration ability compared to the mock control. All experiments were repeated in triplicate. Statistical significance was calculated using the Student t test (*P < 0.05, **P < 0.01, ***P < 0.001).
To confirm these effects, three independent, stable, MAN1A1‐overexpressing 293T cell lines and one stable MAN1C1‐overexpressing Hep3B cell line were generated. The qPCR analysis revealed that these cell lines exhibited significantly increased expression of MAN1A1 or MAN1C1 compared to the control cells (Fig. 2D, left). The cell migration ability of the three stable MAN1A1‐overexpressing 293T stable cell lines was enhanced compared to control cells (Fig. 2D, right). In contrast, the cell migration ability of the MAN1C1‐overexpressing Hep3B stable cell line was decreased by 80% compared to the control (Fig. 2D, right). Together, the data showed that overexpression of MAN1A1 increased cell migration ability whereas overexpression of MAN1C1 reduced cell migration ability.
Recently, the xenotransplantation in vivo migration assay has been used in zebrafish embryos as a tool to investigate metastasis.23 Using the stable clones generated above, the effects of MAN1A1 and MAN1C1 on cell migration and proliferation were examined in vivo. The zebrafish metastasis model was developed using fluorescent DiI‐labeled tumor cells.29 Stable DsRed or MAN1A1‐overexpressing 293T cells were implanted into the yolks of 2‐day‐postfertilization zebrafish embryos, and the in vivo migration ability was observed in multiple embryos possessing the transplanted cells. The migration rate of DsRed stably transfected 293T cells at 1 and 3 days postinjection was 6% and 19%, respectively, whereas following MAN1A1 overexpression, the migration rate at 1 and 3 days postinjection increased to 27% and 57%, respectively (Fig. 3A, left). Conversely, the migration rate of DsRed stably transfected Hep3B cells at 1 and 3 days postinjection was 15% and 36%, respectively, while with MAN1C1 overexpression, the migration rate at 1 and 3 days postinjection decreased to 7% and 19%, respectively (Fig. 3B, left). The representative pictures of zebrafish embryos show the cells migrated from the original sites of injection when overexpressing MAN1A1 in 293T cells but not in the mock/293T control cells (Supporting Fig. S1D, left); migration also occurred in the mock/Hep3B but not in the MAN1C1‐overexpressing Hep3B cells (Supporting Fig. S1D, right).
Figure 3.
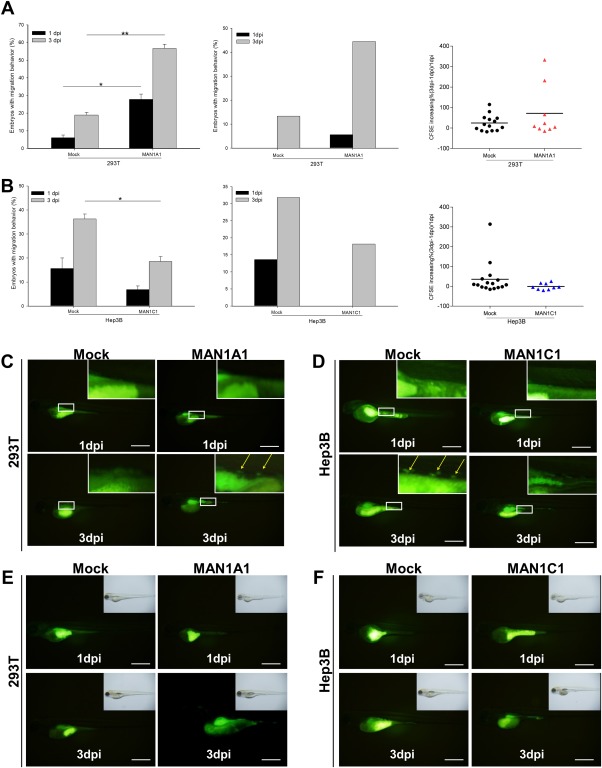
In vivo xenotransplantation assay for different cell lines and MAN1A1‐ or MAN1C1‐overexpressing cells. The cell lines were labeled with a DiI or CFSE fluorescent dye and injected into the yolks of 2‐day‐old zebrafish embryos. (A) Left: In vivo migration ability of embryos injected with MAN1A1 stably overexpressing or control 293T cells. DiI‐labeled MAN1A1 stably overexpressing cells increased the migration ability compared to the DsRed control cells (Mock) at 1 and 3 days postinjection (MAN1A1, n = 37; Mock, n = 48). Middle: Transient CFSE‐labeled MAN1A1‐overexpressing cells increased the migration ability (MAN1A1, n = 18; Mock, n = 15). Right: Proliferation ability of embryos injected with MAN1A1‐overexpressing 293T cells analyzed by area using Image J; MAN1A1‐overexpressing 293T cells increased the proliferation ability compared to the Mock 293T cells at 1 and 3 days postinjection. The data are presented as dot plots with a horizontal line for the mean. (B) Left: In vivo migration ability from the embryos injected with stably MAN1C1‐overexpressing Hep3B cells. MAN1C1 stably overexpressing cells decreased the migration ability compared to the Mock Hep3B cells. (MAN1C1, n = 95; Mock, n = 155). Middle: CFSE‐labeled MAN1C1 transiently overexpressing cells decreased the migration ability (MAN1C1, n = 11; Mock, n = 22). Right: Proliferation ability of embryos injected with MAN1C1‐overexpressing Hep3B cells decreased the proliferation ability compared to the Mock Hep3B cells at 1 and 3 days postinjection. The data are presented as dot plots with a horizontal line for the mean. (C) Representative images of MAN1A1 transiently overexpressing 293T cells increased the migration ability compared to the Mock 293T cells at 1 and 3 days postinjection. (D) Representative images of MAN1C1 transiently overexpressing Hep3B cells decreased the migration ability compared to the Mock Hep3B cells. (E) Representative images of MAN1A1 transiently overexpressing 293T cells increased the proliferation ability compared to the Mock 293T cells at 1 and 3 days postinjection. (F) Representative images of MAN1C1 transiently overexpressing Hep3B cells decreased the proliferation ability compared to the Mock Hep3B cells at 1 and 3 days postinjection. The xenotransplantation experiments were repeated 4 times. The images were taken at magnification ×48; scale shown is for 1 mm. Abbreviation: dpi, dots per inch.
