Abstract
About 40 years ago, the large and small tumor antigens (LT-Ag and sT-Ag) of the polyomavirus (PyVs) simian vacuolating virus 40 have been identified and characterized. To date, it is well known that all the discovered human PyVs (HPyVs) encode these 2 multifunctional and tumorigenic proteins, expressed at viral replication early stage. The 2 T-Ags are able to transform cells both in vitro and in vivo and seem to play a distinct role in the pathogenesis of some tumors in humans. In addition, they are involved in viral DNA replication, transcription, and virion assembly. This short review focuses on the structural and functional features of the HPyVs’ LT-Ag and sT-Ag, with special attention to their transforming properties.
Keywords: Polyomavirus, large T antigen, small T antigen, neoplasia
Introduction
The Polyomaviridae family has been considered capable of transforming mammals’ cells in vitro. In fact, since the first polyomaviruses were discovered in the 1950s, their tumorigenic properties observed in cell culture and animal models rendered the family name: “polyoma”-multiple tumors.1-5 Despite the transformative abilities of simian vacuolating virus 40 (SV40) and murine polyomavirus (MPyV), the human BK polyomavirus (BKPyV)6 and human JC polyomavirus (JCPyV)7 infections often are asymptomatic and mostly related to nonneoplastic diseases in immunocompromised individuals. For instance, BKPyV has been associated with nephropathy (polyomavirus-associated nephropathy) in renal transplant recipients and hemorrhagic cystitis in allogeneic hematopoietic stem cell transplantation,8-10 whereas JCPyV is the causative agent of progressive multifocal leukoencephalopathy in HIV/AIDS and oncologic patients, along with those under immunomodulatory therapies.11,12
Almost 40 years have elapsed since the discovery of the first 2 human polyomaviruses (HPyVs) until the third and fourth HPyVs identification took place, in 2007. The Washington University polyomavirus (WUPyV) and the Karolinska Institute polyomavirus (KIPyV) were detected in children with acute respiratory tract illness through molecular techniques, although their pathogenicity is still controversial.5,13-18 In 2008, Feng et al,19 using digital transcriptome subtraction, identified a polyomavirus clonally integrated in about 80% of Merkel cell carcinoma (MCC), a rare and aggressive neuroendocrine neoplasia and hence named the virus after the cancer. Several other research groups confirmed the Merkel cell polyomavirus (MCPyV) presence in MCC, although the viral prevalence in MCC varies around the world.20-34 This HPyV was the first and, so far, the only HPyV to be etiologically related to a neoplasia in humans. Since its discovery, the scientific curiosity on polyomaviruses, especially regarding their relationship to human cancer, has been reignited. This resulted in new researches aiming for the identification of HPyVs in different biological samples through new sequencing technologies.35-41 Consequently, there are 14 proposed HPyV species, but only 6 of them (BKPyV, JCPyV, MCPyV, HPyV-6, HPyV-7, and the trichodysplasia spinulosa – associated HPyV [TSPyV]) have been associated with human diseases.39,42,43
To accommodate the fast growing number of new PyVs, an update in the family taxonomy was published in 2016. So far, the International Committee on Taxonomy of Viruses (ICTV) acknowledges 73 polyomavirus species, 13 of which are HPyVs, classified in 4 genera, according to their genomic properties and natural host.44 A new nomenclature was proposed, grouping HPyV in 3 genera and using the official term “Human polyomavirus” followed by the viral discovery order, as follows: Alphapolyomavirus genus: MCPyV or HPyV-5, TSPyV or HPyV-8, HPyV-9, HPyV-12, NJPyV or HPyV-13; Betapolyomavirus genus: BKPyV or HPyV-1, JCPyV or HPyV-2, KIPyV or HPyV-3, and WUPyV or HPyV-4; and Gammapolyomavirus genus: HPyV-6, HPyV-7, HPyV-10, or MWPyV and HPyV-11 or TSPyV.44 Despite the taxonomical effort made to encompass all described polyomaviruses, 3 unclassified viruses and a recently described HPyV have not been classified yet.41,44
Human PyVs have a small, nonenveloped icosahedral capsid, presenting 72 pentameric capsomers formed mainly by VP1 associated with minor structural proteins such as VP2 and VP3.45-50 Viral genome is formed by a double-stranded, histone-associated circular DNA functionally divided into 3 main regions: a noncoding control region, an early coding region, and a late coding region. Polyomaviruses have bidirectional genomes, and therefore, the early and late coding regions are in opposite strands.19,51-55 Most, if not all, of the PyVs’ transforming capacities are derived from the expression of the early coding region, also known as tumor antigen locus. This region codes for multiple spliced transcripts that, despite the variations observed among HPyVs, generate 2 main proteins: the large tumor antigen (LT-Ag) and small tumor antigen (sT-Ag).56-60 These 2 T-Ags are involved in the coordination of viral replication and gene expression, as well as in the cell cycle progress and malignant transformation both in vivo and in vitro.57-65 Hence, this review presents the principal interactions of HPyVs’ T-Ag involved in tumorigenesis.
Human Polyomaviruses’ Large Tumor Antigen
The LT-Ag is a complex, multifunctional protein with many roles in viral replication and cell cycle progression. Due to its ability to replicate in and transform cell cultures, most of the knowledge about T antigens was obtained from SV40 studies,66 which can be extrapolated to other PyVs.55 Indeed, genetic and protein analyses have shown that LT-Ag virtually belonging to all HPyVs have several common domains: DnaJ domain, origin-binding domain (OBD), zinc (Zn)-binding domain, and helicase/ATPase (adenosine triphosphatase) domain55,66-68 (Figure 1, panels A to C). For viral DNA replication, 6 LT-Ag molecules interact as an hexameric structure through the recognition of 3 out of 4 pentanucleotide G(A/G)GGC (P1, P2, and P4) motifs in viral replication origin core, which also presents a highly conserved AT-rich region. This recognition is allowed by multiple and complex interactions between OBD, pentanucleotide motifs, GpC dinucleotides, Zn-binding domain, AT-rich region, and histidine residues in the helicase/ATPase.69-72 Two hexameric LT-Ag molecules unwind the viral genome in both directions through ATP hydrolysis, followed by the binding of the eukaryotic replication protein A at the single-stranded viral DNA. The cellular topoisomerase I then releases the torsional stress, whereas α-primase and DNA polymerase initialize the viral DNA synthesis.55,72-76 This replicative role of LT-Ag is frequently seen in lytic infections and can be abrogated in neoplastic tissues.
Figure 1.
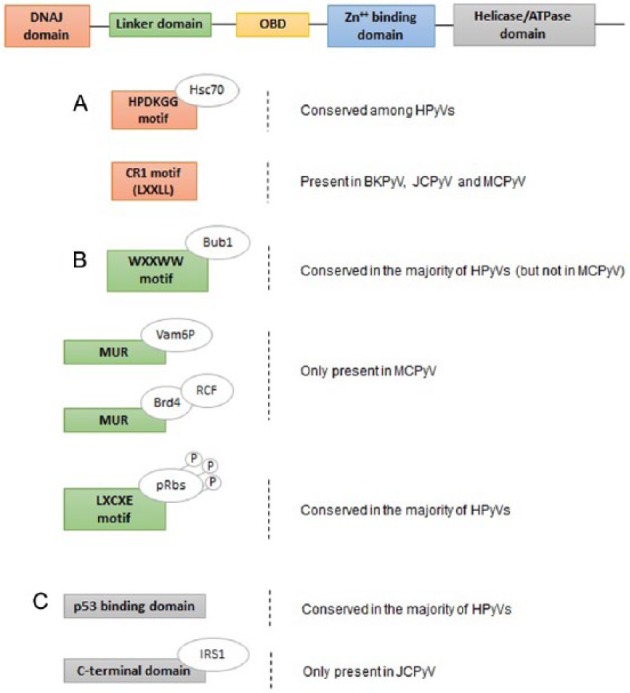
Scheme of the functional domains of the HPyVs’ large tumor antigen (LT-Ag). The LT-Ag consists of several functional domains: DnaJ domain, linker domain, origin-binding domain (OBD), zinc (Zn++)-binding domain, and helicase/ATPase domain. (A) The DnaJ domain contains a HPDKGG motif, conserved among HPyVs, which is able to bind to the Hsc70, a cellular chaperone and transcriptional repressor; a CR1 motif (LXXLL), present only in BKPyV, JCPyV, and MCPyV. (B) The linker domain contains WXXWW motif, conserved in most of the HPyVs but not in MCPyV, for binding Bub1, a mitotic checkpoint serine-threonine protein kinase; a unique region (MUR), only present in MCPyV, that binds the Vamp6P-protein and the Bromodomain protein 4 for the recruitment of cellular protein factor C (RFC); a LXCXE motif, conserved in most of the HPyVs, crucial for the interaction with the retinoblastoma protein (pRb) family. (C) The helicase/ATPase domain comprises a p53-binding domain, conserved in most of the HPyVs; a C-terminal domain, only present in JCPyV, that binds the insulin receptor substrate 1 (IRF1). ATPase indicates adenosine triphosphatase; BKPyV, human BK polyomavirus; HPyVs, human polyomaviruses; JCPyV, human JC polyomavirus; MCPyV, Merkel cell polyomavirus; MUR, MCPyV T antigen unique region.
