Abstract
Respiratory syncytial virus (RSV) is the leading cause of lower respiratory tract infection (LRTI) in children from infancy up to early childhood. Recently, we demonstrated that RSV infection alters cellular small non-coding RNA (sncRNA) expression, most notably the tRNA-derived RNA fragments (tRFs). However, the functions of the tRFs in virus–host interaction are largely unknown. Herein, we examined the role of three RSV-induced tRFs derived from the 5-end of mature tRNAs decoding GlyCCC, LysCTT and CysGCA (named tRF5-GlyCCC, tRF5-LysCTT and tRF5-CysGCA, respectively) in controlling RSV replication. We found that tRF5-GlyCCC and tRF5-LysCTT, but not tRF5-CysGCA, promote RSV replication, demonstrating the functional specificity of tRFs. The associated molecular mechanisms underlying the functions of tRF5-GlyCCC and tRF5-LysCTT were also investigated. Regulating the expression and/or activity of these tRFs may provide new insights into preventive and therapeutic strategies for RSV infection. The study also accumulated data for future development of a tRF targeting algorithm.
Keywords: RSV, tRNA-derived RNA fragments, viral replication
Abbreviations
LRTI, lower respiratory tract infection; miRNAs, microRNAs; piRNA, piwi-Interacting RNAs; RSV, respiratory syncytial virus; siRNA, small interfering RNA; sncRNAs, small non-coding RNAs; snoRNA, small nucleolar RNA; tRFs, tRNA-derived RNA Fragments.
Introduction
Respiratory syncytial virus (RSV) is an enveloped virus which used to belong to the family Paramyxoviridae and the genus Pneumovirus [1]. Pneumovirus has recently been reclassified as a separate family; RSV is now in the family Pneumoviridae [2]. As the most commonly identified virus in young children with acute lower respiratory tract infection (LRTI), RSV contributed to an estimated 33.8 million new cases globally in 2005 [3]. Furthermore, 3.4 million children underwent severe RSV-associated LRTI, requiring hospitalization and nearly 199 000 children younger than 5 years old died that year. Preventive strategies are limited to active paediatric and passive maternal immunization [4]. However, the treatment or vaccine for RSV infection is not effective, which suggests the need for a comprehensive understanding of host–RSV interaction for the development of better treatments and vaccines.
During the last decade, significant attention has been directed towards the identification of small non-coding RNAs (sncRNAs; <200 nt long) and their diverse regulatory functions, which has revolutionized biomedicine. Some well-studied sncRNAs, such as small interfering RNA (siRNA), microRNA (miRNA) and piwi-interacting RNA (piRNA), have emerged as a key regulatory group of antiviral innate immune responses (reviewed in [5]). With the development of high-throughput sequencing technologies, more classes of sncRNAs, including those derived from mRNA (messenger), rRNA (ribosomal), tRNA (transfer) and snoRNA (small nucleolar RNA) were discovered; however, with little information on their biogenesis and functions [6–8].
sncRNAs derived from tRNAs, called tRNA-derived RNA fragments (tRFs), have been recently identified by several research groups, including us [9–11]. There is increasing evidence that some tRFs are not by-products of random degradation, but functional molecules [5, 9, 10, 12]. We recently found that RSV specifically led to an abundant induction of tRFs [10, 13]. We believe these tRFs are functional based on: (1) not all tRNAs being cleaved by RSV infection; (2) the cleavage is virus-specific, as a close member of RSV called human metapneumovirus does not induce tRFs; (3) the cleavage seems dependent on a specific RNase angiogenin (ANG); and (4) the cleavage happens right at the anticodon region of tRNAs. However, the functions of most RSV- induced tRFs, except the one derived from the 5′-end of mature tRNA GluCTC (tRF5-GluCTC), have not been investigated.
In this study, we examined the role of three RSV inducible tRFs derived from the 5′-end of mature tRNAs decoding GlyCCC, LysCTT and CysGCA (named tRF5-GlyCCC, LysCTT and CysGCA, respectively) in controlling RSV replication. Unlike the previously described tRF5-GluCTC, these tRFs seem specific in viral infection, as they have not been reported in other biological settings including cancer and cellular stress [9, 14, 15]. We found that tRF5-GlyCCC and tRF5-LysCTT, but not tRF5-CysGCA, play a significant role in promoting RSV replication, demonstrating the functional specificity of these tRFs. The molecular mechanisms underlying the functions/targeting of tRF5-GlyCCC and tRF5-LysCTT were also investigated. This is the first study to report that two additional novel tRFs are functionally important for RSV replication, therefore providing potential therapeutic targets to be used to control RSV replication. In addition, this study investigated targeting mechanisms of tRF5-GlyCCC and tRF5-LysCTT, generating more data for the development of a tRF-targeting algorithm in the future. Therefore, the results of the study will not only help the virology community to understand the effect of this new family of sncRNAs on virus–host interaction, but also the RNA research community in general regarding methods for tRF function studies and the targeting mechanisms of tRFs.