DiI is a lipophilic carbocyanine fluorescent dye for membrane labeling, so the fluorescent intensity could be cell debris. To observe the proliferation and migration ability of cells in vivo, cells were labeled with carboxyfluorescein succinimidyl ester (CFSE), an amine‐reactive fluorescent dye, and followed in embryos.30 The MAN1A1‐overexpressing 293T cell line and the MAN1C1‐overexpressing Hep3B cell line were labeled with CFSE and implanted in the yolks of 2‐day‐postfertilization zebrafish embryos. The migration rate of the 293T mock control cells at 1 and 3 days postinjection was 0% and 13%, respectively, whereas the migration rate of MAN1A1‐overexpressing 293T cells at 1 and 3 days postinjection was increased to 11% and 44.4%, respectively (Fig. 3A, middle, C). Conversely, the migration rate of the Hep3B mock control cells at 1 and 3 days postinjection was 13.6% and 31.8%, respectively, whereas the migration rate of MAN1C1‐overexpressing Hep3B cells at 1 and 3 days postinjection was reduced to 0% and 18.1%, respectively (Fig. 3B, middle, D). Using Image J to compare the average fluorescence differences between 3 days postinjection and 1 day postinjection, we found that the intensity increased from 24% in the control cells to 72% in the MAN1A1‐overexpressing 293T cells (Fig. 3A, right, 3E). Conversely, the average fluorescent differences between 3 days postinjection and 1 day postinjection from MAN1C1‐expressing Hep3B cells was decreased to –1% compared to 36% in the mock control cells (Fig. 3B, right, 3F). These data demonstrate that MAN1A1 enhances while MAN1C1 decreases cell migration and proliferation ability in vivo.
IN COLONY FORMATION ASSAYS, THE OVEREXPRESSION OF MAN1A1 AND MAN1C1 HAVE OPPOSITE EFFECTS ON CELLULAR TRANSFORMATION
The colony formation assay was used to examine the transforming ability of cell lines on overexpression of either MAN1A1 or MAN1C1. Stable overexpression of MAN1A1 in 293T cells resulted in significantly increased numbers and sizes of colonies relative to the control cells (Fig. 4A; Supporting Fig. S1E). In contrast, stable overexpression of MAN1C1 in Hep3B cells produced fewer and smaller colonies compared to the control cells (Fig. 4B; Supporting Fig. S1E). In conjunction with the results from the cell migration assays, these data suggest that MAN1A1 activates the transforming ability while MAN1C1 represses the transforming ability.
Figure 4.
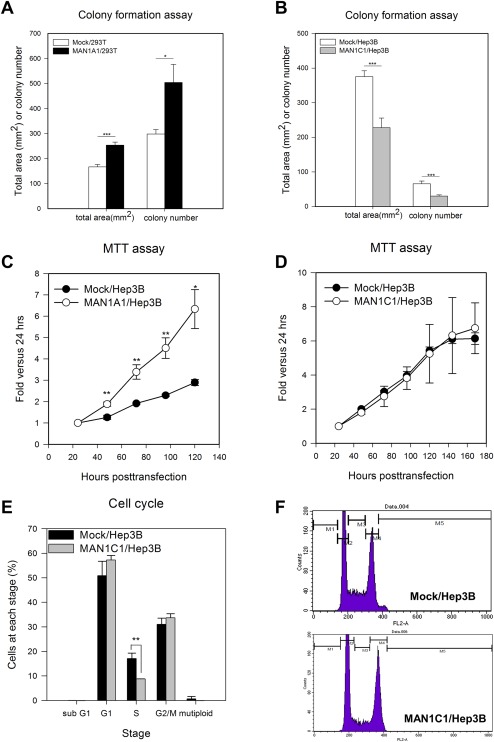
Colony formation, proliferation ability and cell cycle of MAN1A1‐ or MAN1C1‐overexpressing cell lines. (A) Stable overexpression of MAN1A1 in 293T cells resulted in more colonies and a larger colony size. (B) Stable overexpression of MAN1C1 in Hep3B cells resulted in fewer colonies and smaller colony size compared to the Hep3B control. (C) Cell proliferation was analyzed using the MTT assay after the overexpression of MAN1A1 in the Hep3B cell line. The result of transient expression of MAN1A1 in the Hep3B cells showed increased cell proliferation up to 6‐fold at 120 hours posttransection compared to 24 hours posttransection; this result was 3‐fold higher than the mock control. (D) Cell proliferation as assessed by the MTT assay after the transient overexpression of MAN1C1 in the Hep3B cell line displayed no difference. (E) Cell cycle analysis was performed using flow cytometry with a FACSCalibur (BD Biosciences). The result showed that overexpression of MAN1C1 in Hep3B cells resulted in increased cell number in the G1 and G2‐M phases by approximately 6.33% and 2.69%, respectively, compared to parental Hep3B cells; however, the cells in S phase were decreased by approximately 8.33% compared to the Hep3B mock cells. Overexpression of MAN1C1 might have acted on regulators of the S phase and caused fewer cells to enter the S phase, resulting in cell cycle arrest. (F) Flow cytometry profile of mock Hep3B cells versus overexpression of MAN1C1 in Hep3B cells (M1 = sub G1, M2 = G1, M3 = S, M4 = G2/M, M5 = mutiploid phase). All experiments were repeated 3 times. The statistical significance was calculated using the Student t test (*P < 0.05, **P < 0.01, ***P < 0.001). Abbreviations: MTT, 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (a tetrazole).
