Abstract
Metabolomics is a relatively new and rapidly growing area of post-genomic biological research. As use of metabolomics technology grows throughout the spectrum of drug discovery and development, and its applications broaden, its impact is expanding dramatically. This review seeks to provide the reader with a brief history of the development of metabolomics, its significance and strategies for conducting metabolomics studies. The most widely used analytical tools for metabolomics: NMR, LC–MS and GC–MS, are discussed along with considerations for their use. Herein, we will show how metabolomics can assist in pharmaceutical research studies, such as pharmacology and toxicology, and discuss some examples of the importance of metabolomics analysis in research and development.
Keywords: Metabolomics, drug discovery and development, ultraperformance liquid chromatography, mass spectrometry
History
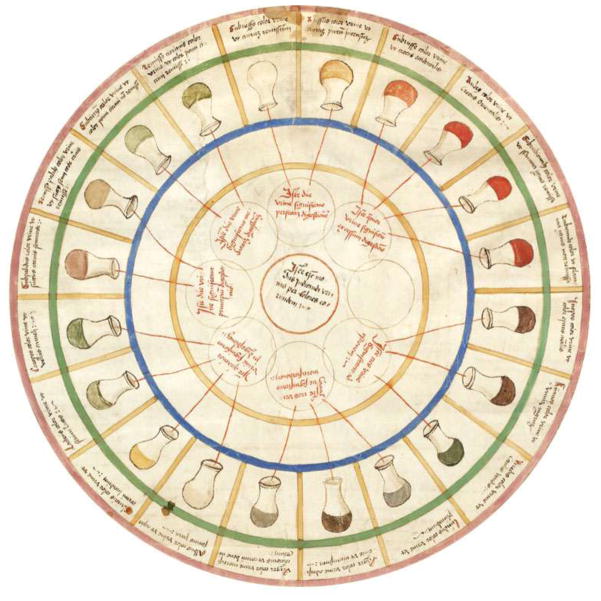
Metabolic profiling is not new; it has been used for clinical detection of human disease using blood and urine samples for centuries. In the days when people consulted with astrologers, or were diagnosed via combinations of their ‘humours’, the medical analysis of urine was the gold standard. The urine wheel from Epiphanie Medicorum by Ullrich Pinder in 1506 was the first application of crude science in medicine [1]. The urine wheel was used for diagnosing diseases based on the color, smell and taste of a patient’s urine in the early 16th century (Figure 1). The different colors and tastes of urine are due to mixtures of different inorganic salts and organic compounds that the body excretes as soluble waste. Diabetes mellitus was originally identified by the taste of sweet urine. Diagnostics through taste and smell might be a low-tech metabolomics approach but diabetes mellitus has been firmly linked to glucose metabolism ever since. After this original discovery type 2 diabetes and some subgroups of diabetes mellitus were identified.
Figure 1.
Urine Wheel for diagnosing metabolic diseases, from Epiphanie Medicorum by Ullrich Pinder, 1506. Reproduced, with permission, from Scientific American Blog Network http://blogs.scientificamerican.com/oscillator/the-urine-wheel/.
Ancient Arab, Hindu and Chinese texts also have anecdotal reports of the same sweet taste in urine from patients who displayed the symptoms of what was later termed diabetes. Urine wheels had been a standard inclusion in medical texts for centuries, and were gradually replaced by tables, test strips and microscope slides. Advanced chromatographic separation techniques were developed in the late 1960s. Linus Pauling published quantitative analysis of urine vapor and breath by gas–liquid partition chromatography in 1971 [2]. In the early 1970s gas chromatography–mass spectrometry (GC–MS) technologies were used to analyze steroids, acids and neutral and acidic urinary drug metabolites [3]. Metabolic profiling using GC–MS was reported [4,5]. Metabolic profiling studies were first applied to toxicology, pharmacology, inborn metabolic errors and nutritional function.
During the past two decades, great interest has developed in the application of metabolomics to characterize different pathological states of human diseases like cancer, diabetes, autoimmune and coronary diseases, among others. Metabolomics can provide valuable tools in a wide range of applications, such as pharmacology, toxicology, enzyme discovery and systems biology. As early as 2000, metabolomics was offered as a new technique with the potential to provide faster toxicity screening in pharmaceuticals; by 2005, five pharmaceutical firms and the Imperial College of Science, Technology and Medicine in London formed the Consortium for Metabonomic Toxicology (COMET) [6]. Their work established a metabolomics database that has expanded over the years, and is invaluable to their research, and its use has expanded to other areas such as chemical industry toxicology testing. The Metabolomics Society provides lists of available databases such as comprehensive metabolomic, metabolic pathway, compound or compound-specific, drug, spectral and disease and physiology databases (http://metabolomicssociety.org/resources/metabolomics-databases).
Significance of metabolomics
As these databases are expanded and refined, as well as our techniques and tools, this rapidly developing discipline is becoming a vital force, with the potential to transform pharmaceutical research. The value of metabolomics arises from the wide diversity of small molecules which, individually or in combinations, gives specific profiles for different human conditions including infectious, neoplastic, cardiovascular, neurological, metabolic and inflammatory diseases [7–9]. Metabolomics studies can help us to enhance our understanding of disease mechanisms and drug effects, as well as improving our ability to predict individual variation in drug response phenotypes [10–12]. Owing to its noninvasive nature, and close link with the phenotype, metabolomics is ideal for pharmaceutical research. For example, drug safety screens and biomarker discovery have already enhanced informed decision making. Personalization of analysis and treatment promises to allow us to track our own metabolome, with potential to target specific biomarkers, which will improve treatment strategies.