Results
Expression validation of RSV-induced tRFs
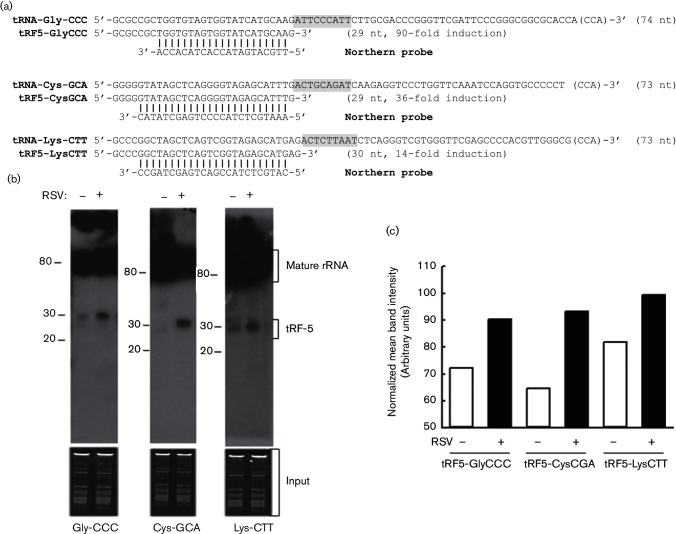
A global change in sncRNA expression in response to RSV infection was recently investigated by Illumina ultra-high-throughput sequencing and sequence mapping. Upon RSV infection, tRFs are robustly induced. The induction of tRFs is virus specific as the infection of hMPV, a closely related respiratory virus, does not result in such a significant change in tRF induction [10]. We scrutinized one of the most abundantly induced tRFs, namely tRF5-GluCTC, to establish the methods to study its gene regulatory mechanisms [10, 13]. However, the functions of other inducible tRFs have not been explored and the experimental methods established from the tRF5-GluCTC study have not been validated. Here, we selected three tRFs with different ranges of induction to confirm the importance of tRFs in RSV infection. They are tRF5-GlyCCC (90-fold induction), tRF5-CysGCA (36-fold induction) and tRF5-LysCTT (14-fold induction), whose induction were confirmed by Northern blot (Fig. 1). Similar to the cleavages of previously studied tRF5-GluCTC, these tRFs are products of the anticodon cleavage of mature tRNAs, supporting that the RNA cleavage is structurally dependent.
Fig. 1.
Experimental validation of tRF-5s. (a) Sequence alignment of tRF-5s with their parental mature tRNAs and Northern probes. The length of each sequence and CCA, post-transcriptionally added to the 3′-end of tRNA, are indicated in parentheses and brackets, respectively. (b) Total RNA from indicated treatments in A549 cells was loaded to a denaturing polyacrylamide gel for Northern hybridization using probes indicated in panel A. Total RNA is shown for equal loading. The positions of tRF-5 and mature tRNA are indicated on the right; molecular size markers are indicated on the left. The blot was exposed for between 8 and 48 h. Data are representative of two–three independent experiments. (c) Densitometric analysis of the tRF bands was performed for Fig. 1(b), using the histogram function of Adobe Photoshop. Basically, the mean tRF intensity was normalized by the corresponding mean intensity of total RNA.
Role of tRF5-GlyCCC and tRF5-LysCTT in viral replication
sncRNAs, if they regulate viral replication, would be ideal tools for controlling viral replication, as they are nonimmunogenic. Therefore, the role of these RSV-induced tRFs in RSV replication was investigated by comparing viral infectious particles produced from the cells treated with antisense oligonucleotide against a specific tRF5 or scrambled control oligo. As shown in Fig. 2(a), suppression of tRF5-GlyCCC and tRF5-LyCTT led to decreased RSV replication by a log. However, the contribution of tRF5-CysCGA seemed insignificant. Western blot for viral protein expression also demonstrated less viral protein expression in infected cells treated with antisense oligo specifically against tRF5-GlyCCC and tRF5-LysCTT, but not against tRF5-CysCGA (Fig. 2b), supporting the result shown in Fig. 2(a). We also used a combination of antisense oligos against tRF5-GlyCCC and tRF5-LyCTT at a 1 : 1 ratio with a final total concentration at 100 nM, and found that treated cells had a about 1.7 log less virus replication than cells treated with control oligos. Conversely, overexpression of mimic for tRF5-GlyCCC or tRF5-LysCTT significantly enhanced RSV replication, further confirming the role of these two tRFs in RSV replication (Fig. 2c, d).
Fig. 2.
Effects of tRFs on RSV replication. (a, b) 100 nM anti-sense oligos (‘anti-tRF5-GlyCCC’, ‘anti-tRF5-LysCTT’, ‘anti-tRF5-CysCGA’ or ‘anti-CN’) were transfected into A549 cells. Two hours post-transfection, cells were mock- or RSV-infected, followed by washing using FK-12 with 2 % FBS for three times and total virus harvesting at 15 h p.i. (hours post-infection) Viral infectious particles were quantified by immunostaining (a). Viral protein synthesis was investigated by Western blot (b). (c, d) A549 cells were transfected with 100 nM of indicated mimic oligos. Two hours post-transfection, cells were mock- or RSV-infected. Two hours post-infection, the supernatant was removed and cells were incubated for additional 15 h, viral titration (c) and viral protein assays (d) were determined. (a, c) Values at the y-axis are a representative of three to four independent experiments and are expressed as mean±standard error (se). *On bars as indicated in the panel denotes a P-value <0.05, relative to the first plain bar [RSV-infected and CN for antisense oligo (α-CN shown in a) or mimic oligo (CN shown in c)]. (e, f) The histogram function of Adobe Photoshop was used to quantify viral bands in (b, d). The intensity sum of viral proteins was normalized by the corresponding β-actin intensity.
