ABSTRACT
Drosophila melanogaster chromosome 4 is an anomaly because of its small size, chromatin structure, and most notably its lack of crossing over during meiosis. Earlier ideas about the absence of crossovers on 4 hypothesize that these unique characteristics function to prevent crossovers. Here, we explore hypotheses about the absence of crossovers on 4, how these have been addressed, and new insights into the mechanism behind this suppression. We review recently published results that indicate that global crossover patterning, in particular the centromere effect, make a major contribution to the prevention of crossovers on 4.
KEYWORDS: Bloom syndrome helicase, chromosome 4, crossover patterning, meiosis, recombination
Preface: Opposing views of Drosophila geneticists on chromosome 4
As a graduate student, I (JS) did my graduate research under the direction of the late Bill Gelbart at Harvard University. My rotation project was to inject a P element transgene construct and then screen for and map any integrants. One integration did not map to the X, 2, or 3, so I told Bill it must have landed on 4. Bill instructed me to autoclave the stock immediately so as not to contaminate the laboratory with something associated with the fourth chromosome. “God gave flies the fourth chromosome so they wouldn't be perfect,” he said. Bill's position was based on the absence of crossovers on 4, which prevented one from doing “real” genetics involving that chromosome. My postdoctoral advisor, Scott Hawley, has the opposite relationship with 4 and has made numerous contributions to understanding unique aspects of the biology of this chromosome, particularly how it segregates in meiosis in the absence of chiasmata.1 Intentionally or not, Bill and Scott's positions helped spark my own interest in chromosome 4.
The absence (and presence) of crossovers on 4
Much of the attraction to chromosome 4 stems from its lack of crossovers, which has been a puzzle for 90 y. In his influential book The Theory of the Gene, T.H. Morgan presented a map of 3 chromosome 4 genes in the order bent (bt) - shaven (sv) - eyeless (ey).2 Both the order and relative distances were wrong; bt is adjacent to ey in the middle of 4, and sv is toward the distal end. The errors occurred because the presumed recombinants were actually cases of nondisjunction.3,4 True crossovers on 4 have been observed, but only under special conditions. Perhaps most notable were the studies of Sturtevant, who found that crossovers are “greatly elevated” in diplo-4 triploid females.5 He used this finding to build a genetic map of 4, reporting 3.0 map units between the most proximal and distal genes known (ci and sv).6 Additionally, it has been reported that heat shock results in crossovers on chromosome 4,7 but it is unknown if these are true meiotic events. Although these cases support the possibility of crossover formation on 4, they do not seem to provide insight into the mechanisms regulating crossover inhibition on 4 in a normal meiosis.
Previous hypotheses for why 4 lacks crossovers have focused on the unusual physical characteristics of this chromosome, including its small size, repetitive sequence, and heterochromatic structure, but studies reported recently by Hatkevich et al. have contributed new insights regarding the regulation of recombination on 4.8 Hatkevich et al. provide support for the idea that the meiotic crossover patterning processes that establish crossover distributions characteristic of chromosomes X, 2, and 3 also prevent crossovers on 4. Here, we review and assess the idea that the absence of crossovers on 4 stems from its physical characteristics and how crossover patterning processes may play a role.
Can unique physical properties of 4 explain the absence of crossovers?
The fourth chromosome in Drosophila melanogaster is much smaller than the other chromosomes, and is often referred to as the “dot chromosome” due to its observed small size in metaphase spreads. It has been suggested that crossovers on chromosome 4 in Drosophila melanogaster do not occur due its physical size. Interestingly, Chino and Kikkawa observed that the small chromosome in Drosophila virilis, which is similar in size to Drosophila melanogaster chromosome 4, does have meiotic crossovers.9 This discrepancy between the 2 species may be explained by the fact that D. virilis has a much higher rate of crossing over on other chromosomes compared with D. melanogaster. For example, the X is the same physical size in D. virilis and D. melanogaster, but D. virilis has about 3 times as many crossovers on the X.10 These data lead Chino and Kikkawa to hypothesize that a combination of the fourth's small size and the overall low crossover rate in D. melanogaster results in such a low probability of crossovers that they are undetectable.
