Abstract
Determining the exact pathogenesis of chronic gastrointestinal diseases remains difficult due to the complex in vivo environment. In this review we give an overview of the available epithelial cell culture systems developed to investigate pathophysiology of gastrointestinal diseases. Traditionally used two-dimensional (2D) immortalised (tumour) cell lines survive long-term, but are not genetically stable nor represent any human in particular. In contrast, primary cultures are patient unique, but short-lived. Three-dimensional (3D) organoid cultures resemble the crypt-villus domain and contain all cell lineages, are long-lived and genetically stable. Unfortunately, manipulation of the 3D organoid system is more challenging. Combining the 3D and 2D technologies may overcome limitations and offer the formation of monolayers on permeable membranes or flow-chambers. Determining the right model to use will depend on the pathology of interest and the focus of the research, defining which cell types need to be included in the model.
Keywords: Organoids, epithelial models, 2D cultures
Introduction
The upper lining of the gastrointestinal tract is covered by a single layer of epithelial cells that primarily function in nutrient absorption and secretion of mucus, antimicrobial peptides, hormones, ions and other factors. Moreover, the intestinal epithelium is exposed to many commensal and potential pathogenic microbes and microbial patterns, which brings the risk of infections but also inappropriate immune responses towards more prevalent commensal antigens. The intestinal epithelium therefore needs to act as a physical barrier against the antigen-rich lumen, separating it from the immune cell containing lamina propria.1 Furthermore, the intestinal immune system has evolved to allow a certain level of tolerance towards antigens of commensal or dietary origin. Many diseases, such as inflammatory bowel disease (IBD), coeliac disease, and colitis-associated neoplasia, result from a disruption of the gut immune homeostasis. The exact pathophysiology of these diseases is not fully understood and curative therapies are lacking. Much effort is being made to uncover the underlying disease pathways and in identifying therapeutic targets. Unfortunately, results from animal studies do not always match results obtained in clinical trials. Thus, developing pre-clinical models which better reflect the in vivo situation and efficiently predict the effect of an experimental therapeutic compound in a patient-specific manner is crucial.2
The complexity of many chronic gastrointestinal diseases makes them difficult to model. In this review we focus on available human intestinal epithelial models, their applications and future perspectives.
Two-dimensional (2D) models
The first used intestinal epithelial cell lines were culture-adapted derivatives of cancer. Several intestinal epithelial cell lines exist, such as the colonic adenocarcinoma cell line Caco-2, which can form differentiated and polarised monolayers containing small intestinal enterocyte-like cells. In contrast to Caco-2 cells, T84 and HT-29 cells can have a goblet cell-like phenotype but they are also derived from malignant cells.3 Culturing these highly proliferative cell lines is relatively easy and cheap, which makes them useful for mechanistic studies or high-throughput screening approaches. A major limitation is their reduced complexity and low physiological relevance. This has been partially overcome by growing these epithelial monolayers on permeable membrane supports instead of culture dishes. A confluent, polarised monolayer will have a differentiated and distinguishable apical and basolateral side that can be targeted independently by adding molecules or bacteria to a specific side of the transwell-insert. Caco-2 cells are mainly being used in this fashion for transport and permeability studies,4,5 but also Caco-2/HT-29-methotrexate co-cultures exist which contain both enterocytes and goblet cells.6
Recently, alternative support structures have been developed to better reflect the intestinal environment, for example seeding the cells in the inside of a hollow silk-based porous scaffold that has an intestine-like topography and can be loaded with human intestinal myofibroblasts.7 Such artificial scaffold structures are also used in the gut-on-chip technique where cells are grown on a porous membrane in a microfluidic flow cell.8 This approach allows a continuous and modifiable flow of nutrients on both sides of the epithelial layer mimicking apical flow forces. Furthermore, cyclic contractions can be applied which resemble gut peristalsis and result in altered gene expression and cell function/morphology. A final layer of complexity can be added in these in vitro approaches by co-culturing epithelial cell lines with immune cells and thereby mimicking the interactions that occur in the intestinal mucosa.9 In this setting the epithelial cells should be plated on the inverted transwell, in order to achieve migration of immune cells from the basolateral side.10
