Abstract
Background
Bovine babesiosis is one of the most economically important tick-borne diseases threatening the livestock industry globally including South Africa. This disease is induced by members of Babesia bovis species. Antigenic variations among geographical strains of B. bovis, and these heterogeneities are cited as the mechanism by which parasites evade from host immune system and they hamper the successful development of a single vaccine that could confer absolute protection. Given the economic importance of livestock industry in South Africa, the extent of genetic diversity among field isolates of B. bovis merits extensive investigation.
In this study, we genetically characterized partial genes of B. bovis and studied the phylogenetic relationship among B. bovis isolates of South African origin. The genes, which were PCR-amplified from bovine samples collected from different locations across South Africa, coded for rhoptry-associated protein 1 (BbRAP-1), cysteine peptidase 2 (BbCP2), spherical body protein 4 (BbSBP-4) and β-tubulin (BbβTUB). Phylogenies were inferred from newly determined sequences using the neighbour-joining approach.
Results
Nested PCR assays with gene-specific primers indicated that, of the 54 bovine samples tested, 59.3% (32/54; 95% CI = 46.0–71.3%), 27.8% (15/54; 95% CI = 17.6–40.9%), 37.0% (20/54; 95% CI = 25.4–50.4%) and 29.6% (16/54; 95% CI = 19.1–42.8%) possessed BbRAP-1, BbCP2, BbSBP-4 and BbβTUB fragments, respectively. Sequencing of PCR-generated fragments revealed that nucleotide sequences of each of the four genes were highly conserved among the B. bovis isolates examined. Phylogenetic analyses of BbCP2, BbSBP-4 and BbβTUB sequences indicated a close phylogenetic relatedness among South African-derived sequences and those of global B. bovis strains.
Conclusion
The data reported in this study indicated that there is a high conservation among the genes of B. bovis isolates from cattle in South Africa. These findings give an indication that immunologically important proteins encoded by these genes could potentially be considered for exploitation as viable candidates for inclusion in recombinant subunit vaccines.
Keywords: Babesia bovis, Cattle, South Africa, Nested PCR assays, Phylogenies
Background
Babesia bovis is an intraerythrocytic protozoan parasite and an important aetiological agent of bovine babesiosis. This tick-borne disease is considered an economically important constraint to livestock production in tropical and subtropical regions of the world [1]. While B. bigemina (another agent of babesiosis) can be transmitted by both Rhipicephalus microplus and R. decoloratus, the confirmed tick vector of B. bovis in southern Africa is R. microplus [2].
Cattle infected by B. bovis are characterized by high fever, ataxia, anorexia and sometimes nervous signs [3]. Further, B. bovis remains the most pathogenic microorganism compared to other Babesia species and induces a more severe disease due to the sequestration of infected red blood cells in cerebral capillaries, thus resulting in parasitaemia levels of less than 1% in the circulating blood [4, 5]. On the other hand, the disease caused by B. bigemina is milder and is characterized by anaemia, fever and haemoglobinuria. Infections by B. bigemina do not involve sequestration of infected red blood cells and the levels of parasitaemia in blood often exceed 10% [3]. Therefore, it remains pivotal that effective control measures are implemented in an endeavour to curtail cattle mortalities resulting from Babesia infections.
In South Africa, vaccination against B. bovis is a method that is widely exploited to immunize cattle against babesiosis. This follows that the cattle which recover from primary acute infection remain persistently infected and serve as reservoirs for parasite transmission to other animals [4]. Nevertheless, it is worth noting that the available vaccines are based on bovine blood infected with live attenuated strains of the parasite [6, 7]. Although these vaccines confer protective immunity, the major limitation associated with the use of attenuated vaccines relate to the possible transmission of other blood-borne pathogens [6]. In addition, the production of live vaccines requires artificial infection of cattle in order to attain high parasitaemia levels in the blood, and this often constitutes important ethical implications [8].
According to Torina et al. [9], the success in the development of vaccines against babesial infections is impeded by the presence of heterogeneities in some parasite proteins. In particular, studies are focused on surface proteins that can confer high level of protection and better safety in comparison to currently used vaccines [10]. Many of these proteins might play a crucial role in erythrocyte invasion and are therefore the targets for vaccine development [9]. Several genes encoding these B. bovis proteins with immunogenic potential are known, and thanks to the complete genome sequencing of an American B. bovis T2Bo strain that these surface proteins were identified [11]. However, it must be acknowledged that there is still a dearth of information regarding the conservation of genes encoding potential immunogenic proteins in South African isolates of B. bovis.
In this context, the present study was borne out of the need to characterize the genes encoding rhoptry-associated protein 1 (BbRAP-1), cysteine peptidase 2 (BbCP2), spherical body protein 4 (BbSBP-4) and β-tubulin (BbβTUB) in B. bovis isolates from South African field bovine samples.
Methods
Blood samples
Field bovine samples (n = 54) were randomly selected to validate the nested PCR assays developed in this study. These samples form part of the sample collection of the Veterinary Parasitology Programme (National Zoological Gardens of South Africa: NZG, South Africa), and the collection of these samples was approved by the NZG Research Ethics and Scientific Committee. Animal owners gave verbal informed consent for the collection of the samples for the epidemiological survey, and animals were not involved in any clinical trials or treatments. All blood samples were maintained at −20°C prior to DNA extraction.
DNA extraction
Genomic DNA was extracted using ZR Genomic DNA™-Tissue MiniPrep kit (Zymo Research Corporation, Irvine, CA, USA) according to the manufacturer’s instructions. DNA quantification was performed using a NanoDrop® ND-1000 (NanoDrop Technologies Inc., Wilmington, USA).