OVEREXPRESSION OF MAN1A1 INCREASES CELL PROLIFERATION WHILE OVEREXPRESSION OF MAN1C1 LEADS TO CELL CYCLE ARREST
To further study the effects of class I α‐1,2‐mannosidases on liver cancer cell proliferation, we performed a 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (a tetrazole) assay, which is a colorimetric assay to assess cell viability. Overexpression of MAN1A1 significantly increased the cell proliferation rate compared to the mock control in 293T cells (Fig. 4C). However, MAN1C1 overexpression did not influence cellular proliferation in Hep3B cells (Fig. 4D). To determine if the overexpression of MAN1C1 might affect the cell cycle, flow cytometry was used to observe various cellular aspects in the Hep3B cell lines. Overexpression of MAN1C1 in Hep3B cells resulted in a significantly decreased cell number in the S phase, about 8.8% compared to 17.1% in the mock Hep3B control, but no significant differences in the G1 and G2/M phases were observed (Fig. 4E,F). This cell cycle analysis suggests that overexpression of MAN1C1 acts on regulators of the S phase, causing fewer cells to enter the S phase and thereby resulting in cell cycle arrest.
MAN1A1 AND MAN1C1 HAVE OPPOSITE EFFECTS ON MMP9, PCNA, AND CCNA2 EXPRESSION IN CELL MIGRATION, PROLIFERATION, AND CELL CYCLE REGULATION
Matrix metalloproteinases (MMPs) are proteases that promote cancer cell growth, migration, invasion, and metastasis.31 MMP9, a member of the MMP family, is known to induce cancer32 and is highly correlated with liver cancer and metastasis in patients with HCC.33 To investigate how MAN1A1 and MAN1C1 affect cell migration, the mRNA levels of MMP9 were measured in MAN1A1‐overexpressing 293T stable cells and MAN1C1‐overexpressing Hep3B stable cells. Overexpression of MAN1A1 significantly induced MMP9 expression, while the overexpression of MAN1C1 significantly repressed MMP9 expression relative to the controls (Fig. 5A).
Figure 5.
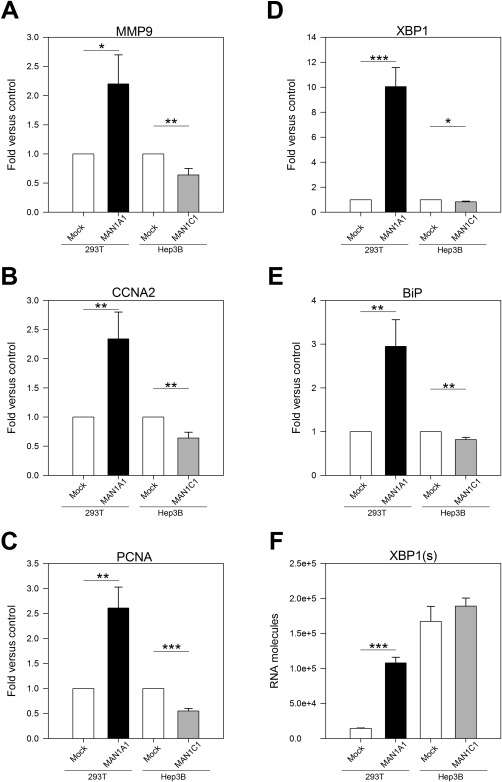
Expression patterns of MMP9, CCNA2, PCNA, and UPR regulator in MAN1A1 or MAN1C1‐overexpressing stable cell lines. (A) Overexpression of MAN1A1 induced MMP9 expression to 2.3‐fold, whereas overexpression of MAN1C1 repressed MMP9 expression to 0.33‐fold compared to the DsRed control (Mock). (B) CCNA2 (cell cycle marker) was up‐regulated in MAN1A1‐overexpressing stable cells and down‐regulated in MAN1C1‐overexpressing cells. (C) PCNA (cell proliferation marker) was up‐regulated in MAN1A1‐overexpressing cells and down‐regulated in MAN1C1‐overexpressing cells. (D) MAN1A1‐overexpressing cells displayed significantly enhanced expression of XBP1, and MAN1C1‐overexpressing cells displayed down‐regulated expression of XBP1. (E) MAN1A1‐overexpressing cells displayed significantly enhanced expression of BiP, and MAN1C1‐overexpressing cells displayed down‐regulated expression of BiP. (F) MAN1A1‐overexpressing cells exhibited higher levels of spliced XBP1 mRNA than mock; however, MAN1C1 overexpression did not appear to decrease XBP1(s) expression. Statistical significance was calculated using the Student t test (*P < 0.05, **P < 0.01, ***P < 0.001). All figures are results of triplicate experiments.
To examine whether MAN1A1 and MAN1C1 influence the expression of cell proliferation and cell cycle regulators, the mRNA levels of the cell cycle marker CCNA2 and the cell proliferation marker PCNA were measured in the MAN1A1‐overexpressing and MAN1C1‐overexpressing stable cells by qPCR. The results showed that CCNA2 (Fig. 5B) and PCNA (Fig. 5C) were significantly up‐regulated in MAN1A1‐overexpressing stable cells and down‐regulated in MAN1C1‐overexpressing stable cells.
UPR REGULATORS ARE ACTIVATED IN MAN1A1‐OVEREXPRESSING CELLS AND REPRESSED IN MAN1C1‐OVEREXPRESSING CELLS
α‐1,2‐mannosidases are mannose‐trimming enzymes that are involved in protein folding. MAN1B1 plays a critical role in the ERAD pathway, although studies have indicated that other mannosidases also affect this pathway.12 Thus, we hypothesized that not only MAN1B1 but also MAN1A1, MAN1A2, and MAN1C1 would affect ER stress. Unfolded or improperly folded proteins initiate the UPR through the activation of three proteins (IRE1, ATF6, and PERK), which in turn activate many downstream transcription factors and signal transduction machinery.34 As the UPR regulators are also transcriptionally regulated by the UPR8, the expression of three key UPR regulators, X‐box binding protein 1 (XBP1), IRE1, and BiP, were also analyzed in four different cell lines.