HPyV integration and LT-Ag truncation
As mentioned previously, the first HPyV associated with human cancer, MCPyV, was discovered clonally integrated in about 80% of Merkel cell carcinomas. The viral integration into the cellular genome is a well-known event that may lead to cancer through several pathways, such as functional loss or gain in cell cycle regulation genes, viral activation of human gene promoters, expression of viral oncogenes, and acquisition of mutational profile both in human and viral genes.77 In fact, LT-Ag premature truncation in MCC samples is considered as a MCPyV tumor-specific signature because the LT-Ag from nontumoral samples does not harbor LT-Ag–truncated forms. Hence, it is suggested that after MCPyV integration into the cellular genome, the infected cells undergo selection of LT-Ag mutations to prevent viral replication and the viral Ori bidirectional unwinding, which would lead to collision with cellular replication forks.57 In 2013, Li et al78 showed that the intact helicase/ATPase domain activates DNA damage response and enhances p53 phosphorylation and cell cycle arrest, which corroborates the proposed explanation for the need of LT-Ag truncation for MCPyV tumorigenesis. Moreover, LT-Ag truncation mutations are unequivocally observed downstream the retinoblastoma protein (pRb)-binding domain, occurring near to the OBD and helicase/ATPase domains. Therefore, a critical role for either pRb-binding domain or sT-Ag preservation in MCC pathogenesis has been suggested.62
Although a strong causal association between other HPyVs with human tumors has not been confirmed yet, circumstantial data suggesting their transformation properties are growing. In fact, a recent meta-analysis revealed that the BKPyV prevalence was significantly higher in prostate cancer (PCa) tissues than in the control, considering as “PCa tissues” samples ranging from very early onset of PCa to well-defined tumors and, as controls, non-PCa samples from patients with PCa or benign prostate hyperplasia.79 There are also evidence linking BKPyV to bladder carcinoma in both immunocompetent80 and renal transplant recipients, as well as in urothelial and renal carcinomas.5,77,81-83
Furthermore, BKPyV DNA has been found integrated into a limited number of brain tumor,84 urothelial carcinoma,85 and, more recently, a case of renal allograft tumor.77 The BKPyV genome was found linearized and inserted into human chromosome 12 in a high-grade urothelial carcinoma from the allograft after almost 10 years of uneventful renal transplant. Viral genome sequencing revealed a new, undescribed variant of BKPyV subtype 1A, which was named Chapel Hill tumor-associated polyomavirus 1 (or CH-1).77 Similarly, the same group reported another undescribed variant of BKPyV 1A subtype, named Chapel Hill 2 (or CH-2) integrated into human chromosome 2 in a poorly differentiated renal cell carcinoma.83 Kenan et al83 suggest that BKPyV integration into host genome after linearization disrupts the expression of late viral transcripts, which would impair LT-Ag downregulation and could eventually lead to tumorigenesis.
Moreover, JCPyV genome has been found integrated into various brain tumors in animal models such as mice and owl monkeys.11,86,87 Viral integration might also happen in human neoplasia, especially in colorectal cancer,88,89 in which JCPyV is considered a cofactor for chromosomal instability.90,91 Interestingly, most of the BKPyV and JCPyV integration studies did not report a truncated LT-Ag. Instead, the viral genome was disrupted at the VP1 region, although replicative impairment has also been observed.77,82,83 Nonetheless, unregulated LT-Ag expression, loss of late protein expression, and replicative arrest occurred following viral integration.83
Although the role of BKPyV integration in human oncogenesis is still hazy, MCPyV integration into host genomes and its involvement at least in MCC carcinogenesis have been better characterized. Still, the low frequency of MCPyV integration in non-MCC tumor has raised attention. Pantulu et al92 identified LT-Ag truncation mutations in 4 chronic lymphocytic leukemia cases, which might indirectly testify the viral integration. In addition, the deletion of a 90-pb fragment in the MCPyV VP1 gene in nonmelanoma skin cancer may also indicate incomplete viral integration.93 Thus, the real role of low-frequency HPyVs’ integration into human cancers is yet to be established.
HPyV LT-Ag domains for cellular protein interaction
Large tumor antigen contains several interaction domains with cellular proteins involved in cell cycle regulation. Some of these motifs are well conserved and most of them facilitate viral replication through activation of cellular proliferation and phase S entry, resulting in upregulation of enzymes involved in cellular DNA replication, DNA damage response, and accessory replicative enzymes.55,65,66 Indeed, LT-Ag expression itself induces viral replication and cellular transformation in animal models and in cell cultures, under specific conditions.62,64,66,79,91-96 The first exon of LT-Ag (approximately 1-70 amino acids, shared with sT-Ag) contains the DnaJ domain, which has a HPDKGG (His-Pro-Asp-Lys-Gly-Gly) motif conserved among HPyV. The DnaJ domain is able to bind to the Hsc70, a cellular chaperone and transcriptional repressor. Human PyVs’ ability to replicate viral DNA in vivo and to promote cell transformation depends on the intact DnaJ domain, as well as capsid assembling in vivo.55,68,97-99 In fact, the MCPyV LT-Ag interaction between its DnaJ domain and Hsc70 has been shown to be necessary for viral replication in vitro.65,68 Moreover, DnaJ domain binding to Hsc70 promotes the chaperone ATPase activity, generating enough energy to dissociate pRb/E2F.55,100-102 The HPyVs, BKPyV, JCPyV, and MCPyV, also contain a CR1 motif, formed by the pentapeptide LXXLL (Leu-X-X-Leu-Leu), which apparently has an auxiliary role in pRb/E2F disruption and cell proliferation.100,103
The LT-Ag linker domain is located downstream to the DnaJ domain. Almost all HPyVs (JCPyV, BKPyV, WUPyV, KIPyV, TSPyV, HPyV-6, HPyV-7, HPyV-9), but not MCPyV, present a conserved motif WXXWW (Trp-X-X-Trp-Trp) that can bind to the Bub1, as demonstrated for SV40 LT-Ag.66,104 Bub1 is a mitotic checkpoint serine-threonine protein kinase that, when functionally impaired, may result in chromosomal instability, as observed in cells expressing SV40 LT-Ag.105 Although MCPyV is oncogenic, it does not present the Bub1-binding motif. Thus, it is not conclusive whether Bub1:LT-Ag interaction is relevant for HPyVs’ transformation in vivo. Instead, at the nucleotide position, where other HPyVs encode Bub1-binding domain, MCPyV has a sequence with little similarity with other PyVs, denominated unique region (MUR [MCPyV T antigen unique region]).65,103 The MUR contains a minimal fragment (171-218 nucleotides), the Vam6P-binding domain, that sequestrates this cytoplasmic protein to a nuclear location, resulting in lysosome clustering impairment, without disruption of transforming growth factor β or mTOR (mammalian target of rapamycin) signaling pathways.103 The LT-Ag interaction with Vam6P seems to be regulatory, as its loss enhances viral replication; consequently, it is considered a mechanism for persistent infection establishment.65,94,95
Another interesting MUR interaction was observed for other known oncoviruses, such as the bovine papillomaviruses (BPVs). Wang et al106 investigated the interaction between LT-Ag and bromodomain protein 4 (Brd4), a member of bromodomain and extra-terminal family involved in cellular growth control, cell cycle progression, and cancer development. The Brd4 interacts with BPV’s early protein 2 (E2) promoting the rightful partition of viral episomes to daughter cells during mitosis.106,107 During the MCPyV infection, a similar interaction between LT-Ag and Brd4 is observed, promoting the MCPyV DNA replication through the cellular protein factor C (RFC) recruitment to viral Ori.106 In addition, recent studies have shown that the human papillomavirus (HPV) are able to interact with Brd4, which is implicated in viral replication and E2 transcriptional activation function as the use of Brd4 inhibitor reduces HPV transcription.108,109 Likewise, the use of a specific peptide, named Brd4 410 to 730, which is homologous to the same region of the Brd4 but without other domains, successfully inhibited MCPyV replication in vitro as it disrupts Brd4:LT-Ag interaction.106
The JCPyV LT-Ag is also able to bind the cellular β-catenin, a cellular protein belonging to the Wnt pathway, important for tissue development, polarity, differentiation, and cell cycle control through cell-cell contact. When hypophosphorylated, β-catenin complexes with LEF-1/TCF-4 transcription factors and migrates to the nucleus, where it promotes cell cycle progression through c-myc and cyclin D1 expression. Phosphorylated β-catenin undergoes degradation via ubiquitin-dependent proteasome through the activation of a complex formed mainly by the glycogen synthase kinase-3 (GSK-3), Axin scaffold proteins, and adenomatous polyposis coli (APC) protein, which phosphorylates β-catenin.91,110-112 In this context, JCPyV promotes β-catenin stabilization through an LT-Ag central domain, which comprehends amino acid residues from 82 to 628. Besides, this interaction increases β-catenin levels and promotes its nucleic localization, with subsequent enhancement of c-myc expression.61,113 Although the JCPyV LT-Ag:β-catenin interaction was also described in mouse medulloblastoma114 and in glioblastoma cell lines,115 it has been recently described in human colorectal carcinoma (CRC), in which β-catenin and Wtn pathway are frequently increased (Figure 2, panel C). Co-localization of JCPyV LT-Ag and β-catenin into the tumor cell nucleus, as well as c-myc and cyclin D1 activation in a subset of JCPyV-positive CRC has been described. This suggests a role of JCPyV in CRC pathogenesis, although JCPyV detection is frequent in both normal colorectal and CRC tissues.91,116 The JCPyV LT-Ag also interacts with the insulinlike growth factor/insulin receptor substrate 1 (IGF/IRS1) pathway, which has been proposed as a mechanism for malign transformation in medulloblastoma. It has been shown that JCPyV LT-Ag binds IRS1 through its C-terminal domain.117 The LT-Ag:IRS1 promotes the IRS1 nuclear translocation, followed by IRS1 interaction with Rad51, which indirectly and eventually prevents DNA damage response90 (Figure 1, panels A to C).
Figure 2.
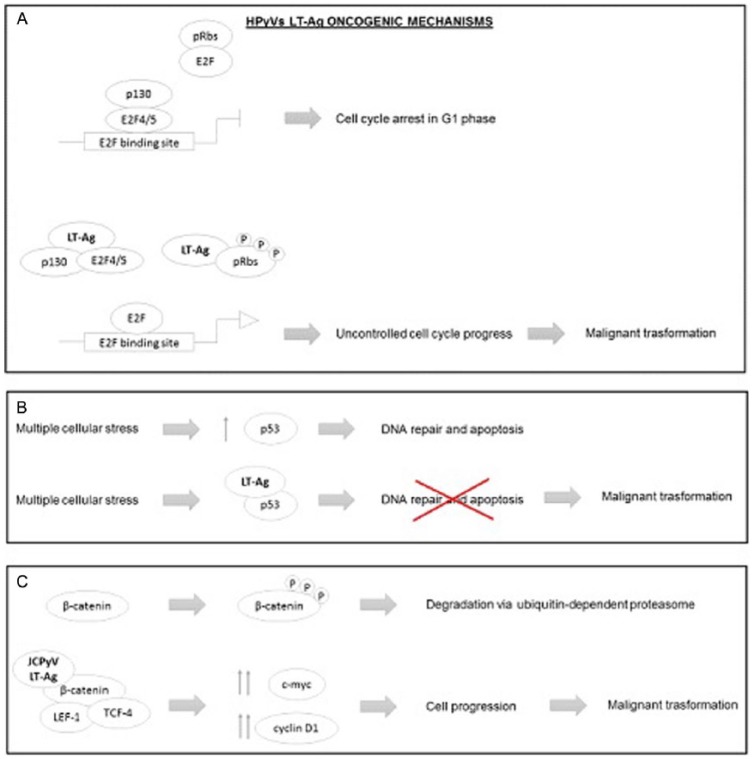
HPyVs’ large tumor antigen (LT-Ag) oncogenic mechanisms. (A) In physiological conditions, the retinoblastoma proteins (pRbs) are in a hypophosphorylated state, which allows them to bind and inhibit the E2F transcription factors, preventing the E2F-mediated gene expression and consequently the transition from G1 to S phase. HPyVs’ LT-Ag is able to bind the pRbs promoting their hyperphosphorylation, thus pRb is unable to bind E2F, leading to its transcriptional activity; at the same time, the hyperphosphorylation of p130 disrupts the transcriptional repressor complex (p130-E2F4/5), leading to uncontrolled cell cycle progression and sometimes to malignant transformation. (B) Multiple cellular stress, normally, raises the levels of p53, which promotes the DNA repair and cell cycle arrest. HPyVs’ LT-Ag is able to bind and block the activity of p53 protein, preventing apoptosis and cell cycle arrest induced by DNA damage. (C) In physiological conditions, the phosphorylated β-catenin undergoes degradation via ubiquitin-dependent proteasome. JCPyVs’ LT-Ag binds the β-catenin protein promoting its hypophosphorylation, thus β-catenin complexes with LEF-1/TCF-4 transcription factors, promoting the cell cycle progression by c-myc and cyclin D1 expression. HPyVs indicate human polyomaviruses; JCPyV, human JC polyomavirus.