Gene regulatory function of tRF5-GlyCCC and tRF5-LysCTT
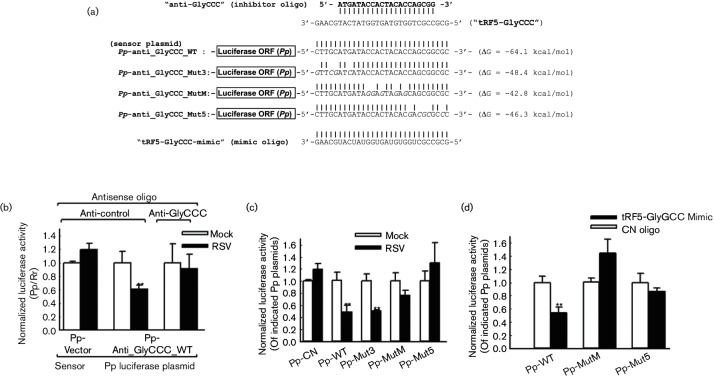
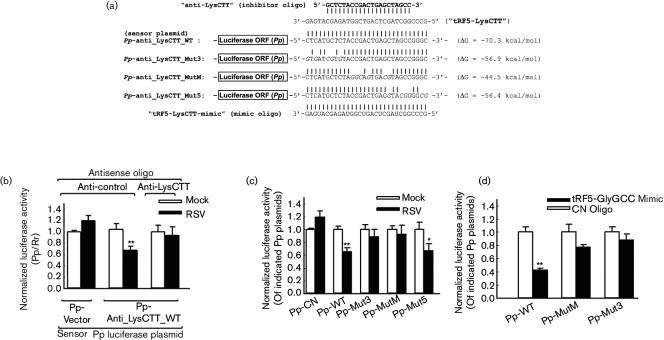
We have previously shown that tRF5-GluCTC promotes RSV replication through its gene trans-silencing function at the post-transcriptional level. tRF5-GlyCCC and tRF5-LysCTT were detected restrictively in the cytosol compartment (Fig. S1, available with the online Supplementary Material). Therefore, if they regulate gene expression, such regulation is likely to be at the level of post-transcription. Here, we first tested whether tRF5-GlyCCC and tRF5-LysCTT also have a gene regulatory capability [13]. If these tRFs have a gene trans-silencing function like tRF5-GluCTC, they would recognize the complementary sequence as a target site, therefore downregulating the expression of Pp harbouring a reverse complementary sequence of tRF5-GlyCCC or tRF5-LysCTT, namely the relative luciferase activity of ‘Pp-anti_GlyCCC_WT’ and ‘Pp-anti_LycCTT_WT’, respectively [10, 16].
As shown in Figs 3(b) and 4(b), compared to mock-infection, RSV infection significantly decreased the relative luciferase activity of ‘Pp-anti_GlyCCC_WT’ and ‘Pp-anti_LycCTT_WT’, but did not affect that of ‘Pp-control’ (left two bars versus middle two bars), suggesting that both induced tRF5-GlyCCC and tRF5-LysCTT have a trans-silencing activity like tRF5-GluCTC. To determine that the observed decrease of luciferase activity was tRF5-GlyCCC- or tRF5-LysCTT-specific, we inhibited the individual tRF by transfecting cells with 20 nt antisense oligonucleotides whose sequences were complementary to tRF5-GlyCCC/tRF5-LysCTT (see Figs 3(a) and 4(a) for their sequence), as previously described [10, 13, 17, 18]. Compared to a non-targeting control oligoribonucleotide (designated as ‘anti-control’), transfection of either of these two tRFs efficiently restored the luciferase activity that was decreased by RSV infection (right two bars versus middle two bars in Figs 3(b) and 4(b)). These data suggest that both tRF5-GlyCCC and tRF5-LysCTT have a gene trans-silencing function.
Fig. 3.
Gene trans-silencing function of tRF5-GlyCCC. (a) The sensor plasmid ‘Pp-anti_GlyCCC’ was constructed by inserting an oligonucleotide, which was complementary to tRF5-GlyCCC, into EcoRI/XhoI sites of pcDNA3.1-Zeo(+)-Pp, as described previously [41]. An empty pcDNA3.1-Zeo (+)-Pp vector was used as a control (designated as ‘Pp-control’). To dissect the target interaction regions of tRF5-GlyCCC, the mutated oligonucleotides were inserted into luciferase reported as instructed. (b) To investigate the effect of RSV-induced tRF5-GlyCCC on mRNA expression, A549 cells were co-transfected with Pp-anti_GlyCCC sensor plasmids (firefly plasmids), pRL-CMV plasmids expressing Rr (renilla luciferase) and anti-tRF5-GlyCCC oligonucleotides (anti-GlyCCC), using Lipofetamine 2000 according to the manufacturer’s instructions (Invitrogen). Pp-control plasmids and anti-control oligonucleotides were used as negative controls. After 2 h of transfection, the cells were infected with mock or RSV at an m.o.i. of 1. At 15 h p.i., cells were lysed for assays by using a dual-luciferase kit (Promega, Madison, WI). Data normalization was done by correcting Pp luciferase by the Rr luciferase within the same group and normalized value in the RSV-infected group was further normalized to its corresponding mock-infected group within same oligo treatment. Values at the y-axis are a representative of three to four independent experiments and are expressed as mean±standard error (se). ** On second black bars (RSV infected and Pp-anti_GlyCCC and anti-control oligo-treated) as indicated denotes a P-value<0.01, relative to the second plain bar (mock infected and anti-control for antisense oligo). (c) To identify targeted regions of tRF5-GlyCCC, A549 cells were co-transfected with Pp-anti_GlyCCC sensor plasmids containing various mutation in regions as indicated and Rr expressing plasmids. Pp-anti_GlyCCC sensor (Pp-WT) and Pp-vector (Pp-CN) plasmids were used as positive and negative controls, respectively. The infection and luciferase normalization were done as described in (b). Values at the y-axis are a representative of three to four independent experiments and are expressed as mean±standard error (se). ** On black bars, as indicated, denotes a P-value<0.01, relative to the corresponding open or plain bar in the same group. (d) The gene trans-silencing function of tRF5-GlyCCC was also done in the context of tRF5-GlyCCC-mimic treatment. For ectopic expression of tRF5-GlyCCC, tRF5-GluCCC-mimic in siRNA format was designed and synthesized from Ambion. The same design with a scrambled sequence was used as a control (‘CN Oligo’). A549 cells were cotransfected with mimics (tRF5-GlyCCC specific or CN oligo), Pp plasmids (Pp-anti_GlyCCC WT or containing mutations in the middle and 5′-end of tRF5-GlyCCC) and Rr-expressing plasmids using Lipofectamine 2000. Cells were harvested for dual-luciferase assays at 30 h post-transfection. Data normalization was done by correcting Pp luciferase by the Rr luciferase within the same group and the normalized value of the tRF5-GlyCCC mimic-treated group was further normalized to its corresponding CN-oligo-treated group within the same Pp vector. Values at the y-axis are a representative of three to four independent experiments and are expressed as mean±standard error (se). ** On first black bars (Pp-WT transfected and tRF5-GlyCCC mimic-treated), as indicated, denotes a P-value<0.01, relative to the first open or plain bar (Pp-WT transfected and CN mimic-treated).