MAN1A1‐overexpressing cells showed significantly enhanced expression of XBP1 and BiP, while MAN1C1‐overexpressing cells exhibited down‐regulated expression of XBP1 and BiP (Fig. 5D,E), revealing a correlation between MAN1A1, MAN1C1, and ER stress. XBP1 mRNA is spliced and activated by the endonuclease activity of IRE1. Thus, we designed primers specific to detect the spliced form of XBP1 [XBP1(s)]. MAN1A1‐overexpressing cells exhibited higher levels of XBP1(s) than controls, whereas MAN1C1‐overexpressing cells did not appear to decrease the levels of XBP1(s) (Fig. 5F).
Our previous results showed that MAN1A1 overexpression activates the UPR signaling pathway and the expression of cell proliferation‐ and migration‐related genes. Thus, we treated MAN1A1‐overexpressing cells with ER stress inhibitors, including the chemical chaperone sodium 4‐phenylbutyrate (4‐PBA) and tauroursodeoxycholic acid (TUDCA), both of which ameliorate ER stress,35 to examine whether they can block or revert the expression of cell cycle‐ and migration‐related genes as well as the UPR regulators in the MAN1A1‐overexpressing cells. On treatment with the ER stress inhibitors 4‐PBA and TUDCA, the up‐regulation of these genes was reversed in Hep3B cells, PLC5 cells, HepG2 cells, and 293T cells (Supporting Fig. S2A‐D). Taken together, the data provide evidence that MAN1A1 overexpression induces the expression of proliferation‐ and migration‐related genes by activation of the UPR pathway.
Treatment of MAN1A1‐overexpressing Hep3B cells with the mannosidase inhibitor DMJ reverses the effects of MAN1A1‐overexpression‐mediated up‐regulation of the cell cycle‐ and migration‐related genes as well as the UPR regulators (Supporting Fig. S2E). DMJ treatment on MAN1C1‐overexpressing cells has an opposite effect on gene expression compared to that of MAN1A1‐overexpressing Hep3B cells (Supporting Fig. S2E,F). Taken together, these data provide further evidence that MAN1C1 functions differently than MAN1A1.
To determine whether MAN1A1 and MAN1C1 affect apoptosis, apoptosis protein markers phosphorylated‐ c‐Jun N‐terminal kinase and cleaved caspase 3 were analyzed by western blot in MAN1A1‐ and MAN1C1‐overexpressing cell lines. These results indicate that neither MAN1A1 nor MAN1C1 seem to affect apoptosis effector protein expression (Supporting Fig. S2G).
OVEREXPRESSION OF MAN1A1 INDUCES HEPATOCARCINOGENESIS IN ZEBRAFISH
During human hepatocarcinogenesis progression, the initial stage is steatosis, followed by cirrhosis, and eventually developing into cancer. Although many HBx transgenic mouse models developed HCC, there was no cirrhosis or fibrosis before HCC formation. HBx transgenic zebrafish in p53 mutant, src transgenic fish or edn1 transgenic fish developed steatosis to fibrosis, hyperplasia, and dysplasia prior to developing HCC.24, 26 Therefore, to study the effects of MAN1A1 and MAN1C1 on liver tumorigenesis in vivo, transgenic zebrafish expressing MAN1A1 or MAN1C1 under the control of the liver tissue‐specific promoter fabp10a were generated and examined for the progression of hepatocarcinogenesis.
MAN1A1 transgenic fish developed steatosis at 3 months, inflammation at 5 months, and hyperplasia at 7 months, whereas the MAN1C1 and enhanced green fluorescent protein (EGFP)‐mCherry transgenic control fish were normal (Fig. 6A). Overexpression of MAN1A1 initiated HCC at 7 months, with HCC lasting until 11 months (Fig. 6B,C). However, overexpression of MAN1C1 had no effect on the growth of hepatocytes (Fig. 6D), and the controls were almost normal except for a few fish exhibiting steatosis at 9 and 11 months (Fig. 6E). Staining with Oil Red O and Sirius Red (Supporting Fig. S3A,B) was used to verify the steatosis and fibrosis in the liver of the earlier stages of MAN1A1 transgenic fish, but steatosis and fibrosis were not observed in the MAN1C1 or EGFP‐mCherry transgenic fish. Using qPCR, the expression levels of lipogenic factors (pparg, chrebp, and srebp1; Supporting Fig. S4A), cell cycle/proliferation‐related genes (ccne1, cdk1, and cdk2; Supporting Fig. S4B), and UPR mediators (atf6, ern2, and xbp1; Supporting Fig. S4C) were examined. The lipogenic factors and UPR mediators were up‐regulated in MAN1A1 transgenic fish from 7 to 11 months but not in the MAN1C1 transgenic fish (Supporting Fig. S4A,C). Cell cycle/proliferation genes were up‐regulated in MAN1A1 transgenic fish from 7 to 11 months, which correlated with hyperplasia and HCC formation, but in the MAN1C1 transgenic fish, these genes were also up‐regulated (Supporting Fig. S4B). Because MAN1C1 is down‐regulated at the very early stage of human hepatocarcinogenesis, overexpression of MAN1C1 might be beneficial in the enhancement of hepatic activity, perhaps resulting in an increase in the expression of cell cycle and proliferation genes. Investigations into these effects are currently underway. In line with the previous histopathological data, the up‐regulation of the BiP protein was detected by IHC in the MAN1A1 transgenic fish but little BiP was detected in the MAN1C1 and EGFP‐mCherry transgenic fish (Supporting Fig. S5). This transgenic fish model provides additional evidence that MAN1A1 might act as an oncogene in vivo.
Figure 6.