Primer design
Nested PCR primers used for amplifying B. bovis-specific BbRAP-1 genes from bovine samples were reported previously [12]. Primers targeting BbCP2, BbSBP-4 and BbβTUB genes were created with Primer-BLAST program of the NCBI using reference sequences BBOV_IV007730 (GenBank accession number XM_001610645), BBOV_IV005390 (XM_001610418) and BBOV_III004850 (XM_001611566), respectively. All primers shown in Table 1 were synthesized by Inqaba Biotechnical Industries (Pretoria, South Africa).
Table 1.
Primer sets used to amplify DNA fragments specific for Babesia bovis
| Gene | Assay | Primer name | Oligonucleotide primers (5′ → 3′) | Annealing | Product sizea | Reference |
|---|---|---|---|---|---|---|
| BbRAP-1 | PCR | BoF | F-CACGAGGAAGGAACTACCGATGTTGA | 55 °C | 360 bp | [12] |
| BoR | R-CCAAGGAGCTTCAACGTACGAGGTCA | [12] | ||||
| nPCR | BoFN | F-TCAACAAGGTACTCTATATGGCTACC | 57 °C | 298 bp | [12] | |
| BoRN | R-CTACCGAGCAGAACCTTCTTCACCAT | [12] | ||||
| BbCP2 | PCR | CpBovF | F-TGCATCGGACCTATCCAACC | 57 °C | 960 bp | This study |
| CpBovR | R-TCAGCAGCCAAATAAGGCCA | This study | ||||
| nPCR | CpBov3 | F-ATCGGAAGAAGTCGCCGTTG | 65 °C | 829 bp | This study | |
| CpBov4 | R-AAGCGTAGTCGCTGTAACCA | This study | ||||
| BbSBP-4 | PCR | BbSBP1 | F-AGTTGTTGGAGGAGGCTAAT | 57 °C | 887–905 bp | This study |
| BbSBP2 | R-CTTCTCGGCGTCCTTTTC | This study | ||||
| nPCR | BbSBP3 | F-CCGCATTCTTAAGACTTCTGA | 60 °C | 726–744 bp | This study | |
| BbSBP4 | R-GTTACCATTTCATCGTTGTCA | This study | ||||
| BbβTUB | PCR | BTbovA | F-AGAGCGGTACTTACCACGGA | 61 °C | 1203 bp | This study |
| BTbovB | R-CGTCGTCGATGGTTGCTTCT | This study | ||||
| nPCR | BTbovC | F-GTTCCACGCGCTGTACTCAT | 65 °C | 954 bp | This study | |
| BTbovD | R-CATGTCCTGGATGGCGGTAG | This study |
aTheoretical product sizes based on nucleotide gene sequences of several B. bovis strains used as templates for primer design
Amplification of genes
PCR was performed in a 25-μl reaction containing 2.5 μl of the extracted DNA template, 0.6 μM of each primer and 12.5 μl of 2× DreamTaq Green PCR Master Mix (Inqaba Biotechnical Industries). The reactions were subjected to thermal conditions in a Bio-Rad T100™ thermal cycler (Bio-Rad Laboratories, Johannesburg, South Africa) with the following temperature profiles: 94 °C for 3 min, followed by 35 cycles of 94°C for 30 s, 55–65 °C for 45 s and 72 °C for 1 min. This was followed by a final extension at 72 °C for 10 min. The specific annealing temperatures are shown in Table 1. For nested PCR, 1 μl of the amplified PCR products was used as a template. Nested PCR products were subjected to gel electrophoresis in 1.2% (w/v) agarose gels stained with Biotium GelRed Acid Stain (Anatech Instruments, Johannesburg, South Africa) and visualized under ultraviolet illumination. GeneRuler 100-bp DNA Ladder (Inqaba Biotechnical Industries) was used as the standard molecular weight marker.
DNA sequencing and phylogenetic analysis
Nested PCR products of positive samples, and whose GenBank accession numbers appear in Table 2, were selected for subsequent sequencing of BbCP2, BbSBP-4 and BbβTUB genes. To corroborate correct amplification of BbRAP-1 genes, 12 positive samples were sequenced. Nucleotide sequences were determined with ABI 3130XL Genetic Analyzer (Applied Biosystems, Johannesburg, South Africa) using a Big Dye Terminator Kit (Applied Biosystems). For each gene, DNA sequencing was performed on both strands using the corresponding forward and reverse primers. Multiple sequence alignments were performed using a Biological Sequence Alignment Editor [13]. Neighbour-joining trees were created using MEGA v5.0 software [14]. Molecular distances were estimated with Kimura two-parameter model [15], and the robustness of branches was determined using bootstrapping analysis with 1000 replicates [16]. Pairwise comparisons of nucleotide sequences were performed with EMBOSS Needle program (version 6.3.1) of the European Bioinformatics Institute (http://www.ebi.ac.uk/tools/psa/emboss_needle). Nucleotide sequences were translated to protein sequences using ExPASy translate tool (http://web.expasy.org/translate).
Table 2.
GenBank accession numbers of nucleotide sequences generated in this study
| Province | Sample ID | GenBank accession number | ||
|---|---|---|---|---|
| BbCP2 | BbSBP-4 | BbβTUB | ||
| KwaZulu-Natal | KZN-C1 | – | KF626629 | – |
| KZN-C2 | KF626619 | KF626630 | KF626639 | |
| INDIA | KF626620 | KF626631 | KF626640 | |
| Mpumalanga | MP-C16 | KF626621 | – | – |
| MP-C17 | – | – | KF626641 | |
| MP-C18 | KF626622 | KF626632 | KF626642 | |
| Western Cape | WC-10270 | KF626623 | KF626633 | KF626643 |
| Gauteng | GP-C7 | KF626624 | KF626634 | KF626644 |
| GP-C15 | – | KF626635 | – | |
| GP-C17 | KF626625 | KF626636 | KF626645 | |
| Eastern Cape | EC-23A | KF626626 | – | – |
| North West | NW-C2 | KF626627 | KF626637 | KF626646 |
| NW-C4 | KF626628 | KF626638 | KF626647 | |
(−-) No sequences generated
Nucleotide sequence accession numbers
Nucleotide sequences determined in this study were deposited in GenBank under the accession numbers KF626619 – KF626647.
