Abstract
The crystal structure of the trans-acyltrans-ferase (AT) from the disorazole polyketide synthase (PKS) was determined at room temperature to a resolution of 2.5 Å using a new method for the direct delivery of the sample into an X-ray free-electron laser. A novel sample extractor efficiently delivered limited quantities of microcrystals directly from the native crystallization solution into the X-ray beam at room temperature. The AT structure revealed important catalytic features of this core PKS enzyme, including the occurrence of conformational changes around the active site. The implications of these conformational changes for polyketide synthase reaction dynamics are discussed.
Polyketide synthases (PKSs) work in a highly modular fashion to synthesize medicinally important natural products with a variety of therapeutic properties.1,2 The basic unit of the module consists of a β-ketosynthase (KS), an acyltransferase (AT), and an acyl carrier protein (ACP). The KS domain receives the growing polyketide chain from the upstream module and subsequently catalyzes chain elongation with an ACP-bound extender unit as the co-substrate (Figure 1). The AT transacylates the extender unit onto the pantetheinyl arm of the ACP (Figure 1). The AT domain is responsible for selection of an appropriate building block (malonyl- or methylmalonyl-CoA) in each chain elongation cycle that is transferred to a dedicated acyl carrier protein (ACP) domain in the same module. Substitution for domains loading other extender units allows introduction of a broadened chemistry into the polyketide backbone.3–5 Considerable research has focused on engineering the PKS system to produce new compounds.6 The observed inefficiency underscores the importance of a deeper understanding of PKS structure-function relationships. The structures of individual PKS enzymes7,8 and multidomain fragments9,10 revealed some mechanistic insights into catalysis; together with recent small angle X-ray scattering analysis, these structural studies have enabled modeling of the entire assembly. However, the lack of high-resolution structures of intact PKS modules limits the ability to improve their synthetic efficiency via engineering approaches. In particular, the difficulty in growing larger crystals and the sensitivity of crystals to X-rays make it difficult to employ conventional crystallographic methods for atomic analysis.
Figure 1.
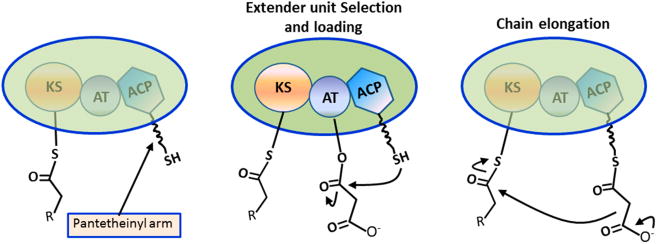
PKS reaction. The AT selects the extender unit and transfers the extender unit onto the pantetheinyl arm of the ACP (middle). The KS domain catalyzes the chain elongation with the ACP-bound extender unit (right).
The ultrashort femtosecond X-ray pulses of X-ray free-electron laser (XFEL) sources can produce diffraction data on time scales that mitigate radiation damage processes. This enables structural investigations of extremely small, weakly diffracting, and radiation-sensitive crystals at ambient temperatures. Because the exposed crystals are destroyed in the process, serial femtosecond crystallography (SFX) data sets are comprised of still diffraction images collected from many individual crystals delivered serially. The XFEL at the SLAC Linac Coherent Light Source (LCLS) produces hard X-ray pulses at a maximum frequency of 120 Hz, and protein crystals for SFX experiments are most often delivered rapidly into the X-ray beam using a liquid jet injector. Significant improvements in liquid jet technology have recently been made, including the use of high-viscosity carrier solutions to minimize sample consumption and to improve reliability.14–17 However, challenges remain, such as the need to alter crystallization conditions and/or transfer crystals into carrier solutions that are compatible with the specific injector technology, a process that often adversely affects diffraction quality. Moreover, to prevent clogging of the injector nozzle, crystal slurries may need to be filtered to limit crystal size. In such circumstances, delicate crystals like the PKS subcomplexes may not survive the shear forces associated with filtration or the injection process. Acoustic injectors that deliver drops containing crystals “on demand” show promise as an alternative way to reduce sample consumption.19 However, compatibility requirements of carrier solutions and limitations in drop size continue to pose technical challenges. Therefore, these techniques are not well suited for AT crystals, which are difficult to produce in large quantities and are sensitive to changes in the crystallization solution.