To address the argument that we have not seen crossovers on 4 due its small size and low rate of crossing over, we can make comparisons with data from another chromosome (Fig. 1). The assembled sequences of proximal 2L and 4 have similar chromatin domains based on ChIP studies from several Drosophila cell lines.11 In the GBrowse chromatin tracks on Flybase,12 most of chromosome 4 is classified as heterochromatin. Proximal 2L is similarly classified as heterochromatin from approximately 22 Mb to the end of the assembly. Chromosome 4 is about 4–5 Mb, of which 1.2 Mb is assembled in the genome sequence.13-15 The pericentric heterochromatin on 2L makes up approximately 5.4 Mb. As on 4, most of this is composed of highly repeated tandem (satellite) sequences, but 1.5 Mb adjacent to proximal 2L euchromatin has been assembled in the genome sequence.15,16 A common interval on 4 where crossover events are scored is from ci to sv, which spans 1.03 Mb. The ci to sv interval and the sequenced region of proximal 2L are approximately the same distance from the centromere, providing a good comparison between 2L proximal crossovers and crossovers on 4.
Figure 1.
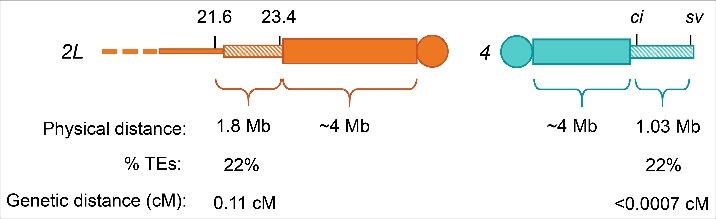
Comparison of proximal 2L and 4. Heterochromatin distances are as reported in Adams et al.16 and Sun et al..14 Distance of euchromatin and percentage transposable elements are from the v6.0 Drosophila melanogaster assembly.15 Genetic distance for 4 is from Sandler and Szauter.17 The distance for proximal 2L interval was calculated from our unpublished described in the text; flies with crossovers between pr and cn were collected and crossover sites were more finely mapped by genotyping with the SNP/indel markers shown (numbers represent positions of on the v6.0 assembly. Chromosomes not drawn to scale.
We have identified SNPs and indels from the genome assembly that span the assembled heterochromatin proximal to the centromere to more finely map crossovers near the centromere. Figure 1 shows the location of 2 of these SNPs/indels, at 21.6 Mb and 23.4 Mb (v6.0 assembly). We collected crossovers between pr and cn and then genotyped them for these 2 genetic markers. We recovered 8 crossovers in the 21.6Mb – 23.4Mb interval region from 7,399 flies scored (unpublished). This gives a genetic distance of 0.11 cM (“map units” are traditionally used to describe recombination frequencies in Drosophila; we use the equivalent but more widely used centiMorgan, cM, here). Using the comparison between proximal 2L and 4, the ci – sv interval on 4 would have an expected genetic distance of 0.06 cM. Based on the number of flies scored for crossovers on 4, it would be very unlikely that the crossovers were simply missed. In one notable example, Sandler and Szauter17 found no crossovers among 58,702 flies, yielding an upper limit of 0.0007 cM. From this comparison, we can infer that the small size of chromosome 4 and rate of recombination are not the only factors preventing crossovers.
