Abstract
Dicentric chromosomes are products of genomic rearrangements that place two centromeres on the same chromosome. Due to the presence of two primary constrictions, they are inherently unstable and overcome their instability by epigenetically inactivating and/or deleting one of the two centromeres, thus resulting in functionally monocentric chromosomes that segregate normally during cell division. Our understanding to date of dicentric chromosome formation, behavior and fate has been largely inferred from observational studies in plants and humans as well as artificially produced de novo dicentrics in yeast and in human cells. We investigate the most recent product of a chromosome fusion event fixed in the human lineage, human chromosome 2, whose stability was acquired by the suppression of one centromere, resulting in a unique difference in chromosome number between humans (46 chromosomes) and our most closely related ape relatives (48 chromosomes). Using molecular cytogenetics, sequencing, and comparative sequence data, we deeply characterize the relicts of the chromosome 2q ancestral centromere and its flanking regions, gaining insight into the ancestral organization that can be easily broadened to all acrocentric chromosome centromeres. Moreover, our analyses offered the opportunity to trace the evolutionary history of rDNA and satellite III sequences among great apes, thus suggesting a new hypothesis for the preferential inactivation of some human centromeres, including IIq. Our results suggest two possible centromere inactivation models to explain the evolutionarily stabilization of human chromosome 2 over the last 5–6 million years. Our results strongly favor centromere excision through a one-step process.
Keywords: dicentric chromosomal stability, satellite III and rDNA evolution, centromere evolution and inactivation
Introduction
Dicentric chromosomes are the result of genomic rearrangements placing two active centromeres on the same chromosome. Although dicentrics can occur between any two chromosomes, in human they are most frequently generated by Robertsonian translocations (ROBs). ROBs involve any two of the ten acrocentric chromosomes (13, 14, 15, 21, and 22), although rob(13; 14) (75.6%) and rob(14; 21) (9.9%) account for nearly 86% of dicentric ROBs ascertained from patients (Page et al. 1996). Moreover, dicentric chromosomes have been identified in hematological malignancies, including the recurrent dic(17; 20) (Watson et al. 2000; Patsouris et al. 2002) and dic(5; 17) (Wang et al. 1997) in myelodysplastic syndromes and acute myeloid leukemia and dic(9; 20) (Heerema et al. 1996) in acute lymphoblastic leukemia, as well as a range of other abnormalities.
Although there are cases in human showing that dicentrics may exist as functionally dicentric chromosomes primarily due to the short distance between the two centromeres (Sullivan and Willard 1998; Ewers et al. 2010), the majority are inherently unstable and they overcome their instability by epigenetically inactivating one centromere or, alternatively, by deleting one of the two centromeres, resulting in functionally monocentric chromosomes that segregate normally during cell division (Sullivan et al. 1994; Sullivan and Schwartz 1995; Page and Shaffer 1998; Stimpson et al. 2012).
Centromere deletion from dicentric chromosomes has been reported in human, in constitutional dicentric Y chromosomes (Tyler-Smith et al. 1993), in several hundreds of dicentrics produced in in vitro assays using human cell transiently expressing a TRF2-mutated protein (Stimpson et al. 2010), in cancer (Berger and Busson-Le Coniat 1999; Andersen and Pedersen-Bjergaard 2000; Andersen et al. 2005; Mackinnon and Campbell 2011) as well as in yeast (Pennaneach and Kolodner 2009). In particular, the deletion is usually restricted to the functional CENP-A domain in human and CDEI, CDEII, and CDEIII domains in yeast (Kramer et al. 1994). In human, the excised section, including the centromere, is usually preserved in a small stably transmitted marker chromosome (Rivera et al. 1993; Wandall et al. 1998; Rivera et al. 2003; Ventura et al. 2003; Stimpson et al. 2010).
From an evolutionary point of view, ROBs are thought to represent one of the most frequent recurring mechanisms by which chromosome evolution in mammals has occurred and chromosome number has been reduced (Slijepcevic 1998). These chromosomal rearrangements have been reported for several species such as lemurs and other mammals, including the Indian muntjac deer (Hartmann and Scherthan 2004) and house mice (Gimenez et al. 2017). A Robertsonian evolutionary fusion led to the formation of human chromosome 2, explaining the only chromosome number difference between humans (46 chromosomes) and great apes (48 chromosomes) (Lejeune et al. 1973; Yunis and Prakash 1982; IJdo et al. 1991). Following the fusion, the 2p centromere remained active whereas the 2q centromere was inactivated leaving relicts of centromeric alphoid DNA (Avarello et al. 1992; Baldini et al. 1993).
Comparative evolutionary analyses of human chromosome 2 have shown that the homolog of 2q, which fused in the human line (about 5–6 million years ago, mya), was identical in marker order and centromere position to those of chimpanzee and gorilla (Stanyon et al. 2008). In contrast, a pericentric inversion in the Homininae ancestor and a centromere repositioning event are needed to explain marker order differences in the orangutan and macaque lineages, respectively (Roberto et al. 2008).
Using molecular cytogenetics, sequencing and in silico data, we deeply characterize the relicts of the ancestral centromere and its flanking regions on chromosome 2q, providing new insights into the ancestral organization. Our results led us to propose two possible centromere inactivation models representing how dicentric chromosomes overcome their instability.
Results
Molecular Cytogenetic Characterization
We precisely located the ancestral centromere of human chromosome 2 (2qAC) in the human assembly NCBI36/hg18 (chr2:132,698,453-132,714,748) using the clone pαH21 as a query (M64321) (Baldini et al. 1993). In order to study the evolutionary origin and define the genomic organization of this region, we constructed a 2 Mbp fosmid-contig of 63 partially overlapping human clones spanning the genomic region chr2:131,655,215-133,759,260 around 2qAC (see supplementary table S1, Supplementary Material online and fig. 1) and performed comparative and reiterative fluorescence in situ hybridization (FISH) experiments on metaphase chromosomal spreads of four hominid species: Homo sapiens (HSA; human), Pan troglodytes (PTR; chimpanzee), Gorilla gorilla (GGO; gorilla), and Pongo pygmaeus (PPY; orangutan) as well as one representative Old World monkey: Macaca mulatta (MMU; macaque).