Despite its recent discovery, it is already known that MCPyV is involved in different molecular regulatory mechanisms. For example, MCPyV detection in non–small-cell lung cancer (NSCLC) was found to deregulate BRAF and Bcl2: the first is involved in cell cycle progression through mitogen-activated protein kinase (MAPK) signaling pathway and was upregulated in MCPyV-positive samples; the second is an antiapoptotic mitochondrial protein that was downregulated in MCPyV infection. These findings were suggestive of MCPyV-mediated deregulation in NSCLC, mainly in smoker patients.118 Furthermore, another NSCLC study reported a significant association between MCPyV DNA detection and LT-Ag expression with mutation on epithelial growth factor receptor mainly in nonsmoker patients, supporting the hypothesis of MCPyV’s participation in a subgroup of NSCLC oncogenesis.119
A similar study investigating PIK3’s mutational profile showed a higher frequency of PIK3 mutations in MCPyV-positive MCC than in MCPyV-negative MCC, although the Akt/mTOR expression was higher in MCPyV-negative tumor, supporting the employment of different mechanisms for MCC oncogenesis in distinct MCPyV backgrounds.120 Furthermore, the expression of tumor-derived MCPyV T-Ag in mice-stratified epithelium promotes gross epithelial phenotypes, consistent with neoplastic progression comparable with those observed for high-grade HPV-16 E6 and E7 oncoproteins. Further molecular studies demonstrated the LT-Ag interaction with BIRC5 survivin and increased E2F target gene expression, being both the pathways mediated by LT-Ag LXCXE motif.121
Finally, BKPyV LT-Ag and other HPyV LT-Ag have been recently found in the upregulation of the DNA cytosine deaminase APOBEC3, an important innate immune system enzyme responsible for suppressing viral replication and cancer. Evidence suggest that APOBEC3 may be correlated with cytosine-based mutation patterns and in coincident sites of DNA rearrangement. As APOBEC3B was also upregulated in HPV-related cancers, it is suggested that HPyV LT-Ag upregulation may also contribute to carcinogenesis through host genomic mutations.122
HPyV LT-Ag binding with pRb
Indubitably, one of the most important LT-Ag interactions is that with pRb family55,66 (Figure 2, panel A). This tumor suppressor family comprises 3 main proteins, pRb, p107, and p130, encoded by the RB1, RBL1, and RBL2 genes, respectively. Although the pRb can regulate the cell cycle through many pathways, its interaction with the E2F transcription factors family is one of the most important, as its loss potentiates tumorigenesis.123,124 In a hypophosphorylated state, pRb binds the E2F transcription factor, preventing the E2F-mediated gene expression and thus promoting cell cycle arrest. During normal cell cycle progress, activation of cyclin-dependent kinases (CDKs) leads to hyperphosphorylation of serine and threonine residues at pRb phosphorylation-specific sites resulting in the release of the E2F transcriptional factors; their transcriptional activity promotes the cell cycle progress from G1 to S phase.125 The components of the pRb family are also known as “pocket proteins” because of a conserved “pocket” domain that interacts with viral oncoproteins, notoriously the SV40 LT-Ag and HPV E7.126
The HPyV LT-Ag, as observed in SV40 LT-Ag prototype, has a highly conserved pRb-binding domain in its linker region. The LXCXE (Leu-X-Cis-X-Glu) motif is responsible for the LT-Ag:pRb family interaction. It has been hypothesized that the energy provided by the LT-Ag DnaJ domain binding to Hsc70 is used to disassociate pRb/E2F through pRb phosphorylation, as corroborated by studies showing the DnaJ domain requirement for the SV40 LT-Ag growth activities.127-129 Large tumor antigen is also capable of disrupting the transcriptional repressor complex formed by p130 and the repressive E2F4/5 proteins through p130 dephosphorylation in a DnaJ domain–dependent pathway.66,91,130 The overall pRb:E2F disruption leads to uncontrolled cell cycle progress and, eventually, to malignant transformation. In fact, the pRb pathway deregulation is frequently observed in many tumors.127,128,131
The SV40 LT-Ag interactions with pRb, p107, and p130 cause well-known transformative effects in cell culture and animal models and have been recently reviewed.66 However, HPyV LT-Ag also mediated cell transformation through pRb family binding. For instance, JCPyV LT-Ag binds the hypophosphorylated pRb in hamster glioblastoma cells and this interaction suppression successfully disables cell transformation, although JCPyV LT-Ag might be less efficient in cell transformation than SV40 LT-Ag.132 Moreover, JCPyV LT-Ag might alter pRb expression pattern and cellular distribution in mouse primitive neuroectodermal tumors (PNET) and human medulloblastoma (HMB) cell lines, suggesting a cell-type–specific LT-Ag–mediated tumorigenesis.133
The BKPyV LT-Ag can significantly reduce pRb, p107, and p130 levels and increase transcriptionally active E2F, requiring intact LT-Ag DnaJ and pRb-binding domains to induce cell cycle deregulation.101,134 Furthermore, BKPyV LT-Ag interaction with pRb and consequent E2F release leads to DNA methyltransferase 1 (DNMT1) gene expression, one of the E2F target genes.135 This enzyme adds methyl residues on cytosine located 5′ to guanosine and is involved in cellular expression control, as methylation significantly reduces gene transcription.135 Thus, DNMT1 expression promoted by BKPyV LT-Ag probably induces the inheritable silencing of tumor suppressor genes and might contribute to carcinogenesis.135
Likewise, MCPyV LT-Ag contains LXCXE motif and interacts with pRb family proteins.57 In fact, this natural interaction is the decisive evidence for LT-Ag tumorigenesis pathway through pRb family, as pRb-binding domain is systematically maintained in MCPyV-positive MCC while the LT-Ag C-terminal is truncated.57 Moreover, truncated LT-Ag is more efficient in transforming cells then full-length LT-Ag, mainly due to its LXCXE motif and to the disruption of the C-terminal region inhibitory activity on cell growth.94 The MCPyV LT-Ag C-terminal inhibitory activity on cell growth might be mediated by the phosphorylation of an LT-Ag serine residue at the position 816, stimulated by the ATM pathway activation. This phosphorylation site might represent a negative selection mechanism to eliminate functional and growth-inhibitory C-terminal during tumorigenesis, which also preserves pRb domain.136 In vitro studies showed that the ablation of MCPyV LT-Ag:pRb would lead to tumor regression, suggesting that the LT-Ag:pRb interaction is critical for cell proliferation and sustained tumor growth.137 Also, a new interaction for MCPyV LT-Ag was observed when Arora et al138 found that the cellular oncoprotein survivin BIRC5a, an antiapoptotic protein, was 7-fold more expressed in MCPyV-positive MCC than in MCPyV-negative MCC. This study also identified the need for an intact pRb-binding domain for the direct LT-Ag binding to BIRC5a gene promoter and hence proposed the survivin as a therapeutic target for MCC.137 However, it was already known that the HPyV LT-Ag:survivin interaction might promote cell survival, as observed in oligodendrocyte and astrocyte cell lines infected in vitro and expressing LT-Ag.91
The LT-Ag:pRb interaction has also been studied in the newly discovered HPyVs. The TSPyV LT-Ag mediates cells cycle progression in trichodysplasia spinulosa cases through pRb interaction. Kassem et al139 showed evidence of TSPyV LT-Ag clusters with phosphorylated pRb by histologic immunofluorescence and concluded that LT-Ag:pRb may induce cell proliferation as a potential driver of papule and spicule formation. In contrast, the MWPyV LT-Ag is less stable and has not been yet associated with transforming abilities in vitro, although it is able to bind to pRb, p107, and p130.139,140 Likely, HPyV-7 LT-Ag expression in thymic epithelial tumors does not correlate with pRb expression,141 although HPyV detection and expression in thymomas were relatively frequent (62%).142
HPyV LT-Ag binding with p53
Finally, another important LT-Ag interaction in HPyV-mediated tumorigenesis is that with p53, a tumor-suppressing protein that regulates the gene expression in response to stressful events such as DNA damage, leading to cell apoptosis, cell cycle arrest, or senescence and is usually deregulated in cancer.143 Large tumor antigen of the PyVs contains a p53-binding site on the helicase/ATPase domain. During SV40 carcinogenesis, LT-Ag binds and blocks p53 activity, preventing apoptosis and cell cycle arrest, induced by DNA damage, derived from pRb:E2F disruption. Instead of inducing p53 degradation through ubiquitinase, as seen for HPV E6 protein, SV40 LT-Ag stabilizes p53, which is sufficient for the abrogation of its transcriptional activities66 (Figure 2, panel B).
The BKPyV LT-Ag also complexes with p53 and, in BKPyV-positive renourinary tumors, such association is considered an important oncogenic mechanism. The complex formation also promotes p53 kidnapping to cytoplasm, which is observed in both infected and transformed cell lines. Even if the sequestration is not a requirement for p53 inhibition, the p53 cytoplasmic accumulation correlates with an increased cellular mutational profile in LT-Ag–expressing cells and is considered the main hallmark of BKPyV involvement in PCa development.143 In addition, the preservation of the LT-Ag:p53–binding domain observed in tumors with BKPyV integration is suggestive for the LT-Ag:p53–mediated oncogenesis.