If size alone does not account for the lack of crossovers on 4, perhaps the sequence makeup of 4 contributes to crossover prevention. A large fraction of the region that has been assembled in the genome sequence consists of transposable elements (TEs): 22% of the ci – sv interval in the v6.0 assembly.18,19 In some organisms, recombination rates are lower in regions of high TE density and absent within TEs themselves.20 Miller et al. demonstrated that only one of 541 Drosophila crossovers they mapped through whole-genome sequencing was within a TE,21 suggesting crossovers are reduced within TEs but not completely absent. The 2L region described above, from 21.6 Mb to 23.4 Mb, is 22% TE in the reference genome, similar to the ci – sv interval on 4 (but it should be noted that we do not know TE structure and density on the chromosomes used in the experiments reported here). Since these 2 intervals are comparable in size and transposable element density, we would expect them to have a similar recombination rate and, thus, genetic distance. Therefore, it is unlikely that TE density alone is responsible for preventing 4 crossovers.
Another aspect of the makeup of chromosome 4, and closely related to the factor of heterochromatic sequences, is chromatin structure. Chromatin structure modifications have been studied in the context of chromosome 4, although there are data on the other autosomes that suggest chromatin structure could play a role in preventing crossovers. For example, in suppressors of variegation (Su(var) mutants), chromatin structure is modified so that heterochromatin is in a more open state, and found that Su(var) mutants resulted in an increase in crossovers proximal to the centromere on both chromosomes 2 and 3.22 However, these studies did not look at crossovers on chromosome 4. It would be interesting to see if Su(var) mutations resulted in crossovers on 4, which would support a role for chromatin structure in the prevention of crossovers on 4.
In summary, the physical properties of 4, including size, TE content, and chromatin structure could potentially play a role in preventing crossovers on 4, but these are not likely the only factors involved, and perhaps not even major factors.
The centromere effect and the absence of crossovers on 4
A different perspective on the reasons for the absence of crossovers on 4 was investigated by Hatkevich et al. –that the absence of crossovers on 4 is a result of meiotic crossover patterning. Meiotic recombination begins with the introduction of DNA double-strand breaks (DSBs) in the DNA. Some DSBs are repaired as crossovers, but most become non-crossovers. Pathway choice is very highly regulated, but the mechanisms involved are poorly understood.
The major meiotic crossover patterning phenomena are interference, assurance, and the centromere effect. Sturtevant, when he first demonstrated the use of meiotic recombination frequencies to make a map of genes on the Drosophila melanogaster X chromsome, also noted that a crossover on one chromosome reduces the likelihood of another crossover in an adjacent interval, a phenomenon he and Muller later termed interference.23,24 While interference applies to crossover distribution along a chromosome, assurance describes distribution among chromosomes, in that there is a tendency to have at least one crossover on every chromosome pair, regardless of size. Assurance was first noticed by Darlington and Dark25 in studies of chiasmata in the grasshopper Stenobothrus parallelus. From these and similar studies, Owen26 suggested that each bivalent has an “obligatory chiasma.” Since interference and assurance were discovered, they have been described in many other organisms, including plants, fungi, flies, nematodes, and mammals.27-29 Since there are no crossovers on 4, neither interference nor assurance is applicable to explain the lack of crossovers; however, the third patterning phenomenon – the centromere effect – might contribute to this absence. The centromere effect is the suppression of crossovers in the centromere-proximal euchromatin. Like interference, the centromere effect was first described in Drosophila,30 and its mechanism remains mysterious.
To determine whether crossover suppression on 4 is due to proximity to the centromere, Osborne31 used T(1;4)wm5, which swaps the distal portions of the X and 4 (Fig. 2). The 4PXD element of this translocation has the centromere and proximal heterochromatin of chromosome 4, a block of heterochromatin thought to be derived from the X, and the distal end of the X through the white (w) gene.32 The XP4D element has most or all of the 4 gene-containing region (Bolen33 thought the 4 break was between bt and ey, but Hawley32 says it is within the pericentric heterochromatin of 4) attached to the X at 3C2, placing chromosome 4 gene sequences far from the X centromere. Osborne asked whether crossovers were able to occur in the chromosome 4 sequences that are now further from the centromere, and, conversely, if crossing over was abolished on the portion of the X translocated onto the centromere of 4. To generate heterozygous markers, He first induced mutations in y and w on the 4PXD chromosome and in ey and sv on the XP4D chromosome. This allowed him to score crossing over in flies homozygous for T(1;4)wm5 but heterozygous for mutations in these 4 genes. Interestingly, Osborne observed crossing over between ey and sv when they were on the end of the truncated X, but there were no detectable crossovers between y and w when they were translocated adjacent to the centromere of 4 (Fig. 2). These results are consistent with the centromere effect largely contributing to crossover prevention on 4, and argues against the hypothesis that size, sequence content, and chromatin structure are the main barriers to crossovers.