We developed a new device, the sample extractor, to efficiently deliver microcrystals in native crystallization solutions for serial crystallography (SX) and SFX experiments (Figure 2 and Figure S1). The sample extractor uses a variety of mesh- or thin film-based substrates to transport crystals directly from their mother liquor to the X-ray interaction region. To mitigate dehydration during X-ray diffraction data collection, both the crystalline sample and the substrate are enclosed in a small vial. A small opening in the vial allows the X-ray beam to enter, and a larger opening on the opposite side covered with a thin X-ray transparent film allows the diffracted X-rays to pass through (Figure S1). The substrate is connected through a small slot on top of the vial to a small solenoid that can rapidly move the substrate in and out of the crystal solution. A small stainless steel weight at the end of the substrate provides positional stability and also agitates the solution, thus preventing the crystals from settling to the bottom of the reservoir.
Figure 2.
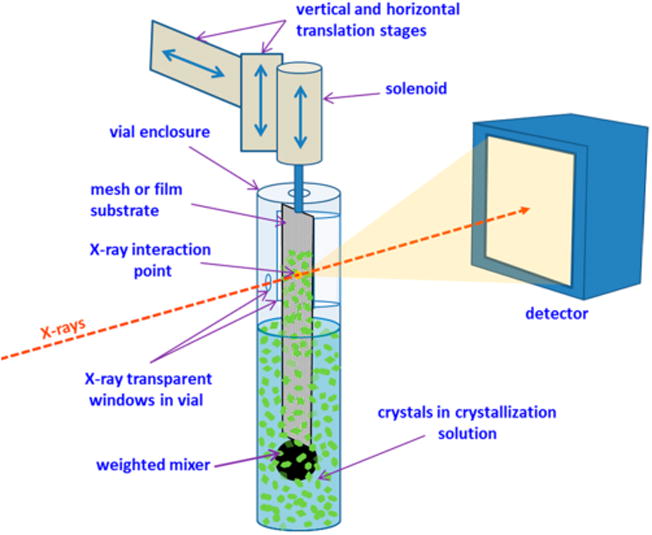
Sample extractor. Schematic view showing the different parts of the sample extractor. Crystals are trapped on a crystal carrier substrate (mesh or film) that is rapidly inserted and removed from the crystal containing solution by a solenoid. Crystals on different areas of the substrate are positioned in the X-ray interaction region by translation stages. A weighted mixer connected to the bottom of the carrier substrate agitates the crystals, keeping them from settling to the bottom of the vial.
During operation, the substrate is first submerged into a solution containing crystals. Crystals are trapped on the substrate as it is rapidly removed from the solution. The substrate is then translated in a zigzag pattern to expose a “grid” of regularly spaced sites, some of which contain crystals. The substrate can be translated rapidly up to 2.5 mm/s using the current device. After a rastering sweep is completed, the substrate is resubmerged, transferring unexposed crystals back into solution to keep them hydrated and to make them available for subsequent exposures. This crystal recycling approach, currently not available in any other sample delivery method, minimizes the loss of valuable crystalline sample during data collection. Furthermore, the sample extractor does not appear to be sensitive to crystal size or shape, and because it operates at ambient temperature and pressure, there is essentially no mechanical stress on the crystals during delivery. The sample extractor device is compact and easily mounted at the LCLS and SSRL crystallography beamlines. Furthermore, the existing collimation, beam stop, and on-axis sample visualization systems can be used for beam conditioning and sample extractor alignment (see Figures S1–S3).