In the same way, as observed for the p53 cytoplasmic sequestration mediated by BKPyV LT-Ag, JCPyV LT-Ag is able to alter p53 expression and its cellular distribution in PNET and HMB cell lines.133 A recent study conducted on paraffin-embedded specimens reported a dramatic decrease in p53 expression in JCPyV-positive glioblastomas when compared with JCPyV-negative glioblastomas, suggesting a potential role of JCPyV LT-Ag in p53 expression modulation and tumorigenesis.144
Nevertheless, MCPyV-mediated MCC tumorigenesis does not require p53 inactivation. In fact, p53 disruption in MCC is MCPyV LT-Ag independent, as demonstrated by Houben et al.145 This experimental study corroborates with other previously published papers, showing that the C-terminal region of MCPyV LT-Ag mediates cell cycle progress inhibition,94 and that the truncated LT-Ag has a higher transformation potential compared with the full-length LT-Ag.62 Finally, these findings are consistent with the MCPyV tumor-specific signature in MCC, where the truncated LT-Ag lacks the helicase/ATPase and p53-binding domain.57
Human Polyomaviruses’ Small Tumor Antigen
The sT-Ag is another PyV protein with cell transformation properties. Although sT-Ag expression is not always a condition for viral replication or transformation, it is required for PyVs’ optimal functioning.68,146 The SV40 sT-Ag is functionally comparable with HPV E7, in such a way that E7 is capable of successfully replacing the functions of sT-Ag in vitro.147 The PyV sT-Ags are encoded in the early region of PyVs’ genomes, superposed to the LT-Ag coding sequence. Produced by an alternative splicing of this region, sT-Ag shares about 80 amino acids in its N-terminus with LT-Ag.55 Therefore, the sT-Ag presents the DnaJ domain with a conserved HPDKGG motif among all HPyVs, which potentially affects viral replication, cell proliferation, and transformation, although the functional importance of this domain for sT-Ag activity is unknown.65,148-150 The BKPyV, JCPyV, and SV40 sT-Ag share high amino acid similarities with each other, especially on the N-terminus, although the sT-Ag middle region is more divergent among HPyVs,151 and its hydrophobicity and flexibility are important for sT-Ag functionality.152
The sT-Ag transcript also contains a unique region, removed from LT-Ag during the intronic processing, which may have distinct features among HPyVs. For instance, JCPyV sT-Ag contains a LYCKE and a LHCWE motif and BKPyV has only one LYCKE motif. Thus, these LXCXE motifs are potentially able to interact with pRb family proteins, whereas MCPyV sT-Ag does not present any LXCXE motif, as well as the other HPyVs.55,153 Regardless, all HPyVs’ sT-Ags have 2 conserved domains at their unique C-terminus region: 2 zinc-binding domains (Cys-X-Cys-X-X-Cys motif), which provide structural and functional stabilities for sT-Ag, and 2 domains for phosphatase 2A (PP2A), rich in cysteine and proline residues responsible for sT-Ag:PP2A interaction, that require the sT-Ag DnaJ domain and the second zinc-binding motif55,151 (Figure 3, panels A to C).
Figure 3.
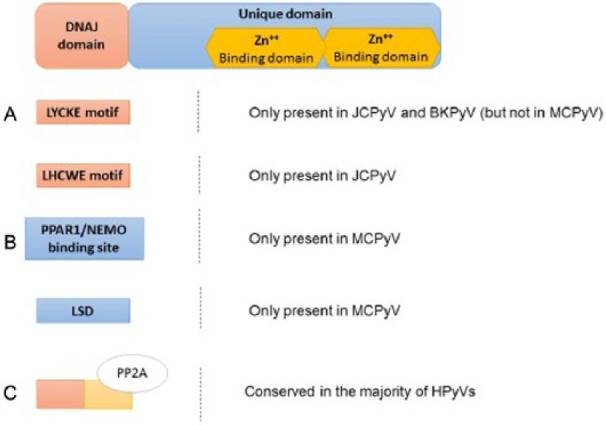
Scheme of the functional domains of the HPyVs’ small tumor antigen (sT-Ag). The sT-Ag presents a DnaJ domain, followed by a unique domain, that is removed from LT-Ag during the splicing process. (A) The DnaJ domain contains a LYCKE motif (JCPyV and BKPyV) and a LHCWE motif (only JCPYV), able to interact with pRb family proteins. (B) The unique domains contain 2 Zn++-binding sites and, additionally and only in MCPyV, a PPAR1/NEMO-binding site and a large T stabilization domain (LSD) that are involved in the oncogenesis process. (C) The binding site for the PP2A is conserved in most of the HPyVs and triggers several pathways related to cellular transformation. BKPyV indicates human BK polyomavirus; HPyVs, human polyomaviruses; JCPyV, human JC polyomavirus; MCPyV, Merkel cell polyomavirus; NEMO, NF-κB essential modulator.
HPyV sT-Ag binding with PP2A
The PP2A is a serine-threonine phosphatase that regulates, among many other important cellular processes, the cell cycle progression and apoptosis by dephosphorylating protein targets such as Akt, p53, c-Myc, and β-catenin and hence is considered a tumor-suppressing protein. Phosphatase 2A is composed of catalytic (PP2Ac), scaffold (PP2AA), and regulatory subunits (PP2AB) that interact to form an active enzymatic complex.154 The SV40 sT-Ag prototype has a cysteine-proline–rich and conserved domain (Cys-X-X-X-Pro-X-Cys) that interacts with PP2A, mainly through the sT-Ag cysteine residues.55,151 In fact, the ablation of only one cysteine from CXXXPXC motif dramatically reduces sT-Ag:PP2A–binding rate.155 The sT-Ag interacts with the PP2AA (scaffold) subunit by the HEAT repeats 3 to 7, displacing the PP2AB (regulatory) subunit that also binds HEAT repeats 3 to 7, and thus inhibiting PP2AAc phosphatase activity152 (Figure 4, panels A and B).
Figure 4.
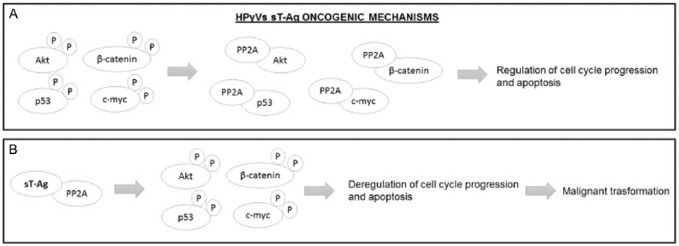
HPyVs’ small tumor antigen (sT-Ag) oncogenic mechanisms. (A) In physiological conditions, the Akt, p53, c-Myc, and β-catenin proteins are in a phosphorylated state; the subsequent dephosphorylation due to the PP2A serine-threonine phosphatase regulates the cell cycle progression and the apoptosis process. (B) The binding between sT-Ag and PP2A avoids the dephosphorylation of Akt, p53, c-Myc, and β-catenin proteins, and the subsequent deregulation of the cell cycle progression and apoptosis process drives the cell to a malignant transformation. HPyVs indicate human polyomaviruses; PP2A, phosphatase 2A.
The sT-Ag:PP2A interaction triggers several pathways related to cellular transformation. The entry on cell cycle S phase may be stimulated by sT-Ag:PP2A, as PP2A no longer dephosphorylates the CDK inhibitor p27, leading to its degradation. In addition, sT-Ag:PP2A also induces the S phase entry by promoting cyclin A/CDK2 and cyclin E/CDK2 expression through a cell cycle–regulated E2F site.156,157 Furthermore, sT-Ag:PP2A has been linked to human fibroblast transformation by CDK2 activation and DNA synthesis induction.158
The SV40 sT-Ag:PP2A interaction might induce oncogenesis in vitro through the activation of the MAPK cascade.159 The MAPKs are involved in cell proliferation via the ERK1 and ERK2 (extracellular signal–related kinases 1 and 2) upregulation of cyclin D1, leading to phosphorylation of pRb and the subsequent releasing of the E2F transcription factor. The MAPK pathway activation has been linked to increased BKPyV replication in cell cultures, although induced by LT-Ag.160 However, Wu et al161 demonstrated that TSPyV sT-Ag overexpression activates the MAPK pathway by enhancing the MEK/ERK phosphorylation. Once more, it is suggested that sT-Ag:PP2A interaction prevents the PP2A dephosphorylation activity and ultimately the suppression of the MAPK cascade.
It has been demonstrated that PyV sT-Ag are capable of upregulating and stabilizing the myelocytomatosis transcription factor (Myc), which requires intact DnaJ and PP2AsT-Ag domains. Studies have demonstrated that sT-Ag:c-Myc upregulation is related to later increase in human telomerase and cyclin D1, suggestive for Myc-targeted expression.162 The c-Myc and cyclin D1 promoter activity was also increased by JCPyV sT-Ag and β-catenin, both separated and associated, the latter found to potentiate the effect.116 Moreover, MCPyV sT-Ag is capable of increasing c-Myc activity. Normally, the Fbw7 ubiquitin ligase protein complex promotes the degradation of proto-oncogene products such as cyclin E, c-Myc, c-Jun, mTOR, and nuclear factor κB (NF-κB) by a phosphorylation-dependent mechanism. Recent findings support the idea that the sT-Ag can inhibit the LT-Ag, c-Myc, and cyclin E proteasomal degradation through their stabilization, thus regulating several cell cycle proliferation pathways.163
Furthermore, human cells expressing SV40 sT-Ag have shown upregulation of antiapoptotic targets of NF-κB,164 despite most viral proteic interactions with NF-κB pathway have been shown to have inhibitory effects. For instance, MCPyV sT-Ag downregulates NF-κB by targeting the NF-κB essential modulator (NEMO) adaptor protein and, thus, disrupting the inflammatory pathway.165 Moreover, NF-κB regulation pathway by MCPyV sT-Ag involves the regulatory subunit 1 of the protein phosphatase 4 (PP4R1), which is required for NEMO adaptor protein interaction.166 Nuclear factor κB has also been described as a promoter of JCPyV replication during DNA damage response induced by JCPyV itself.167 These sT-Ag inhibitory activities on NF-κB pathway, responsible for innate immune response, might be related to host immune response evasion, whose impact on tumorigenesis still remains unclear.