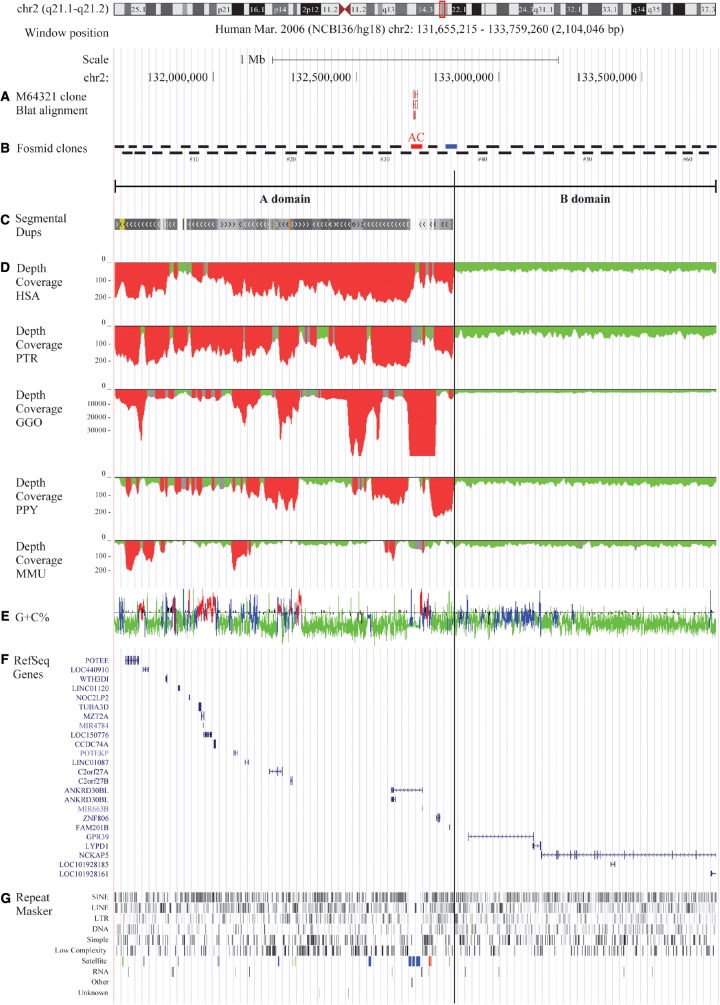
Fig. 1.
Overview of the 2qAC region and the A and B domains. (a) Blat results with the M64321 clone; (b) overlapping fosmid clones spanning the investigated area with fosmid #33 highlighted in red and #37 highlighted in blue; (c) segmental duplication content; (d) depth of coverage on HSA, PTR, GGO, PPY, MMU; (e) GC content and isochores distribution; (f) RefSeq genes composition; and (g) RepeatMasker results and the satellite elements highlighted in blue (centromeric family), green (telomeric family) and red (acrocentric family).
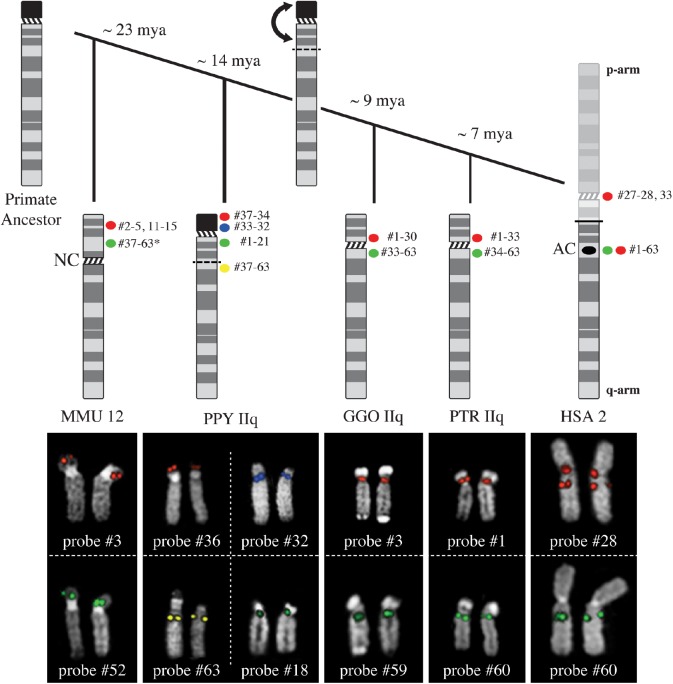
FISH experiments on human metaphases revealed intra and interchromosomal duplications (see supplementary table S2, Supplementary Material online). With respect to the functional centromeres in chimpanzee and gorilla, we detected two different patterns: probes #1–33 hybridized to the p-arm pericentromere of chimpanzee chromosome IIq, whereas probes #34–63 were on the q-arm pericentromere. The same patterns were detected on gorilla chromosome IIq, although signals corresponding to probes #31 and #32 could not be detected in gorilla. In contrast to chimpanzee and gorilla, where all probes mapped to the pericentromeric region consistent with human orientation, for orangutan chromosome IIq some probes did not. Remarkably, four different FISH patterns were observed on PPY IIq, using the centromere as landmark: probes #1–21 and #37–63 localized to the q-arm, with the first cluster proximal and the second distal; probes #32–33 and #34–37 localized to the p-arm, with the first cluster proximal and the second distal. Moreover, probes #22–31 gave signals on chromosomes not syntenic to HSA 2q, in particular on PPY chromosomes III, VII, X, XIII, XV, XX, XXI, and XXII, whereas probe #37, showed double signals both on the PPY IIq p-arm (tip) and q-arm (see supplementary tables S3–S5, Supplementary Material online and fig. 2).
Fig. 2.