Fig. 4.
Gene trans-silencing function of tRF5-LysCTT. (a) To identify whether tRF5-LysCTT has the gene trans-silencing function and associated regulatory mechanism, the sensor plasmid ‘Pp-anti_LysCTT’ or its mutants were constructed as described in Fig. 3(a). (b). To define the gene trans-silencing function of tRF5-LysCTT, dual-luciferase assays were done as described in Fig. 3(b), except Pp-anti_LysCTT sensor plasmids (firefly plasmids) and anti-LysCTT oligonucleotides were used. ** On second black bars (RSV infected, Pp-anti_LysCTT and anti-control oligo-treated), as indicated, denotes a P-value<0.01, relative to the second open or plain bar (mock-infected and Pp-anti_LysCTT and anti-control oligo-treated). (c) To identify targeting mechanisms of tRF5-LysCTT, the experiment was carried out as described in Fig. 3(c) using the plasmids designed in Fig. 4(a). (d) To confirm the interacting regions identified in Fig. 4(c), tRF5-LysCTT mimic was used to investigate its suppression on the luciferase expression of Pp-anti-LysCTT (Pp-WT) and motif mutants identified in Fig. 4(c).
In this study, we also used mutants of ‘Pp-anti_GlyCCC_WT’ and ‘Pp-anti_LycCTT_WT’ to determine the sequence requirements for the trans-silencing capacity of these two tRFs. 3~4 nt mutations at the 3′-, middle- or 5′-region of the tRF5-GlyCCC or tRF5-LysCTT target sites were introduced into the sensor plasmids ‘Pp-anti_GlyCCC’ and ‘Pp-anti_LysCTT’ as depicted in Figs 3(a) and 4(a), respectively. As shown in Fig. 3(c), contrary to Pp-anti_GlyCCC_WT or ‘Mut3’, the luciferase activity of the other two mutants had impaired responsiveness to RSV infection, indicating that the 5′- and the middle regions of tRF5-GlyCCC were critical for its trans-silencing activity. Hence, the sequence requirement for tRF5-GlyCCC was slightly different from that of miRNAs whose 5′-portion (called the ‘seed sequence’) was almost solely responsible for target recognition. The trans-silencing mechanism used by tRF5-GlyCCC was also distinct from that of tRF5-GluCTC whose 3′-portion was the most important one for its trans-silencing function.
The importance of 5′- and middle-portions of tRF5-GlyCCC were also confirmed by investigating the luciferase expression of ‘Pp-anti_GlyCCC’, WT, Mut5 or MutM, in response to the ectopic expression of tRF5-GlyCCC, as described previously [10]. Upon transfection of this mimic into A549 cells, the luciferase activity of ‘Pp-anti_GlyCCC_WT’, was significantly suppressed by ‘tRF5-GlyCCC-mimic’ (Fig. 3d). However, the luciferase expression of Mut-3 and Mut-Middle was not sensitive to the treatment of tRF5-GlyCCC-mimic (Fig. 3d). In summary, our data demonstrated that tRF5-GlyCCC suppressed target gene expression, through a mechanism distinct from the recently characterized tRF5-GluCTC.
Similar strategies were applied to identify functional regions of tRF5-LysCTT. As shown in Fig. 4(c, d), when the 3′- and middle-portions were mutated, the luciferase expression became significantly less responsive to RSV infection or the ectopic expression of tRF5-LysCTT, compared to the luciferase expression controlled by the sequence complementary to WT tRF5-LysCTT, suggesting that these two regions were important to its gene trans-silencing function. This sequence requirement for gene silencing was different from miRNAs but similar to tRF5-GluCTC. Collectively, in our mutant experiments for several tRFs, the degree of trans-silencing activity did not simply correlate to overall °G values and each tRF needed different regions for base pairing to the targets, suggesting that certain specific mechanisms seem to exist for individual tRFs.
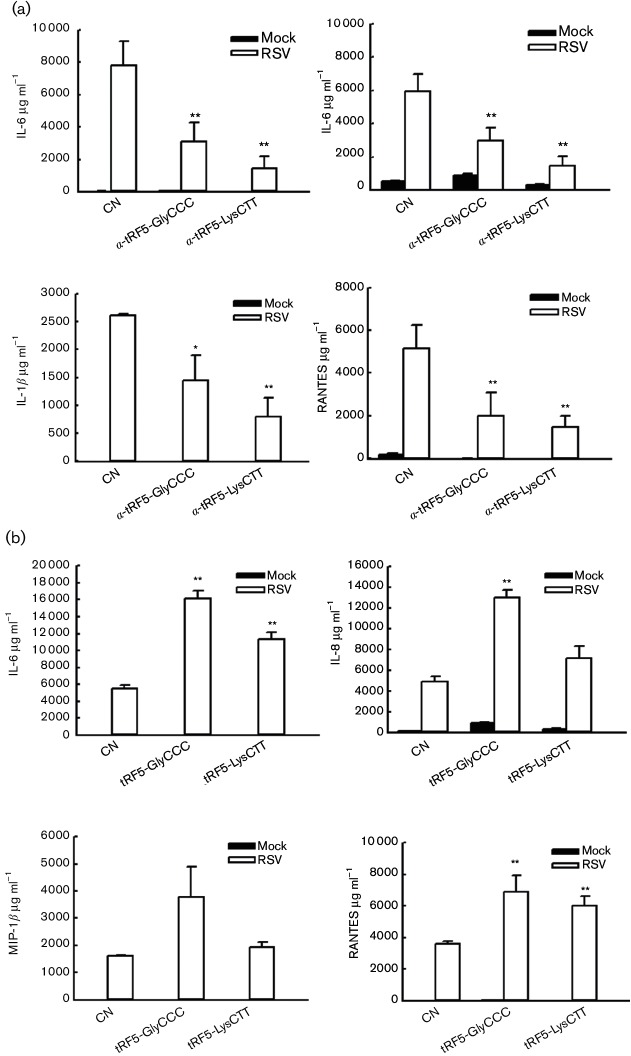
Effects of tRF5-GlyCCC and tRF5-LysCTT on RSV-induced cytokines and chemokines
Inflammation is always associated with viral infection. In this study, we also compared RSV-induced chemokines and cytokines in cells treated with a control antisense oligo or an oligo specifically against tRF5-GlyCCC or tRF5-LysCTT (Fig. 5a). Downregulation of these two tRFs suppressed the induction of IL-6, IL-8, MIP-1b and RANTES by RSV, while the treatment of tRF5-GlyCCC mimic stimulated cells to produce these in response to RSV (Fig. 5b). We also observed that tRF5-LysCTT enhanced the induction of IL-6 and RANTES, but not MIP-β and IL-8 in response to RSV infection (Fig. 5b). Although IL-8 induction in tRF5-LysCTT mimic-treated cells was not significant (P=0.7), there was a tendency of increased induction. Overall, these data suggest that antisense treatment may not only reduce RSV replication, but also associated inflammation, providing a more promising direction for using anti-tRF oligo to control both RSV replication and pathogenesis.