sT-Ag/PP2A–independent pathways
Despite the principal PyVs’ sT-Ag oncogenic pathway is associated with the PP2A deregulation, MCPyV sT-Ag may be able to induce cell transformation in a PP2A-independent way. For instance, it has been shown that MCPyV sT-Ag may induce cell proliferation depending on the Akt-mTOR signaling without PP2A deregulation.168 The mTOR is a serine-threonine kinase that controls cellular functions such as transcription and translation. When activated, mTOR phosphorylates some translational control proteins, such as the initiation factor 4E-binding protein 1 (4E-BP1), preventing its inhibitory activity and releasing the initiation factor 4 (eIF4E), which may then promote translation and further cell cycle progression.151 The PP2A domain of MCPyV sT-Ag domain is not required for epithelial transformation in transgenic mice.169 Furthermore, it has been demonstrated that MCPyV sT-Ag is able to dislocate a restricted number of PP2AB subunits, which lead to a sT-Ag:PP2A interaction insufficient to promote tumorigenesis in vitro. Instead, MCPyV sT-Ag induces oncogenesis through the so-called large T stabilization domain (LSD).163,170 The LSD is located at residues 91 to 95 and inhibits MCPyV LT-Ag proteasomal degradation because LT-Ag is the target for the Fbw7 E3 ubiquitin ligase. Mutations at LSD disrupt LT-Ag stabilization, prevent sT-Ag cell transformation and viral replication, as well as reduce sT-Ag induction of cellular oncoprotein, still in a PP2A-independent manner.65,170
Another MCPyV sT-Ag interaction related to in vitro tumorigenesis is that with c-Jun, a transcription factor that regulates cellular differentiation and proliferation. When hyperphosphorylated, c-Jun is capable of inducing tumorigenesis, especially in keratinocytes.171 A recent study172 observed that c-Jun hyperphosphorylation increased after MCPyV sT-Ag overexpression in HEK293 cell line. In addition, although c-Jun phosphorylation status in in vivo MCC is currently unknown, its activation by MCPyV sT-Ag may contribute to the MCC aggressive pattern, thus demanding further investigations.172 Finally, it has also been demonstrated that TSPyV sT-Ag overexpression is associated with c-Jun phosphorylation and activation, indicating a role for TSPyV sT-Ag in the trichodysplasia spinulosa (TS) pathogenesis.161
Conclusions
Indubitably, among the proteins encoded by the HPyVs, the principal agent involved in cell transformation and tumor development is the LT-Ag, which is also the most studied. Interestingly, the structure and function of this protein are quite conserved among the different HPyVs, testifying that it is indispensable for both viral replication and interaction with the host cells. However, this review underlined also the strategic role of the other early proteins encoded by the HPyVs, the sT-Ag. In addition, it should be taken into account that some of the classic and newly discovered HPyVs are also able to produce agnoprotein, which may have transforming activities itself.91 Most probably, these proteins act synergically, orchestrated by LT-Ag, fighting a battle against the infected host, trying to evade from the immune system, and targeting multiple cellular pathways. To this particular regard, the evasion of the innate immune system by HPyV has been so far studied for MCPyV. Both MCPyV LT-Ag and sT-Ag, and also BKPyV LT-Ag, are able to interact with the toll-like receptor 9 (TLR9) and to inhibit it, causing the subsequent lack of transcription of C/EBPβ. This last transcription factor plays several roles in the suppression activity of the tumor proliferation, ie, it regulates interleukin (IL)-6, IL-8, tumor necrosis factor α, and E2F expressions. Consequently, the suppression of C/EBPβ expression by the HPyVs’ T-Ag induces modification in the immune system reactions against the viruses and also triggers the cell proliferation.173-176
Not all of the involved mechanisms, neither the interactions among the viral proteins have been fully understood, and the continuous discovery of new HPyVs might favor the understanding of cell transformation mediated by the HPyVs.
Acknowledgments
The authors would like to thank Mrs Rosalia Ticozzi for her technical support.
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: CFB wrote the first draft of the manuscript. RBV, SV, and SD contributed to the writing of the manuscript. CFB, RBV, SV, and SD agree with manuscript results and conclusions; jointly developed the structure and arguments for the paper; and made critical revisions and approved final version. All authors reviewed and approved the final manuscript.
Disclosures and Ethics: As a requirement of publication, authors have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality, and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section.
References
- 1. Sweet BH, Hilleman MR. The vacuolating virus, S.V. 40. Proc Soc Exp Biol Med. 1960;105:420-427. [DOI] [PubMed] [Google Scholar]
- 2. Gross L. A filterable agent, recovered from Ak leukemic extracts, causing salivary gland carcinomas in C3H mice. Proc Soc Exp Biol Med. 1953;83:414-421. [DOI] [PubMed] [Google Scholar]
- 3. Stewart SE, Eddy BE, Borgese N. Neoplasms in mice inoculated with a tumor agent carried in tissue culture. J Natl Cancer Inst. 1958;20:1223-1243. [DOI] [PubMed] [Google Scholar]
- 4. Butel JS, Lednicky JA. Cell and molecular biology of simian virus 40: implications for human infections and disease. J Natl Cancer Inst. 1999;91:119-134. [DOI] [PubMed] [Google Scholar]
- 5. Dalianis T, Hirsch HH. Human polyomaviruses in disease and cancer. Virology. 2013;437:63-72. [DOI] [PubMed] [Google Scholar]
- 6. Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet. 1971;1:1253-1257. [DOI] [PubMed] [Google Scholar]
- 7. Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet. 1971;1:1257-1260. [DOI] [PubMed] [Google Scholar]
- 8. Bogdanovic G, Priftakis P, Giraud G, et al. Association between a high BK virus load in urine samples of patients with graft-versus-host disease and development of hemorrhagic cystitis after hematopoietic stem cell transplantation. J Clin Microbiol. 2004;42:5394-5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kato J, Mori T, Suzuki T, et al. Nosocomial BK polyomavirus infection causing hemorrhagic cystitis among patients with hematological malignancies after hematopoietic stem cell transplantation. Am J Transplant. 2017;17:2428-2433. [DOI] [PubMed] [Google Scholar]
- 10. Hirsch HH, Randhawa P. BK polyomavirus in solid organ transplantation. Am J Transplant. 2013;13:179-188. [DOI] [PubMed] [Google Scholar]
- 11. Hirsch HH, Kardas P, Kranz D, Leboeuf C. The human JC polyomavirus (JCPyV): virological background and clinical implications. Apmis. 2013;121:685-727. [DOI] [PubMed] [Google Scholar]
- 12. Wollebo HS, White MK, Gordon J, Berger JR, Khalili K. Persistence and pathogenesis of the neurotropic polyomavirus JC. Ann Neurol. 2015;77:560-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gaynor AM, Nissen MD, Whiley DM, et al. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007;3:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allander T, Andreasson K, Gupta S, et al. Identification of a third human polyomavirus. J Virol. 2007;81:4130-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rao S, Lucero MG, Nohynek H, et al. WU and KI polyomavirus infections in Filipino children with lower respiratory tract disease. J Clin Virol. 2016;82:112-118. [DOI] [PubMed] [Google Scholar]
- 16. Dehority WN, Eickman MM, Schwalm KC, et al. Complete genome sequence of a KI polyomavirus isolated from an otherwise healthy child with severe lower respiratory tract infection. J Med Virol. 2017;89:926-930. [DOI] [PubMed] [Google Scholar]
- 17. Siebrasse EA, Pastrana DV, Nguyen NL, et al. WU polyomavirus in respiratory epithelial cells from lung transplant patient with Job syndrome. Emerg Infect Dis. 2015;21:103-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Siebrasse EA, Nguyen NL, Willby MJ, Erdman DD, Menegus MA, Wang D. Multiorgan WU polyomavirus infection in bone marrow transplant recipient. Emerg Infect Dis. 2016;22:24-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paolini F, Donati P, Amantea A, Bucher S, Migliano E, Venuti A. Merkel cell polyomavirus in Merkel cell carcinoma of Italian patients. Virol J. 2011;8:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jung HS, Choi YL, Choi JS, et al. Detection of Merkel cell polyomavirus in Merkel cell carcinomas and small cell carcinomas by PCR and immunohistochemistry. Histol Histopathol. 2011;26:1231-1241. [DOI] [PubMed] [Google Scholar]
- 22. Mangana J, Dziunycz P, Kerl K, Dummer R, Cozzio A. Prevalence of Merkel cell polyomavirus among Swiss Merkel cell carcinoma patients. Dermatology. 2010;221:184-188. [DOI] [PubMed] [Google Scholar]
- 23. Kassem A, Schopflin A, Diaz C, et al. Frequent detection of Merkel cell polyomavirus in human Merkel cell carcinomas and identification of a unique deletion in the VP1 gene. Cancer Res. 2008;68:5009-5013. [DOI] [PubMed] [Google Scholar]
- 24. Sastre-Garau X, Peter M, Avril MF, et al. Merkel cell carcinoma of the skin: pathological and molecular evidence for a causative role of MCV in oncogenesis. J Pathol. 2009;218:48-56. [DOI] [PubMed] [Google Scholar]
- 25. Santos-Juanes J, Fernandez-Vega I, Fuentes N, et al. Merkel cell carcinoma and Merkel cell polyomavirus: a systematic review and meta-analysis. Br J Dermatol. 2015;173:42-49. [DOI] [PubMed] [Google Scholar]
- 26. Andea AA, Patel R, Ponnazhagan S, Isayeva T, Kumar S, Siegal GP. Detection of Merkel cell polyomavirus in formalin-fixed, paraffin-embedded tissue of Merkel cell carcinoma and correlation with prognosis. Rom J Morphol Embryol. 2014;55:1057-1062. [PubMed] [Google Scholar]
- 27. Mizuno Y, Kato G, Shu E, et al. Merkel cell polyomavirus-positive Merkel cell carcinoma in a patient with epidermodysplasia verruciformis. Acta Derm Venereol. 2015;95:98-99. [DOI] [PubMed] [Google Scholar]
- 28. Vaira F, Nazzaro G, Pesapane F, et al. Detection of polyomavirus in Merkel cell carcinoma by immunohistochemistry: report of three cases. G Ital Dermatol Venereol. 2015;150:617-621. [PubMed] [Google Scholar]
- 29. Erovic BM, Al Habeeb A, Harris L, Goldstein DP, Ghazarian D, Irish JC. Significant overexpression of the Merkel cell polyomavirus (MCPyV) large T antigen in Merkel cell carcinoma. Head Neck. 2013;35:184-189. [DOI] [PubMed] [Google Scholar]