For every species, probes with the same FISH pattern were grouped and representative results for each class are displayed. In HSA, all fosmids mapped to chromosome 2, only three of them mapped both on the ancestral centromere (AC) and on the primary constriction of chromosome 2 (HSA red signals). For the other species, we used the chromosome IIq active centromere as a landmark. In PTR and GGO, we were able to group the FISH results into two classes, since some probes mapped to the p-side of the centromere, whereas others to the q-side. In PPY, we distinguished four clusters of signals, two for each chromosome arm: we observed distal and proximal signals on both the p- and q-arm. Finally, in MMU we detected signals only on the p-arm, where the inactivated centromere is located. The active centromere is a neocentromere (NC). The * indicates that not all probes from #37 to #63 actually mapped on MMU 12.
On macaque metaphases, 14 out of 63 fosmid clones (22%) did not produce any hybridization signals whereas ten probes (probes #16, #17, #21–25, #33, #34, and #36) showed signals on chromosomes that are not syntenic to HSA 2q (MMU 3, 10, 15, and 19). Only 33 out of 63 probes hybridized on chromosome 12 (homologous to human 2q) and they all mapped to the p-arm of MMU 12, with probes from #2 to #5 as well as probes from #11 to #15 distal, and probes from #37 to #63 (except for probes #46, #50, and #57) proximal (see supplementary table S6, Supplementary Material online and fig. 2).
Focusing our analysis on the genomic organization of orangutan chromosome IIq p-arm, we found two Sumatran individuals (PPY 9 and PPY 13) that were heterozygous for the deletion of the terminal part of the studied chromosome whereas three other individuals (PPY 11 (Sumatran); PPY 12 and PPY 19 (Bornean)) were homozygous for the presence of the region encompassed by fosmids #34–37 (see supplementary fig. S1, Supplementary Material online). In order to characterize this region more fully, we decided to investigate the presence of rDNA and satellite III, which are often found on the p-arms of acrocentric chromosomes.
Using cohybridization experiments with fosmid clone #34 and a BAC clone containing rDNA (CH507-159O11), we confirmed the presence of rDNA sequences on the tip of orangutan chromosome IIq p-arm as well as on all the other orangutan acrocentric chromosomes p-arms. In contrast to the rDNA distal localization, we demonstrated that satellite III repeats, known to map to acrocentric chromosomes p-arms in human, displayed proximal signals on orangutan IIq p-arm, as revealed by cohybridization experiments using the fosmid clone #34 and a labeled PCR product of satellite III repeats (see supplementary fig. S2, Supplementary Material online). It is worth noting that PPY 9 and PPY 13 were heterozygous for the deletion of both rDNA and satellite III sequences on chromosome IIq.
Based on these observations, we broadened the investigation of satellite III distribution to include the other great apes. Remarkably no traces were detected on gorilla, chimpanzee, and human IIq homologs. Conversely, chromosome IIp was positive for the satellite III carrying probe in all examined species (table 1 and see supplementary fig. S3, Supplementary Material online).
Table 1.
Distribution on HSA, PTR, GGO, PPY, and MMU chromosomes of β-satellite (Modified from Cardone et al. [2004]), satellite III and rDNA Repeats (Modified from Jarmuz et al. [2007]).
| Chromosomes |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | I | IIp | IIq | IV | IX | XIII | XIV | XV | XVI | XVII | XVIII | XIX | XX | XXI | XXII | Y | |
| HSA | β-satellite | ||||||||||||||||
| rDNA | |||||||||||||||||
| Satellite III | |||||||||||||||||
| PTR | β-satellite | ||||||||||||||||
| rDNA | |||||||||||||||||
| Satellite III | |||||||||||||||||
| GGO | β-satellite | ||||||||||||||||
| rDNA | |||||||||||||||||
| Satellite III | ** | * | |||||||||||||||
| PPY | β-satellite | ||||||||||||||||
| rDNA | |||||||||||||||||
| Satellite III | |||||||||||||||||
| MMU | β-satellite | ||||||||||||||||
| rDNA | |||||||||||||||||
| Satellite III | |||||||||||||||||
Note.—Black indicates the presence of the above-mentioned repeats as reported in the literature and confirmed by our experiments, gray stands for discordant results, with * indicating the distribution reported in literature, whereas ** indicates the distribution inferred exclusively by our results.
Sequence-Based Characterization
In agreement with our FISH results, in silico analyses showed that the investigated area consists of two different regions: a 1.2 Mbp duplicated “A domain” containing the 2qAC region and a “B domain” (0.9 Mbp) totally devoid of segmental duplications in all five examined species (fig. 1). The boundary between these two domains was located at chr2:132,838,104, harbored by the fosmid clone ABC8_2139340_H20 (fosmid clone #37).
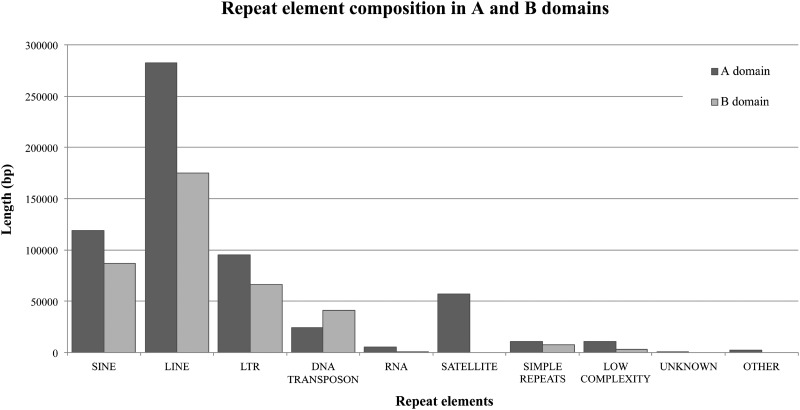
Further comparative analyses between the A and B domains were performed. Repeat elements showed enrichment in the A domain and, in particular, satellite sequences (telomeric, acrocentric, and centromeric) were only detected in this domain (figs. 1 and 3 and see supplementary notes S1, Supplementary Material online).
Fig. 3.
Histograms depicting the different composition in terms of repeat elements in the A (dark gray) and B (light gray) domains.