Results
Amplification of target genes by PCR
The nested PCR primers specifically designed to detect B. bovis DNA in bovine samples yielded single amplicons of 298 bp for BbRAP-1, 829 bp for BbCP2, 726–744 bp for BbSBP-4 and 954 bp for BbβTUB. Nested PCR assays with primers targeting four different genes were able to detect B. bovis DNA from some of the tested bovine samples. For example, BoFN/BoRN primers detected the BbRAP-1 genes in 32 out of 54 samples tested (59.3%). On the other hand, a total of 15 (27.8%), 20 (37.0%) and 16 (29.6%) samples possessed BbCP2, BbSBP-4 and BbβTUB fragments, respectively. In testing the specificity of PCR and nested PCR assays, purified DNA samples of B. bigemina, Anaplasma centrale, Ehrlichia ruminantium and Theileria parva were employed as negative controls, and as such, they yielded no PCR amplifications.
In order to corroborate if the correct PCR fragments were amplified, 12 samples possessing the BbRAP-1 gene and representing seven provinces were sequenced. The resulting nucleotide sequences were subjected to homology searches in GenBank. The BbRAP-1 sequences determined in this study exhibited 99–100% identity with published sequences of B. bovis strains originating from Philippines (JX860283), Brazil (FJ588009 to FJ588013 and AF030057 to AF030058), Uruguay (AF030060 to AF030061), Argentina (AF030053 and AF030056), USA (AF030054 and AF030059) and Cuba (JF279443).
In silico analysis of B. bovis sequences
Among the samples showing positive PCR amplifications, samples whose GenBank accession numbers appear in Table 2 were selected for sequencing. Alignment of 10 BbCP2 gene sequences determined in this study revealed the existence of single nucleotide polymorphisms (SNPs) between the sequences. The sequences from NW-C4, GP-C7, GP-C17 and MP-C16 shared 100% identity, and contained 5 nucleotide differences in relation to MP-C18 sequence. Similarly, the sequences from EC-23A, NW-C2 and WC-10270 shared 100% identity, while the highest identity recorded with sequences from KZN-C2 and INDIA was 98.9% (9 nucleotide differences). A blastn search in GenBank exhibited that the sequences determined in this study were most closely related to B. bovis strains identified in cattle from other parts of the world. In particular, our sequences exhibited 99% identity with sequences of B. bovis strains R1A (GQ412131), M3P (GQ412133), S2P (GQ412136), Veracruz (GQ412135), Uruguay (GQ412134) and Brazil (GQ412132).
Pairwise comparison of the BbSBP-4 sequences using EMBOSS Needle program indicated a close genetic similarity between sequences of South African B. bovis isolates and world strains. As shown in Table 3, the BbSBP-4 sequences from samples designated KZN-C2 and WC-10270 shared 100% identity with sequences of B. bovis strains previously identified in cattle from Ghana (AB569301) and South Africa (AB569303). The BbSBP-4 sequence of GP-C17 indicated 99.9% identity with the sequence of INDIA sample and 100% identity with that of KZN-C1. In an in silico analysis of BbSBP-4 gene sequences derived from MP-C18, NW-C4, reference strain (XM_001610418) and other world strains (Fig. 1a), it was worth noting that these sequences possessed a gap with 18 nucleotides missing in relation to sequences originating from KZN-C2, WC-10270 and two other published sequences (AB569301 and AB569303). A similar trend was observed after translating nucleotide sequences to protein sequences; the latter strains also possessed a gap with six amino acid residues missing (Fig. 1b). Similarly, the BbSBP-4 sequences from NW-C2, GP-C7, GP-C15, GP-C17, INDIA and KZN-C1 had 9 nucleotides missing (Fig. 1a), sharing between 98.1 and 98.8% sequence identities. Despite the high percentage identities observed among closely related sequences, there were SNPs occurring at different positions across the length of BbSBP-4 gene sequences (Fig. 1a).
Table 3.