Figure 2.
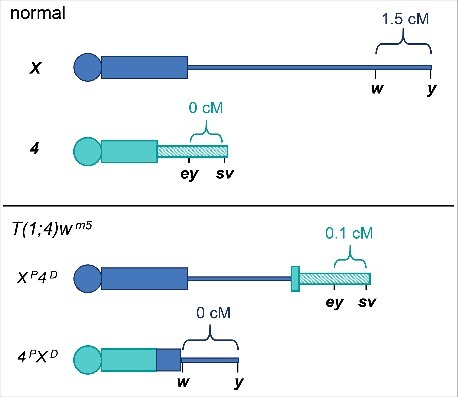
Representation of T(1;4)wm5 with markers that Osborne used to measure recombination. The y – w distance on the wild-type X chromosome is the standard value based on recombination maps.40 The values in the T(1;4)wm5 experiment are from Osborne's data.31 For clarity, we flipped the orientation of the X chromosome from the standard map. Not drawn to scale.
Eliminating crossover patterning allows crossovers on 4
Crossovers have a characteristic distribution, forming mainly in the middle of each chromosome arm. In contrast, non-crossover gene conversion events detected in whole-genome sequencing are distributed more evenly along each major chromosome arm.21,34 Chromosome 4 at first seems to be an exception to this meiotic crossover patterning because it has no crossovers; however, Comeron34 reported finding non-crossover gene conversion events on 4, so we can infer that DSBs are made on 4; these DSBs on 4 are actively prevented from becoming crossovers through meiotic patterning processes. How could the absence of crossovers on 4 result from meiotic patterning? Since 4 is very small, the euchromatin is located very near the centromere, so perhaps the entirety of the chromosome is under the influence of the centromere effect. In our mapping of crossovers on 2L (Fig. 1), crossovers do occur in the one Mb interval adjacent to the proximal heterochromatin. However, this does not necessarily mean we would expect crossovers to occur in the euchromatic regions of the fourth for 2 reasons. First, the telomere effect (a crossover suppression at the distal ends of each chromosome, much weaker than the centromere effect) could be acting together with the centromere effect. Second, the centromere effect differs among chromosome arms,21 and is possibly stronger on 4.
Hatkevich et al.8 showed that crossover patterning is lost in the absence of Blm, a helicase involved in multiple DNA repair pathways.35,36 In studying meiotic phenotypes of Blm mutants, Hatkevich et al. found that interference, assurance, and the centromere effect were all absent or severely reduced. Interestingly, they also reported the occurrence of crossovers on 4 in a Blm mutant.
Why does loss of Blm lead to loss of crossover patterning and the presence of crossovers on 4? In many organisms, there are 2 pathways that can generate meiotic crossovers37 (Fig. 3). The major pathway produces “class I” crossovers that exhibit meiotic patterning. The second pathway is minor, perhaps mostly resolving problems that arise during repair by the major pathway. The “class II” crossovers produced by this minor pathway are not patterned: They do not experience interference, assurance, or the centromere effect.8 In Blm mutants, all crossovers appear to be class II, suggesting that Blm is required to chaperone DSBs into the pathway that produces class I crossovers under the influence of crossover patterning. Hatkevich et al. concluded that the absence of crossovers on 4 is due to meiotic patterning processes, with the centromere as the major contributor to this absence.
Figure 3.