Similar to the common repeats, the 23 genes mapping to this region were asymmetrically distributed between the two domains with the A domain showing higher gene density than the B domain. Interestingly, average gene length was lower in the A than B domain (A domain: ∼20 kbp, B domain: ∼384 kbp). This ∼20-fold difference in the average length could be due to the presence of two long genes in the B domain (fig. 1). We also investigated gene expression differences between human and chimpanzee by querying the RNA-seq gene expression profile database across 16 selected tissues. Notably, we found a gene, ANKRD30BL, that was ∼25-fold overexpressed in a large variety of chimpanzee tissues, although the database we queried did not differentiate between the two isoforms of this gene, one of which fully retained the relicts of the ancestral centromere in the longest intron (www.ncbi.nlm.nih.gov/IEB/Research/Acembly) (see supplementary notes S1, Supplementary Material online).
Of the 23 genes, 12 were analyzed for their evolutionary history, revealing that all of them were conserved in the common chordate ancestor with the exception of C2orf27A and C2orf27B protein-coding genes and MIR663B, an ncRNA that emerged in the common ancestor of human and chimpanzee (see supplementary table S7 and supplementary notes S1, Supplementary Material online).
Additionally, we characterized the α-satellite relicts and identified four α-satellite arrays, spanning about 40 kbp, showing the overall organization that has been recently reported by Miga (Miga 2016). Indeed, the first block (11417 bp, block A) was separated from the second (8446 bp, block B) by a (TCTCCC)n simple repeat and three SVA elements, whereas L1PA3 (about 6 kbp) separated the second block from the third (11215 bp). Since no additional repetitive elements mapped between the third and fourth (1901 bp) blocks but they were only 12 bp apart from each other, we considered them as a single unit (block C). Initially, we divided the alphoid arrays into ∼171 bp monomers (n = 198), using the human α-satellite consensus sequence (X07685) as a guide to identify the start position of each monomer, obtaining 68 monomers for block A, 51 for block B, and 79 for block C, with the last 19 monomers containing reverse-complement sequences (fig. 4). Base-level investigations revealed that the last monomer of block A composed of 86 bp lacked its terminal part (85 bp) that was actually located at the beginning of block B. Indeed, the juxtaposition of these two monomers perfectly reconstitutes a complete monomer unit (171 bp). On the other hand, the analysis of boundary monomers between blocks B and C revealed an imperfect juxtaposition due to the presence of a 5 bp deletion (148–154 bp) (see supplementary fig. S4, Supplementary Material online).
Fig. 4.
(a) Fosmid clones encompassing the relicts of the ancestral centromere (fosmid #32–34) and the repeat elements mapping to this genomic area. There are three α-satellite blocks: block A (red), block B (green), and block C (blue). The number of monomers is shown above each rectangle. The third block is actually made up of two separated units (a 12 bp interruption occurred inside the 68th monomer, as indicated by a light blue arrow). All the monomers are directly oriented except for the last 19 monomers of block C (the boundary is highlight by a purple arrow). (b) Dot matrix view showing regions of similarity (based upon the BLAST results) obtained after plotting self-comparison of the sequence harboring the relicts of the ancient centromere. The presence of inverted sequences was visually revealed by the creation of gray streaks running diagonally from top left to bottom right. Dotted purple lines show the boundary between directly oriented and inverted sequences.
We also explored the main α-satellite DNA features in terms of primary sequence (the content of A-type and B-type monomers). First, all monomers were evaluated for sites of homology to the reported pJα (CTAPyGGTGPuAAAAGGAA) and CENP-B (PyTTCGTTGGAAPuCGGGA) box motifs (essential positions or “core” underlined) in forward and reverse complement orientations. Except for 16 monomers that could not be confidently assigned and two without the box motif, 63% (124/198) were pJα motif-positive, with 65% (80/124) of them revealing a perfect conservation with the “pJα-core;” 28% (56/198) of monomers possessed the CENP-B box, with 29% (16/56) of them displaying all essential positions completely unaltered.
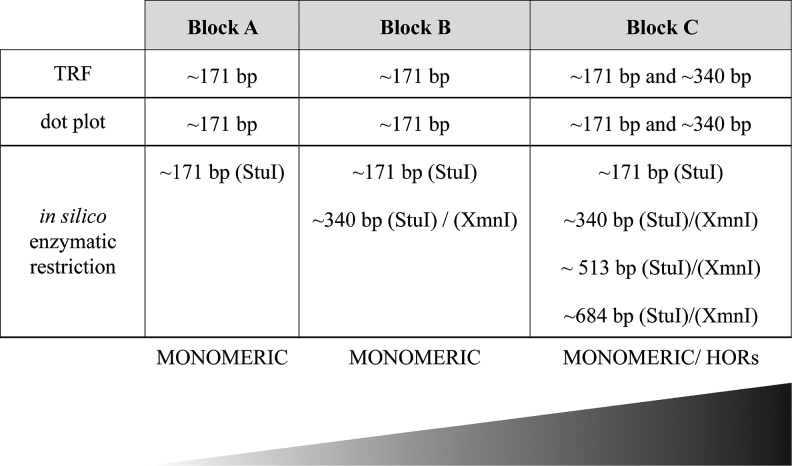
Next, to shed light on the mutation rates of alphoid sequences with respect to their counterpart on active human centromeres, we used 12 human alphoid consensus sequences: the J1-J2 and D1-D2 monomers regularly distributed in the human dimeric suprachromosomal families SF1 and SF2, respectively; the W1-W2-W3-W4-W5 forming the pentameric SF3 (Willard and Waye 1987), the M1 shaping the monomeric SF4 (Alexandrov et al. 1993); and the R1-R2 monomers irregularly alternating in the human dimeric SF5 (Jorgensen et al. 1986; Thompson et al. 1989). By constructing multiple sequence alignments and p-distance matrices, we determined that all 2q alphoid monomers showed the greatest sequence identity with SF4-SF5 consensus sequences except for four monomers in block C that showed the highest identity to SF2 consensus sequences. We performed additional analyses to investigate if 2qAC alphoid sequences were organized into a higher-order repeat (HOR) structure, like all human centromeres. In particular, our sequences were examined for higher-order periodicities by Tandem Repeats Finder (TRF), dot plot (JDotter) and in vitro enzymatic restriction, and whereas the majority of the monomers were composed of monomeric arrays, the presence of ∼340 bp arrays was interestingly displayed for some monomers of block C, pointing out a dimeric organization (fig. 5, see supplementary fig. S5, Supplementary Material online).