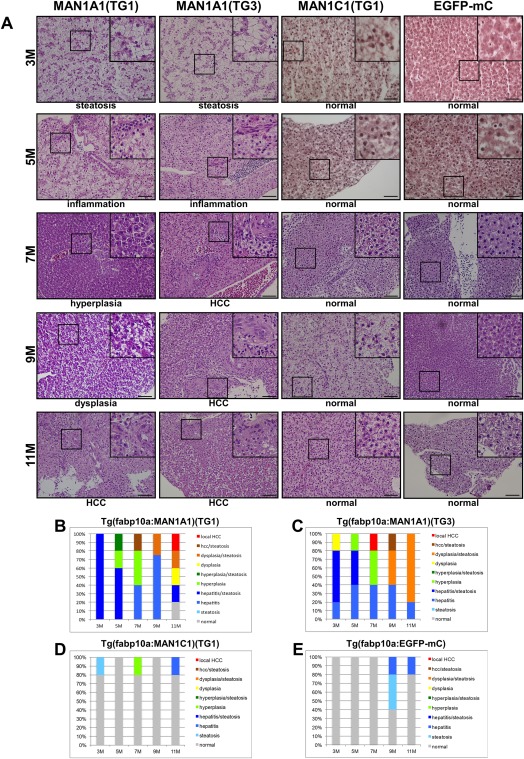
Histopathological analysis of hepatocytes in MAN1A1 and MAN1C1 transgenic fish at 3, 5, 7, 9, and 11 months of age. (A) Hematoxylin and eosin staining of liver sections from the TG1 and TG3 independent lines of MAN1A1 and MAN1C1(TG1) transgenic fish. Hematoxylin and eosin staining of the liver sections from MAN1A1 transgenic fish showed various pathological features, such as steatosis, inflammation, hyperplasia, dysplasia, and HCC. The MAN1C1 transgenic fish and EGFP‐mCherry control fish revealed normal phenotypes. The images were taken at magnification ×400, and scale shown is for 30 μm. (B) Statistical analysis of MAN1A1 (TG1), (C) MAN1A1 (TG3), (D) MAN1C1 (TG1), and (E) Tg(fabp10a:EGFP‐mCherry) is shown; n = 5 for each stage of transgenic fish.
TREATMENT WITH A MANNOSIDASE INHIBITOR, ER STRESS INHIBITORS, OR MAN1A1 shRNA REDUCED MAN1A1‐OVEREXPRESSION‐MEDIATED CELL PROLIFERATION IN THE XENOTRANSPLANTATION ASSAY
Our previous results indicated that the ER stress inhibitors 4‐PBA and TUDCA reversed the expression of cell cycle‐ and migration‐related genes as well as UPR regulators in the MAN1A1‐overexpressing cells. Using xenotransplantation, we verified the inhibitory function of these reagents in vivo. Figure 7A shows representative images of 1 day postinjection versus 3 days postinjection of embryos carrying MAN1A1‐overexpressing Hep3B cells with corresponding drug treatments (Fig. 7A). The treatment of cells with DMJ, 4‐PBA, and TUDCA, or shRNA knockdown of MAN1A1, significantly reduced the proliferation ability when assayed in the xenograft model (Fig. 7B). Quantification of CFSE intensity also showed the same phenomena (Fig. 7C). Taken together, these data provide in vivo animal model evidence that MAN1A1 overexpression promotes cellular proliferation through the activation of the UPR pathway in an in vivo animal model.
Figure 7.
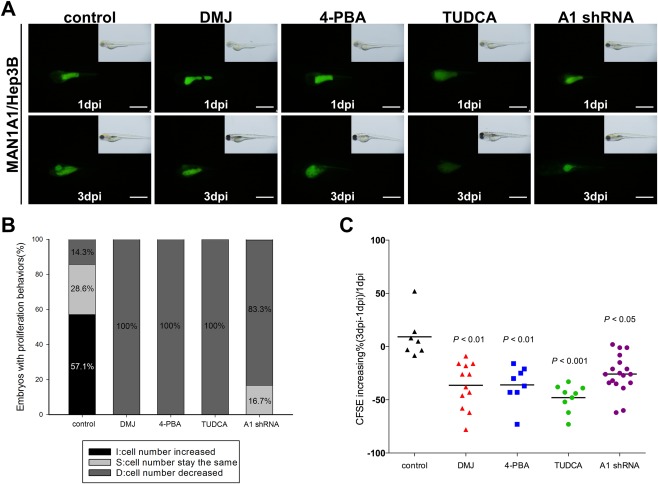
In vivo xenotransplantation assay for different inhibitors or MAN1A1 shRNA treatment in MAN1A1‐overexpressing cells. The cell lines were labeled with a CFSE florescent dye and injected into the yolks of 2‐day‐old zebrafish embryos. (A) Representative images of 1 day postinjection versus 3 days postinjection of MAN1A1 transiently overexpressing Hep3B cells treated with sterile water, DMJ, 4‐PBA, TUDCA, and MAN1A1 shRNA are shown from the same fish at 1 day postinjection and 3 days postinjection. (B) The percentage of embryos with increased, no change, or decreased number of injected CFSE‐dyed cells was analyzed using Image J and compared between 3 days postinjection and 1 day postinjection. (C) The proliferation ability of MAN1A1 transiently overexpressing Hep3B cells was effectively inhibited by the mannosidase inhibitors, ER stress inhibitor, and MAN1A1 shRNA. The data are presented as dot plots with a horizontal line for the mean. Experiments were repeated 3 times. Statistical significance was calculated using the Student t test (*P < 0.05, **P < 0.01, ***P < 0.001). Images were taken at magnification ×48, and the scale shown is for 1 mm. Abbreviation: dpi, dots per inch.
Discussion
HCC is the most common primary malignancy and the second leading cause of mortality worldwide,36, 37 with deaths caused by primary liver cancer affecting 0.8 million people annually worldwide.38 Traditionally, AFP was used as a marker for the detection of liver cancer, cirrhosis, and hepatitis; however, due to its low specificity and frequently obtained false‐positive results, AFP has been excluded as a diagnostic criterion.39 Therefore, the identification of reliable biomarkers for early diagnosis and the detection of metastasis is essential for saving lives as well as understanding the molecular mechanisms of hepatocarcinogenesis. We report here the identification of potential biomarkers for the diagnosis of HCC, including MAN1A1, MAN1A2, and MAN1B1, molecules that are up‐regulated at the later stages of HCC, and conversely, MAN1C1, which is down‐regulated at stage I of HCC.