Nevertheless, findings might not always be physiologically relevant because of the tumour-like nature of cell lines and their discrepancies (e.g. karyotype, gene/protein expression) compared to epithelial tissue in/ex vivo. Efforts have therefore been made to combine the assets of the patient-specific and physiologically relevant, three-dimensional (3D) model (organoids, discussed later) with the practical (and financial) advantages of 2D epithelial cell monolayers. Just like organoids, growing patient-derived intestinal epithelial cells as a monolayer has been proven to be challenging. One of the major difficulties was to obtain enough donor material in order to perform multiple experiments on cells from a single patient. Perreault and Beaulieu were able to isolate and grow epithelial cells derived from whole small intestines from legally aborted human foetuses.11 Likewise, whole and partial organ resection tissue has been processed to isolate and grow larger amounts of intestinal epithelial cells.12,13 Although larger amounts of cells can be acquired, the requirement of bowel resection samples is a major limitation as these samples represent only patients with severe inflammation or disease-related complications. Furthermore, growing foetal or resection tissue-derived cells is of little use for personalised medicine approaches. In contrast, an ideal tool for personalised medicine and biomarker studies would be to culture intestinal biopsy-derived epithelial cells as monolayers. Pedersen and colleagues were able to isolate and grow colonic epithelial cells from mucosal biopsies from healthy individuals on collagen.14 Their reported viability was only 24% after 48 hours, clearly limiting applications. Our group has recently developed and characterised another biopsy-derived epithelial cell culture system with an improved proliferation/apoptosis ratio. In a first attempt to use this system for patient stratification, we showed that increasing number of IBD-associated mutations in endoplasmic reticulum (ER) stress and autophagy genes led to functionally increased ER stress responses in the colonic epithelium.15
3D models
The discovery of Leucin-rich repeat containing G-protein coupled receptor 5 (LGR5) as a robust intestinal stem cell marker, and consequently the formation of the 3D organoid model, has led to a better understanding of the intestinal epithelium.16–18 When isolated single LGR5-positive intestinal stem cells (ISCs) or crypts fractions are embedded in Matrigel (an extracellular matrix-mimicking substance)17 and are overlaid with a rich culture medium, cells undergo unlimited proliferation while remaining genetically stable.19 The majority of them form 3D structures with a clear crypt-villus domain that maintain polarisation towards a lumen, and can differentiate into all downstream epithelial cell lineages. Others will be more spherical. Two ways to form organoids currently exist: either from adult mucosal tissue,17 or by differentiation of induced pluripotent stem cells (iPSC).20 In contrast to the first type, iPSC-derived organoids have a more foetal phenotype and do contain a mesenchymal cell compartment as a result from differentiation procedures.20,21 Despite initiatives to name organoids according to the tissues from which they were generated (enteroids, colonoids), the term organoid is still used in a broad and unspecific manner. Here we refer to organoids as those from a purely epithelial origin.17 This purity is both a strength and a weakness of the model: It allows the pinpointing of epithelial causality, but does not mimic the complexity of the in vivo situation since organoids lack for example an immune system compartment, a nerve system, or nutrients providing mesenchymal niche. This has recently been overcome by co-culture systems, e.g. with intraepithelial lymphoid cells (IELs).22 The most recent advance in making the organoid model more representative was the addition of a functional enteric nervous system.23
The organoid model has already been used to study an extensive range of diseases but can also be used to perform mechanistic studies in normal human tissue. While we focus on human disease modelling, some studies referred to here were performed in organoids from mice. Yet, these studies are great proof of principles and are highly likely to be recapitulated in the human condition.
Organoids retain basic physiological principles of the intestinal epithelium: Zietek and colleagues showed that organoids derived from mice lacking specific nutrient transporters (SGLT1, GLUT5) have impaired transport of glucose and fructose respectively, and consequently there was also a decrease in hormone release in these organoids.24 Validation of intestinal organoids as an efficient ion transport-model was performed by Foulke-Abel and colleagues, who demonstrated that organoids have, among others, steady Na+/H+ exchanger 3 activity which can be modulated by inhibitors, as well as bacterial enterotoxins.25 Furthermore, dietary fat absorption and the synthesis of chylomicron may be linked to the pathophysiology of cardiovascular risk factors, as shown by apoC-III overexpression leading to smaller chylomicrons with less dietary triacylglycerol.26
Disease modelling with organoids
Many diseases are being modelled with organoids. Here we discuss the most applicable and promising possibilities.