Fig. 5.
Effects of tRF5-GlyCCC and tRF5-LysCTT on RSV-induced cytokines and chemokines. A549 cells were transfected with 100 nM of indicated anti-oligos (a) or tRF5 mimic (b). Two hours post-transfection, cells were mock- or RSV-infected. Fifteen hours post-infection, supernatant was harvested to measure secretion of cytokines/chemokines by Bioplex or ELISA, as described previously [42]. ELISA kits for IL-8/RANTES and IFN-β were from R and D (R and D system, Minneapolis, MN) and PBL (PBL Interferon Source, Piscataway, NJ), respectively. Data shown are representative of three independent experiments. * P<0.05 relative to CN-treated and RSV-infected cells (black bars).
Biogenesis of tRF-GlyCCC and tRF5-LysCTT
We have previously shown that tRF5-GluCTC is generated by an endonuclease angiogenin (ANG). Close inspection of tRFs in the high-throughput data revealed several consensus features of induced tRFs, including the enrichment of 5′-end, but not 3′-end fragments and the preferential cleavage sites at the 5′-side of the anti-codon loop, suggesting ANG may be also responsible for the cleavage of other RSV-induced tRFs.
To test this hypothesis, we artificially suppressed ANG by a siRNA and measured tRF5-GlyCCC and tRF5-LysCTT in RSV-infected A549 cells. When the siRNA led to the successful knockdown of ANG (Fig. 6a), the induction of tRF5-GlyCCC and tRF5-LysCTT by RSV infection was significantly (>50 %) attenuated (Fig. 6b). For comparison, we did similar knockdown experiments for Dicer, also a cytosol endonuclease, but responsible for the biogenesis of miRNAs, another sncRNA family. Interesting, although these two tRFs have gene trans-silencing activity, their induction by RSV seems to be Dicer-independent. Suppressing siRNAs specific against Dicer did not affect the induction of tRF5-GlyCCC and tRF5-LysCTT upon RSV infection (Fig. 6b). Collectively, our data support our hypothesis that the production of tRF-5s upon RSV infection specifically required ANG.
Fig. 6.
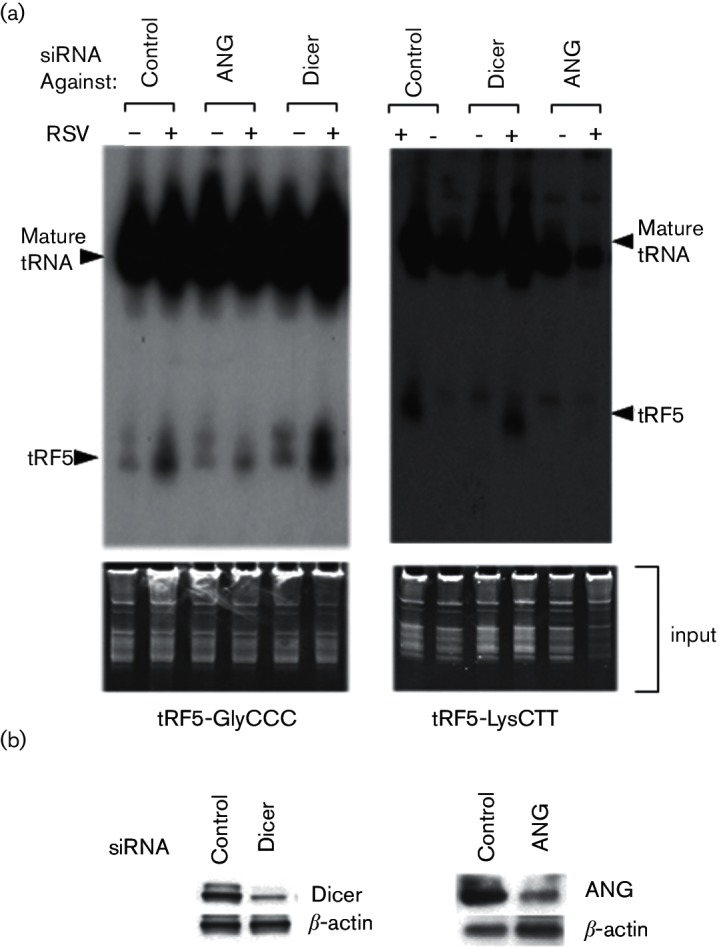
tRF5-GlyCCC and tRF5-LysCTT are generated by Angiogenin. (a) A549 cells were transfected with 100 nM of siRNA against indicated proteins or scrambled siRNA as a negative control. At 40 h post-transfection, the cells were mock- or RSV-infected for 15 h. Total RNAs were then subjected to Northern hybridization as described in Fig. 1(b) (top panel). EtBr staining is shown for equal loading (bottom panel). (b) The suppression of target proteins by each siRNA was confirmed by Western blot.