It has been recently reported that ambient-temperature X-ray crystallography can access protein conformational dynamics that are locked out in low-temperature structures.20 The sample extractor device allowed us to collect an ambient-temperature X-ray diffraction data set of an AT complex to explore these potential conformations. The serial X-ray diffraction data for AT crystals were collected during protein crystal screening (PCS) time at the LCLS X-ray pump probe (XPP) station using 3.3 mg of protein in 300 μL of native crystallization solution (approximately 75 crystals of ∼50 × ∼10 × ∼2 μM3 per microliter) (Figure S4). The XFEL operated at a wavelength of 9.5 keV with a pulse length of 40 fs. Still diffraction images were recorded using a Rayonix MX170-HS X-ray detector operating at a frame rate of 10 Hz. A nylon mesh with diamond-shaped holes (1.0 mm × 0.9 mm) was used as the sample extractor carrier substrate. After extracting the substrate from the crystal solution and transferring it into the X-ray beam path, we recorded ∼40 exposures while the substrate was translated by 60 μm between each exposure. The detector operated in a continuous mode, yielding five images without diffraction each time the substrate was submerged and the crystals were reloaded. Thus, ∼12% of the total 22985 detector images collected in ∼40 min lacked diffraction data because the sample was being regenerated. A total of 2075 images contained diffraction patterns with at least 50 peaks [with resolution as high as 2.1 Å (Figure S4) ], yielding a hit rate of ∼10%. Of these, 1482 images could be successfully indexed and integrated. The quality of the merged data set was improved by applying stringent rejection criteria. As a result, the final data set contained 771 of the filtered images and was 98.6% complete to a resolution of 2.5 Å with a redundancy of 14 (space group P212121) and a CC1/2 of 91.7% (Table S1). The structure was refined to yield Rwork and Rfree values of 23.1 and 27.1%, respectively. Despite the thin plate morphology of the AT crystals (Figure S4), the data set was complete, indicating crystals were not preferentially orienting on the substrate. In comparison, previous experiments using the gas dynamic virtual nozzle liquid injector13 at the LCLS XFEL required 6 mL of AT crystals to produce 66 images with diffraction, the highest observed resolution being only 2.6 Å. The sample extractor described here holds 300 μL of the crystal suspension, and smaller vial designs are in progress. After continuous diffraction data had been collected for ∼40 min using the same sample, there was no observable deterioration in diffraction quality. Because crystals are typically destroyed upon exposure to the X-ray beam, the chance of repeat exposure of the same crystal is very low. For the few crystals that survive the beam, the repeat diffraction image will be substantially poorer and will therefore be excluded from the final data set by standard data processing methods.
The resulting AT structure represents the first ambient-temperature structure of a PKS enzyme to date. Comparison of the ambient-temperature structure with the recently published 100 K structure7 reveals conformational flexibility at ambient temperature that is not observed at low temperatures. A notable difference was found in the active site comprised of Ser86 and His191 (which form the catalytic dyad), Arg111 (which stabilizes the C3 carboxylate of the malonyl extender unit through a salt bridge), and Phe190 and Gln9 (which are believed to control substrate specificity by interacting with C2 and any attendant substituent of the extender unit) (Figure 3).7 In particular, the Gln9 side chain has undergone a reorientation, resulting in disruption of the hydrogen bond between the Gln9 carboxamide and the side chain of Thr57 (Figure 3). This observed widening of the substrate binding cavity comprised of the Phe190 and Gln9 side chains is intriguing, as it suggests that active site dynamics play an important role in controlling the high substrate specificity of the AT family of enzymes. To test this hypothesis, we compared structures of AT crystals soaked in malonate and citrate solutions, collected using crystals held at 100 K. As shown in Figure 3, the orientation of Gln9 in the room-temperature LCLS structure is analogous to that observed in the malonate- and citrate-bound complexes. The larger substrates are accommodated by the movement of the Phe190 side chain (Figure 3b). Thus, it appears that room-temperature X-ray diffraction analysis of the disorazole apo-AT has allowed the visualization of a higher-energy conformation of this enzyme that may be critical for accommodating its cognate substrate with high specificity.
Figure 3.