In the cystic fibrosis (CF) field, patient-derived organoids are being used to screen therapeutic compounds using the forskolin swelling assay to determine their potential to rescue cystic fibrosis transmembrane conductance regulator (CFTR) function in a personalised manner.27 Moreover, using CRISPR/Cas9 gene editing, CFTR function was restored in organoids from patients with CF.28
Salas’ group was the first to use organoids investigating IL-1R2 in ulcerative colitis (UC), confirming the observation from biopsy data that differentiation leads to IL1R2 expression.29 They, as well as our group, have also used the organoid system to study stem cells from patients with IBD, and found differentially expressed genes in organoids from patients with UC, compared to controls, indicating a genetic imprinting which is recapitulated in vitro.30,31
Hackam’s group used the organoid system to dissect the molecular basis of necrotising enterocolitis (NEC) and showed that Toll-like receptor 4 activation in ISCs led to ER stress-mediated apoptosis in crypts.32 Microvillus inclusion disease (MVID) has also been modelled in organoids from patients, showing many of the typical epithelial defects observed in vivo.33 Mechanisms behind colon cancer were investigated by both the groups of Toshiro Sato and Hans Clevers. After consecutive induction of mutations in genes involved in tumour suppression or oncogenesis (APC, SMAD4, TP53, KRAS, PIK3CA), mutated organoids became independent of proliferative niche factors and also showed to be able to proliferate in vivo.34,35 Lastly, differentiation of ISCs can be easily manipulated ex vivo, which for instance allowed the induction and study of M cells, which are normally very scarce in vivo.36
Dissecting the effect of microbes on the intestinal epithelium
Luminal exposure studies require micro-injection of agents or microbes into the lumen of the organoid, which is technically challenging. This technique is put forward to study host-microbe interactions. Indeed, many studies have already showed the potency of this model.
One such study showed an interplay of epithelial cells with Salmonella enterica, and the effect of genetics: Micro-injection into the lumen of murine ileal organoids showed that infection is limited by α-defensins, and interestingly this was impaired in MMP7–/–, but not in NOD2–/– organoids.37
Other novel studies showed the culture of viruses, such as rotavirus.38,39 Bacteria have been micro-injected into the lumen of organoids, such as Helicobacter pylori in gastric organoids, which induced a strong inflammatory reaction in gastric gland cells, but not in gastric pit cells.40 H. pylori is a capnophile, and some strains grow best in microaerobic conditions such as in the stomach. Many other bacterial species are strictly anaerobe, thus limiting experiments in which cells are kept under normoxic conditions. Furthermore, many bacteria are unculturable, although this is still improving.41 Advanced technical setups will be required to overcome these limitations.
Peck and colleagues demonstrated a direct effect of microbiota on the expression of miRNA in murine ISCs, and upon knockdown of miR-375, organoids showed increased proliferation.42 Moreover, bacterial components also have (long-lasting) effects on organoids, as Hibiya and colleagues found decreased differentiation and increased NFκB signalling in organoids at 11 weeks after a 60-week exposure to a cocktail of cytokines and bacterial cell wall components.43 These advancements illustrate how organoid cultures are an excellent model for studying many diseases: homing of immune cells towards the epithelium, pathologies linked to the enteric nervous system, barrier defects, and the genetic causality in these disease phenotypes, and like in CF, test compounds on their therapeutic potential. Organoids can now be formed from all parts of the gastrointestinal tract, and additionally from liver,44 pancreas,45 and many other non-intestinal tissues. While these organoids have many growth factors in common, they retain their lineage and location specificity.46,47 For organoids formed from LGR5+ ISCs this is less well described. Interestingly, organoids established from murine foetal small intestine can engraft on colon, attaining corresponding colonic expression patterns.48
Explants
Whole mucosal biopsies may also be used ex vivo to recapitulate a more realistic situation as these biopsies contain both epithelial and immune cells. For example, whole biopsies can be placed in culture medium and exposed to cytokines.49 Alternatively they can be mounted in Ussing chambers for mucosal barrier studies.50,51 In short, barrier function is assessed by monitoring the passage of fluorescent molecules or by measuring the transepithelial electrical resistance (TEER).