Discussion
Unbiased deep sequencing of human sncRNAs has shown the changes of tRFs in response to various types of diseases, including cancer, neuro-developmental disorders and infections; however, the significance of tRFs in diseases and the associated molecular mechanisms remain largely unknown, mainly due to the lack of methods for the functional studies [9, 10, 19–21]. Following our report on the function of tRF5-GluCTC in regulating RSV replication [10, 22–24], many recent studies suggest the role of tRFs in infectious diseases; however, other important tRFs in infectious diseases have not been identified. Here, we identified two novel tRF(s), other than tRF5-GluCTC, that play a critical role in regulating RSV replication and the associated host responses. We found that tRF5-GlyCCC and tRF5-LysCTT have such functions, based on their regulation of RSV replication, their ability in gene trans-silencing and their impact on RSV-induced cytokines/chemokines.
In an effort to develop a tRF targeting algorithm for future use, we first attempted to determine a critical region(s) for gene trans-silencing functions. We found that, like tRF5-GluCTC, tRF5-LysCTT used its 3′- region to recognize the target site ([10] and Fig. 4c), while the 5′-end of tRF5-GlyCCC was critical in gene targeting (Fig. 3c). Interestingly, the middle region of these ~30 nt tRFs were also important in gene trans-silencing in all the three tRFs. Although there were too few cases to draw a general conclusion, it appears that some degree of base pairing interaction at the middle portion is required for anchoring tRFs to target mRNAs and then additional interaction at the 5′ or 3′-portions is also needed for trans-silencing action. For a consensus conclusion, more mutant data will be needed. In addition, identification/characterization of protein components should be included to elucidate the target recognition rule.
It has been shown that the cytosine methylation of tRFs may play an important role in neuro-developmental disorders, suggesting that the modification of tRFs is critical for their function [20, 21]. However, the methylation of tRFs may not affect their interaction with target regions, as the cytosine-rich region of all three tRFs is located in the 5′-end. In addition, the 5′-end regions of tRF5-GlyCCC and tRF5-LysCTT are quite similar, yet their roles in gene trans-silencing are quite different (Fig. 1). The different targeting mechanisms of tRFs are an interesting research focus for the future. In addition, to finalize the targeting algorithm, more function and targeting mechanism assays are needed for other induced tRFs. Nevertheless, based on the cytosolic location (Fig. S1), we believe that the gene trans-silencing function of all three tRFs is at the protein translation and/or RNA stability level, as with miRNAs.
Previously described tRNA halves identified in bacteria, fungi and plants have the cleavage sites throughout the anticodon loop and the TψC arm [25–27], while the cleavage sites of RSV-induced tRFs are biased towards the 5′-side of the anticodon ([10] and Fig. 1a). Species-specific RNases, nuclease isoforms and cellular stimuli are key factors responsible for the difference in tRNA cleavage [15, 25, 28, 29]. We have previously shown that, in response to infection, mammalian cells use ANG for tRNA cleavage, which results in ~30 nt 5′-end derived tRFs (Fig. 1a and [10, 22]). In addition to ANG, Dicer is also responsible for producing tRF5s, but a 19 mer in mammalian cells under stress [28]. In response to RSV infection, we confirmed the importance of ANG, but not Dicer, in producing 30 mer tRF5s (Fig. 6a). Overall, the stimuli- and endonuclease-specific tRNA cleavage supports that tRFs are biologically meaningful [9, 15, 28, 29].
Despite the early ignorance of the presence of abundant tRFs in next generation sequencing data, our data and the accumulating evidence from others support tRFs as being an important class of functional molecules. In addition to their fragmentation patterns as discussed, we found tRFs have gene silencing functions. The gene silencing functions of tRFs are consistent with studies showing some tRFs being bound to Argonaute/Piwi proteins, well-known components of the RNA-induced silencing complex [30–35]. In this study, we are particularly interested in the role of tRFs in the replication of RSV and the associated host response, as the pulmonary RSV load is correlated to the disease severity [36–38]. We found that both tRF5-GlyCCC and tRF5-LysCTT play significant roles in controlling inflammation and RSV replication, suggesting a translational potential or conceptual framework for designing therapeutic molecules. Although tRF5-CysGCA seems unimportant in controlling RSV replication, this does not exclude it from being critical in the infection. We will continue to investigate its biological function(s), including but not limited to its role in growth factor induction and the host mRNA modification. Given the fact that only a subset of tRNAs are cleaved by RSV infection, we believe there is a reason for tRNA-CysCGA to be cleaved. Overall, we have validated our experimental methods to study the functions of tRFs. These methods are helpful to define the roles of tRFs in other biological settings, including stress responses, cell proliferation and tissue development. The feature of endonuclease-specific tRNA cleavage also provides new insights into the disease control, for example, by modifying the activity or the expression of the endonuclease. We are accumulating valuable data to develop the targeting algorithm of tRFs, which, we hope, will also accelerate functional studies of tRFs in a near future.
Methods
Cell lines and virus
HEp-2 (Human epithelial type 2), A549 (human alveolar type II-like epithelial cells) and 293 (human embryonic kidney epithelial cells) cells were from ATCC, Manassas, VA, and maintained as described previously [13, 39, 40]. RSV A2 strain was grown in HEp-2 cells and purified by sucrose-gradient as we previously described [10, 13, 40]. Viral titre was determined by immunostaining in HEp-2 cells using polyclonal biotin-conjugated goat anti-RSV antibody (Ad direct, Barberton, OH) and streptavidin peroxidase polymer (Sigma, St. Louis, MO) sequentially, as described previously [10, 13].
Viral infection
A549 cells, treated with/without oligonucleotides against or mimicking an interested tRF, were infected with RSV, at a multiplicity of infection (m.o.i.) of 1. An equivalent amount of fresh FK-12 medium with 2 % FBS was added to uninfected cells, as control (mock infection). After initial absorption, viral inoculum was removed and cells were supplied with fresh medium with 2 % FBS. Fifteen hours later, infected cells were scrapped into the medium, followed by sonication to release intracellular viruses and centrifugation. Viruses in the supernatant were titrated by immunostaining in HEp-2 cells, as described above.