- 30. Leitz M, Stieler K, Grundhoff A, Moll I, Brandner JM, Fischer N. Merkel cell polyomavirus detection in Merkel cell cancer tumors in Northern Germany using PCR and protein expression. J Med Virol. 2014;86:1813-1819. [DOI] [PubMed] [Google Scholar]
- 31. de Biase D, Ragazzi M, Asioli S, Eusebi V. Extracutaneous Merkel cell carcinomas harbor polyomavirus DNA. Hum Pathol. 2012;43:980-985. [DOI] [PubMed] [Google Scholar]
- 32. Matsushita M, Iwasaki T, Kuwamoto S, et al. Merkel cell polyomavirus (MCPyV) strains in Japanese Merkel cell carcinomas (MCC) are distinct from Caucasian type MCPyVs: genetic variability and phylogeny of MCPyV genomes obtained from Japanese MCPyV-infected MCCs. Virus Genes. 2014;48:233-242. [DOI] [PubMed] [Google Scholar]
- 33. Garneski KM, Warcola AH, Feng Q, Kiviat NB, Leonard JH, Nghiem P. Merkel cell polyomavirus is more frequently present in North American than Australian Merkel cell carcinoma tumors. J Invest Dermatol. 2009;129:246-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Becker JC, Houben R, Ugurel S, Trefzer U, Pfohler C, Schrama D. MC polyomavirus is frequently present in Merkel cell carcinoma of European patients. J Invest Dermatol. 2009;129:248-250. [DOI] [PubMed] [Google Scholar]
- 35. Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scuda N, Hofmann J, Calvignac-Spencer S, et al. A novel human polyomavirus closely related to the African green monkey-derived lymphotropic polyomavirus. J Virol. 2011;85:4586-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Siebrasse EA, Reyes A, Lim ES, et al. Identification of MW polyomavirus, a novel polyomavirus in human stool. J Virol. 2012;86:10321-10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lim ES, Reyes A, Antonio M, et al. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology. 2013;436:295-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Korup S, Rietscher J, Calvignac-Spencer S, et al. Identification of a novel human polyomavirus in organs of the gastrointestinal tract. PLoS ONE. 2013;8:e58021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mishra N, Pereira M, Rhodes RH, et al. Identification of a novel polyomavirus in a pancreatic transplant recipient with retinal blindness and vasculitic myopathy. J Infect Dis. 2014;210:1595-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gheit T, Dutta S, Oliver J, et al. Isolation and characterization of a novel putative human polyomavirus. Virology. 2017;506:45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ho J, Jedrych JJ, Feng H, et al. Human polyomavirus 7-associated pruritic rash and viremia in transplant recipients. J Infect Dis. 2015;211:1560-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nguyen KD, Lee EE, Yue Y, et al. Human polyomavirus 6 and 7 are associated with pruritic and dyskeratotic dermatoses. J Am Acad Dermatol. 2017;76:932.e933-940.e933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Calvignac-Spencer S, Feltkamp MC, Daugherty MD, et al. A taxonomy update for the family Polyomaviridae. Arch Virol. 2016;161:1739-1750. [DOI] [PubMed] [Google Scholar]
- 45. Stehle T, Gamblin SJ, Yan Y, Harrison SC. The structure of simian virus 40 refined at 3.1 A resolution. Structure. 1996;4:165-182. [DOI] [PubMed] [Google Scholar]
- 46. Liddington RC, Yan Y, Moulai J, Sahli R, Benjamin TL, Harrison SC. Structure of simian virus 40 at 3.8-A resolution. Nature. 1991;354:278-284. [DOI] [PubMed] [Google Scholar]
- 47. Li TC, Takeda N, Kato K, et al. Characterization of self-assembled virus-like particles of human polyomavirus BK generated by recombinant baculoviruses. Virology. 2003;311:115-124. [DOI] [PubMed] [Google Scholar]
- 48. Stroh LJ, Neu U, Blaum BS, Buch MH, Garcea RL, Stehle T. Structure analysis of the major capsid proteins of human polyomaviruses 6 and 7 reveals an obstructed sialic acid binding site. J Virol. 2014;88:10831-10839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li TC, Iwasaki K, Katano H, et al. Characterization of self-assembled virus-like particles of Merkel cell polyomavirus. PLoS ONE. 2015;10:e0115646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hurdiss DL, Morgan EL, Thompson RF, et al. New structural insights into the genome and minor capsid proteins of BK polyomavirus using cryo-electron microscopy. Structure. 2016;24:528-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fiers W, Contreras R, Haegemann G, et al. Complete nucleotide sequence of SV40 DNA. Nature. 1978;273:113-120. [DOI] [PubMed] [Google Scholar]
- 52. Reddy VB, Thimmappaya B, Dhar R, et al. The genome of simian virus 40. Science. 1978;200:494-502. [DOI] [PubMed] [Google Scholar]
- 53. Frisque RJ, Bream GL, Cannella MT. Human polyomavirus JC virus genome. J Virol. 1984;51:458-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hirsch HH, Steiger J. Polyomavirus BK. Lancet Infect Dis. 2003;3:611-623. [DOI] [PubMed] [Google Scholar]
- 55. Van Ghelue M, Khan MT, Ehlers B, Moens U. Genome analysis of the new human polyomaviruses. Rev Med Virol. 2012;22:354-377. [DOI] [PubMed] [Google Scholar]
- 56. Prins C, Frisque RJ. JC virus T′ proteins encoded by alternatively spliced early mRNAs enhance T antigen-mediated viral DNA replication in human cells. J Neurovirol. 2001;7:250-264. [DOI] [PubMed] [Google Scholar]
- 57. Shuda M, Feng H, Kwun HJ, et al. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci U S A. 2008;105:16272-16277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Abend JR, Joseph AE, Das D, Campbell-Cecen DB, Imperiale MJ. A truncated T antigen expressed from an alternatively spliced BK virus early mRNA. J Gen Virol. 2009;90:1238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. van der Meijden E, Kazem S, Dargel CA, van Vuren N, Hensbergen PJ, Feltkamp MC. Characterization of T antigens, including middle T and alternative T, expressed by the human polyomavirus associated with trichodysplasia spinulosa. J Virol. 2015;89:9427-9439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Carter JJ, Daugherty MD, Qi X, et al. Identification of an overprinting gene in Merkel cell polyomavirus provides evolutionary insight into the birth of viral genes. Proc Natl Acad Sci U S A. 2013;110:12744-12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gan DD, Khalili K. Interaction between JCV large T-antigen and beta-catenin. Oncogene. 2004;23:483-490. [DOI] [PubMed] [Google Scholar]
- 62. Borchert S, Czech-Sioli M, Neumann F, et al. High-affinity Rb binding, p53 inhibition, subcellular localization, and transformation by wild-type or tumor-derived shortened Merkel cell polyomavirus large T antigens. J Virol. 2014;88:3144-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gou WF, Zhao S, Shen DF, et al. The oncogenic role of JC virus T antigen in lens tumors without cell specificity of alternative splicing of its intron. Oncotarget. 2015;6:8036-8045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Houben R, Angermeyer S, Haferkamp S, et al. Characterization of functional domains in the Merkel cell polyoma virus Large T antigen. Int J Cancer. 2015;136:E290-E300. [DOI] [PubMed] [Google Scholar]
- 65. Wendzicki JA, Moore PS, Chang Y. Large T and small T antigens of Merkel cell polyomavirus. Curr Opin Virol. 2015;11:38-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. An P, Saenz Robles MT, Pipas JM. Large T antigens of polyomaviruses: amazing molecular machines. Annu Rev Microbiol. 2012;66:213-236. [DOI] [PubMed] [Google Scholar]
- 67. Topalis D, Andrei G, Snoeck R. The large tumor antigen: a “Swiss Army knife” protein possessing the functions required for the polyomavirus life cycle. Antiviral Res. 2013;97:122-136. [DOI] [PubMed] [Google Scholar]
- 68. Kwun HJ, Guastafierro A, Shuda M, et al. The minimum replication origin of Merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication. J Virol. 2009;83:12118-12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Harrison CJ, Meinke G, Kwun HJ, et al. Asymmetric assembly of Merkel cell polyomavirus large T-antigen origin binding domains at the viral origin. J Mol Biol. 2011;409:529-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Harrison C, Jiang T, Banerjee P, et al. Polyomavirus large T antigen binds symmetrical repeats at the viral origin in an asymmetrical manner. J Virol. 2013;87:13751-13759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chang YP, Xu M, Machado AC, Yu XJ, Rohs R, Chen XS. Mechanism of origin DNA recognition and assembly of an initiator-helicase complex by SV40 large tumor antigen. Cell Rep. 2013;3:1117-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Meinke G, Phelan PJ, Kalekar R, et al. Insights into the initiation of JC virus DNA replication derived from the crystal structure of the T-antigen origin binding domain. PLoS Pathog. 2014;10:e1003966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mahon C, Liang B, Tikhanovich I, et al. Restriction of human polyomavirus BK virus DNA replication in murine cells and extracts. J Virol. 2009;83:5708-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tikhanovich I, Nasheuer HP. Host-specific replication of BK virus DNA in mouse cell extracts is independently controlled by DNA polymerase alpha-primase and inhibitory activities. J Virol. 2010;84:6636-6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Deyerle KL, Sajjadi FG, Subramani S. Analysis of origin of DNA replication of human papovavirus BK. J Virol. 1989;63:356-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Liang B, Tikhanovich I, Nasheuer HP, Folk WR. Stimulation of BK virus DNA replication by NFI family transcription factors. J Virol. 2012;86:3264-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kenan DJ, Mieczkowski PA, Burger-Calderon R, Singh HK, Nickeleit V. The oncogenic potential of BK-polyomavirus is linked to viral integration into the human genome. J Pathol. 2015;237:379-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li J, Wang X, Diaz J, Tsang SH, Buck CB, You J. Merkel cell polyomavirus large T antigen disrupts host genomic integrity and inhibits cellular proliferation. J Virol. 2013;87:9173-9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Delbue S, Ferrante P, Provenzano M. Polyomavirus BK and prostate cancer: an unworthy scientific effort? Oncoscience. 2014;1:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Weinreb DB, Desman GT, Amolat-Apiado MJ, Burstein DE, Godbold JH, Johnson EM. Polyoma virus infection is a prominent risk factor for bladder carcinoma in immunocompetent individuals. Diagn Cytopathol. 2006;34:201-203. [DOI] [PubMed] [Google Scholar]