Investigators have also used surgically removed parts of intestine to study intestinal pathophysiology.52 In this model, tissue was placed in a cylinder and exposed to Salmonella typhimurium with or without Lactobacillus paracasei supernatant. The authors demonstrated a protective effect of L. paracasei, as determined by tumour necrosis factor (TNF) and interleukin (IL)-10 secretion whereas L. paracasei exposure to tissues from IBD patients showed negative effects, indicating that even anti-inflammatory microbes have detrimental effects in predisposed individuals. More recently, an elegant study showed a more advanced model of a murine colon explant system in which the colon was excised and placed into a chamber where flow of medium was controlled, as well as the luminal flow.53 The enteric nervous system was involved in pro- or anti-inflammatory responses by modulating induction of RORg+ T-regulatory cells, proving that this model allows a far more detailed investigation than was previously possible. Unfortunately, explant tissues’ viability is in the range of days, with most experiments being performed within 24 hours.
Comparison of models
Organoids are without doubt an excellent model to study intestinal epithelium and its proliferative capacity. Due to the 3D nature, however, luminal exposure or transport studies require organoid injection, which is a challenging technique. Immune cells can be easily added to the Matrigel or medium for migration and interaction studies, same as in 2D models, as long as the cells are seeded on an inverted transwell. Cost is a drawback of organoid experiments: Matrigel is expensive, especially in high-throughput approaches, making fast-dividing cancer cell lines a more economical but a less realistic model. Explant also require some investment in materials, while their advantage is the multi-cellular composition. Most assays are feasible for all discussed models, with limitations for some (Table 1). The main limitations are because of cell numbers in the case of western blotting of organoids. In general, 2D models are accessible, while organoids and more advanced explant models require training and more work, for usage as well as downstream procedures.
Table 1.
Intestinal epithelial culture methods and potential applications.
| Culture | Source | Applications | Read-outs | Future applications | Cost |
|---|---|---|---|---|---|
| Cancer cell lines (CaCo-2, HT-29, SW480, T84, etc.) | Cancer tissues, adapted to cell culture | – Basic physiology (limited) – Host-microbe interactions | – RNA levels (+) – Western blot (+) – Immunostainings (+) – Transport of molecules (+) – TEER measurement (+) – Multiple cell types (−/+) – Cell viability/proliferation (+) | – Limited | Low |
| Primary 2D monolayers | – Crypts from mucosal biopsies – Resected tissue | – Basic physiology – Host-microbe interactions | – RNA levels (+) – Western blot (+) – Immunostainings (+) – Transport of molecules (+) – TEER measurement (+) – Multiple cell types (−/+) – Cell viability/proliferation (+) | – Pathway-based patient stratification – Gut-on-a-chip – Co-culture with other patient-derived cell types, microbiota | Low–Moderate |
| Primary 3D organoids | – Crypts or LGR5+ stem cells from mucosal biopsies – Resected tissue – iPSC-derived | – Basic physiology – Drug efficacy (e.g. CF) – Host-microbe interactions | – RNA levels (+) – Western blot (–/+) – Immunostainings (+) – Transport of molecules (+/−) – TEER measurement (+) – Multiple cell types (+) – Cell viability/proliferation (+) | – Co-culture systems (enteric nervous system, immune cells, myofibroblasts, microbiota) – Drug efficacy/toxicity screening | Expensive |
| Primary explants | – Mucosal biopsies – Resected tissue | – Ussing chambers (barrier function) – Effect on epithelium – Immune system and CNS in one system | – RNA levels (+) – Western blot (+) – Immunostainings (+) – Transport of molecules (+) – TEER measurement (+) – Multiple cell types (+) – Cell viability/proliferation (+) | – Limited/to be explored | Moderate |
TEER: transepithelial electrical resistance; 2D: two-dimensional; 3D: three-dimensional; LGR5: Leucin-rich repeat containing G-protein coupled receptor 5; CF: cystic fibrosis; CNS: central nervous system.