Increased expression of α‐1,2 mannosidases has been associated with many cancers, including larynx cancer.19 More specifically, MAN1A1 is associated with breast cancer,40 MAN1A2 is a prognostic indicator in B cell lymphoma,41 and MAN1B1 is up‐regulated and promotes transformation phenotypes in HCC.18 In our study, the expression levels of MAN1A1, MAN1A2, and MAN1B1 were increased during the late stages of human liver cancer progression, whereas MAN1C1 was dramatically repressed at an early stage of liver cancer development (stage I). These distinct expression patterns of MAN1 genes in liver cancer represent novel findings. We demonstrated that MAN1A1 acts as an oncogenic factor in cell lines and in zebrafish and that MAN1C1 can suppress proliferation and migration in cell lines.
Our functional studies indicate that the overexpression of MAN1A1 promotes cell migration, cell proliferation, and enhances colony‐formation ability. We provide further in vivo evidence that, the overexpression of MAN1A1 in the liver results in HCC formation in MAN1A1 transgenic zebrafish. These data support the role of MAN1A1 in oncogenicity. The observations that MAN1C1 overexpression reduced cellular migration (both in vitro and in vivo), decreased colony‐formation ability, and resulted in a shortened S phase suggest that MAN1C1 may function as a tumor suppressor in the liver.
It has been reported that MAN1A1 is down‐regulated while MAN1C1 is up‐regulated in human HCC cell lines with different metastatic potentials (MHCC97L, MHCC97H, and HCCLM3) versus Hep3B, a nonmetastatic HCC cell line.42 However, patients with HCC do not exhibit a similar trend. The comparison between metastatic versus nonmetastatic cells was different from our study when comparing the absolute molecules of α‐1,2 mannosidases mRNA from different hepatoma cell lines (PLC5, Hep3B, and HepG2). MAN1A1, MAN1A2, and MAN1B1 were highly expressed in all three cell lines, while conversely, the expression of MAN1C1 was significantly low to undetectable in all three cell lines; the expression pattern of the cell lines correlated to patients with HCC. Moreover, our clinicopathology data showed that higher MAN1A1 levels were associated with tumors of a higher grade, larger size, and invasiveness (Supporting Table S1).
In this report, we propose that the aberrant expression of four α‐1,2 mannosidase subtypes activates UPR signaling during hepatocarcinogenesis. Abnormal glycosylation is associated with malignance, tumor progression, metastasis, and also results in UPR activation.40, 42, 43, 44 UPR is an important tightly regulated response required for cellular homeostasis, and it intersects with many other pathways that are critical for glucose metabolism, glycogen synthesis, lipid metabolism, inflammation, and metabolic disease.45 Recent studies have shown that the UPR plays a role in liver disease,46, 47 that its activation is associated with hepatic insulin resistance and fatty acid flux,48, 49 and that the UPR mediator XBP1 can up‐regulate hepatic lipogenic factor.47, 50 Our study suggests that overexpression of MAN1A1 leads to steatosis, inflammation, and HCC formation, possibly through the UPR, perhaps evoking the transcriptional networks involved in hepatocarcinogenesis.
Interesting results from this study include the differential expression patterns and functions of the α‐1,2‐mannosidases genes. MAN1B1 is a mannosidase found within the ER and trims residues from Man9GlcNAc2 to produce Man8GlcNAc2. MAN1A1, MAN1A2, and MAN1C1 are Golgi mannosidases that trim residues from Man8GlcNAc2 to produce Man5GlcNAc2. We discovered that the expression levels of α‐1,2 mannosidase subtypes correlate with the severity of liver cancer. In particular, our in vitro and in vivo functional studies provide evidence that MAN1A1 and MAN1C1 possess oncogenic‐ and tumor‐suppressor functions, respectively. Furthermore, we found that the cellular aspects affected by the dysregulation of α‐1,2 mannosidase genes are mediated through the UPR pathway. MAN1A1‐mediated cleavage of Man8GlcNAc2 to Man6GlcNAc2 occurs rapidly in vitro; however, MAN1A1 slowly cleaves Man6GlcNAc2 to Man5GlcNAc2.2, 43 We hypothesize that MAN1C1 is responsible for trimming the mannose from Man6GlcNAc2 to Man5GlcNAc2 in HCC. During the earlier stages of HCC, the down‐regulation of MAN1C1 causes modest ER stress due to the accumulation of uncleaved glycoprotein precursors. However, at the later stages the up‐regulation of MAN1A1, MAN1A2, or MAN1B1 results in more Man6GlcNAc2. Without MAN1C1, N‐glycosylation is dysregulated, which may lead to the accumulation of greater ER stress and eventually cancer. Active experiments are currently underway testing this hypothesis. We have performed the comparative glycomic analysis comparing MAN1A1 and MAN1C1 overexpression versus their knockdown. Our preliminary results indicated that MAN1A1 overexpression indeed increases the all mannose and Man6+7 as well as Man6GlcNAc2 but not Man5GlcNAc2 glycoproteins; shRNA knockdown MAN1A1 reverses the effect. Our preliminary discovery supports our hypothesis.
Our data also provide insight for future therapeutic interventions. Previous clinical trials using mannosidase inhibitors to treat cancer have been unsuccessful. However, our data suggest that for the effective treatment of liver cancer, specific inhibitors for MAN1A1, MAN1A2, and MAN1B1 must be generated while the introduction of a functional MAN1C1 protein may be important in rescuing a MAN1C1 deficiency. Experiments using synthetic MAN1C1 protein as a potential cancer therapy are currently underway.
Supporting information
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1002/hep4.1032/suppinfo.
Supporting Information.
Acknowledgment
We thank Dr. Micheline Laurent and Dr. Theodore Brummel for editing the manuscript. The fellowship to Miss Hsiao‐Chen Tu from the Liver Disease Prevention Research Foundation is acknowledged. We thank the Taiwan Zebrafish Core Facility at NTHU‐NHRI for providing fish lines and resources and the Taiwan Liver Cancer Network (TLCN) for providing the hepatocellular carcinoma tissue samples and related clinical data (all anonymously) for our research.
Potential conflict of interest: Nothing to report.