RNA extraction and expression confirmation of tRFs by Northern blot
Total cellular RNA was extracted by TRIzol Reagents from Thermo Fisher Scientific, Waltham, MA. Northern hybridization for sncRNAs were performed, as described previously [10, 13]. Briefly, RNA was separated in an 8 % denaturing polyacrylamide gel with 7M urea and then transferred to a positively charged nylon membrane (Amersham Biosciences, Piscataway, NJ). The membrane was hybridized with 32P-lablled probes in ULTRAhyb-Oligo solution (Life Technologies, Grand Island, NY), followed by washing according to the manufacturer’s instructions.
Construction of luciferase sensor plasmids and luciferase assays
The sensor plasmid ‘Pp-anti_GlyCCC_WT’ and ‘Pp-anti_LysCTT_WT’ were constructed to investigate whether tRF5-GlyCCC and/or tRF5-LysCTT have gene silencing functions. Briefly, an oligonucleotide which was complementary to tRF5-GlyCCC or tRF5-LysCTT, respectively, was inserted into EcoRI/XhoI sites of pcDNA3.1-Zeo(+)-Pp, as we previously described [10]. The paired primers used for insertion were: 5′-AATTCTTGCATGATACCACTACACCAGCGGCGC-3′ and 5′-TCGAGCCCCGCTGGTGTAGTGGTATCATGCAAG-3′ (bold letters represent extra nts to generate EcoRI/XhoI overhangs) for tRF5-GlyCCC, and 5′-AATTCTCATGCTCTACCGACTGAGCTACCCGGGC-3′ and 5′-TCGACCCCGGCTAGCTCAGTCGGTAGAGCATGAG-3′ for tRF5-LysCTT. An empty pcDNA3.1-Zeo (+)-Pp vector was used as a control (designated as ‘Pp-vector’). Three mutant plasmids were constructed in the same manner (their mutated sites shown in Figs 3(a) and 4(a)) to identify which domain(s) are sensitive to RSV-induced tRF5-GlyCCC and/or tRF5-LysCTT.
To investigate whether tRF5-GlyCCC and tRF5-LysCTT on the luciferase expression are controlled specifically by the complimentary sequences of tRFs, A549 cells were co-transfected with Pp-anti_GlyCCC or Pp-anti_LysCTT sensor plasmids (firefly plasmids), pRL-CMV plasmids expressing Rr (renilla luciferase) and anti-GlyCCC or anti-LysCTT oligonucleotides, using Lipofetamine 2000 according to the manufacturer’s instructions (Invitrogen). Pp-vector plasmids and/or anti-control oligonucleotides were used as negative controls. After 2 h of transfection, the cells were infected with mock or RSV. At 15 h post infection (p.i.), cells were lysed for luciferase assays using a dual-luciferase kit (Promega, Madison, WI) to assess the effect of antisense oligos on the luciferase expression. The sequences of ‘anti-GlyCCC’ and ‘anti-LysCTT’ are shown in Figs 3(a) and 4(a). Data processing and normalization are described in the legends of Figs 3 and 4. Synthetic anti-tRF5-GlyCCC or anti-tRF5-LysCTT oligonucleotides, purchased from Sigma, were 20 nt mixed oligonucleotides that contain a backbone phosphorothioate and have 5 nts on each end substituted with 2′-O-methyl ribonucleotides [10, 17, 18]. We also used mutant Pp plasmids to confirm the inhibition specificity by interested tRFs and to identify the domains of tRFs responsible for the gene inhibition.
The gene regulatory function of tRF5-GlyCCC or tRF5-LysCTT was also performed in ectopic expression conditions, where mimic dsRNAs were transfected. ‘tRF-specific-mimic’ and ‘Control-mimic’ were purchased also from Sigma. A549 cells were co-transfected with the mimics and luciferase sensor plasmids using Lipofectamine 2000. Cells were harvested for dual-luciferase assays at 40 h post-transfection. All other details, such as the concentrations of oligonucleotides and plasmids, are described in Figs 3(a) and 4(a).
RNA interference
siRNAs against angiogenin or Dicer were purchased from Sigma or Invitrogen. 100 nM of siRNA was transfected into A549 cells, by using Mirus (Madison, WI) according to the manufacturer’s recommendations. Forty hours later, A549 cells were mock- or RSV-infected for 15 h at an m.o.i. of 1.
Western blot analysis
The total cell lysate after the infection was prepared using RIPA buffer. The lysates were collected and quantified with a protein quantification kit from Bio-Rad, followed by fractionation by SDS-PAGE. The separated proteins were then transferred to polyvinylidene difluoride membranes. Membranes were blocked with 5 % milk in Tris-buffered saline (TBS)-Tween 20 and incubated with the proper primary antibodies according to manufacturer's instructions.
Cytokine and chemokine quantification
The levels of chemokines and cytokines shown in Fig. 5 were quantified by using a Multi-Analytic Profiling Human Cytokine/Chemokine Kit (Bio-Rad, Hercules, CA), according to the manufacturer's instructions. Data were analysed using the Multiplex Analyst Software from Bio-Rad.
Statistical analysis
Statistical significance was analysed using analysis of variance (ANOVA). P value of less than 0.05 was considered significant. Mean±standard error (se) is shown.
Funding information
This work was supported by grants from the National Institutes of Health-National Institute of Allergy and Infectious Diseases 1R01AI107033-01 and R21AI113771-01A1, and American Thoracic Grant unrestricted Research Grant to X. B.