It is well known that 2D models are easy to manipulate and cost friendly, though their biological relevance is often disputed. Modifications towards chip-based microfluidics systems increases biological relevance but also cost, while decreasing ease of use and decreasing cell numbers, thereby limiting analyses. The next step will be to apply the aforementioned improvements of 2D cell lines to patient-derived epithelial monolayers combined with the proliferative advantage of the 3D models. A crucial leap forward in science originating from organoids was the ability to finally culture norovirus in a 2D layer formed from organoids.54 VanDussen et al. demonstrated how biopsy-derived epithelial cells from different regions of the intestine can be grown as confluent monolayers on membrane supports after enriching the number through initial expansion as organoids.55 This adaptation allows patient-specific assessment of the epithelial barrier function and possibly, in the future, also co-cultures with (patient-derived) immune or neuronal cells. Patient-derived epithelial monolayers can thus be generated from intestinal organoids allowing storage of isolated cells from a specific patient as a long-term organoid culture and even the performance of complementary experiments in both models in parallel. To achieve co-culture with patients’ own cells, and given the time and effort it takes to establish organoids, storing additional patient material such as immune cells and faecal samples will allow the recreation of the patient-specific microenvironment.
Conclusions and future perspectives
It is clear that many valid models for the intestinal epithelium exist, choosing the right model for a specific research question is essential as each model has its specific benefits and limitations as discussed earlier (Table 1).
Models that better mimic the complexity of the intestinal tissue, combining all the different cells types that may influence the response of these tissues, are still lacking. But the recent advances with organoids and 2D co-cultures brings hope that more intricate systems will be generated in the near future (also see Figure 1). In fact, the organoid model is continuously being adapted, and can now be extended to include other cellular components that we find in vivo, such as nerve, muscle, and immune cells.22,23 Adding immune cells may be difficult, but using iPSCs to create immune cells from the same individual may overcome this issue. The interaction of the epithelium with bacteria is one of the most interesting aspects to study. In the last decade, interest in microbiota has increased tremendously.56 Still, it is unclear what exactly a healthy microbiota is, and how it is related to homeostasis, inflammation and disasese.57
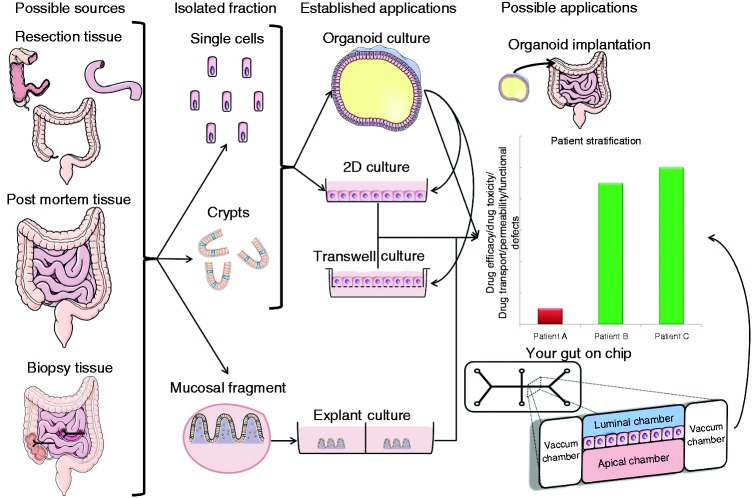
Figure 1.
Overview of intestinal epithelial culture possibilities, applications and the tissues of origin. Left: sample types available. Middle: tissue-derivatives which are cultured. Right: culture systems. Either crypts or single cells may be cultured as organoids under the right conditions. Organoids can be exposed to any environmental stimulus or through micro-injection be luminally exposed to microbes. Primary cells may be cultured on collagen either directly from isolated crypts or from organoids. This facilitates the direct exposure to stimuli and may be used (like organoids) in assays to investigate patients' response to drugs. 2D: two-dimensional.
The systems discussed here may prove invaluable models to further our understanding of disease processes, such as the importance of microbiota in intestinal cancer and IBD.58 Indeed, the future is looking bright for the advancement through human cell culture models, which are directly linked to the patient from whom the tissue was derived, also allowing personalised medicine approaches.