Pairwise comparisons of the BbSBP-4 nucleotide gene sequences of South African Babesia bovis isolates and GenBank strains
| Isolate origin | 01 | 02 | 03 | 04 | 05 | 06 | 07 | 08 | 09 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| South Africa | 01 | 100 | 100 | 94.5 | 94.5 | 94.2 | 94.4 | 94.4 | 93.7 | 94.4 | 94.5 | 94.5 | 94.5 | 94.8 | 94.8 | 98.3 | 98.1 | 98.3 | 98.1 | 98.3 | 98.8 | 100 | 100 |
| Ghana | 02 | 100 | 94.5 | 94.5 | 94.2 | 94.4 | 94.4 | 93.7 | 94.4 | 94.5 | 94.5 | 94.5 | 94.8 | 94.8 | 98.3 | 98.1 | 98.3 | 98.1 | 98.3 | 98.8 | 100 | 100 | |
| Brazil | 03 | 100 | 100 | 99.7 | 99.9 | 99.9 | 99.2 | 99.9 | 100 | 100 | 100 | 98.6 | 98.6 | 96.1 | 95.9 | 96.1 | 96.2 | 96.2 | 95.6 | 94.5 | 94.5 | ||
| Japan | 04 | 100 | 99.7 | 99.9 | 99.9 | 99.2 | 99.9 | 100 | 100 | 100 | 98.6 | 98.6 | 96.1 | 95.9 | 96.1 | 96.2 | 96.2 | 95.6 | 94.5 | 94.5 | |||
| Mongolia | 05 | 100 | 99.6 | 99.6 | 99.2 | 99.6 | 99.7 | 99.7 | 99.7 | 98.3 | 98.3 | 95.8 | 95.6 | 95.8 | 95.9 | 95.9 | 95.4 | 94.2 | 94.2 | ||||
| Syria | 06 | 100 | 99.7 | 99.0 | 99.7 | 99.9 | 99.9 | 99.9 | 98.5 | 98.5 | 95.9 | 95.8 | 95.9 | 96.1 | 96.1 | 95.5 | 94.4 | 94.4 | |||||
| Syria | 07 | 100 | 99.0 | 99.7 | 99.9 | 99.9 | 99.9 | 98.5 | 98.5 | 95.9 | 95.8 | 95.9 | 96.1 | 96.1 | 95.5 | 94.4 | 94.4 | ||||||
| Thailand | 08 | 100 | 99.0 | 99.2 | 99.2 | 99.2 | 97.9 | 97.9 | 95.2 | 95.1 | 95.2 | 95.4 | 95.4 | 94.8 | 93.7 | 93.7 | |||||||
| Thailand | 09 | 100 | 99.9 | 99.9 | 99.9 | 98.5 | 98.5 | 95.9 | 95.8 | 95.9 | 96.1 | 96.1 | 95.5 | 94.4 | 94.4 | ||||||||
| Thailand | 10 | 100 | 100 | 100 | 98.6 | 98.6 | 96.1 | 95.9 | 96.1 | 96.2 | 96.2 | 95.6 | 94.5 | 94.5 | |||||||||
| USA | 11 | 100 | 100 | 98.6 | 98.6 | 96.1 | 95.9 | 96.1 | 96.2 | 96.2 | 95.6 | 94.5 | 94.5 | ||||||||||
| USA | 12 | 100 | 98.6 | 98.6 | 96.1 | 95.9 | 96.1 | 96.2 | 96.2 | 95.6 | 94.5 | 94.5 | |||||||||||
| MP-C18 | 13 | 100 | 100 | 96.3 | 96.2 | 96.3 | 96.5 | 96.5 | 95.9 | 94.8 | 94.8 | ||||||||||||
| NW-C4 | 14 | 100 | 96.3 | 96.2 | 96.3 | 96.5 | 96.5 | 95.9 | 94.8 | 94.8 | |||||||||||||
| GP-C17 | 15 | 100 | 99.9 | 100 | 99.9 | 99.7 | 99.5 | 98.3 | 98.3 | ||||||||||||||
| INDIA | 16 | 100 | 99.9 | 99.7 | 99.6 | 99.3 | 98.1 | 98.1 | |||||||||||||||
| KZN-C1 | 17 | 100 | 99.9 | 99.7 | 99.5 | 98.3 | 98.3 | ||||||||||||||||
| NW-C2 | 18 | 100 | 99.9 | 99.3 | 98.1 | 98.1 | |||||||||||||||||
| GP-C7 | 19 | 100 | 99.5 | 98.3 | 98.3 | ||||||||||||||||||
| GP-C15 | 20 | 100 | 98.8 | 98.8 | |||||||||||||||||||
| KZN-C2 | 21 | 100 | 100 | ||||||||||||||||||||
| WC-10270 | 22 | 100 |
Accession numbers of BbSBP-4 sequences extracted from GenBank are (01 through 12): AB569303, AB569301, AB569300, AB594813, AB569302, AB617639, AB617641, AB571871, AB586125, AB594814, AF486506 and AF486507, respectively. Accession numbers of sequences generated in this study are (13 through 22): KF626632, KF626638, KF626636, KF626631, KF626629, KF626637, KF626634, KF626635, KF626630, KF626633
Fig. 1.
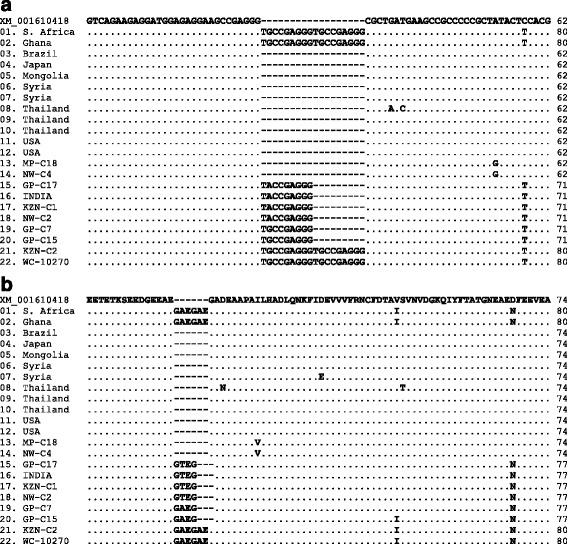
Multiple sequence alignments of partial BbSBP-4 nucleotide gene sequences (a) and deduced amino acid sequences (b) of 12 GenBank strains and 10 South African isolates of Babesia bovis. The dots and dashes respectively indicate sequence identities and gaps in relation to a reference sequence (accession number XM_001610418). Accession numbers of sequences extracted from GenBank (01 to 14) correspond to those given in Table 3
A multiple sequence alignment of BbβTUB sequences revealed the presence of SNPs among the South African B. bovis isolates and GenBank strains. The BbβTUB sequence from MP-C17 sample showed 97% identity with corresponding sequences from Texas (AK440534) and Samford (L00978) strains. MP-C17 sequence was also compared to other BbβTUB sequences determined in this study and the highest sequence identities recorded were between 97.2 and 97.5%. Sequences derived from other South African bovine samples (GP-C7, GP-C17, KZN-C2, INDIA, MP-C18, NW-C2, NW-C4 and WC-10270) showed 99% sequence identity when compared to those of Texas (AK440534) and Samford (L00978) strains.