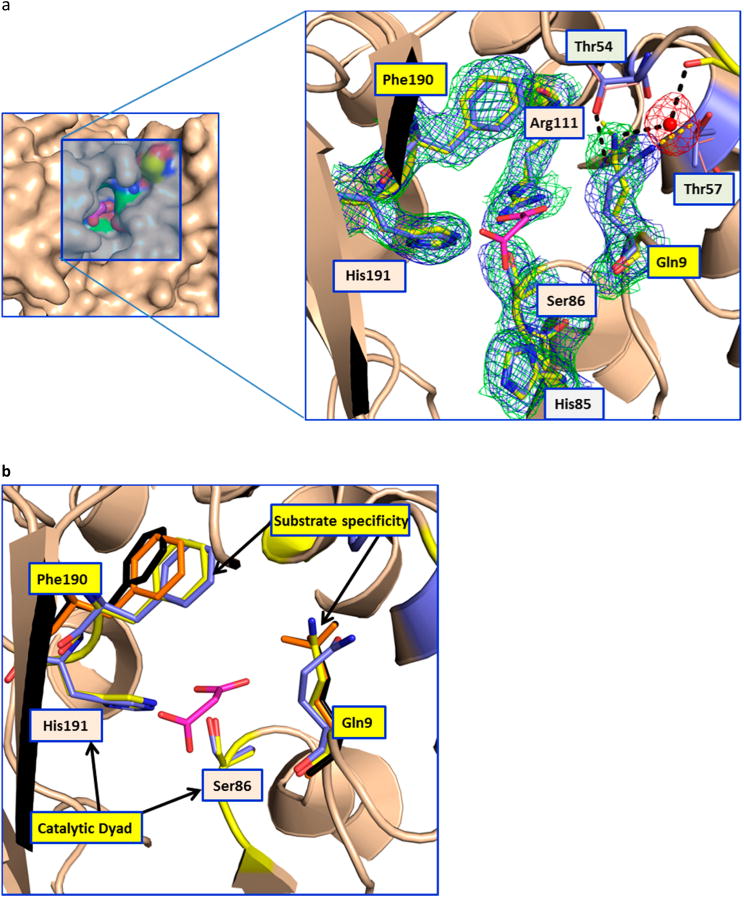
Conformational changes in the AT. (a) The left panel shows the surface representation of the AT active site (apo-LCLS structure), highlighting its deep narrow pocket. The close-up view of the malonate binding site is shown at the right. In the close-up view, the catalytic dyad residues (Ser86 and His191) are labeled with orange backgrounds and the gatekeeper residues (Gln9 and Phe190) are labeled with yellow backgrounds. Thr54 and Thr57, which are involved in the stabilizing Gln9, are also shown. The hydrogen bonds in the room-temperature and 100 K structures are shown as black and yellow dashed lines, respectively. Gln9 in the room-temperature structure is stabilized by a water molecule that bridges to the main chain carbonyl of Ser52. The 2Fo – Fc maps contoured at 1σ for the room-temperature and 100 K structures are shown as green and blue meshes, respectively (Phenix was used to superpose the electron density maps, ref 5 in the Supporting Information). In the low-temperature structure of the apoenzyme, access to the binding site of the malonate extender unit is blocked off. In contrast, the room-temperature structure shows a higher-energy conformation that appears to be critical for substrate access. The residue equivalent to Gln9 in the AT from the bacillaene PKS (unpublished structure, Protein Data Bank entry 5dz6) shows hydrogen bonding to a glycerol molecule in the malonate binding site. (b) The catalytic dyad and the residues involved in the substrate specificity are shown as sticks. The residues involved in substrate specificity (Phe190 and Gln9) are colored orange and black for the malonate and citrate complexes, respectively. This picture shows the movement of the Phe190 side chain to accommodate larger substrates in the binding pocket.
Additional conformational changes are observed near the active site in the vicinity of Gln156. Gln156 shows alternate conformations in the room-temperature structure and forms a direct hydrogen bond with Asn152 (Figure 4a). While the published structures of the native and acetate complexes had four water molecules connecting these residues (Figure 4b), the structures of the malonate complex and the citrate complex show hydrogen bonds involving only one water molecule (Figure 4c). Flipping of the side chain of Gln156 moves the side chain of nearby Gln276. This reorganization results in a new long-range interaction (16 Å from Cα of Gln276 to His85) in the room-temperature structure. This interaction connects the solvent-exposed surface to the internal active site (Figure 4d). More generally, our findings demonstrate that room-temperature diffraction is capable of uncovering conformational flexibility that may otherwise be locked out at lower temperatures.