Acknowledgements
Author contributions are as follows: MN: writing of manuscript; WV: writing of manuscript; KA: writing of manuscript; ASR: manuscript corrections; GVA: manuscript corrections; SV: manuscript corrections; CV: manuscript corrections; MF: manuscript corrections and final approval.
Declaration of conflicting interests
Manuel Noben: PhD fellowship, Research Foundation – Flanders (FWO). Wiebe Vanhove: Nothing to declare. Kaline Arnauts: Nothing to declare. Anabela Santa Ramalho: Nothing to declare. Gert Van Assche: Financial support for research: Abbvie, MSD; lecture fees: Abbvie, Ferring, MSD, Janssen, and Takeda; consultancy: Abbvie, MSD, and Takeda. Catherine Verfaillie: Nothing to declare. Severine Vermeire: Grant support: AbbVie, MSD, Pfizer, and Takeda; speaker fees: AbbVie, MSD, Takeda, Ferring, Dr. Falk Pharma, Hospira, Pfizer Inc, and Tillots; consultancy: AbbVie, MSD, Takeda, Ferring, Genentech/Roche, Shire, Pfizer Inc, Galapagos, Mundipharma, Hospira, Celgene, Second Genome, and Janssen. Marc Ferrante: Research grant: Takeda; speakers fee: Abbvie, Boehringer-Ingelheim, Chiesi, Falk, Ferring, Janssen, Mitsubishi Tanabe, MSD, Takeda, Tillotts, and Zeria; consultancy: Abbvie, Boehringer-Ingelheim, Ferring, Janssen, and MSD.
Funding
Manuel Noben is a doctoral fellow, and Gert Van Assche, Séverine Vermeire, and Marc Ferrante are senior clinical investigators of the Research Foundation – Flanders (FWO). This study was partially funded by a CREA research grant of the KU Leuven (CREA/12/031), a research grant by the European Crohn’s and Colitis Organization (ZKC4621/10143459), a grant from the Flemish Gastroenterology Association (VVGE), and a grant from Belgian Inflammatory Bowel Disease Research and Development (BIRD).
Ethics approval
Not required.
Informed consent
Not required.
References
- 1.Ahmad R, Sorrell MF, Batra SK, et al. Gut permeability and mucosal inflammation: Bad, good or context dependent. Mucosal Immunol 2017; 10: 307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hay M, Thomas DW, Craighead JL, et al. Clinical development success rates for investigational drugs. Nat Biotechnol 2014; 32: 40–51. [DOI] [PubMed] [Google Scholar]
- 3.Liévin-Le Moal V, Servin AL. Pathogenesis of human enterovirulent bacteria: Lessons from cultured, fully differentiated human colon cancer cell lines. Microbiol Mol Biol Rev 2013; 77: 380–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varadi J, Harazin A, Fenyvesi F, et al. Alpha-melanocyte stimulating hormone protects against cytokine-induced barrier damage in Caco-2 intestinal epithelial monolayers. PLoS One 2017; 12: e0170537–e0170537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spinks CB, Zidan AS, Khan MA, et al. Pharmaceutical characterization of novel tenofovir liposomal formulations for enhanced oral drug delivery: In vitro pharmaceutics and Caco-2 permeability investigations. Clin Pharmacol 2017; 9: 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walter E, Janich S, Roessler BJ, et al. HT29-MTX/Caco-2 cocultures as an in vitro model for the intestinal epithelium: In vitro-in vivo correlation with permeability data from rats and humans. J Pharm Sci 1996; 85: 1070–1076. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Lin Y, Davis KM, et al. Robust bioengineered 3D functional human intestinal epithelium. Sci Rep 2015; 5: 13708–13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim HJ, Li H, Collins JJ, et al. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci U S A 2016; 113: E7–E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lozoya-Agullo I, Araújo F, González-Álvarez I, et al. Usefulness of Caco-2/HT29-MTX and Caco-2/HT29-MTX/Raji B coculture models to predict intestinal and colonic permeability compared to Caco-2 monoculture. Mol Pharm 2017; 14: 1264–1270. [DOI] [PubMed] [Google Scholar]
- 10.Kusek ME, Pazos MA, Pirzai W, et al. In vitro coculture assay to assess pathogen induced neutrophil trans-epithelial migration. J Vis Exp 2014, pp. 83: e50823–83: e50823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perreault N, Beaulieu JF. Primary cultures of fully differentiated and pure human intestinal epithelial cells. Exp Cell Res 1998; 245: 34–42. [DOI] [PubMed] [Google Scholar]