Phylogenetic tree analyses
In order to study the phylogenetic relationship among the sequences of tested B. bovis isolates, nested PCR-amplified fragments of selected samples were sequenced in both strands. A neighbour-joining tree constructed with the BbCP2 sequences showed a clear phylogenetic separation of sequences derived from geographical isolates of B. bovis (Fig. 2). The BbCP2 sequences determined in this study were found in four different clusters. The first cluster grouped the reference strain (XM_001610645) with sequences from NW-C4, GP-C7, GP-C17 and MP-C16. Sequences of EC-23A, NW-C2 and WC-10270 formed a cluster clearly distinct from that of other sequences, with a high bootstrap support of 97%. While MP-C18 sequence clustered with that of R1A strain (GQ412131), the BbCP2 sequences of INDIA and KZN-C2 exhibited a close phylogenetic relationship with the sequence of S2P strain (GQ412136).
Fig. 2.
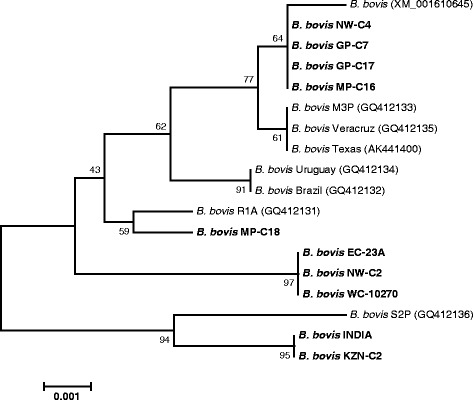
Phylogenetic tree inferred with BbCP2 gene sequences (809 nt) of several B. bovis isolates from bovine samples collected at various geographical regions in South Africa, together with corresponding sequences retrieved from GenBank. Accession numbers are given in parentheses. Bootstrap values calculated as percentages of 1000 replicates are indicated at branching points. The horizontal scale bar shows the number of base substitutions per site
A phylogram created with neighbour-joining method based on BbSBP-4 nucleotide gene sequences is shown in Fig. 3. The sequences obtained in this study fell into three clades that resulted from the existence of SNPs across the lengths of BbSBP-4 genes. The sequences of MP-C18 and NW-C4 were clearly distinct from other sequences. The BbSBP-4 gene sequences from GP-C7, GP-C17, KZN-C1, INDIA and NW-C2 all formed a separate grouping. Similarly, GP-C15, WC-10270 and KZN-C2 sequences showed a very close phylogenetic relationship with sequences of B. bovis strains published in GenBank (AB569301 and AB569303), and were supported by a high bootstrap value.
Fig. 3.
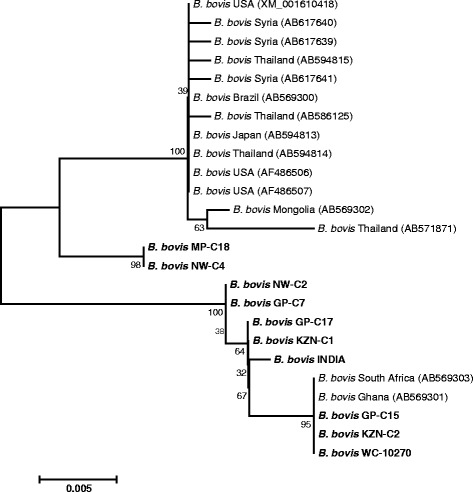
A neighbour-joining tree generated with BbSBP-4 nucleotide gene sequences (726–744 nt) of South African B. bovis isolates (in bold type), together with sequences previously published in GenBank. Accession numbers are given in parentheses. Bootstrap values calculated as percentages of 1000 replicates are indicated at branching points. The horizontal scale bar shows the number of base substitutions per site
The phylogenetic tree inferred with BbβTUB sequences is reflected in Fig. 4. In the phylogeny, it was observed that the sequences from GP-C7, GP-C17, INDIA, MP-C18, NW-C2 and NW-C4 clustered together. The DNA sequences derived from KZN-C2 and WC-10270 showed a grouping with Texas (AK440534) and T2Bo (XM_001611566) strains. Furthermore, it was noted that the BbβTUB sequence of MP-C17 isolate formed a separate cluster clearly distinct from that of other South African isolates, with a bootstrap support of 100%. Phylogenetic analysis of BbβTUB gene also indicated that Babesia ovata (AB634844) is genetically related to B. bigemina Argentina strain (AB634846).
Fig. 4.
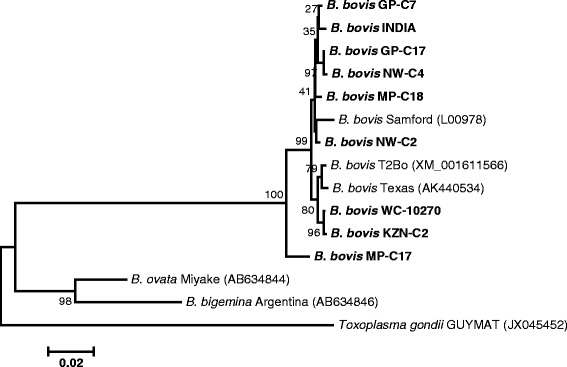
Phylogenetic tree based on BbβTUB nucleotide gene sequences (920 nt) derived from blood samples collected from South African cattle (in bold type), together with sequences previously published in GenBank (accession numbers in parentheses). Bootstrap values calculated as percentages of 1000 replicates are indicated at branching points. The horizontal scale bar shows the number of base substitutions per site. Toxoplasma gondii GUYMAT (accession no. JX045452) was used as an outgroup
Discussion
The search for B. bovis vaccine that could confer absolute protective immunity against bovine babesiosis has been the subject of many studies over the last decades. To date, no recombinant subunit vaccines are available to eliminate babesial infections in cattle globally because of extensive antigenic variations displayed by heterologous parasitic strains. As a result, many research groups have intensified their efforts in an endeavour to search for novel vaccine candidates with potential to offer complete protective immune response against a challenge by heterologous strains.