Figure 4.
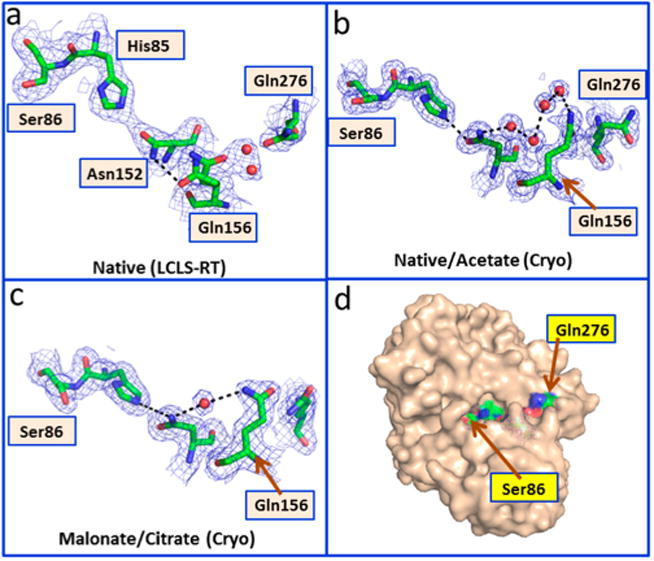
Conformational changes in the AT at room temperature. The panels show conformational changes in the vicinity of Gln156. All residues are labeled in the room-temperature structure; the other structures are drawn in an analogous orientation. Panel A shows the interactions observed in the XFEL room-temperature structure. Panel B shows the interactions observed in the 100 K structures of the native enzyme and the acetate complex. Panel C shows the interactions observed in the 100 K structures of the malonate and citrate complexes. Panel D shows the surface representation highlighting the pathway. The 2Fo — Fc maps are contoured at 1.0σ
In addition, these experiments have established the efficient operation and utility of the sample extractor device to study fragile crystals in limited supply. To broaden the utility of the sample extractor for room-temperature structural analysis, we also tested the device at the Stanford Synchrotron Radiation Lightsource (SSRL) and successfully collected a 2.0 Å data set of hen egg white lysozyme (HEWL) microcrystals (Table S2 and Figures S5 and S6). Use of a film rather than a mesh substrate helped to restrict crystal motion during the relatively long X-ray exposure time of 0.2 s.
In summary, the first room-temperature structure of a polyketide enzyme using the novel sample extractor device was determined to 2.5 Å resolution. Structural comparison with the low-temperature structures revealed conformational flexibility in this complex and a dynamic PKS enzyme. Further experiments are in progress to collect serial crystallography data sets for microcrystals of PKS complexes stabilized by antibodies.
Supplementary Material
Acknowledgments
Funding: T.R. and C.K. were supported by a grant from the National Institutes of Health (R01 GM087934).
We thank Scott Mitchell and Ronald Reyes for their part in the design and the machining of mechanical components. We thank Paul Ehrensberger, Renato Avelar, and Vladimir Vinetskiy for assistance with the design and installation of the electronic components and Daniel DePonte and Alex Wallace for assistance with the remote delivery setup. We thank Roberto Alonso Mori and Silke Nelson and LCLS staff for help with XFEL data collection. Portions of this research were performed at the LCLS at the SLAC National Accelerator Laboratory. The LCLS is supported by the U.S. Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences (OBES), under Contract DE-AC02-76SF00515. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (including Grant P41GM103393). A.T.B. acknowledges a Howard Hughes Collaborative Innovation Award.