Animal models are an essential tool for researchers hoping to learn more about metabolic disease. In many cases, data cannot be collected from living patients with a metabolic disease, because this sometimes calls for organ dissection or other highly invasive procedures. Model animals can be engineered to express the disease phenotype and can be euthanized to collect data. This overcomes a major difficulty in collecting data from living patients; there is no need for dangerous and destructive invasive procedures such as organ dissection. As an example, one research group successfully developed a means of monitoring asthma status by metabolomics and urine sampling. They hypothesized NMR could be utilized to detect and analyze the unique patterns of urine metabolites produced during various states of airway dysfunction and inflammation, and subsequently measured by multivariate statistical analysis. Five groups of guinea pigs were studied: control, control treated with dexamethasone, sensitized (ovalbumin, administered intraperitoneally), sensitized and challenged (ovalbumin, administered intraperitoneally, plus ovalbumin aerosol), and sensitized–challenged with dexamethasone. For each group, the researchers measured airway hyper-reactivity (AHR) to histamine (administered intravenously) and inflammation. Their work showed that the challenged guinea pigs developed AHR and increased inflammation compared with sensitized or control animals, and that dexamethasone significantly improved AHR. This study demonstrates that urine metabolites correlate with airway dysfunction in an asthma model, making urine NMR analysis a promising, noninvasive technique for monitoring asthma in humans [13].
Study workflow design
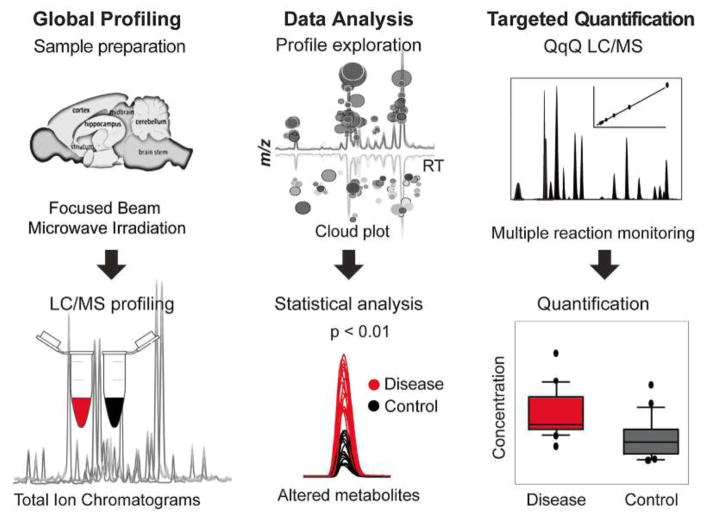
Figure 1 shows a proposed metabolomic strategy. After metabolism quenching, LC–MS data are acquired, followed by retention time correction data for chromatogram alignment and visualization of dysregulated metabolite features. Metabolite features whose levels were significantly changed in disease vs control samples are then filtered out and identified by MS/MS matching. The identified metabolites are quantified by targeted multiple reaction monitoring (MRM) analysis using standard compounds [14].
Sample preparation is one of the most important factors that determine the quality of metabolomics data [15]. Sampling affects the observed metabolite content and biological interpretation of the data. An ideal sample preparation method for global metabolomics should be as nonselective as possible to ensure adequate depth of metabolite coverage. Also, the best method will avoid metabolite loss and degradation during preparation, facilitate high throughput and be highly reproducible. A procedure that requires a minimum of steps, as well as the shortest practicable time, will help the researcher achieve these goals. One step that cannot be overlooked, in order to provide the most accurate metabolomics composition at the time of sampling, is that of metabolic quenching. Sample preparation methods must be able to facilitate the analysis of a variety of biological fluids, such as blood, urine, sputum, cerebrospinal fluid (CSF) and exhaled breath condensate in humans, or bacterial sources such as culture media. Urine is a more common matrix that requires relatively little sample preparation. For MS analysis, this involves dilution followed by centrifugation to remove particulates. Serum and plasma are two other relevant and highly valuable matrices that have long been used in diagnostic, health and drug monitoring studies. Preparation for ultra performance liquid chromatography (UPLC)–MS metabolomics analysis involves a methanolic protein precipitation technique, which is simple and effective in extracting all classes of metabolites present in serum or plasma samples. One advantage of using urine or plasma in drug toxicology experiments is that the sample is collected noninvasively so that it can be applied in clinical studies. Another advantage is that multiple biofluid samples from a single subject can be collected over a time course, which allows the determination of a metabolic trajectory that describes the toxic response and recovery period. The analysis of metabolites in biofluids permits a toxicological evaluation of the health of many different organs within the same animal over time and could allow simultaneous evaluation of drug efficacy from the same biofluid sample.
Thus, a successful protocol will allow removal of unwanted proteins and other particulates while retaining hydrophobic and hydrophilic metabolites, and quenching of the metabolism, at which point the samples are ready for analysis. Although the majority of the work done in pharmaceutical labs is on peripheral biofluids that can be obtained noninvasively, there are times when tissue metabolomics is essential to complete understanding of the system under study. There are a number of methods that are routinely employed to physically lyse cells, including mechanical disruption, liquid homogenization, high frequency sound waves, freeze/thaw cycles and manual grinding.