Supported by grants from the Ministry of Science and Technology (MOST) of the Republic of China (102‐2311‐B‐007‐008‐MY3, 105‐2311‐B‐007‐006) to H‐D.W. and (103‐2321‐B400‐022‐MY2, 105‐2314‐B‐400‐031‐MY2) to C‐H.Y. and from the National Health Research Institutes (NHRI) (MG‐105‐PP‐06) to C‐H.Y. The Taiwan Zebrafish Core Facility is supported by grant 105‐2319‐B‐400‐001 from the MOST. The Taiwan Liver Cancer Network is supported by grants from the National Science Council (NSC 94‐3112‐B‐182‐002, NSC 97‐3112‐B‐182‐004) and the NHRI.
Contributor Information
Horng‐Dar Wang, Email: hdwang@life.nthu.edu.tw.
Chiou‐Hwa Yuh, Email: chyuh@nhri.org.tw.
REFERENCES
- 1. Roth J, Zuber C, Park S, Jang I, Lee Y, Kysela KG, et al. Protein N‐glycosylation, protein folding, and protein quality control. Mol Cells 2010;30:497–506. [DOI] [PubMed] [Google Scholar]
- 2. Tempel W, Karaveg K, Liu ZJ, Rose J, Wang BC, Moremen KW. Structure of mouse Golgi alpha‐mannosidase IA reveals the molecular basis for substrate specificity among class 1 (family 47 glycosylhydrolase) alpha1,2‐mannosidases. J Biol Chem 2004;279:29774–29786. [DOI] [PubMed] [Google Scholar]
- 3. Barlowe CK, Miller EA. Secretory protein biogenesis and traffic in the early secretory pathway. Genetics 2013;193:383–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Herscovics A. Structure and function of class I alpha 1,2‐mannosidases involved in glycoprotein synthesis and endoplasmic reticulum quality control. Biochimie 2001;83:757–762. [DOI] [PubMed] [Google Scholar]
- 5. Molinari M. N‐glycan structure dictates extension of protein folding or onset of disposal. Nat Chem Biol 2007;3:313–320. [DOI] [PubMed] [Google Scholar]
- 6. Pan S, Cheng X, Sifers RN. Golgi‐situated endoplasmic reticulum alpha‐1, 2‐mannosidase contributes to the retrieval of ERAD substrates through a direct interaction with gamma‐COP. Mol Biol Cell 2013;24:1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rab A, Bartoszewski R, Jurkuvenaite A, Wakefield J, Collawn JF, Bebok Z. Endoplasmic reticulum stress and the unfolded protein response regulate genomic cystic fibrosis transmembrane conductance regulator expression. Am J Physiol Cell Physiol 2007;292:C756–766. [DOI] [PubMed] [Google Scholar]
- 8. Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 2011;334:1081–1086. [DOI] [PubMed] [Google Scholar]
- 9. Lu W, Hagiwara D, Morishita Y, Tochiya M, Azuma Y, Suga H, et al. Unfolded protein response in hypothalamic cultures of wild‐type and ATF6alpha‐knockout mice. Neurosci Lett 2016;612:199–203. [DOI] [PubMed] [Google Scholar]
- 10. Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 2016;529:326–335. [DOI] [PubMed] [Google Scholar]
- 11. Hazari YM, Bashir A, Haq EU, Fazili KM. Emerging tale of UPR and cancer: an essentiality for malignancy. Tumour Biol 2016;37:14381–14390. [DOI] [PubMed] [Google Scholar]
- 12. Hosokawa N, You Z, Tremblay LO, Nagata K, Herscovics A. Stimulation of ERAD of misfolded null Hong Kong alpha1‐antitrypsin by Golgi alpha1,2‐mannosidases. Biochem Biophys Res Commun 2007;362:626–632. [DOI] [PubMed] [Google Scholar]
- 13. Ogen‐Shtern N, Avezov E, Shenkman M, Benyair R, Lederkremer GZ. Mannosidase IA is in quality control vesicles and participates in glycoprotein targeting to ERAD. J Mol Biol 2016;428:3194–3205. [DOI] [PubMed] [Google Scholar]
- 14. Goss PE, Baker MA, Carver JP, Dennis JW. Inhibitors of carbohydrate processing: a new class of anticancer agents. Clin Cancer Res 1995;1:935–944. [PubMed] [Google Scholar]
- 15. Dennis JW, White SL, Freer AM, Dime D. Carbonoyloxy analogs of the anti‐metastatic drug swainsonine. Activation in tumor cells by esterases. Biochem Pharmacol 1993;46:1459–1466. [DOI] [PubMed] [Google Scholar]
- 16. Zhang Y, Zhao JH, Zhang XY, Guo HB, Liu F, Chen HL. Relations of the type and branch of surface N‐glycans to cell adhesion, migration and integrin expressions. Mol Cell Biochem 2004;260:137–146. [DOI] [PubMed] [Google Scholar]
- 17. Lu Y, Xu YY, Fan KY, Shen ZH. 1‐Deoxymannojirimycin, the alpha1,2‐mannosidase inhibitor, induced cellular endoplasmic reticulum stress in human hepatocarcinoma cell 7721. Biochem Biophys Res Commun 2006;344:221–225. [DOI] [PubMed] [Google Scholar]
- 18. Pan S, Cheng X, Chen H, Castro PD, Ittmann MM, Hutson AW, et al. ERManI is a target of miR‐125b and promotes transformation phenotypes in hepatocellular carcinoma (HCC). PLoS One 2013;8:e72829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Olszewska E, Borzym‐Kluczyk M, Rzewnicki I, Wojtowicz J, Rogowski M, Pietruski JK, et al. Possible role of alpha‐mannosidase and beta‐galactosidase in larynx cancer. Contemp Oncol (Pozn) 2012;16:154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gerber‐Lemaire S, Juillerat‐Jeanneret L. Studies toward new anti‐cancer strategies based on alpha‐mannosidase inhibition. Chimia (Aarau) 2010;64:634–639. [DOI] [PubMed] [Google Scholar]
- 21. Lu JW, Hsia Y, Yang WY, Lin YI, Li CC, Tsai TF, et al. Identification of the common regulators for hepatocellular carcinoma induced by hepatitis B virus X antigen in a mouse model. Carcinogenesis 2012;33:209–219. [DOI] [PubMed] [Google Scholar]
- 22. Ciou SC, Chou YT, Liu YL, Nieh YC, Lu JW, Huang SF, et al. Ribose‐5‐phosphate isomerase A regulates hepatocarcinogenesis via PP2A and ERK signaling. Int J Cancer 2015;137:104–115. [DOI] [PubMed] [Google Scholar]