Fig. 5.
Summary of the differences among monomers belonging to blocks A, B, and C in terms of higher-order organization. The gradient displays the degeneration of the monomers (light gray: monomeric, black: evidence of HOR structure). TRF: Tandem Repeats Finder.
Finally, we compared our sequences with other centromeres of the African great ape chromosome sequences, exploiting gorilla α-satellite sequences retrieved by Catacchio et al. (2015) since chimpanzee centromeric sequences were, unfortunately, not available. Specifically, the first comparisons were performed using 2qAC monomers and some gorilla sequences from monomeric α-satellite arrays mapping at junctions of the pericentromeric region. Interestingly, all the 2qAC monomers somehow intermingled with the gorilla sequences without forming any lineage-specific clusters (see supplementary fig. S6, Supplementary Material online). Next, we compared 2qAC monomers with sequences representative of gorilla HOR alphoid DNA families SF1 and SF2, known to be part of the centromere bulk; of note, the phylogenic trees we obtained revealed that 2qAC block A and B monomers clustered independently from gorilla sequences, thus suggesting the absence of any hierarchical organization, whereas block C monomers formed a clade with the gorilla SF1 and SF2 sequences, suggesting conservation. Taken together, these results suggest that block C monomers were organized more similarly to the ancestral centromere still active in GGO than either block A and B monomers (fig. 5 and see supplementary fig. S6, Supplementary Material online).
Discussion
Studying the evolutionary history of chromosomes can facilitate our understanding of biological and molecular mechanisms, such as the process underlying dicentric chromosome stabilization. We used human chromosome 2 as an example for the investigation of dicentric chromosome fate, since it is the most recent product of a chromosome fusion event fixed in the human lineage. It is largely known that the stability of the ancestrally dicentric chromosome 2 was gained by centromere inactivation and progressive degradation, as demonstrated by the detection of α-satellite DNA remains corresponding to the IIq centromere (2qAC). For this reason, we analyzed by FISH a 2 Mbp area around the 2qAC region. We compared human to the homologous regions in chimpanzee, gorilla, orangutan and macaque, thus refining the 2qAC evolutionary history. This approach allowed us to shed light on the processes of chromosome dicentric stabilization and centromere inactivation—two phenomena still unclear at the molecular level.
Previous studies on the evolution of chromosome 2, excluding the human-specific IIp-IIq fusion, reported that a pericentric inversion occurred after the divergence of orangutan from other great apes. Using partially overlapping fosmid clones, we accurately characterized this inversion as well as the entire content of this genomic area. We were able to pinpoint one breakpoint of this inversion spanned by fosmid #37, whose hybridization pattern in orangutan showed a split signal on the q-arm and on the p-arm tip of chromosome IIq, whereas human, chimpanzee and gorilla signals were uniquely observed on the q-arm of the homologous chromosome. In addition, our in silico analyses revealed that fosmid #37 demarcated the boundary between two distinguishable domains that formed the 2qAC regions. It is noteworthy that this architecture was not unique to humans but was found in all the other examined species.
Architecture of the 2qAC Region
The 2qAC region consists of two considerably different domains: a 1.2 Mbp domain, named the A domain, which was a patchwork of intra and interchromosomal duplications, and the B domain (0.9 Mbp), which is completely devoid of chromosome duplications. In detail, the human euchromatic A domain shared high sequence similarity to other chromosome pericentromeric regions, with more than 80% of the region being duplicated at least once to another chromosome (see supplementary fig. S7, Supplementary Material online). The A domain typifies the enrichment in segmental duplications observed in great apes and is consistent with the burst of duplications that occurred after the separation of Old World monkeys and the hominoid lineage (∼25 mya) (Marques-Bonet et al. 2009), although this evidence may be biased since the centromere was repositioned in macaque. And although our human-centric approach may have caused an underestimation of species-specific duplications, we similarly documented a different duplication amount among great apes. For example, using probes #22–26 we detected hybridization signals on chromosome IIq exclusively in African great apes whereas no signals were observed in PPY and MMU.
This confirmed that a second wave of duplications occurred after the divergence of orangutan from other great apes (∼14 mya) in agreement with previous studies (Golfier et al. 2003; Marques-Bonet et al. 2009). Examining the map locations of A domain segmental duplications, we find ∼73% are shared between human and orangutan. We propose two rounds of duplication events from chromosome 2 to other chromosomes—one prior to the PPY-specific inversion, followed by locus-specific and species-specific duplications in the African great ape lineage.
In addition, the higher gene density in the A domain compared with the B domain may be associated with the presence of large duplication blocks because of its enrichment in paralogous copies of human genes (Bailey and Eichler 2006) (see supplementary table S7, Supplementary Material online). Indeed, although the majority of these genes were transcriptionally active, we also found evidence of pseudogenes (e.g., WTH3DI and POTEKP) as evidence that some paralogs originated by duplicative events, which in many cases degenerated into nonfunctional genes (Fan et al. 2002). In stark contrast, the B domain harbored a limited number of genes, although such genes were, on average, larger than those in the A domain. Remarkably, we documented the presence of a gene, ANKRD30BL, highly differentially expressed between human and chimpanzee that may represent a fascinating subject to study in the future, since one of its two isoforms fully retained the relicts of the ancestral centromere in the longest intron.
Further, we analyzed the distribution of repeat elements and observed a remarkable difference between the A and B domains. In particular, we documented the presence of a TAR1 element (telomere-associated repeat sequence) in fosmid #37 (A domain), likely ascribable to the ancestral localization of this genomic segment on the telomere of chromosome IIq p-arm in orangutan. Similarly, we found evidence of relicts of rDNA elements, including an LSU_rRNA, an SSU_rRNA, and an ACRO1 satellite element, consistent with the observation that the ape ancestral chromosome IIq is acrocentric and carries rDNA. Overall, our data suggest that ancestrally the 2qAC A domain was organized as subtelomeric/pericentromeric heterochromatin of orangutan chromosome IIq, thus resembling the structure of the common ancestor to the chromosome IIq of Hominoidea (Stanyon et al. 2008).