There are two approaches to metabolomics studies: targeted and untargeted. Target analysis has been applied for many decades and includes the determination and quantification of a small but rapidly expanding set of known metabolites using one particular analytical technique of best performance for the compounds of interest. Authentic standards are available in these studies. The untargeted approach is effectively used to identify a number of unknown metabolites by comparing the relative abundances of metabolites in multiple samples without prior identification [16,20]. The goals of untargeted metabolomics are to determine the metabolomic profiles of phenotypic diseases and healthy states, involving a complex set of genomic and proteomic changes. Untargeted metabolomics makes it possible to analyze hundreds of individual metabolites. Some applications of untargeted metabolomics include biomarkers, cardiovascular, diabetes and obesity disease research and drug discovery.
Platforms for metabolomics analysis
The most widely used analytical tools for metabolomic studies to identify large numbers of metabolites are proton NMR (1H-NMR) spectroscopy, GC–MS and LC–MS [16,20,21]. Hundreds of metabolites can be separated and measured in samples of interest such as plasma, CSF, urine or cell extracts using a diversity of commonly used metabolomics tools such as NMR, GC–MS and LC–MS detection. Each of these analytical tools carries its own advantages, disadvantages and applications as shown in Table 1 [22]. Table 1 shows the most common analytical tools for analysis of the various classes of compounds, such as lipids, carbohydrates, amino acids, organic acids, sugars, sugar phosphates, biogenic amines, nucleotides, vitamins, purines, fatty acids and steroids. It shows that no single tool is fit to analyze all compounds; generally, a combination of two or three analytical platforms is needed to identify the various stages of a disease and to differentiate diseases. To have the desired statistical power, it is proposed that a minimum of three biological replicates is required and five replicates are preferred to obtain reliable data. Overall, NMR has advantages such as reproducibility, the nondestructive nature of sample preparation and the comprehensive coverage of chemical species, whereas MS possesses much better sensitivity and resolution as well as high-throughput capacity. NMR scores well in terms of reproducibility, whereas MS techniques have the advantage of significantly improved sensitivity (Table 1).
Table 1.
Tools used in metabolomics studies
| Information/key point | NMR | LC MS | GC MS |
|---|---|---|---|
| Detected metabolites | Universal detector | Universal detector for ionization molecules | Universal detector for ionization molecules |
| Reproducibility | High | Lower than NMR | Lower than NMR |
| Sensitivity | Low sensitivity (mg) | High sensitivity (ng) | High sensitivity (pg) |
| Resolution | Low | High | High |
| Quantitativity | Yes | Yes, with IS | Yes, with IS |
| Sample preparation | Minimal | Extensive sample preparation steps | Extensive sample preparation steps |
| Destructive method | No | Yes | Yes |
| Structural information | Yes | Yes | Yes |
| No. of metabolites | 50–200 | 100–800 | 100–500 |
| Type of metabolite |
|
|
|
| Application | Metabolite profiling Metabolite flux analysis Metabolic fingerprinting |
Metabolite target analysis Metabolite profiling Metabolomics |
Metabolite target analysis Metabolomics |
| Cost | Expensive | Expensive | Non expensive |
Abbreviations: GC–MS, gas chromatography mass spectrometry.
Considerations when using NMR, LC–MS or GC–MS
Collection time for basic data: NMR 5 min; MS 10 min for UPLC–MS. More-comprehensive LC–MS methods involve run times of ~30 min, with the potential to detect 200 metabolites. Run times for NMR are about the same, with the ability to quantify 50 metabolites in samples.
The detection limits are in mg, ng, pg for NMR (600 MHz), LC–MS and GC–MS, respectively.
Metabolite detection: with NMR, if the metabolite contains hydrogen it will be detected assuming the concentration is sufficient and protein binding does not cause marked line-broadening. MS usually requires a more targeted approach. Although there can be problems with loss of metabolites in void volume with ion suppression, this is reduced when using UPLC. The ability to run negative and positive ion detection gives extra information. NMR cannot detect or identify salts and inorganic ions, whereas LC–MS and GC–MS can detect most organic and some inorganic molecules.
Sample handling and analysis: using NMR, the whole sample is analyzed in one measurement with no destruction of the samples during analysis. LC–MS requires samples to be extracted into a suitable solvent, as well as specific LC conditions for different classes of metabolites, whereas GC–MS requires derivatization. For GC–MS and LC–MS the samples are not recoverable.
Volume of sample required: NMR typically needs 200–400 μl; MS uses ~10 μl.
Reproducibility: NMR and GC–MS are excellent for separation reproducibility whereas LC–MS is fair.
Robustness: NMR and GC–MS are both robust and mature technologies; LC–MS is less so.
Analysis of tissue samples: NMR is compatible with liquids and solids, LC–MS can be used in metabolite imaging [matrix-assisted laser desorption/ionization (MALDI) or desorption electrospray ionization (DESI)], and GC–MS is compatible with gases and liquids.
Quantitative: NMR can quantify a wide range of organic compounds; GC–MS and LC–MS need calibration standards for quantitative analysis.