- 81. Alexiev BA, Randhawa P, Vazquez Martul E, et al. BK virus-associated urinary bladder carcinoma in transplant recipients: report of 2 cases, review of the literature, and proposed pathogenetic model. Hum Pathol. 2013;44:908-917. [DOI] [PubMed] [Google Scholar]
- 82. Roberts IS, Besarani D, Mason P, Turner G, Friend PJ, Newton R. Polyoma virus infection and urothelial carcinoma of the bladder following renal transplantation. Br J Cancer. 2008;99:1383-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kenan DJ, Mieczkowski PA, Latulippe E, Cote I, Singh HK, Nickeleit V. BK polyomavirus genomic integration and large T antigen expression: evolving paradigms in human oncogenesis. Am J Transplant. 2017;17:1674-1680. [DOI] [PubMed] [Google Scholar]
- 84. Dorries K, Loeber G, Meixensberger J. Association of polyomaviruses JC, SV40, and BK with human brain tumors. Virology. 1987;160:268-270. [DOI] [PubMed] [Google Scholar]
- 85. Bialasiewicz S, Cho Y, Rockett R, et al. Association of micropapillary urothelial carcinoma of the bladder and BK viruria in kidney transplant recipients. Transpl Infect Dis. 2013;15:283-289. [DOI] [PubMed] [Google Scholar]
- 86. Wold WS, Green M, Mackey JK, Martin JD, Padgett BL, Walker DL. Integration pattern of human JC virus sequences in two clones of a cell line established from a JC virus-induced hamster brain tumor. J Virol. 1980;33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Del Valle L, White MK, Khalili K. Potential mechanisms of the human polyomavirus JC in neural oncogenesis. J Neuropathol Exp Neurol. 2008;67:729-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hori R, Murai Y, Tsuneyama K, et al. Detection of JC virus DNA sequences in colorectal cancers in Japan. Virchows Arch. 2005;447:723-730. [DOI] [PubMed] [Google Scholar]
- 89. Coelho TR, Almeida L, Lazo PA. JC virus in the pathogenesis of colorectal cancer, an etiological agent or another component in a multistep process? Virol J. 2010;7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Theodoropoulos G, Panoussopoulos D, Papaconstantinou I, et al. Assessment of JC polyoma virus in colon neoplasms. Dis Colon Rectum. 2005;48:86-91. [DOI] [PubMed] [Google Scholar]
- 91. Delbue S, Comar M, Ferrante P. Review on the role of the human Polyomavirus JC in the development of tumors. Infect Agent Cancer. 2017;12:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Pantulu ND, Pallasch CP, Kurz AK, et al. Detection of a novel truncating Merkel cell polyomavirus large T antigen deletion in chronic lymphocytic leukemia cells. Blood. 2010;116:5280-5284. [DOI] [PubMed] [Google Scholar]
- 93. Kassem A, Technau K, Kurz AK, et al. Merkel cell polyomavirus sequences are frequently detected in nonmelanoma skin cancer of immunosuppressed patients. Int J Cancer. 2009;125:356-361. [DOI] [PubMed] [Google Scholar]
- 94. Cheng J, Rozenblatt-Rosen O, Paulson KG, Nghiem P, DeCaprio JA. Merkel cell polyomavirus large T antigen has growth-promoting and inhibitory activities. J Virol. 2013;87:6118-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Stakaityte G, Wood JJ, Knight LM, et al. Merkel cell polyomavirus: molecular insights into the most recently discovered human tumour virus. Cancers (Basel). 2014;6:1267-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Trabanelli C, Corallini A, Gruppioni R, et al. Chromosomal aberrations induced by BK virus T antigen in human fibroblasts. Virology. 1998;243:492-496. [DOI] [PubMed] [Google Scholar]
- 97. Whalen KA, de Jesus R, Kean JA, Schaffhausen BS. Genetic analysis of the polyomavirus DnaJ domain. J Virol. 2005;79:9982-9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Berjanskii MV, Riley MI, Xie A, Semenchenko V, Folk WR, Van Doren SR. NMR structure of the N-terminal J domain of murine polyomavirus T antigens. Implications for DnaJ-like domains and for mutations of T antigens. J Biol Chem. 2000;275:36094-36103. [DOI] [PubMed] [Google Scholar]
- 99. Chromy LR, Pipas JM, Garcea RL. Chaperone-mediated in vitro assembly of Polyomavirus capsids. Proc Natl Acad Sci U S A. 2003;100:10477-10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sheng Q, Denis D, Ratnofsky M, Roberts TM, DeCaprio JA, Schaffhausen B. The DnaJ domain of polyomavirus large T antigen is required to regulate Rb family tumor suppressor function. J Virol. 1997;71:9410-9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Harris KF, Christensen JB, Radany EH, Imperiale MJ. Novel mechanisms of E2F induction by BK virus large-T antigen: requirement of both the pRb-binding and the J domains. Mol Cell Biol. 1998;18:1746-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bollag B, Prins C, Snyder EL, Frisque RJ. Purified JC virus T and T′ proteins differentially interact with the retinoblastoma family of tumor suppressor proteins. Virology. 2000;274:165-178. [DOI] [PubMed] [Google Scholar]
- 103. Spurgeon ME, Lambert PF. Merkel cell polyomavirus: a newly discovered human virus with oncogenic potential. Virology. 2013;435:118-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Cotsiki M, Lock RL, Cheng Y, et al. Simian virus 40 large T antigen targets the spindle assembly checkpoint protein Bub1. Proc Natl Acad Sci U S A. 2004;101:947-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hu L, Filippakis H, Huang H, Yen TJ, Gjoerup OV. Replication stress and mitotic dysfunction in cells expressing simian virus 40 large T antigen. J Virol. 2013;87:13179-13192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wang X, Helfer CM, Pancholi N, Bradner JE, You J. Recruitment of Brd4 to the human papillomavirus type 16 DNA replication complex is essential for replication of viral DNA. J Virol. 2013;87:3871-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. You J, Croyle JL, Nishimura A, Ozato K, Howley PM. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117:349-360. [DOI] [PubMed] [Google Scholar]
- 108. Gauson EJ, Donaldson MM, Dornan ES, et al. Evidence supporting a role for TopBP1 and Brd4 in the initiation but not continuation of human papillomavirus 16 E1/E2-mediated DNA replication. J Virol. 2015;89:4980-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Helfer CM, Yan J, You J. The cellular bromodomain protein Brd4 has multiple functions in E2-mediated papillomavirus transcription activation. Viruses. 2014;6:3228-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Voronkov A, Krauss S. Wnt/beta-catenin signaling and small molecule inhibitors. Curr Pharm Des. 2013;19:634-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. Embo J. 1998;17:1371-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Xing Y, Clements WK, Kimelman D, Xu W. Crystal structure of a beta-catenin/axin complex suggests a mechanism for the beta-catenin destruction complex. Genes Dev. 2003;17:2753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Enam S, Del Valle L, Lara C, et al. Association of human polyomavirus JCV with colon cancer: evidence for interaction of viral T-antigen and beta-catenin. Cancer Res. 2002;62:7093-7101. [PubMed] [Google Scholar]
- 114. Krynska B, Del Valle L, Gordon J, Otte J, Croul S, Khalili K. Identification of a novel p53 mutation in JCV-induced mouse medulloblastoma. Virology. 2000;274:65-74. [DOI] [PubMed] [Google Scholar]
- 115. Bhattacharyya R, Noch EK, Khalili K. A novel role of Rac1 GTPase in JCV T-antigen-mediated beta-catenin stabilization. Oncogene. 2007;26:7628-7636. [DOI] [PubMed] [Google Scholar]
- 116. Ripple MJ, Parker Struckhoff A, Trillo-Tinoco J, et al. Activation of c-Myc and Cyclin D1 by JCV T-antigen and beta-catenin in colon cancer. PLoS ONE. 2014;9:e106257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Khalili K, Del Valle L, Wang JY, et al. T-antigen of human polyomavirus JC cooperates with IGF-IR signaling system in cerebellar tumors of the childhood-medulloblastomas. Anticancer Res. 2003;23:2035-2041. [PubMed] [Google Scholar]
- 118. Lasithiotaki I, Antoniou KM, Derdas SP, et al. The presence of Merkel cell polyomavirus is associated with deregulated expression of BRAF and Bcl-2 genes in non-small cell lung cancer. Int J Cancer. 2013;133:604-611. [DOI] [PubMed] [Google Scholar]
- 119. Xu S, Jiang J, Yu X, Sheng D, Zhu T, Jin M. Association of Merkel cell polyomavirus infection with EGFR mutation status in Chinese non-small cell lung cancer patients. Lung Cancer. 2014;83:341-346. [DOI] [PubMed] [Google Scholar]
- 120. Iwasaki T, Matsushita M, Nonaka D, et al. Comparison of Akt/mTOR/4E-BP1 pathway signal activation and mutations of PIK3CA in Merkel cell polyomavirus-positive and Merkel cell polyomavirus-negative carcinomas. Hum Pathol. 2015;46:210-216. [DOI] [PubMed] [Google Scholar]
- 121. Spurgeon ME, Cheng J, Bronson RT, Lambert PF, DeCaprio JA. Tumorigenic activity of Merkel cell polyomavirus T antigens expressed in the stratified epithelium of mice. Cancer Res. 2015;75:1068-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Verhalen B, Starrett GJ, Harris RS, Jiang M. Functional upregulation of the DNA cytosine deaminase APOBEC3B by polyomaviruses. J Virol. 2016;90:6379-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Di Fiore R, D’Anneo A, Tesoriere G, Vento R. RB1 in cancer: different mechanisms of RB1 inactivation and alterations of pRb pathway in tumorigenesis. J Cell Physiol. 2013;228:1676-1687. [DOI] [PubMed] [Google Scholar]
- 124. Chinnam M, Goodrich DW. RB1, development, and cancer. Curr Top Dev Biol. 2011;94:129-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14:518-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Chellappan S, Kraus VB, Kroger B, et al. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc Natl Acad Sci U S A. 1992;89:4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Stubdal H, Zalvide J, Campbell KS, Schweitzer C, Roberts TM, DeCaprio JA. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol. 1997;17:4979-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Brown VD, Gallie BL. The B-domain lysine patch of pRB is required for binding to large T antigen and release of E2F by phosphorylation. Mol Cell Biol. 2002;22:1390-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Sullivan CS, Cantalupo P, Pipas JM. The molecular chaperone activity of simian virus 40 large T antigen is required to disrupt Rb-E2F family complexes by an ATP-dependent mechanism. Mol Cell Biol. 2000;20:6233-6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Lin JY, DeCaprio JA. SV40 large T antigen promotes dephosphorylation of p130. J Biol Chem. 2003;278:46482-46487. [DOI] [PubMed] [Google Scholar]