In particular, many studies have focused on functionally important proteins that are believed to play a pivotal role in parasite survival and growth [17, 18]. For example, the RAP-1 protein is involved in the process of invasion of bovine red blood cells by merozoites [19, 20]. Peptidases, such as cysteine peptidase 2, possess enzymatic activities identified as virulence factors for Apicomplexan parasites [21]. The BbSBP is involved in stabilizing the environment after parasite invasion and plays a role in parasite growth [22]. In addition, BbSBP has been documented as a new serological antigen for global epidemiological research [23]. Likewise, the β-tubulin gene of Babesia parasites has been used as an informative marker for species discrimination [24].
The fight against bovine babesiosis in southern Africa has always relied on the use of live attenuated vaccines which are more prone to cross-contamination [6]. Furthermore, the practical application of these vaccines is hampered by genetic heterogeneities observed among field isolates [25]. Therefore, it remains imperative to document the genetic profiles of B. bovis parasitic field isolates in an endeavour to develop effective control strategies with potential to curtail infection of susceptible bovines by Babesia parasites.
In the present study, we describe the successful development and application of three nested PCR assays for the specific detection of B. bovis DNA in field bovine samples. The BbRAP-1 nested PCR assay developed previously [12] was employed as the control for the newly developed nested PCR assays. Due to the high number of samples tested positive with BbRAP-1 nested PCR-based assay, it appears that this assay was more sensitive than BbCP2, BbSBP-4 and BbβTUB assays developed in the present study. However, it is reported in the literature that the high number of positive samples detected by BbRAP-1 nested PCR may be attributed to the presence of several nearly identical copies of B. bovis BbRAP-1 genes in the genome [26, 27].
To corroborate the specificities of nested PCR assays, PCR-generated amplicons from randomly selected positive samples were sequenced. Sequence analysis revealed high genetic conservation among the BbRAP-1 DNA fragments of South African B. bovis isolates and those published in GenBank, with identities ranging between 99 and 100%. The high conservation of BbRAP-1 genes in B. bovis field isolates is comparable to previous studies that observed significant similarities in BbRAP-1 sequences of isolates from countries other than South Africa [10, 28].
Accordingly, in silico analysis of BbCP2 sequences indicated a high degree of sequence conservation between South African B. bovis isolates and GenBank strains. In the BLAST search for homologous sequences in GenBank, the highest BbCP2 sequence identities (99%) were recorded with B. bovis strains originating from Argentina, Mexico, Uruguay and Brazil. This suggested high conservation of BbCP2 gene sequences among B. bovis strains from geographically distinct regions of the world. Although cysteine peptidases of B. bovis have not been well characterized [29], the evidence of the importance of peptidases for parasite growth and survival was obtained when specific inhibitors of these enzymes impaired merozoite growth in vitro [30]. Recent findings on in vitro and in vivo expression of bovipain-2, a cysteine protease, in B. bovis also suggest that peptidases might potentially be considered as important targets for the development of effective control strategies against bovine babesiosis [29].
Sequencing of BbSBP-4 genes from B. bovis isolates examined in this study demonstrated the existence of gaps in some isolates, a feature commensurate with base-pair insertion or deletion events. These insertions and deletions of nucleotide bases within the open reading frames (ORFs) may result in frame shift mutations, depending on whether the nucleotides that are added to (insertion event) or deleted from (deletion event) the ORF are a multiple of three or not. To further investigate whether there were any significant differences between the protein sequences as well as the possibility of frame shift mutations among B. bovis isolates possessing gaps, the determined nucleotide sequences were translated to protein sequences in silico and aligned with corresponding sequences of B. bovis strains published in GenBank. Analysis of aligned protein sequences revealed high conservation between sequences, albeit there were amino acid differences observed along the stretch of the protein sequences. The conservation of BbSBP sequences among geographical isolates of B. bovis has also been observed elsewhere [31, 32].
In studying the phylogenetic relationship among B. bovis isolates based on BbCP2, BbSBP-4 and BbβTUB gene sequences, neighbour-joining trees were inferred. The analyses of phylogenies indicated a close phylogenetic relationship between the South African B. bovis isolates and geographical strains originating from cattle in other countries, albeit the sequences showed varying phylogenetic groupings because of the existence of SNPs across the lengths of nucleotide gene sequences. However, the observed clustering of BbCP2 and BbSBP-4 sequences incorporated in the phylogenies should be interpreted with caution given that the trees were not rooted. Furthermore, the current dearth of B. bovis BbβTUB sequences in GenBank could not allow for a better discrimination of the phylogenetic relatedness between B. bovis isolates of South African origin and those of countries other than South Africa. Given the importance of β-tubulin protein-encoding gene as the molecular marker for species identification and discrimination [24], it is of paramount importance that more BbβTUB genes of geographical B. bovis strains are sequenced and made available in GenBank.