Identification of novel chemicals: NMR works well and most spectral features are identifiable. For LC–MS and GC–MS novel compound identification is difficult.
Instrument cost and lifetime: NMR has high costs >US$1 million; LC–MS is ~US$500 000; and GC–MS is ~US$250 000. NMR instruments have a life time of over 20 years, LC–MS ~8 years and GC–MS >10 years.
Metabolomics in drug discovery
Metabolomics has been applied in preclinical drug development studies such as pharmacology, toxicology and ADME [23–25]. One example of the use of metabolomics involves investigation of the lipid expression patterns of six anti-inflammatory compounds using principal component discriminant analysis [26]. The research group studied the effects on the transcriptome, proteome and metabolome of macrophage-like U937 cells when they were exposed to various anti-inflammatory drugs, including β2-adrenergic agonists. Using LC–MS, profiles of choline derivatives were also determined, including lysophosphocholines, phosphocholines, triglycerides and cholesterol esters. The study showed that three β2-adrenergic agonists employed: zilpaterol, clenbuterol and salbutamol, produced similar choline derivative changes in U937 cells compared to dexamethasone and SB203580 (a specific inhibitor of the p38/MAPK pathway). Subsequently, the data were used to characterize the pharmacological effects of a group of anti-inflammatory drugs in terms of their effects on target cell transcriptome, proteome and metabolome. The study concluded that the tested drugs produced no important marker molecules [26]. Another group studied the changes in lysophospholipids noted in patients with nonalcoholic steatohepatitis (NASH), which were consistent with changes that have been noted in hepatocellular carcinoma, showing a potential mechanistic progression pathway from NASH to carcinoma [27].
The pharmacology phase is one of the key components of the drug discovery process. Drugs fail in clinical trials because they either do not work or are not safe to patients. The prediction of therapeutic efficacy and adverse event risk on an individual patient basis through systems pharmacology is believed to have promise in reducing failure rates in Phase II and III clinical trials. In one such study, NMR was used to detect metabolic changes in cells and provide information about the mechanism of action of antimicrobial agents. In this study it was reported that, by using the mutant strain Aspergillus nidulans uaZ14 mutant and comparative NMR metabolomics data, 8-azaxanthine inhibits A. nidulans hyphal growth by in vivo inactivation of urate oxidase [28]. As another example, metabolomics research provided more-comprehensive information on pharmacogenetic metabolic phenotypes, such as the urinary metabolic pattern obtained after the administration of debrisoquine, as well as the phenotype for CYP2D6 polymorphism [29,30]. Metabolomic studies were employed to investigate urine of wild-type and Farnesoid X receptor (Fxr)-null mice fed cholic acid, an FXR ligand, using UPLC–MS. The studies revealed that several metabolites of corticosterone and cholic acid had been highly elevated in Fxr-null mice by cholic acid loading but not in wild-type mice [31].
Toxicity is a leading cause of attrition at all stages of the drug development process. Early indicators of drug safety are of intense interest to the pharmaceutical industry, which set up the Predictive Safety Testing Consortium comprising over 120 scientists from 16 drug companies who examine preclinical biomarkers for drug safety [32]. It has been estimated that the drug industry spends US$30 million a year in research to validate biomarkers for preclinical toxicity [33]. Also, it has been stated that predictive safety tests currently developed by individual companies are not useful to the agency because they have not been validated by an independent party. Metabolic profiling has the potential to identify toxicity early in the drug discovery process, which can save time and money for pharmaceutical companies [34].
Metabonomics methods were used in a preclinical study on a compound known to cause hepatotoxicity in several species [35]. NMR spectra were acquired on urine and liver tissue samples obtained from rats administered either a vehicle or a developmental compound (MrkA) that was shown to induce hepatotoxicity in several animal species. Results showed that urinary dicarboxylic aciduria is associated with defective metabolism of fatty acids; subsequent in vitro experiments confirmed that MrkA impairs fatty acid metabolism. Characterization of the urinary metabonome was the key step in rapidly ascertaining the probable mechanism of toxicity and led to the development of appropriate in vivo and in vitro experiments confirming that MrkA impaired fatty acid metabolism. This is a case where metabonomics was able to provide mechanistic insight to the hepatotoxicity of a drug [35].
Examples of the value of metabolomics analysis
An early diagnosis is crucial to reduce cancer mortality rates. In a recent study, a total of 58 human serum metabolites were collected from 15 esophageal cancer patients, 11 gastric cancer patients, 12 colorectal cancer patients and 12 healthy volunteers. Of these metabolites the resulting GC–MS chromatograms showed differences in levels for each type of cancer compared with those of the healthy group [36]. In another study [37], an untargeted approach was used to analyze the relative metabolite concentrations between pancreatic cancer patients and healthy controls by high-resolution, flow-injection Fourier transform ion cyclotron MS. Several metabolites including serum phospholipids and novel ultra-long-chain fatty acids showed significantly altered levels between diseased and healthy volunteers. Both of these studies are examples of detecting biomarker panels instead of a single target, which represents a systems approach in solving complex diseases.