Abbreviations
- PKS
polyketide synthase
- AT
acyltransferase
- CoA
coenzyme A
- ACP
acyl carrier protein
- DEBS
6-deoxyerythronolide B synthase
- DSZS
disorazole synthase
- KS
ketosynthase
- SFX
serial femtosecond crystallography
- SX
serial crystallography
- XFEL
X-ray free-electron laser
- LCLS
Linac Coherent Light Source
- XPP
X-ray pump-probe
Footnotes
Supporting Information: The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-chem.7b00711. Methods, Figures S1—S8, and Tables S1 and S2 (PDF)
Author Contributions: I.I.M. contributed to design, crystallization, data collection, structure refinement, and structure analysis. A.E.C. and M.S. contributed to design, data collection, and structure analysis. K.A. contributed to design, data collection, and SEM data collection. M.H. contributed to design. T.R. and C.K. contributed to acyltransferase protein and structure analysis. A.Y.L., M.U., and A.T.B. contributed to data processing, refinement, and structure analysis. The manuscript was prepared by A.E.C., S.E.M., I.I.M., A.Y.L., C.K., and M.U. with input from all authors.
Notes: The authors declare no competing financial interest.
References
- 1.Khosla C. J Org Chem. 2009;74:6416–6420. doi: 10.1021/jo9012089. [DOI] [PubMed] [Google Scholar]
- 2.Robbins T, Liu YC, Cane DE, Khosla C. Curr Opin Struct Biol. 2016;41:10–18. doi: 10.1016/j.sbi.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Hazzard C, Eustaquio AS, Reynolds KA, Moore BS. J Am Chem Soc. 2009;131:10376–10377. doi: 10.1021/ja9042824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson MC, Nam SJ, Gulder TA, Kauffman CA, Jensen PR, Fenical W, Moore BS. J Am Chem Soc. 2011;133:1971–1977. doi: 10.1021/ja109226s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cummings M, Breitling R, Takano E. FEMS Microbiol Lett. 2014;351:116–125. doi: 10.1111/1574-6968.12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menzella HG, Reid R, Carney JR, Chandran SS, Reisinger SJ, Patel KG, Hopwood DA, Santi DV. Nat Biotechnol. 2005;23:1171–1176. doi: 10.1038/nbt1128. [DOI] [PubMed] [Google Scholar]
- 7.Wong FT, Jin X, Mathews II, Cane DE, Khosla C. Biochemistry. 2011;50:6539–6548. doi: 10.1021/bi200632j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gay DC, Spear PJ, Keatinge-Clay AT. ACS Chem Biol. 2014;9:2374–2381. doi: 10.1021/cb500459b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. Proc Natl Acad Sci U S A. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gay DC, Gay G, Axelrod AJ, Jenner M, Kohlhaas C, Kampa A, Oldham NJ, Piel J, Keatinge-Clay AT. Structure. 2014;22:444–451. doi: 10.1016/j.str.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards AL, Matsui T, Weiss TM, Khosla C. J Mol Biol. 2014;426:2229–2245. doi: 10.1016/j.jmb.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapman HN, Fromme P, Barty A, White TA, Kirian RA, Aquila A, Hunter MS, Schulz J, DePonte DP, Weierstall U, Doak RB, Maia FR, Martin AV, Schlichting I, Lomb L, Coppola N, Shoeman RL, Epp SW, Hartmann R, Rolles D, Rudenko A, Foucar L, Kimmel N, Weidenspointner G, Holl P, Liang M, Barthelmess M, Caleman C, Boutet S, Bogan MJ, Krzywinski J, Bostedt C, Bajt S, Gumprecht L, Rudek B, Erk B, Schmidt C, Homke A, Reich C, Pietschner D, Struder L, Hauser G, Gorke H, Ullrich J, Herrmann S, Schaller G, Schopper F, Soltau H, Kuhnel KU, Messerschmidt M, Bozek JD, Hau-Riege SP, Frank M, Hampton CY, Sierra RG, Starodub D, Williams GJ, Hajdu J, Timneanu N, Seibert MM, Andreasson J, Rocker A, Jonsson O, Svenda M, Stern S, Nass K, Andritschke R, Schroter CD, Krasniqi F, Bott M, Schmidt KE, Wang X, Grotjohann I, Holton JM, Barends TR, Neutze R, Marchesini S, Fromme R, Schorb S, Rupp D, Adolph M, Gorkhover T, Andersson I, Hirsemann H, Potdevin G, Graafsma H, Nilsson B, Spence JC. Nature. 2011;470:73–77. [Google Scholar]