- 23. Marques IJ, Weiss FU, Vlecken DH, Nitsche C, Bakkers J, Lagendijk AK, et al. Metastatic behaviour of primary human tumours in a zebrafish xenotransplantation model. BMC Cancer 2009;9:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu JW, Liao CY, Yang WY, Lin YM, Jin SL, Wang HD, et al. Overexpression of endothelin 1 triggers hepatocarcinogenesis in zebrafish and promotes cell proliferation and migration through the AKT pathway. PLoS One 2014;9:e85318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu JW, Yang WY, Lin YM, Jin SL, Yuh CH. Hepatitis B virus X antigen and aflatoxin B1 synergistically cause hepatitis, steatosis and liver hyperplasia in transgenic zebrafish. Acta Histochem 2013;115:728–739. [DOI] [PubMed] [Google Scholar]
- 26. Lu JW, Yang WY, Tsai SM, Lin YM, Chang PH, Chen JR, et al. Liver‐specific expressions of HBx and src in the p53 mutant trigger hepatocarcinogenesis in zebrafish. PLoS One 2013;8:e76951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu YL, Lu WC, Brummel TJ, Yuh CH, Lin PT, Kao TY, et al. Reduced expression of alpha‐1,2‐mannosidase I extends lifespan in Drosophila melanogaster and Caenorhabditis elegans. Aging Cell 2009;8:370–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blagosklonny MV. Prevention of cancer by inhibiting aging. Cancer Biol Ther 2008;7:1520–1524. [DOI] [PubMed] [Google Scholar]
- 29. Rouhi P, Jensen LD, Cao Z, Hosaka K, Lanne T, Wahlberg E, et al. Hypoxia‐induced metastasis model in embryonic zebrafish. Nat Protoc 2010;5:1911–1918. [DOI] [PubMed] [Google Scholar]
- 30. Progatzky F, Dallman MJ, Lo Celso C. From seeing to believing: labelling strategies for in vivo cell‐tracking experiments. Interface Focus 2013;3:20130001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Na Rev Cancer 2002;2:161–174. [DOI] [PubMed] [Google Scholar]
- 32. Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer 2003;3:401–410. [DOI] [PubMed] [Google Scholar]
- 33. Tien YW, Lee PH, Hu RH, Hsu SM, Chang KJ. The role of gelatinase in hepatic metastasis of colorectal cancer. Clin Cancer Res 2003;9:4891–4896. [PubMed] [Google Scholar]
- 34. Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 2007;8:519–529. [DOI] [PubMed] [Google Scholar]
- 35. de Almeida SF, Picarote G, Fleming JV, Carmo‐Fonseca M, Azevedo JE, de Sousa M. Chemical chaperones reduce endoplasmic reticulum stress and prevent mutant HFE aggregate formation. J Biol Chem 2007;282:27905–27912. [DOI] [PubMed] [Google Scholar]
- 36. Mazzanti R, Arena U, Tassi R. Hepatocellular carcinoma: where are we? World J Exp Med 2016;6:21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127:2893–2917. [DOI] [PubMed] [Google Scholar]
- 38. McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr 2016;7:418–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Song PP, Xia JF, Inagaki Y, Hasegawa K, Sakamoto Y, Kokudo N, et al. Controversies regarding and perspectives on clinical utility of biomarkers in hepatocellular carcinoma. World J Gastroenterol 2016;22:262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Milde‐Langosch K, Karn T, Schmidt M, zu Eulenburg C, Oliveira‐Ferrer L, Wirtz RM, et al. Prognostic relevance of glycosylation‐associated genes in breast cancer. Breast Cancer Res Treat 2014;145:295–305. [DOI] [PubMed] [Google Scholar]
- 41. Kim SJ, Sohn I, Do IG, Jung SH, Ko YH, Yoo HY, et al. Gene expression profiles for the prediction of progression‐free survival in diffuse large B cell lymphoma: results of a DASL assay. Ann Hematol 2014;93:437–447. [DOI] [PubMed] [Google Scholar]
- 42. Liu T, Zhang S, Chen J, Jiang K, Zhang Q, Guo K, et al. The transcriptional profiling of glycogenes associated with hepatocellular carcinoma metastasis. PLoS One 2014;9:e107941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vojta A, Samarzija I, Bockor L, Zoldos V. Glyco‐genes change expression in cancer through aberrant methylation. Biochim Biophys Acta 2016;1860:1776–1785. [DOI] [PubMed] [Google Scholar]
- 44. Liu Y, Ren S, Xie L, Cui C, Xing Y, Liu C, et al. Mutation of N‐linked glycosylation at Asn548 in CD133 decreases its ability to promote hepatoma cell growth. Oncotarget 2015;6:20650–20660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 2010;140:900–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vandewynckel YP, Laukens D, Devisscher L, Paridaens A, Bogaerts E, Verhelst X, et al. Tauroursodeoxycholic acid dampens oncogenic apoptosis induced by endoplasmic reticulum stress during hepatocarcinogen exposure. Oncotarget 2015;6:28011–28025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 2008;320:1492–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science 2006;313:1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yoshiuchi K, Kaneto H, Matsuoka TA, Kasami R, Kohno K, Iwawaki T, et al. Pioglitazone reduces ER stress in the liver: direct monitoring of in vivo ER stress using ER stress‐activated indicator transgenic mice. Endocr J 2009;56:1103–1111. [DOI] [PubMed] [Google Scholar]
- 50. Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab 2008;7:520–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1002/hep4.1032/suppinfo.
Supporting Information.