Use of the pathway that generates class I crossovers requires Blm. In wild type flies, crossover patterning processes contribute to designating which DSBs become crossovers, resulting in observance of the centromere effect, interference, and assurance, as well as the absence of crossovers on 4. In a Blm mutant, crossovers arise from a backup pathway and are not patterned, resulting in a random distribution of COs and NCOs across the genome, including on chromosome 4.
Conclusions and future directions
There are many factors that could potentially contribute to the absence of crossovers on chromosome 4, but until recently, only a few of the most basic physical characteristics of 4 have been studied in the context of crossover prevention. Based on recent studies, Hatkevich et al. suggest that it may not be the physical properties of 4 that prevent crossovers, but that the key regulator of this crossover suppression is likely the meiotic patterning of crossovers. Hatkevich et al. show that Blm directs double-strand break repair down the class I pathway toward crossovers that experience meiotic patterning. Without Blm, meiotic patterning is abolished and thus crossovers are permitted on 4. There are still lingering questions such as whether any of the physical features of 4, such as the small size, repetitive sequence content, or heterochromatic chromatin states, play a role in meiotic patterning. Additionally, the Hatkevich et al. paper raises questions about what Blm is actually doing in meiosis and how interference, assurance and the centromere effect are enforced. Finally, although the data from Osbourne and Hatkevich et al. reviewed above make a strong case for the hypothesis that crossover prevention on 4 results largely from the centromere effect, we know essentially nothing about how the centromere effect is conferred. In considering mechanism, however, Sturtevant's classic mapping of 4 crossovers in triploids may provide a clue.6 Perhaps the increased number of centromeres (11 in diplo-4 triploids vs. 8 in normal diploids) dilutes the strength of the effect on each centromere. Redfield studied crossing over on chromosomes 2 and 3 in diploid and triploid females.38,39 She reported that triploids had elevated crossovers in the middle of each chromosome (i.e., where the centromere is located) and reduced crossovers away from the central region (crossovers were also elevated near each end of chromosome 2). Thus, it appears that the centromere effect is sensitive to the number of centromeres. It will be interesting to confirm with conclusion through other manipulations and to further investigate the nature of this interesting phenomenon.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Hawley RS, Theurkauf WE. Requiem for distributive segregation: achiasmate segregation in Drosophila females. Trends Genet 1993; 9:310-7; PMID:8236460; http://dx.doi.org/ 10.1016/0168-9525(93)90249-H [DOI] [PubMed] [Google Scholar]
- [2].Morgan TH. The Theory of the Gene. New Haven, CT: Yale University Press, 1928 [Google Scholar]
- [3].Morgan TH, Sturtevant AH, Bridges CB. Constitution of the germinal material in relation to heredity. Yrbk Carn Inst Wash 1926; 25:308-12 [Google Scholar]
- [4].Patterson JT, Muller HJ. Are “progressive” mutations produced by X-rays? Genetics 1930; 15:495-577; PMID:17246607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Morgan TH, Sturtevant AH, Morgan LV. Maintenance of a Drosophila stock center, in connection with investigations on the constitution of the germinal material in relation to heredity. Yrbk Carn Inst Wash 1945; 55:157-60 [Google Scholar]