- 13.DePonte DP, Doak RB, Hunter M, Liu Z, Weierstall U, Spence JC. Micron. 2009;40:507–509. doi: 10.1016/j.micron.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Sierra RG, Laksmono H, Kern J, Tran R, Hattne J, Alonso-Mori R, Lassalle-Kaiser B, Glockner C, Hellmich J, Schafer DW, Echols N, Gildea RJ, Grosse-Kunstleve RW, Sellberg J, McQueen TA, Fry AR, Messerschmidt MM, Miahnahri A, Seibert MM, Hampton CY, Starodub D, Loh ND, Sokaras D, Weng TC, Zwart PH, Glatzel P, Milathianaki D, White WE, Adams PD, Williams GJ, Boutet S, Zouni A, Messinger J, Sauter NK, Bergmann U, Yano J, Yachandra VK, Bogan MJ. Acta Crystallogr Sect D Biol Crystallogr. 2012;68:1584–1587. doi: 10.1107/S0907444912038152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sierra RG, Gati C, Laksmono H, Dao EH, Gul S, Fuller F, Kern J, Chatterjee R, Ibrahim M, Brewster AS, Young ID, Michels-Clark T, Aquila A, Liang M, Hunter MS, Koglin JE, Boutet S, Junco EA, Hayes B, Bogan MJ, Hampton CY, Puglisi EV, Sauter NK, Stan CA, Zouni A, Yano J, Yachandra VK, Soltis SM, Puglisi JD, DeMirci D. Nat Methods. 2016;13:59–62. doi: 10.1038/nmeth.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugahara ME, Mizohata E, Nango E, Suzuki M, Tanaka T, Masuda T, Tanaka R, Shimamura T, Tanaka Y, Suno C, Ihara K, Pan D, Kakinouchi K, Sugiyama S, Murata M, Inoue T, Tono K, Song C, Park J, Kameshima T, Hatsui T, Joti Y, Yabashi M, Iwata S. Nat Methods. 2015;12:61–63. doi: 10.1038/nmeth.3172. [DOI] [PubMed] [Google Scholar]
- 17.Weierstall D, James D, Wang C, White TA, Wang D, Liu W, Spence JC, Doak RB, Nelson G, Fromme P, Fromme R, Grotjohann I, Kupitz C, Zatsepin NA, Liu H, Basu S, Wacker D, Won Han G, Katritch V, Boutet S, Messerschmidt M, Williams GJ, Koglin JE, Seibert MM, Klinker M, Gati C, Shoeman RL, Barty A, Chapman HN, Kirian RA, Beyerlein KR, Stevens RC, Li D, Shah ST, Howe N, Caffrey M, Cherezov V. Nat Commun. 2014;5:3309. doi: 10.1038/ncomms4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stevenson HP, DePonte DP, Makhov AM, Conway JF, Zeldin OB, Boutet S, Calero G, Cohen AE. Philos Trans R Soc B. 2014;369:20130322. doi: 10.1098/rstb.2013.0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roessler CG, Agarwal R, Allaire M, Alonso-Mori R, Andi B, Bachega JF, Bommer M, Brewster AS, Browne MC, Chatterjee R, Cho E, Cohen AE, Cowan M, Datwani S, Davidson VL, Defever J, Eaton B, Ellson R, Feng Y, Ghislain LP, Glownia JM, Han G, Hattne J, Hellmich J, Heroux A, Ibrahim M, Kern J, Kuczewski A, Lemke HT, Liu P, Majlof L, McClintock WM, Myers S, Nelsen S, Olechno J, Orville AM, Sauter NK, Soares AS, Soltis SM, Song H, Stearns RG, Tran R, Tsai Y, Uervirojnangkoorn M, Wilmot CM, Yachandra V, Yano J, Yukl ET, Zhu D, Zouni A. Structure. 2016;24:631–640. doi: 10.1016/j.str.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraser JS, van den Bedem H, Samelson AJ, Lang PT, Holton JM, Echols N, Alber T. Proc Natl Acad Sci U S A. 2011;108:16247–16252. doi: 10.1073/pnas.1111325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