New Insights into PPY Chromosome IIq p-Arm Structure
By further characterizing this region in orangutan, we found two individuals, PPY 9 and 13 (Pongo abelii), heterozygous for the deletion of the whole chromosome IIq p-arm, but we found no correlation between this deletion and the species to which these orangutans belong (see supplementary fig. S1, Supplementary Material online). Furthermore, we reported the presence of satellite III repeats between the α-satellite arrays and the rDNA clusters on the investigated orangutan chromosome, as well as on the other orangutan acrocentric chromosomes, demonstrating that the reciprocal organization of these elements, so far uniquely described in human acrocentric chromosome p-arms, was perfectly identical in orangutan counterparts (Stimpson et al. 2010).
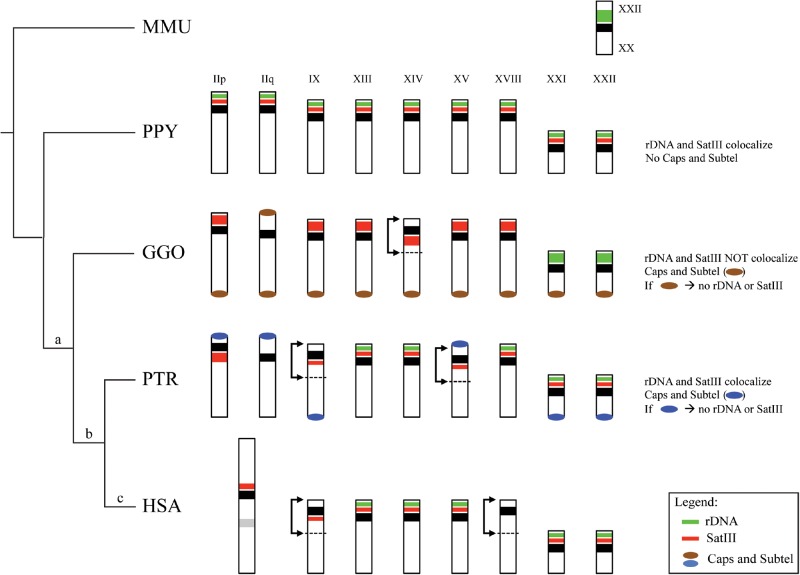
Moreover, our analysis shows that the previous satellite III mapping among great apes performed by Jarmuż et al. (2007) was incorrect for some chromosomes (see supplementary fig. S3, Supplementary Material online). Therefore, we propose a new comprehensive overview of this element distribution with respect to the rDNA localization. Unlike the rDNA and ACRO1 elements whose relicts were still detectable in human within 2qAC, no traces of satellite III DNA were identified in human 2qAC. Its “absence” in the homologous regions of gorilla, chimpanzee, and human allowed us to propose that this satellite was completely lost in conjunction with the IIq pericentric inversion after the divergence of orangutan from other great apes. Interestingly, this loss did not occur on all chromosomes that underwent lineage-specific inversions. Absence of this satellite only occurs on human chromosomes IIq and XVIII (fig. 6), whereas the other satellite III-bearing chromosomes retained and internalized satellite III irrespective of the inversion they underwent (chromosomes IIp and IX were subjected to pericentric inversions after the divergence of gorilla from other great apes, chromosome XVIII was inverted after the divergence of chimpanzee from other great apes, and chromosomes XIV and XV were inverted in gorilla and chimpanzee, respectively).
Fig. 6.
Schematic representation of satellite III, rDNA, and caps and subtelomeric (Ventura et al. 2012) repeat distribution in the MMU rDNA-bearing chromosome (MMU10) as well as in all the great apes acrocentric chromosomes. The genomic rearrangements that chromosome IIp and IIq underwent are reported above branches: (a) pericentric inversion that involved the chromosome IIq, (b) pericentric inversion that involved the chromosome IIp, and (c) fusion event that involved chromosome IIp and IIq. A description of the reciprocal distribution of the investigated repeat elements is reported on the right side. Black bars are centromeres and black arrows indicate other inversion events involving other chromosomes.
We therefore hypothesize a possible correlation between the chromosome IIq centromere suppression and the lack of satellite III. This association may have forced its inactivation, unlike its presence on the nearby chromosome IIp centromere, which remained functional. This hypothesis is further supported by evidence of nonrandom centromere suppression collected from studies of dicentric chromosomes in plants and human ROBs. In particular, in a series of patient-derived ROBs, a functional hierarchy was observed: chromosome 14 remained active most often, irrespective of the other acrocentrics involved in the ROB, whereas the centromere of chromosome 15 was more likely to be inactivated (Sullivan et al. 1994). Bandyopadhyay et al. compared the acrocentric chromosome satellite III subfamilies composition and reported that only a few subfamilies reside close by the chromosome 15 centromere (D15Z1, pR1-2 and 4), whereas several and different satellite III subfamilies compose the p-arm of the chromosome 14 (pE-1 and 2, pK-1, pTRS-63, PTR9-s3, pTRS-47, pR1-2 and 4) (Bandyopadhyay et al. 2001). Taking this into consideration, we propose the hypothesis that the preferential inactivation of the chromosome 15 centromere may be favored by the low content of nearby satellite III DNA subfamilies. On the other hand, the existence of multiple subfamilies close to the chromosome 14 centromere could play a protective role against centromere suppression.
Models for Human Dicentric Chromosome 2 Stabilization
In the light of our results, we propose some possible mechanisms mediating the centromere inactivation responsible for the stabilization of the dicentric chromosome 2 originated in the human lineage. Previous studies showed that dicentric chromosomes overcame their instability via the inactivation of one of the two centromeres. It could be a functional inactivation, if mediated by epigenetic modifications without any changes of the α-satellite DNA in terms of size or sequence, or more often it could consist of the physical excision of the α-satellite DNA (Mackinnon and Campbell 2011). Unlike yeast dicentric chromosomes that undergo stability via the full-size centromere excision, in human it was described that centromeric deletion generally removes only the region of chromatin containing CENP-A that presumably identifies the site of kinetochore assembly and represents the central domain of the centromere known as “core” flanked by HORs (Stimpson et al. 2010; Henikoff et al. 2015).