- 131. Pilon AA, Desjardins P, Hassell JA, Mes-Masson AM. Functional implications of mutations within polyomavirus large T antigen Rb-binding domain: effects on pRb and p107 binding in vitro and immortalization activity in vivo. J Virol. 1996;70:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Caracciolo V, Reiss K, Khalili K, De Falco G, Giordano A. Role of the interaction between large T antigen and Rb family members in the oncogenicity of JC virus. Oncogene. 2006;25:5294-5301. [DOI] [PubMed] [Google Scholar]
- 133. Caracciolo V, Macaluso M, D’Agostino L, et al. Cross-talk between T-Ag presence and pRb family and p53/p73 signaling in mouse and human medulloblastoma. J Cell Biochem. 2010;110:182-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Tognon M, Corallini A, Martini F, Negrini M, Barbanti-Brodano G. Oncogenic transformation by BK virus and association with human tumors. Oncogene. 2003;22:5192-5200. [DOI] [PubMed] [Google Scholar]
- 135. McCabe MT, Low JA, Imperiale MJ, Day ML. Human polyomavirus BKV transcriptionally activates DNA methyltransferase 1 through the pRb/E2F pathway. Oncogene. 2006;25:2727-2735. [DOI] [PubMed] [Google Scholar]
- 136. Li J, Diaz J, Wang X, Tsang SH, You J. Phosphorylation of Merkel cell polyomavirus large tumor antigen at serine 816 by ATM kinase induces apoptosis in host cells. J Biol Chem. 2015;290:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Houben R, Adam C, Baeurle A, et al. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. Int J Cancer. 2012;130:847-856. [DOI] [PubMed] [Google Scholar]
- 138. Arora R, Shuda M, Guastafierro A, et al. Survivin is a therapeutic target in Merkel cell carcinoma. Sci Transl Med. 2012;4:133ra156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Kazem S, van der Meijden E, Wang RC, et al. Polyomavirus-associated trichodysplasia spinulosa involves hyperproliferation, pRB phosphorylation and upregulation of p16 and p21. PLoS ONE. 2014;9:e108947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Berrios C, Jung J, Primi B, et al. Malawi polyomavirus is a prevalent human virus that interacts with known tumor suppressors. J Virol. 2015;89:857-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Keijzers M, Rensspiess D, Pujari S, et al. Expression of pRb and p16INK4 in human thymic epithelial tumors in relation to the presence of human polyomavirus 7. Diagn Pathol. 2015;10:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Rennspiess D, Pujari S, Keijzers M, et al. Detection of human polyomavirus 7 in human thymic epithelial tumors. J Thorac Oncol. 2015;10:360-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Liu J, Zhang C, Hu W, Feng Z. Tumor suppressor p53 and its mutants in cancer metabolism. Cancer Lett. 2015;356:197-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Afshar RM, Mollaei HR, Zandi B, Iranpour M. Evaluation of JC and cytomegalo viruses in glioblastoma tissue. Asian Pac J Cancer Prev. 2016;17:4907-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Houben R, Dreher C, Angermeyer S, et al. Mechanisms of p53 restriction in Merkel cell carcinoma cells are independent of the Merkel cell polyoma virus T antigens. J Invest Dermatol. 2013;133:2453-2460. [DOI] [PubMed] [Google Scholar]
- 146. Tsang SH, Wang R, Nakamaru-Ogiso E, Knight SA, Buck CB, You J. The oncogenic small tumor antigen of Merkel cell polyomavirus is an iron-sulfur cluster protein that enhances viral DNA replication. J Virol. 2015;90:1544-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. White EA, Kramer RE, Hwang JH, et al. Papillomavirus E7 oncoproteins share functions with polyomavirus small T antigens. J Virol. 2015;89:2857-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Srinivasan A, McClellan AJ, Vartikar J, et al. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol Cell Biol. 1997;17:4761-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Whalen B, Laffin J, Friedrich TD, Lehman JM. SV40 small T antigen enhances progression to >G2 during lytic infection. Exp Cell Res. 1999;251:121-127. [DOI] [PubMed] [Google Scholar]
- 150. Kolzau T, Hansen RS, Zahra D, Reddel RR, Braithwaite AW. Inhibition of SV40 large T antigen induced apoptosis by small T antigen. Oncogene. 1999;18:5598-5603. [DOI] [PubMed] [Google Scholar]
- 151. Khalili K, Sariyer IK, Safak M. Small tumor antigen of polyomaviruses: role in viral life cycle and cell transformation. J Cell Physiol. 2008;215:309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Cho US, Morrone S, Sablina AA, Arroyo JD, Hahn WC, Xu W. Structural basis of PP2A inhibition by small t antigen. PLoS Biol. 2007;5:e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Bollag B, Hofstetter CA, Reviriego-Mendoza MM, Frisque RJ. JC virus small T antigen binds phosphatase PP2A and Rb family proteins and is required for efficient viral DNA replication activity. PLoS ONE. 2010;5:e10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Seshacharyulu P, Pandey P, Datta K, Batra SK. Phosphatase: PP2A structural importance, regulation and its aberrant expression in cancer. Cancer Lett. 2013;335:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Mungre S, Enderle K, Turk B, et al. Mutations which affect the inhibition of protein phosphatase 2A by simian virus 40 small-t antigen in vitro decrease viral transformation. J Virol. 1994;68:1675-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Schuchner S, Wintersberger E. Binding of polyomavirus small T antigen to protein phosphatase 2A is required for elimination of p27 and support of S-phase induction in concert with large T antigen. J Virol. 1999;73:9266-9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Skoczylas C, Henglein B, Rundell K. PP2A-dependent transactivation of the cyclin A promoter by SV40 ST is mediated by a cell cycle-regulated E2F site. Virology. 2005;332:596-601. [DOI] [PubMed] [Google Scholar]
- 158. Sotillo E, Garriga J, Kurimchak A, Grana X, Cyclin E. SV40 small T antigen cooperate to bypass quiescence and contribute to transformation by activating CDK2 in human fibroblasts. J Biol Chem. 2008;283:11280-11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Frost JA, Alberts AS, Sontag E, Guan K, Mumby MC, Feramisco JR. Simian virus 40 small t antigen cooperates with mitogen-activated kinases to stimulate AP-1 activity. Mol Cell Biol. 1994;14:6244-6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Seamone ME, Wang W, Acott P, Beck PL, Tibbles LA, Muruve DA. MAP kinase activation increases BK polyomavirus replication and facilitates viral propagation in vitro. J Virol Methods. 2010;170:21-29. [DOI] [PubMed] [Google Scholar]
- 161. Wu JH, Simonette RA, Nguyen HP, Rady PL, Tyring SK. Small T-antigen of the TS-associated polyomavirus activates factors implicated in the MAPK pathway. J Eur Acad Dermatol Venereol. 2016;30:1061-1062. [DOI] [PubMed] [Google Scholar]
- 162. Klucky B, Wintersberger E. Polyomavirus small T antigen transactivates genes by its ability to provoke the synthesis and the stabilization of MYC. Oncogene. 2007;26:6356-6360. [DOI] [PubMed] [Google Scholar]
- 163. Kwun HJ, Shuda M, Feng H, Camacho CJ, Moore PS, Chang Y. Merkel cell polyomavirus small T antigen controls viral replication and oncoprotein expression by targeting the cellular ubiquitin ligase SCFFbw7. Cell Host Microbe. 2013;14:125-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Sablina AA, Hahn WC. SV40 small T antigen and PP2A phosphatase in cell transformation. Cancer Metastasis Rev. 2008;27:137-146. [DOI] [PubMed] [Google Scholar]
- 165. Griffiths DA, Abdul-Sada H, Knight LM, et al. Merkel cell polyomavirus small T antigen targets the NEMO adaptor protein to disrupt inflammatory signaling. J Virol. 2013;87:13853-13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Abdul-Sada H, Muller M, Mehta R, et al. The PP4R1 sub-unit of protein phosphatase PP4 is essential for inhibition of NF-κB by Merkel polyomavirus small tumour antigen. Oncotarget. 2017;8:25418-25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. White MK, Bellizzi A, Ibba G, Pietropaolo V, Palamara AT, Wollebo HS. The DNA damage response promotes polyomavirus JC infection by nucleus to cytoplasm NF-kappaB activation. Virol J. 2017;14:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J Clin Invest. 2011;121:3623-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Verhaegen ME, Mangelberger D, Harms PW, et al. Merkel cell polyomavirus small T antigen is oncogenic in transgenic mice. J Invest Dermatol. 2015;135:1415-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Kwun HJ, Shuda M, Camacho CJ, et al. Restricted protein phosphatase 2A targeting by Merkel cell polyomavirus small T antigen. J Virol. 2015;89:4191-4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Zenz R, Scheuch H, Martin P, et al. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev Cell. 2003;4:879-889. [DOI] [PubMed] [Google Scholar]
- 172. Wu JH, Simonette RA, Nguyen HP, Rady PL, Tyring SK. Merkel cell polyomavirus in Merkel cell carcinogenesis: small T antigen-mediates c-Jun phosphorylation. Virus Genes. 2016;52:397-399. [DOI] [PubMed] [Google Scholar]
- 173. Shahzad N, Shuda M, Gheit T, et al. The T antigen locus of Merkel cell polyomavirus downregulates human Toll-like receptor 9 expression. J Virol. 2013;87:13009-13019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Li H, Gade P, Xiao W, Kalvakolanu DV. The interferon signaling network and transcription factor C/EBP-beta. Cell Mol Immunol. 2007;4:407-418. [PMC free article] [PubMed] [Google Scholar]
- 175. Johnson PF. Molecular stop signs: regulation of cell-cycle arrest by C/EBP transcription factors. J Cell Sci. 2005;118:2545-2555. [DOI] [PubMed] [Google Scholar]
- 176. Nerlov C. The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007;17:318-324. [DOI] [PubMed] [Google Scholar]