- [6].Sturtevant AH. A map of the fourth chromosome of Drosophila melanogaster, based on crossing over in triploid females. Proc Natl Acad Sci USA 1951; 37:405-7; PMID:14853956; http://dx.doi.org/ 10.1073/pnas.37.7.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Grell RF. Heat-induced exchange in the fourth chromosome of diploid females of Drosophila melanogaster. Genetics 1971; 69:523-7; PMID:5003507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hatkevich T, Kohl KP, McMahan S, Hartmann MA, Williams AM, Sekelsky J. Bloom syndrome helicase promotes meiotic crossover patterning and homolog disjunction. Curr Biol 2017; 27:1-5; PMID:27916526; http://dx.doi.org/ 10.1016/j.cub.2016.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chino M, Kikkawa H. Mutants and crossing over in the dot-like chromosome of Drosophila virilis. Genetics 1932; 18:111-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chino M. Genetic studies on the Japanese stock of Drosophila virilis. I. Jpn J Genet 1929; 4:117-31; http://dx.doi.org/ 10.1266/jjg.4.117 [DOI] [Google Scholar]
- [11].Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, et al.. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 2010; 330:1787-97; PMID:21177974; http://dx.doi.org/ 10.1126/science.1198374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gramates LS, Marygold SJ, Santos GD, Urbano JM, Antonazzo G, Matthews BB, Rey AJ, Tabone CJ, Crosby MA, Emmert DB, et al.. FlyBase at 25: looking to the future. Nucleic Acids Res 2017; 45:D663-D71; PMID:27799470; http://dx.doi.org/ 10.1093/nar/gkw1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Locke J, McDermid HE. Analysis of Drosophila chromosome 4 using pulsed field gel electrophoresis. Chromosoma 1993; 102:718-23; PMID:8149812; http://dx.doi.org/ 10.1007/BF00650898 [DOI] [PubMed] [Google Scholar]
- [14].Sun FL, Cuaycong MH, Craig CA, Wallrath LL, Locke J, Elgin SC. The fourth chromosome of Drosophila melanogaster: interspersed euchromatic and heterochromatic domains. Proc Natl Acad Sci USA 2000; 97:5340-5; PMID:10779561; http://dx.doi.org/ 10.1073/pnas.090530797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hoskins RA, Carlson JW, Wan KH, Park S, Mendez I, Galle SE, Booth BW, Pfeiffer BD, George RA, Svirskas R, et al.. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res 2015; 25:445-58; PMID:25589440; http://dx.doi.org/ 10.1101/gr.185579.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al.. The genome sequence of Drosophila melanogaster. Science 2000; 287:2185-96; PMID:10731132; http://dx.doi.org/ 10.1126/science.287.5461.2185 [DOI] [PubMed] [Google Scholar]
- [17].Sandler L, Szauter P. The effect of recombination-defective meiotic mutants on fourth-chromosome crossing over in Drosophila melanogaster. Genetics 1978; 90:699-712; PMID:105965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Slawson EE, Shaffer CD, Malone CD, Leung W, Kellmann E, Shevchek RB, Craig CA, Bloom SM, Bogenpohl J 2nd, Dee J, et al.. Comparison of dot chromosome sequences from D. melanogaster and D. virilis reveals an enrichment of DNA transposon sequences in heterochromatic domains. Genome Biol 2006; 7:R15; PMID:16507169; http://dx.doi.org/ 10.1186/gb-2006-7-2-r15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Riddle NC, Leung W, Haynes KA, Granok H, Wuller J, Elgin SC. An investigation of heterochromatin domains on the fourth chromosome of Drosophila melanogaster. Genetics 2008; 178:1177-91; PMID:18245350; http://dx.doi.org/ 10.1534/genetics.107.081828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dolgin ES, Charlesworth B. The effects of recombination rate on the distribution and abundance of transposable elements. Genetics 2008; 178:2169-77; PMID:18430942; http://dx.doi.org/ 10.1534/genetics.107.082743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Miller DE, Smith CB, Yeganeh Kazemi N, Cockrell AJ, Arvanitakis AV, Blumenstiel JP, et al.. Whole-genome analysis of individual meiotic events in Drosophila melanogaster reveals that noncrossover gene conversions are insensitive to interference and the centromere effect. Genetics 2016; 203:159-71; PMID:26944917; http://dx.doi.org/ 10.1534/genetics.115.186486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Westphal T, Reuter G. Recombinogenic effects of suppressors of position-effect variegation in Drosophila. Genetics 2002; 160:609-21; PMID:11861565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sturtevant AH. The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J Exp Biol 1913; 14:43-59 [Google Scholar]