- 12.Scott A, Rouch JD, Jabaji Z, et al. Long-term renewable human intestinal epithelial stem cells as monolayers: A potential for clinical use. J Pediatr Surg 2016; 51: 995–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graves CL, Harden SW, LaPato M, et al. A method for high purity intestinal epithelial cell culture from adult human and murine tissues for the investigation of innate immune function. J Immunol Methods 2014; 414: 20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedersen G, Saermark T, Giese B, et al. A simple method to establish short-term cultures of normal human colonic epithelial cells from endoscopic biopsy specimens. Comparison of isolation methods, assessment of viability and metabolic activity. Scand J Gastroenterol 2000; 35: 772–780. [DOI] [PubMed] [Google Scholar]
- 15.Vanhove W, Nys K, Arijs I, et al. P095 The genetic risk in ER stress and autophagy translates into quantifiable epithelial ER stress levels in IBD patients. J Crohns Colitis 2017; 11: s124–s125. [Google Scholar]
- 16.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007; 449: 1003–1007. [DOI] [PubMed] [Google Scholar]
- 17.Sato T, Stange DE, Ferrante M, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011; 141: 1762–1772. [DOI] [PubMed] [Google Scholar]
- 18.Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009; 459: 262–265. [DOI] [PubMed] [Google Scholar]
- 19.van de Wetering M, Francies HE, Francis JM, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015; 161: 933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spence JR, Mayhew CN, Rankin SA, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011; 470: 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finkbeiner SR, Hill DR, Altheim CH, et al. Transcriptome-wide analysis reveals hallmarks of human intestine development and maturation in vitro and in vivo. Stem Cell Reports 2015; 4: 1140–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nozaki K, Mochizuki W, Matsumoto Y, et al. Co-culture with intestinal epithelial organoids allows efficient expansion and motility analysis of intraepithelial lymphocytes. J Gastroenterol 2016; 51: 206–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Workman MJ, Mahe MM, Trisno S, et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat Med 2017; 23: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zietek T, Rath E, Haller D, et al. Intestinal organoids for assessing nutrient transport, sensing and incretin secretion. Sci Rep 2015; 5: 16831–16831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foulke-Abel J, In J, Yin J, et al. Human enteroids as a model of upper small intestinal ion transport physiology and pathophysiology. Gastroenterology 2016; 150: 638–649.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jattan J, Rodia C, Li D, et al. Using primary murine intestinal enteroids to study dietary TAG absorption, lipoprotein synthesis, and the role of apoC-III in the intestine. J Lipid Res 2017; 58: 853–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dekkers JF, Wiegerinck CL, de Jonge HR, et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med 2013; 19: 939–945. [DOI] [PubMed] [Google Scholar]
- 28.Schwank G, Koo BK, Sasselli V, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 2013; 13: 653–658. [DOI] [PubMed] [Google Scholar]
- 29.Mora-Buch R, Dotti I, Planell N, et al. Epithelial IL-1R2 acts as a homeostatic regulator during remission of ulcerative colitis. Mucosal Immunol 2016; 9: 950–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dotti I, Mora-Buch R, Ferrer-Picón E, et al. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut. Epub ahead of print 1 November 2016. DOI: 10.1136/gutjnl-2016-312609. [DOI] [PMC free article] [PubMed]
- 31.Noben M, Verstockt B, de Bruyn M, et al. Epithelial organoid cultures from patients with ulcerative colitis and Crohn’s disease: A truly long-term model to study the molecular basis for inflammatory bowel disease? Gut. Epub ahead of print 3 February 2017. DOI: 10.1136/gutjnl-2016-313667. [DOI] [PubMed]
- 32.Neal MD, Sodhi CP, Jia H, et al. Toll-like receptor 4 is expressed on intestinal stem cells and regulates their proliferation and apoptosis via the p53 up-regulated modulator of apoptosis. J Biol Chem 2012; 287: 37296–37308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiegerinck CL, Janecke AR, Schneeberger K, et al. Loss of syntaxin 3 causes variant microvillus inclusion disease. Gastroenterology 2014; 147: 65–68.e10. [DOI] [PubMed] [Google Scholar]
- 34.Drost J, van Jaarsveld RH, Ponsioen B, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015; 521: 43–47. [DOI] [PubMed]