The current state of testing for metabolic diseases like type 2 diabetes mellitus involves prognostic and diagnostic biomarkers, such as the oral glucose tolerance test and blood glucose level, which are well-defined. Insulin deficiency is an indicator that diabetes has probably already started; what is lacking is a set of biomarkers that can predict or detect the onset of such diseases. Metabolomics research has shown that early indications of type 2 diabetes include impaired glucose and amino acid and fatty acid metabolism. Wang et al. [38] studied alterations in amino acids over a 12-year period; their work produced a potential biomarker panel of amino acids which can detect those that were altered when compared with otherwise healthy volunteers. Because this alteration begins well before the onset of insulin depletion, such a panel has the potential to prove invaluable in identifying diabetes long before its onset.
Cancer is one of the leading causes of death in the world. Currently nearly 15 million people in the USA are either living with cancer or are cancer survivors [38]. In 2015 more than 1.65 million Americans will be diagnosed with cancer and 590 000 will die from it. Because cancer is such a widespread, pernicious disease that requires significant, long-term medical intervention, the economic costs are considerable. Current estimates of the cost of cancer care in the USA are pegged at US$150 billion per year and are expected to rise to nearly US$173 billion per year by 2020. Cancer is widely known as a genetic disease arising from mutations in key oncogenes or tumor suppressors. Table 2 outlines cancer and the discovery of oncometabolites. Oncometabolites are endogenous metabolites and their accumulation initiates or sustains tumor growth and metastasis [39]. The first oncometabolite to be discovered was 2-hydroxyglutarate, a natural metabolite that is found in high concentrations in gliomas. This compound seems to indirectly alter histone methylation patterns, which ultimately leads to tumorigenesis. Since the discovery of 2-hydroxyglutarate, many other oncometabolites have been identified. Another example is methionine; the mechanism of its possible antihepatotoxic activity is not entirely clear. High doses of acetaminophen in the liver are thought to lead to decreased levels of hepatic glutathione and increased oxidative stress. Antioxidant activity of L-methionine and some of its metabolites appear to account for its possible antihepatotoxic activity. L-methionine is a precursor to L-cysteine, which by itself can have antioxidant activity. L-cysteine, in turn, is a precursor to the antioxidant glutathione. Recent research also suggests that methionine acts as a free-radical scavenger by virtue of its sulfur, as well as its chelating ability (http://www.drugbank.ca/drugs/DB00134).
Table 2.
Oncometabolites and their roles in cancer
| Metabolitea | Mechanism | Refs |
|---|---|---|
| 2-Hydroxyglutarate |
|
[43,44] |
| Fumarate |
|
[44,45] |
| Succinate |
|
[46] |
| Sarcosine |
|
[47] |
| Glucose |
|
[48,49] |
| Glutamine |
|
[50,51] |
| Asparagine |
|
[52] |
| Choline |
|
[53] |
| Lactate |
|
[54,55] |
Abbreviations: mTOR, mammalian target of rapamycin; TCA, tricarboxylic acid.
All of the listed metabolites are required for tumor survival or tumor propagation. All are locally elevated in tumors but not in surrounding tissues. Most alter important cell signaling and cell division pathways. This table was reproduced, with permission, from [39].
Sarcosine was reported to activate prostate cancer cells and to indicate the malignancy of prostate cancer cells when measured in urine [40]. However, this conclusion has been disputed. A German research team reported a different result in 2010. After measuring sarcosine levels in urine samples from prostate cancer patients, they concluded that measuring sarcosine in urine fails as a marker in prostate cancer detection and identification of aggressive tumors. In addition, another report concluded that serum sarcosine is not a marker for prostate cancer. A review of the literature reached a similar conclusion [40]. When metformin was combined with tamoxifen the concentration of tamoxifen required for growth inhibition was substantially reduced. It shows that metformin and tamoxifen additively inhibited the growth and augmented the apoptosis of estrogen receptor (ER)-positive breast cancer cells. It provides leads for future research on this drug combination for the treatment of ER-positive breast cancer [41].