- [24].Sturtevant AH. The behavior of the chromosomes as studied through linkage. Zeitschrift für Induktive Abstammungs- und Vererbungslehre 1915; 13:234-87 [Google Scholar]
- [25].Darlington CD, Dark SOS. The origin and behaviour of chiasmata, II. Stenobothrus parallelus. Cytologia 1932; 3:169-85; http://dx.doi.org/ 10.1508/cytologia.3.169 [DOI] [Google Scholar]
- [26].Owen ARG. A possible interpretation of the apparent interference across the centromere found by Callan and Montalenti in Culex pipiens. Heredity (Edinb) 1949; 3:357-67; PMID:15408913; http://dx.doi.org/ 10.1038/hdy.1949.26 [DOI] [PubMed] [Google Scholar]
- [27].Jones GH, Franklin FC. Meiotic crossing-over: obligation and interference. Cell 2006; 126:246-8; PMID:16873056; http://dx.doi.org/ 10.1016/j.cell.2006.07.010 [DOI] [PubMed] [Google Scholar]
- [28].Berchowitz LE, Copenhaver GP. Genetic interference: don't stand so close to me. Curr Genomics 2010; 11:91-102; PMID:20885817; http://dx.doi.org/ 10.2174/138920210790886835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang S, Zickler D, Kleckner N, Zhang L. Meiotic crossover patterns: Obligatory crossover, interference and homeostasis in a single process. Cell Cycle 2015; 14:305-14; PMID:25590558; http://dx.doi.org/ 10.4161/15384101.2014.991185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Beadle GW. A possible influence of the spindle fibre on crossing-over in Drosophila. Proc Natl Acad Sci USA 1932; 18:160-5; PMID:16577442; http://dx.doi.org/ 10.1073/pnas.18.2.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Osborne JD. Crossing over in a T(1;4) translocation in Drosophila melanogaster. Department of Biol Sci Edmonton, Alberta, Canada: University of Alberta, 1999:94 [Google Scholar]
- [32].Hawley RS, Irick H, Zitron AE, Haddox DA, Lohe A, New C, Whitley MD, Arbel T, Jang J, McKim K, et al.. There are two mechanisms of achiasmate segregation in Drosophila females, one of which requires heterochromatic homology. Dev Genetics 1992; 13:440-67; PMID:1304424; http://dx.doi.org/ 10.1002/dvg.1020130608 [DOI] [PubMed] [Google Scholar]
- [33].Bolen HR. A mutual translocation involving the fourth and the X-chromosomes of Drosophila. Amer Natur 1931; 65:417-22; http://dx.doi.org/ 10.1086/280386 [DOI] [Google Scholar]
- [34].Comeron JM, Ratnappan R, Bailin S. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet 2012; 8:e1002905; PMID:23071443; http://dx.doi.org/ 10.1371/journal.pgen.1002905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].McVey M, Andersen SL, Broze Y, Sekelsky J. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics 2007; 176:1979-92; PMID:17507683; http://dx.doi.org/ 10.1534/genetics.106.070052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Adams MD, McVey M, Sekelsky J. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 2003; 299:265-7; PMID:12522255; http://dx.doi.org/ 10.1126/science.1077198 [DOI] [PubMed] [Google Scholar]
- [37].Kohl KP, Sekelsky J. Meiotic and mitotic recombination in meiosis. Genetics 2013; 194:327-34; PMID:23733849; http://dx.doi.org/ 10.1534/genetics.113.150581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Redfield H. Crossing over in the third chromosomes of triploids of Drosophila melanogaster. Genetics 1930; 15:205-52; PMID:17246599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Redfield H. A comparison of triploid and diploid crossing over for chromosome II of Drosophila melanogaster. Genetics 1932; 17:137-52; PMID:17246648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lindsley DL, Zimm GG. The Genome of Drosophila melanogaster. San Diego, CA: Academic Press, Inc, 1992 [Google Scholar]