- 35.Matano M, Date S, Shimokawa M, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med 2015; 21: 256–262. [DOI] [PubMed] [Google Scholar]
- 36.Rouch JD, Scott A, Lei NY, et al. Development of functional microfold (M) cells from intestinal stem cells in primary human enteroids. PLoS One 2016; 11: e0148216–e0148216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson SS, Tocchi A, Holly MK, et al. A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal Immunol 2015; 8: 352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finkbeiner SR, Zeng XL, Utama B, et al. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. MBio 2012; 3: e00159–e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin Y, Bijvelds M, Dang W, et al. Modeling rotavirus infection and antiviral therapy using primary intestinal organoids. Antiviral Res 2015; 123: 120–131. [DOI] [PubMed] [Google Scholar]
- 40.Bartfeld S, Bayram T, van de Wetering M, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 2015; 148: 126–136.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Browne HP, Forster SC, Anonye BO, et al. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 2016; 533: 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peck BC, Mah AT, Pitman WA, et al. Functional transcriptomics in diverse intestinal epithelial cell types reveals robust microRNA sensitivity in intestinal stem cells to microbial status. J Biol Chem 2017; 292: 2586–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hibiya S, Tsuchiya K, Hayashi R, et al. Long-term inflammation transforms intestinal epithelial cells of colonic organoids. J Crohns Colitis 2017; 11: 621–630. [DOI] [PubMed] [Google Scholar]
- 44.Huch M, Gehart H, van Boxtel R, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015; 160: 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huch M, Bonfanti P, Boj SF, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J 2013; 32: 2708–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fukuda M, Mizutani T, Mochizuki W, et al. Small intestinal stem cell identity is maintained with functional Paneth cells in heterotopically grafted epithelium onto the colon. Genes Dev 2014; 28: 1752–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Middendorp S, Schneeberger K, Wiegerinck CL, et al. Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells 2014; 32: 1083–1091. [DOI] [PubMed] [Google Scholar]
- 48.Fordham RP, Yui S, Hannan NR, et al. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell 2013; 13: 734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jarry A, Malard F, Bou-Hanna C, et al. Interferon-alpha promotes Th1 response and epithelial apoptosis via inflammasome activation in human intestinal mucosa. Cell Mol Gastroenterol Hepatol 2017; 3: 72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herrmann JR, Turner JR. Beyond Ussing’s chambers: Contemporary thoughts on integration of transepithelial transport. Am J Physiol Cell Physiol 2016; 310: C423–C431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ussing HH. Transport through biological membranes. Annu Rev Physiol 1953; 15: 1–20. [DOI] [PubMed] [Google Scholar]
- 52.Tsilingiri K, Barbosa T, Penna G, et al. Probiotic and postbiotic activity in health and disease: Comparison on a novel polarised ex-vivo organ culture model. Gut 2012; 61: 1007–1015. [DOI] [PubMed] [Google Scholar]
- 53.Yissachar N, Zhou Y, Ung L, et al. An intestinal organ culture system uncovers a role for the nervous system in microbe-immune crosstalk. Cell 2017; 168: 1135–1148.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ettayebi K, Crawford SE, Murakami K, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016; 353: 1387–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.VanDussen KL, Marinshaw JM, Shaikh N, et al. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 2015; 64: 911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levy M, Kolodziejczyk AA, Thaiss CA, et al. Dysbiosis and the immune system. Nat Rev Immunol 2017; 17: 219–232. [DOI] [PubMed] [Google Scholar]
- 57.van den Elsen LW, Poyntz HC, Weyrich LS, et al. Embracing the gut microbiota: The new frontier for inflammatory and infectious diseases. Clin Transl Immunology 2017; 6: e125–e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsilimigras MC, Fodor A, Jobin C. Carcinogenesis and therapeutics: The microbiota perspective. Nat Microbiol 2017; 2: 17008–17008. [DOI] [PMC free article] [PubMed] [Google Scholar]