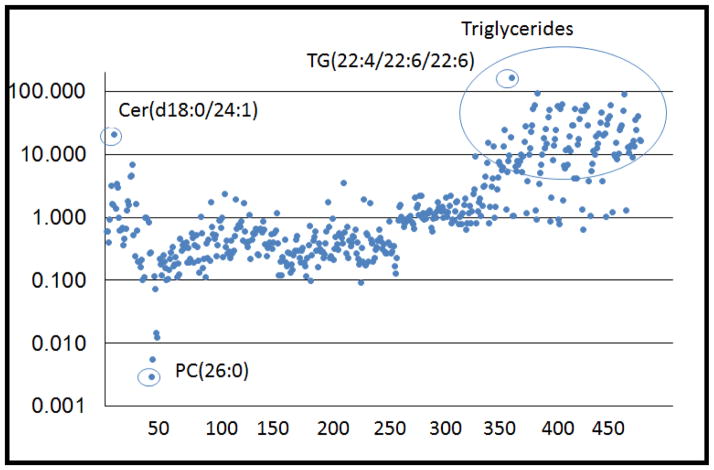
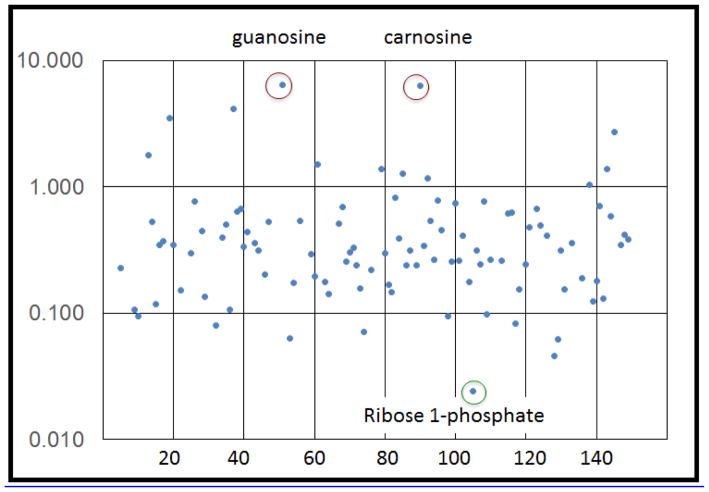
One recent example from our lab involves deciphering the effects of chemical exposures, which could ultimately help identify potential targets for preventing exposure-induced illness or post-exposure therapeutics. As a first step in our investigation, we undertook in vitro studies with the chemical allergen hexemethylene diisocyanate (HDI), which is well-recognized to cause occupational asthma. We assayed for changes in 472 lipid and 148 polar metabolites in the human monocyte-like cell line (U937) following incubation for 48 h in the presence or absence of HDI (pre-reacted with albumin to create antigenic changes) [42]. The relative change in the level of each of the different lipid and polar metabolites is depicted in Figures 3 and 4. As shown, HDI exposure caused substantial increases in trigylcerides and certain ceramides, but not other (e.g., phosho) lipids. Notably, the levels of the specific triglyceride (TG22:4/22:6/22:6) were over 100-fold increased whereas the levels of the phosphocholine (PC 26:0) were over 100-fold downregulated. Changes in polar metabolites included nearly tenfold increases in guanosine and carnosine, concomitant with relative decreases in ribose-1-phosphate. Further analyses of metabolite changes induced by HDI and other chemical exposures should help elucidate the pathways by which these exposures cause disease, and might identify targets for prevention and/or therapy.
Figure 3.
Changes in lipid metabolites induced by occupational allergen. U937 cells were incubated with hexemethylene diisocyanate (HDI)-conjugated human serum albumin for 48 h and the levels of different lipid metabolites were expressed as a ratio relative to control samples. Each dot represents a different lipid metabolite: 1–17 (ceramides); 18–24 (diglycerides); 25–37 (lysophosphatidylcholine); 38 (monoacylglyceride); 39–152 (phosphatidyl choline); 153–222 (phosphoethanolamine); 223–224 (phosphoglycerols); 225–235 (phosphatidylinositol); 236–238 (phosphoserines); 239–257 (phosphosphingolipids); 257–472 (triglycerides).
Figure 4.
Changes in polar metabolites induced by occupational allergen. U937 cells were incubated with hexemethylene diisocyanate (HDI)-conjugated human serum albumin for 48 h and the levels of different polar metabolites were expressed as a ratio relative to control samples. Each dot represents a different polar metabolite. Note the increase in guanosine and carnosine and decrease in ribose-1-phosphate.
Concluding remarks
Metabolomics is an emerging field with tremendous potential to improve our understanding of human health and disease, leading to the development of personalized approaches for disease diagnosis, patient monitoring and treatment response evaluation. Metabolomics has the potential to make a powerful impact in pharmaceutical and clinical research, including the identification of new targets, the elucidation of the mechanism of action of new drugs, the development of safety and efficacy profiles, as well as the discovery of biomarkers for early disease diagnosis and treatment. In addition, the identification and characterization of new biomarkers will allow early diagnosis and prevention of many diseases, as well as the discovery of new drugs.
Figure 2.
Metabolomic workflow. Global profiling summarizes the experimental design with respect to metabolism quenching and global LC–MS profiling of different sample groups. LC–MS data acquisition is followed by retention time correction for chromatogram alignment and visualization of dysregulated metabolite features. Metabolite features where levels were significantly changed in disease vs control samples are then filtered out and identified by MS/MS matching. The identified metabolites are quantified by targeted multiple reaction monitoring (MRM) analysis using standard compounds. Reproduced, with permission, from permission from [14].
Teaser.
The field of metabolomics is continually growing with our knowledge base of metabolites, analytical tools and bioinformatics approaches to analyze data. This rapidly developing new discipline has important potential implications for the pharmaceutical industry.
Highlights.
Metabolomics provides a very powerful tool in pharmaceutical and clinical research
Metabolomic biomarkers can improve personalized treatments
Metabolomics can help us understand drug candidate actions and target selection
Biofluid testing can facilitate the diagnosis and evaluation of treatment response
Acknowledgments
This research is supported by the R01OH0190941/CDC/NIOSH grant.