Focusing on our region, the fine characterization of the α-satellite elements spanned by probes #32–34, revealed the presence of a dramatically low amount of the α-satellite arrays (about 40 kbp) compared with the classical active human centromere ranging from 250 to 5,000 kbp (Aldrup-Macdonald and Sullivan 2014). We hypothesize that the centromere inactivation was triggered by the full deletion of the active centromeric core and the significant reduction of the bulk of higher-ordered centromeric satellite DNA consistent with α-satellite relicts now present on human chromosome 2. The majority of the residual satellite shows exclusively a monomeric organization and, at the nucleotide level, they are more similar to pericentromeric junction monomeric sequences rather than the centromeric core. Few HOR relicts showing a dimeric organization were detected within block C (fig. 5). In particular, eligible excision sites may be represented by the boundary between blocks B and C or, alternatively, the junction between directly oriented monomers and monomers with an inverted orientation (60th monomer of block C). Notably, recent data provided by Miga documented that α-satellite monomers on the chimpanzee p-arm are organized in a forward orientation, whereas monomers that share high identity with sequences on the q-arm are observed in the reverse orientation. This clearly supports that the excision site is at the boundary between the differently oriented monomers within block C (Miga 2016).
Assuming this centromere-deletion model, two potential mechanisms could explain the centromere excision: a one-step excision, consisting of the deletion of the entire centromeric core in a single step, or a step-by-step mechanism, consisting of the gradual removal of HOR monomers. Evidence for the one-step centromere excision has been clearly reported in the literature. In one human clinical case showing neocentromere formation, involving chromosome 3, the old centromere was excised and retained in a supernumerary small marker chromosome (Wandall et al. 1998; Ventura et al. 2004). Similarly, data on engineered dicentric human chromosomes revealed that the presence of functionally stabilized monocentric chromosomes was accompanied by the temporary appearance of small, marker-sized chromosome fragments, shown to contain α-satellite DNA homologous to the inactive centromere (Stimpson et al. 2010). In contrast with the step-by-step model, which has never been supported by experimental evidence, the one-step excision model has already been proposed for dicentric stabilization. For this reason, it may be considered a likely mechanism that may have mediated the 2qAC inactivation.
In this likely scenario, a supernumerary small marker chromosome retaining the chromosome IIq centromeric core might also have been generated after the IIp-IIq fusion but it was subsequently lost by genetic drift, as described for dicentrics analyzed over time in the human cell culture model (Stimpson et al. 2010). Regardless of the IIq centromere excision mechanism timing (one-step or step-by-step), the detailed characterization of the α-satellite relicts revealed that the monomers length was constant (∼171 bp), thereby suggesting that the removal process was not random.
Conclusions
In summary, our data support a model where human chromosome 2 likely overcame its instability by removing the chromosome IIq centromeric core located within the α-satellite block C via a one-step excision mechanism. This model does not exclude the involvement of epigenetic modifications, which may have also played a crucial role in triggering the inactivation before the centromere deletion. This model further supports the preferential inactivation of the chromosome IIq centromere as a result of its lack of satellite III, consistent with the correlation we found between the preferential centromere activity described for human acrocentric chromosome centromeres (Sullivan et al. 1994) and the distribution of satellite III subfamilies (Bandyopadhyay et al. 2001). Indeed, we hypothesize that, like the chromosome IIq centromere, the chromosome 15 centromere is more prone to inactivation because of the relatively low abundance of satellite III sequences. In contrast, the presence of several satellite III subfamilies flanking the chromosome 14 centromere (reflective of the chromosome IIp centromeric architecture) prevents its inactivation during Robertsonian translocation fusions. Our study represents the first investigation of an inactivated evolutionary centromere in mammals and sheds light on the dicentric chromosome stabilization phenomenon that is still not fully understood. Further studies are needed to explore the molecular mechanisms controlling centromere inactivation in dicentric chromosomes.
Materials and Methods
Cell Lines
Metaphase spreads were prepared from lymphoblastoid or fibroblast cell lines of Pan troglodytes (PTR), Gorilla gorilla (GGO), Pongo pygmaeus (PPY), one representative of Old World monkey (Macaca mulatta, MMU), and two representatives of New World monkeys (Callithrix jacchus (CJA) and Callicebus moloch (CMO)). In particular, we used five orangutans as representatives for the two species: three Sumatran individuals (PPY 9, 11, and 13) and two Bornean (PPY 12 and PPY 19). Human spreads were prepared from phytohemagglutinin-stimulated peripheral lymphocytes of normal donors by standard procedures.
FISH Experiments
DNA extraction from BACs and fosmids was performed as previously described (Chiatante et al. 2016). Briefly, DNA probes were directly labeled with Cy3-dUTP, Cy5-dUTP (GE Healthcare), or Fluorescein-dUTP (Invitrogen) by nick translation. Two hundred ng of labeled probe were hybridized on metaphases spreads; hybridization was performed overnight at 37 °C in 2× SSC, 50% (v/v) formamide, 10% (w/v) dextran sulphate, 3 μl C0t-1 DNA (Roche), and 5 μl sonicated salmon sperm DNA, in a volume of 10 μl. Posthybridization washing was performed at 60 °C in 0.1× SSC (three times, high stringency). Washes of interspecies hybridization experiments were performed at lower stringency: 37 °C in 2× SSC, 50% formamide (3×), followed by washes at 42 °C in 2× SSC (3×). Chromosome identification was obtained by DAPI staining, producing a Q-banding pattern. Digital images were obtained using a Leica DMRXA epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). Cy3, Cy5, Fluorescein, and DAPI fluorescence signals, detected with specific filters, were recorded separately as grayscale images. Pseudocoloring and merging of images were performed using Adobe Photoshop software.
rDNA and Satellite III Chromosomal Distribution
To confirm the presence of rDNA relicts embodied by fosmid ABC8_2606640_P13 (#34) and in order to assess rDNA localization in great apes, we performed a cohybridization experiment by using the rDNA-carrying BAC clone CH507-159O11 (Ventura et al. 2012).