Conclusion
In this study, we have described the development and application of three newly developed nested PCR assays for the specific detection and genetic characterization of B. bovis isolates in field bovine samples. Although only few sequences were incorporated in the phylogenetic analyses, it must be acknowledged that the findings from this study provide valuable information regarding the genetic conservation among B. bovis isolates of South African origin in comparison to those previously published in GenBank. Taken together, the data presented in this study has given an indication that immunologically important proteins encoded by the genes examined in this study could potentially be considered for exploitation as viable candidates for inclusion in recombinant subunit vaccines.
Acknowledgements
The authors are indebted to the farmers who participated during bovine blood sample collection. The technical assistance of Lerato Dlamini and Veronica Phetla is gratefully acknowledged. This work is based upon the research supported financially by the National Research Foundation (NRF) and the National Zoological Gardens of South Africa (NZG). Any opinion, finding and conclusion or recommendation expressed in this material is that of the authors and the NRF does not accept any liability in this regard.
Funding
This research was supported by the National Research Foundation (NRF) (Moses Sibusiso Mtshali, grant number 69090) and the National Zoological Gardens of South Africa. The funders played no role in study design, data collection and interpretation, or the decision to submit this work for publication.
Availability of data and materials
All datasets generated and/or analyzed in this current study are available in the manuscript.
Authors’ contributions
PS conceived the study. PS and MS participated in study design and coordination. PS carried out laboratory assays, performed data analysis and interpretation, constructed phylogenetic trees and drafted the manuscript. PS and MS critically reviewed the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Ethics approval was granted by the National Zoological Gardens of South Africa Research Ethics and Scientific Committee, project number NZG/RES/P/001/F/01 for the national research project on the epidemiology and genetic diversity of bovine anaplasmosis in South Africa. The NZG Research Ethics and Scientific Committee approved the collection of blood samples from animals. Animal owners gave verbal informed consent for the collection of the samples for the epidemiological survey, and animals were not involved in any clinical trials or treatments.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Phillip Senzo Mtshali, Email: senzo4me@yahoo.com.
Moses Sibusiso Mtshali, Email: sibusiso.mtshali@nrf.ac.za.
References
- 1.Jongejan F, Uilenberg G. The global importance of ticks. Parasitology. 2004;129:S3–14. doi: 10.1017/S0031182004005967. [DOI] [PubMed] [Google Scholar]
- 2.de Vos AJ, Potgieter FT. Bovine babesiosis. In: JAW C, Thomson GR, Tustin RC, editors. Infectious diseases of livestock. South Africa: Oxford University Press; 1994. pp. 278–294. [Google Scholar]
- 3.OIE. Bovine babesiosis: OIE Terrestrial Manual [Internet]. Available from: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.04.02_bovine_babesiosis.pdf (2010). Accessed 29 Aug 2013.
- 4.Bock R, Jackson L, de Vos AJ, Jorgensen W. Babesiosis of cattle. Parasitology. 2004;129:S247–S269. doi: 10.1017/S0031182004005190. [DOI] [PubMed] [Google Scholar]
- 5.Ristic M. Babesiosis. In: Ristic M, McIntyre I, editors. Disease of cattle in the tropics. The Netherlands: Martinus Nijhof Publishers; 1981. pp. 443–468. [Google Scholar]
- 6.de Vos A, Bock RE. Vaccination against bovine babesiosis. Ann N Y Acad Sci. 2000;916:540–545. doi: 10.1111/j.1749-6632.2000.tb05333.x. [DOI] [PubMed] [Google Scholar]
- 7.Ojeda JJ, Orozco L, Flores R, Figueroa JV, Alvarez JA. Validation of an attenuated live vaccine against babesiosis in native cattle in an endemic area. Transbound Emerg Dis. 2010;57(1–2):84–86. doi: 10.1111/j.1865-1682.2010.01123.x. [DOI] [PubMed] [Google Scholar]
- 8.Callow LL, Dalgliesh RJ, de Vos AJ. Development of effective living vaccines against bovine babesiosis: the longest field trial? Int J Parasitol. 1997;27(7):747–767. doi: 10.1016/S0020-7519(97)00034-9. [DOI] [PubMed] [Google Scholar]
- 9.Torina A, Agnone A, Sireci G, Mosqueda JJ, Blanda V, Albanese I, La Farina M, Cerrone A, Cusumano F, Caracappa S. Characterization of the apical membrane antigen-1 in Italian strains of Babesia bigemina. Transbound Emerg Dis. 2010;57(1–2):52–56. doi: 10.1111/j.1865-1682.2010.01118.x. [DOI] [PubMed] [Google Scholar]
- 10.Ramos CAN, Araújo FR, Alves LC, de Souza IIF, Guedes DS, Jr, Soares CO. Genetic conservation of potentially immunogenic proteins among Brazilian isolates of Babesia bovis. Vet Parasitol. 2012;187(3–4):548–552. doi: 10.1016/j.vetpar.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 11.Brayton KA, Lau AOT, Herndon DR, Hannick L, Kappmeyer LS, Berens SJ, Bidwell SL, Brown W, Crabtree J, Fadrosh D, Feldblum T, Forberger HA, Haas BJ, Howell JM, Khouri H, Koo H, Mann DJ, Norimine J, Paulsen IT, Radune D, Nene VM. Genome sequence of Babesia bovis and comparative analysis of apicomplexan hemoprotozoa. PLoS Pathog. 2007;3(10):1401–1413. doi: 10.1371/journal.ppat.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figueroa JV, Chieves LP, Johnson GS, Buening GM. Multiplex polymerase chain reaction based assay for the detection of Babesia bigemina, Babesia bovis and Anaplasma marginale DNA in bovine blood. Vet Parasitol. 1993;50(1–2):69–81. doi: 10.1016/0304-4017(93)90008-B. [DOI] [PubMed] [Google Scholar]