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nicholson JK, Lindon JC. Systems biology: metabonomics. Nature. 2008;455:1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 2.Pauling L, et al. Quantitative analysis of urine vapor and breath by gas liquid partition chromatography. Proc Natl Acad Sci U S A. 1971;68:2374–2376. doi: 10.1073/pnas.68.10.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devaux PG, et al. Benyzloxime derivative of steroids, a new metabolic profile procedure for human urinary steroids. Anal Lett. 1971;4:151. [Google Scholar]
- 4.Horning BC, Horning MG. Human metabolic profiles obtained by GC and GC/MS. J Chromatogr Sci. 1971;9:129–140. [Google Scholar]
- 5.Gates SC, Sweeley CC. Quantitative metabolic profiling based on gas chromatography. Clin Chem. 1978;24:1663–1673. [PubMed] [Google Scholar]
- 6.Lindon JC, et al. The Consortium for Metabonomic Toxicology (COMET): aims, activities and achievements. Pharmacogenomics. 2005;6:691–699. doi: 10.2217/14622416.6.7.691. [DOI] [PubMed] [Google Scholar]
- 7.Nicholson JK, Lindon JC. Origins of the field; difference metabonomics vs. metabolomics; metanomics beginnings; relation to systems biology; analytical techniques, pros/cons, interpretation; drawbacks; discoveries. Metabonomics. Nature. 2008;455:054–1056. [Google Scholar]
- 8.Kaddurah-Daouk R, et al. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 9.Collino S, et al. Clinical metabolomics paves the way towards future healthcare strategies. Br J Clin Pharmacol. 2013;75:619–629. doi: 10.1111/j.1365-2125.2012.04216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Ravenzwaay B, et al. The use of metabolomics for the discovery of new biomarkers of effect. Toxicol Lett. 2007;172:21–28. doi: 10.1016/j.toxlet.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 11.Mirsaeidi M, et al. Metabolomics: applications and promise in mycobacterial disease. Annal Am Thorac Soc. 2015;12:1278–1287. doi: 10.1513/AnnalsATS.201505-279PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puchades-Carrasco L, Pineda-Lucena A. Metabolomics in pharmaceutical research and development. Curr Opin Biotechnol. 2015;35:73–77. doi: 10.1016/j.copbio.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Adamko DJ, et al. A pilot study differentiating diseases. J Allergy Clin Immunol. 2015;136:571–580. doi: 10.1016/j.jaci.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Vanisevic J, Siuzdak G. The role of metabolomics in brain metabolism research. J Neuroimmune Pharmacol. 2015;10:391–395. doi: 10.1007/s11481-015-9621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vuckovic D. Current trends and challenges in sample preparation for global metabolomics using liquid chromatography–mass spectrometry. Anal Bioanal Chem. 2012;403:1523–1548. doi: 10.1007/s00216-012-6039-y. [DOI] [PubMed] [Google Scholar]
- 16.Reily MD, et al. LC-MS in endogenous metabolite profiling and small molecule biomarker discovery. In: Lee MS, Zhu M, editors. Mass Spectrometry in Drug Metabolism and Disposition: Basic Principles and Applications. John Wiley & Sons; 2011. pp. 685–722. [Google Scholar]
- 17.Guo B, et al. Liquid chromatography-mass spectrometric multiple reaction monitoring-based strategies for expanding targeted profiling towards quantitative metabolomics. Curr Drug Metab. 2012;13:1226–1243. doi: 10.2174/138920012803341401. [DOI] [PubMed] [Google Scholar]
- 18.Lei Z, et al. Mass spectrometry strategies in metabolomics. J Biol Chem. 2011;286:25435–25442. doi: 10.1074/jbc.R111.238691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward JL, et al. Recent applications of NMR spectroscopy in plant metabolomics. FEBS J. 2007;274:1126–1131. doi: 10.1111/j.1742-4658.2007.05675.x. [DOI] [PubMed] [Google Scholar]
- 20.Lu W, et al. Analytical strategies for LC-MS-based targeted metabolomics. J Chromatogr B. 2008;871:236–242. doi: 10.1016/j.jchromb.2008.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mishur RJ, Rea SL. Applications of mass spectrometry to metabolomics and metabonomics: detection of biomarkers of aging and of age-related diseases. Mass Spectrom Rev. 2012;31:70–95. doi: 10.1002/mas.20338. [DOI] [PubMed] [Google Scholar]
- 22.Martin JC, et al. Can we trust untargeted metabolomics? Results of the metaboring initiative, a large-scale, multi-instrument inter-laboratory study. Metabolomics. 2015;11:807–821. doi: 10.1007/s11306-014-0740-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nassar AEF, et al. Learning to improve the decision-making process in structural modification of drug candidates: reduce toxicity. Drug Discov Today. 2004;9:1055–1064. doi: 10.1016/S1359-6446(04)03297-0. [DOI] [PubMed] [Google Scholar]
- 24.Nassar AEF, Talaat R. Strategies for dealing with metabolite elucidation in drug discovery and development. Drug Discov Today. 2004;9:317–327. doi: 10.1016/S1359-6446(03)03018-6. [DOI] [PubMed] [Google Scholar]
- 25.Nassar AEF, Adams PE. Metabolite characterization in drug discovery utilizing robotic liquid-handling, quadrupole time-of-flight mass spectrometry and in silico prediction. Curr Drug Metab. 2003;4:259–271. doi: 10.2174/1389200033489406. [DOI] [PubMed] [Google Scholar]
- 26.Verhoeckx KC, et al. In search of secreted protein biomarkers for the anti-inflammatory effect of beta2-adrenergic receptor agonists: application of DIGE technology in combination with multivariate and univariate data analysis tools. J Proteome Res. 2005;4:2015–2023. doi: 10.1021/pr050183u. [DOI] [PubMed] [Google Scholar]