Acknowledgements
The authors thank Dr Animesh Chandra and Betty Johnson for assistance with manuscript editing and submission supported in part by a Clinical and Translational Science Award (UL1TR001439) from the National Center for Advancing Translational Sciences, National Institutes of Health.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
References
- 1.Lay MK, Bueno SM, Gálvez N, Riedel CA, Kalergis AM. New insights on the viral and host factors contributing to the airway pathogenesis caused by the respiratory syncytial virus. Crit Rev Microbiol. 2015;42:1–13. doi: 10.3109/1040841X.2015.1055711. [DOI] [PubMed] [Google Scholar]
- 2.Afonso CL, Amarasinghe GK, Bányai K, Bào Y, Basler CF, et al. Taxonomy of the order Mononegavirales: update 2016. Arch Virol. 2016;161:2351–2360. doi: 10.1007/s00705-016-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazur NI, Martinon-Torres F, Baraldi E, Fauroux B, Greenough A, et al. Lower respiratory tract infection caused by respiratory Syncytial Virus: current management and new therapeutics. Lancet Respir Med. 2015;3:888–900. doi: 10.1016/S2213-2600(15)00255-6. [DOI] [PubMed] [Google Scholar]
- 5.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomes AQ, Nolasco S, Soares H. Non-coding RNAs: multi-tasking molecules in the cell. Int J Mol Sci. 2013;14:16010–16039. doi: 10.3390/ijms140816010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q, Lee I, Ren J, Ajay SS, Lee YS, et al. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther. 2013;21:368–379. doi: 10.1038/mt.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saikia M, Krokowski D, Guan BJ, Ivanov P, Parisien M, et al. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem. 2012;287:42708–42725. doi: 10.1074/jbc.M112.371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng J, Ptashkin RN, Chen Y, Cheng Z, Liu G, et al. Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol Ther. 2015;23:1622–1629. doi: 10.1038/mt.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pekarsky Y, Balatti V, Palamarchuk A, Rizzotto L, Veneziano D, et al. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci USA. 2016;113:5071–5076. doi: 10.1073/pnas.1604266113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YS, Nakahara K, Pham JW, Kim K, He Z, et al. Distinct roles for drosophila dicer-1 and dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/S0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 17.Yoo BH, Bochkareva E, Bochkarev A, Mou TC, Gray DM. 2'-O-methyl-modified phosphorothioate antisense oligonucleotides have reduced non-specific effects in vitro. Nucleic Acids Res. 2004;32:2008–2016. doi: 10.1093/nar/gkh516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ideue T, Hino K, Kitao S, Yokoi T, Hirose T. Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. RNA. 2009;15:1578–1587. doi: 10.1261/rna.1657609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olvedy M, Scaravilli M, Hoogstrate Y, Visakorpi T, Jenster G, et al. A comprehensive repertoire of tRNA-derived fragments in prostate cancer. Oncotarget. 2016;7:24766–24777. doi: 10.18632/oncotarget.8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–2039. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanco S, Frye M. Role of RNA methyltransferases in tissue renewal and pathology. Curr Opin Cell Biol. 2014;31:1–7. doi: 10.1016/j.ceb.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong B, Lee YS, Lee I, Shelite TR, Kunkeaw N, et al. Compartmentalized, functional role of angiogenin during spotted fever group rickettsia-induced endothelial barrier dysfunction: evidence of possible mediation by host tRNA-derived small noncoding RNAs. BMC Infect Dis. 2013;13:285. doi: 10.1186/1471-2334-13-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selitsky SR, Baran-Gale J, Honda M, Yamane D, Masaki T, et al. Small tRNA-derived RNAs are increased and more abundant than microRNAs in chronic hepatitis B and C. Sci Rep. 2015;5:7675. doi: 10.1038/srep07675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Zhang Y, Shi J, Zhang H, Cao Z, et al. Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J Mol Cell Biol. 2014;6:172–174. doi: 10.1093/jmcb/mjt052. [DOI] [PubMed] [Google Scholar]
- 25.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem. 2005;280:42744–42749. doi: 10.1074/jbc.M510356200. [DOI] [PubMed] [Google Scholar]
- 27.Haiser HJ, Karginov FV, Hannon GJ, Elliot MA. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 2008;36:732–741. doi: 10.1093/nar/gkm1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raina M, Ibba M. tRNAs as regulators of biological processes. Front Genet. 2014;5:171. doi: 10.3389/fgene.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10:1798–1806. doi: 10.4161/rna.27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Silva MR, Cabrera-Cabrera F, Güida MC, Cayota A. Hints of tRNA-derived small RNAs role in RNA silencing mechanisms. Genes. 2012;3:603–614. doi: 10.3390/genes3040603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sobala A, Hutvagner G. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip Rev RNA. 2011;2:853–862. doi: 10.1002/wrna.96. [DOI] [PubMed] [Google Scholar]
- 35.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Devincenzo JP, El Saleeby CM, Bush AJ. Respiratory syncytial virus load predicts disease severity in previously healthy infants. J Infect Dis. 2005;191:1861–1868. doi: 10.1086/430008. [DOI] [PubMed] [Google Scholar]
- 37.Devincenzo JP, Wilkinson T, Vaishnaw A, Cehelsky J, Meyers R, et al. Viral load drives disease in humans experimentally infected with respiratory syncytial virus. Am J Respir Crit Care Med. 2010;182:1305–1314. doi: 10.1164/rccm.201002-0221OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El Saleeby CM, Bush AJ, Harrison LM, Aitken JA, Devincenzo JP. Respiratory syncytial virus load, viral dynamics, and disease severity in previously healthy naturally infected children. J Infect Dis. 2011;204:996–1002. doi: 10.1093/infdis/jir494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bao X, Indukuri H, Liu T, Liao SL, Tian B, et al. IKKε modulates RSV-induced NF-κB-dependent gene transcription. Virology. 2010;408:224–231. doi: 10.1016/j.virol.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren J, Liu T, Pang L, Li K, Garofalo RP, et al. A novel mechanism for the inhibition of interferon regulatory factor-3-dependent gene expression by human respiratory syncytial virus NS1 protein. J Gen Virol. 2011;92:2153–2159. doi: 10.1099/vir.0.032987-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee K, Kunkeaw N, Jeon SH, Lee I, Johnson BH, et al. Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA. 2011;17:1076–1089. doi: 10.1261/rna.2701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bao X, Liu T, Spetch L, Kolli D, Garofalo RP, et al. Airway epithelial cell response to human metapneumovirus infection. Virology. 2007;368:91–101. doi: 10.1016/j.virol.2007.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.