- 13.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 14.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 17.Yokoyama N, Okamura M, Igarashi I. Erythrocyte invasion by Babesia parasites: current advances in the elucidation of the molecular interactions between the protozoan ligands and host receptors in the invasion stage. Vet Parasitol. 2006;138(1–2):22–32. doi: 10.1016/j.vetpar.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 18.Florin-Christensen M, Schnittger L, Dominguez M, Mesplet M, Rodriguez A, Ferreri L, Asenzo G, Wilkowsky S, Farber M, Echaide I, Suarez C. Search for Babesia bovis vaccine candidates. Parassitologia. 2007;1:9–12. [PubMed] [Google Scholar]
- 19.Suarez CE, Palmer GH, Hӧtzel I, Hines SA, McElwain TF. Sequence and functional analysis of the intergenic regions separating babesial rhoptry-associated protein-1 (rap-1) genes. Exp Parasitol. 1998;90(2):189–194. doi: 10.1006/expr.1998.4321. [DOI] [PubMed] [Google Scholar]
- 20.Gaffar FR, Yatsuda AP, Franssen FF, Vries EA. A Babesia bovis merozoite protein with a domain architecture highly similar to the thrombospondin-related anonymous protein (TRAP) present in Plasmodium sporozoites. Mol Biochem Parasitol. 2004;136(1):25–34. doi: 10.1016/j.molbiopara.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Mesplet M, Palmer GH, Pedroni MJ, Echaide I, Florin-Christensen M, Schnittger L, Lau AOT. Genome-wide analysis of peptidase content and expression in a virulent and attenuated Babesia bovis strain pair. Mol Biochem Parasitol. 2011;179(2–2):111–113. doi: 10.1016/j.molbiopara.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goo YK, Jia H, Aboge GO, Terkawi MA, Kuriki K, Nakamura C, Kumagai A, Zhou J, Lee EG, Nishikawa Y, Igarashi I, Fujisaki K, Xuan X. Babesia gibsoni: serodiagnosis of infection in dogs by an enzyme-linked immunosorbent assay with recombinant BgTRAP. Exp Parasitol. 2008;118(4):555–560. doi: 10.1016/j.exppara.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Terkawi MA, Huyen NX, Wibowo PE, Seuseu FJ, AbouLaila M, Ueno A, Goo YK, Yokoyama N, Xuan X, Igarashi I. Spherical body protein 4 is a new serological antigen for global detection of Babesia bovis infection in cattle. Clin Vaccine Immunol. 2011;18(2):337–342. doi: 10.1128/CVI.00388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cacciò S, Cammà C, Onuma M, Severini C. The β-tubulin gene of Babesia and Theileria parasites is an informative marker for species discrimination. Int J Parasitol. 2000;30(11):1181–1185. doi: 10.1016/S0020-7519(00)00105-3. [DOI] [PubMed] [Google Scholar]
- 25.Berens SJ, Brayton KA, Molloy JB, Bock RE, Lew AE, McElwain TF. Merozoite surface antigen 2 proteins of Babesia bovis vaccine breakthrough isolates contain a unique hypervariable region composed of degenerate repeats. Infect Immun. 2005;73(11):7180–7189. doi: 10.1128/IAI.73.11.7180-7189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norimine J, Suarez CE, McElwain TF, Florin-Christensen M, Brown WC. Immunodominant epitopes in Babesia bovis rhoptry-associated protein 1 that elicit memory CD4+-T-lymphocyte responses in B. bovis-immune individuals are located in the amino-terminal domain. Infect Immun. 2002;70(4):2039–2048. doi: 10.1128/IAI.70.4.2039-2048.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.AbouLaila M, Yokoyama N, Igarashi I. Development and evaluation of two nested PCR assays for the detection of Babesia bovis from cattle blood. Vet Parasitol. 2010;172(1–2):65–70. doi: 10.1016/j.vetpar.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 28.Silva MG, Henriques G, Sánchez C, Marques PX, Suarez CE, Oova A. First survey for Babesia bovis and Babesia bigemina infection in cattle from central and southern regions of Portugal using serological and DNA detection methods. Vet Parasitol. 2009;166(1–2):66–72. doi: 10.1016/j.vetpar.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 29.Mesplet M, Echaide I, Dominguez M, Mosqueda JJ, Suarez CE, Schnittger L, Florin-Christensen M. Bovipain-2, the falcipain-2 ortholog, is expressed in intraerythrocytic stages of the tick-transmitted hemoparasite Babesia bovis. Parasit Vectors. 2010;3:113. doi: 10.1186/1756-3305-3-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okubo K, Yokoyama N, Govind Y, Alhassan A, Igarashi I. Babesia bovis: effects of cysteine protease inhibitors on in vitro growth. Exp Parasitol. 2007;117(2):214–217. doi: 10.1016/j.exppara.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 31.Ruef BJ, Dowling SC, Conley PG, Perryman LE, Brown WC, Jasmer DP, Rice-Ficht AC. A unique Babesia bovis spherical body protein is conserved among geographical isolates and localizes to the infected erythrocyte membrane. Mol Biochem Parasitol. 2000;105(1):1–12. doi: 10.1016/S0166-6851(99)00167-X. [DOI] [PubMed] [Google Scholar]
- 32.Terkawi MA, Seuseu FJ, Eko-Wibowo P, Huyen NX, Minoda Y, AbouLaila M, Kawai S, Yokoyama N, Xuan X, Igarashi I. Secretion of a new spherical body protein of Babesia bovis into the cytoplasm of infected erythrocytes. Mol Biochem Parasitol. 2011;178(1–2):40–45. doi: 10.1016/j.molbiopara.2011.02.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets generated and/or analyzed in this current study are available in the manuscript.