- 27.Clarke J, et al. Characterization of hepatocellular carcinoma-related genes and metabolites in human nonalcoholic fatty liver disease. Dig Dis Sci. 2014;59:365–374. doi: 10.1007/s10620-013-2873-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forgue P, et al. NMR metabolic profiling of Aspergillus nidulans to monitor drug and protein activity. J Proteome Res. 2006;5:1916–1923. doi: 10.1021/pr060114v. [DOI] [PubMed] [Google Scholar]
- 29.Mahgoub A, et al. Polymorphic hydroxylation of debrisoquine in man. Lancet. 1977;17:584–586. doi: 10.1016/s0140-6736(77)91430-1. [DOI] [PubMed] [Google Scholar]
- 30.Dorado P, et al. Dermination of debrisoquine and 4-hydroxydebrisoquine by high-performance liquid chromatography: application to the evaluation of CYP2D6 genotype and debrisoquine metabolic ratio relationship. Clin Chem Lab Med. 2005;43:275–259. doi: 10.1515/CCLM.2005.046. [DOI] [PubMed] [Google Scholar]
- 31.Cho JY, et al. Urinary metabolomics in Fxr-null mice reveals activated adaptive metabolic pathways upon bile acid challenge. J Lipid Res. 2010;51:1063–1074. doi: 10.1194/jlr.M002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodsaid FM, et al. The Predictive Safety Testing Consortium: a synthesis of the goals, challenges and accomplishments of the Critical Path. Drug Discov Today: Technol. 2007;2:47–50. doi: 10.1016/j.ddtec.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Finkelstein JB. FDA starts moving on Critical Path, but initiative running out of steam. JNCI. 2007;99:426–427. doi: 10.1093/jnci/djk142. [DOI] [PubMed] [Google Scholar]
- 34.Altar CA. The Biomarkers Consortium: on the critical path of drug discovery. Clin Pharmacol Ther. 2008;83:361–364. doi: 10.1038/sj.clpt.6100471. [DOI] [PubMed] [Google Scholar]
- 35.Mortshire-Smith RJ, et al. Use of metabonomics to identify impaired fatty acid metabolism as the mechanism of a drug-induced toxicity. Chem Res Toxicol. 2004;17:165–173. doi: 10.1021/tx034123j. [DOI] [PubMed] [Google Scholar]
- 36.Ritchie SA, et al. Metabolic system alterations in pancreatic cancer patient serum: potential for early detection. BMC Cancer. 2013;13:416. doi: 10.1186/1471-2407-13-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tabak AG, et al. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet. 2009;373:2215–2221. doi: 10.1016/S0140-6736(09)60619-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang TJ, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wishart DS. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Drug Discov. 2016;15:473–484. doi: 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- 40.Ma J, et al. Metformin enhances tamoxifen-mediated tumor growth inhibition in ER-positive breast carcinoma. BMC Cancer. 2014;14:172. doi: 10.1186/1471-2407-14-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Struys EA, et al. Serum sarcosine is not a marker for prostate cancer. Annal Clin Biochem. 2010;47:282. doi: 10.1258/acb.2010.009270. [DOI] [PubMed] [Google Scholar]
- 42.Wisnewski AV, et al. Isocyanate vapor-induced antigenicity of human albumin. J Allergy Clin Immunol. 2004;113:1178–1184. doi: 10.1016/j.jaci.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 43.Yang M, et al. Oncometabolites: linking altered metabolism with cancer. J Clin Invest. 2013;123:3652–3658. doi: 10.1172/JCI67228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang M. The emerging role of fumarate as an oncometabolite. Front Oncol. 2012;2:85. doi: 10.3389/fonc.2012.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morin A. Oncometabolites-driven tumorigenesis: from genetics to targeted therapy. Int J Cancer. 2014;135:2237–2248. doi: 10.1002/ijc.29080. [DOI] [PubMed] [Google Scholar]
- 46.Khan AP, et al. The role of sarcosine metabolism in prostate cancer progression. Neoplasia. 2013;15:491–501. doi: 10.1593/neo.13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen KT, et al. AMPA receptor–mTOR activation is required for the antidepressant-like effects of sarcosine during swim tests in rats: insertion of AMPA receptor may play a role. Front Behav Neurosci. 2015;9:162. doi: 10.3389/fnbeh.2015.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee AS. Glucose-regulated proteins in cancer: molecular mechanisms and therapeutic potential. Nat Rev Cancer. 2014;14:263–276. doi: 10.1038/nrc3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolf A, et al. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med. 2011;208:313–326. doi: 10.1084/jem.20101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller DM, et al. c-Myc and cancer metabolism. Clin Cancer Res. 2012;18:5546–5553. doi: 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J, et al. Asparagine plays a critical role in regulating cellular adaptation to glutamine depletion. Mol Cell. 2014;56:205–218. doi: 10.1016/j.molcel.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Awwad HM, et al. The role of choline in prostate cancer. Clin Biochem. 2012;45:1548–1553. doi: 10.1016/j.clinbiochem.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 54.Choi SY, et al. Cancer-generated lactic acid: a regulatory, immunosuppressive metabolite? J Pathol. 2013;230:350–355. doi: 10.1002/path.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gillies RJ, Gatenby RA. Metabolism and its sequelae in cancer evolution and therapy. Cancer J. 2015;21:88–96. doi: 10.1097/PPO.0000000000000102. [DOI] [PMC free article] [PubMed] [Google Scholar]