Furthermore, to determine the chromosomal distribution of satellite III sequences, we amplified human genomic DNA (NA18507) by polymerase chain reactions (PCR). Primer sets were designed from the known sequences of two satellite III subfamilies: pE-1 (GATTCGATTCCATTGCACTCG, forward and GGACTGAAACAAAATGGAGACC, reverse) and pW-1 (AATGGGATGGAACCGAGTGG, forward and CCTTTCATTTCAAGTCCCTTCGC, reverse) (Bandyopadhyay et al. 2001). Amplified products were then directly labeled with Cy3-dUTP by PCR labeling. PCR labeling was carried out in a final volume of 20 μl that contained 100 ng PCR product, 2 μl 10× reaction buffer (Invitrogen), 2 μl 50 mM MgCl2, 0.5 μl each primer (10 μM), 2,5 2 mM μl dACG, 2.5 μl 1 mM Cy3-dUTP, 5 μl 2% BSA, and 0.3 μl Taq polymerase (5 U/μl). For both amplification reactions, the cycling parameters used were as follows: 3 min initial denaturation at 94 °C, followed by 30 cycles of: 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s. Final extension was at 72 °C for 10 min. PCR labeling products were used as probes in FISH assays in cohybridization with a chromosome IIq-specific BAC probe (RP11-449J2) and a chromosome IIp-specific BAC probe (RP11-496P1).
Sequence Composition and Repeats
The UCSC Genome Browser (human assembly NCBI36/hg18) provided information about the segmental duplication content in the investigated region visualized in a circular layout by using the software package Circos (Krzywinski et al. 2009), as well as the presence of repetitive elements. In particular, the Alu subfamilies’ distribution was analyzed in detail by calculating their content after dividing the whole region into 43 windows (50 kbp each). The GC content of the entire region was estimated by using the GESTALT program (Glusman and Lancet 2000). The same results were obtained using the most recent release (GRCh38/hg38).
Gene Content
The characterization of the gene content of the region of interest in human was achieved by collecting information from different browsers and databases: the UCSC Genome Browser (http://genome.ucsc.edu, RefSeq genes track), AceView (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/), the NCBI gene database (http://www.ncbi.nlm.nih.gov/gene/) and GeneCards (http://www.genecards.org).
Detailed information about expression levels of the genes harbored in the investigated region was available for 16 human tissues (cerebellum, brain, pituitary gland, ovary, testis, kidney, liver, heart, skeletal muscle, lung, colon, bone marrow, spleen, lymph node, thymus, and whole blood) on http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/. On the same browser, RNA-seq gene expression profiles data across the same 16 tissues were available for the orthologous genes in chimpanzee (Nonhuman Primate Reference Transcriptome Resource). Human and chimpanzee gene expression results were finally compared. qPCR experiments were performed to validate the bioinformatics predictions for some randomly selected genes (see supplementary notes S1, Supplementary Material online).
Alphoid Relict Characterization
The DNA sequence of the four α-satellite blocks detected within the 2qAC region spanned by fosmid clone #32–33 was available on the UCSC Genome Browser (http://genome.ucsc.edu). The third and the fourth blocks were subsequently collapsed and considered as a single unit because they were only interrupted by a 12 bp insertion. The last 19 monomers of block C showed an inverted orientation whereby reverse complement sequences were obtained using the free web tool available at http://www.bioinformatics.org/sms/rev_comp.html. Blocks were indicated as Block A, B, and C. Each block was manually divided into ∼171 bp monomers using the human α-satellite consensus sequence X07685 as a guide to identify the start position of each monomer, and then checked by using the ClustalW algorithm (http://www.ebi.ac.uk/Tools/msa/clustalw2), a multi-alignment program.
The 2qAC monomers were evaluated for sites of homology to the reported pJα and CENP-B box motif and compared with the 12 human alphoid consensus sequences representative for the J1 (AJ130753.1), J2 (AJ130754.1), D1 (AJ130751.1), D2 (AJ130752.1), W1 (AJ130758.1), W2 (AJ130759.1), W3 (AJ130760.1), W4 (AJ130761.1), W5 (AJ130762.1), M1 (AJ130755.1), R1 (AJ130756.1), and R2 (AJ130757.1) by calculating pairwise distances (bootstrap 1000, substitution to include: transitions and transversions) using the MEGA (Molecular Evolutionary Genetic Analysis, version 6.0) program (Tamura et al. 2013). Monomers were investigated for the presence of HOR structure by performing in silico enzyme digestion with NEBcutter (http://tools.neb.com/REBsites/index.php) using specific enzymes able to identify dimeric arrays of α-satellite (EcoRI, XmnI, StuI, BamHI, and XbaI). We found evidence to support the higher organization of monomers by using Tandem Repeats Finder (TRF, http://tandem.bu.edu/trf/trf.html, basic option with the default settings) and Java Dot Plot Alignments or JDotter (http://athena.bioc.uvic.ca/virology-ca-tools/jdotter/). We finally compared the 2qAC monomers and two different sets of the gorilla α-satellite (Catacchio et al. 2015). The first set (CABD02041326, CABD02043063, CABD02043744, CABD02043767, CABD02340093, CABD02340102, CABD02062800, CABD02042655, CABD02043786, CABD02399170) was made up of gorilla monomeric α-satellite arrays known to be found at the junctions to the pericentromeric regions, whereas the second set (CABD02219203.1, CABD02196967.1, CABD02043194.1, CABD02043380.1, CABD02162145.1, JQ685209.1, JQ685208.1, JQ685198.1, JQ685199.1, JQ685218.1, JQ685200.1) was composed of sequences representative of gorilla HOR alphoid DNA families SF1 and SF2, known to be part of the centromere bulk. Multiple sequence alignments were performed using Multiple Alignment using Fast Fourier Transform (MAFFT) software (Katoh and Standley 2013). Multiple alignment editing was performed with the multiple alignment editing program Jalview (Waterhouse et al. 2009). Phylogenetic analyses were carried out using the Approximate Maximum Likelihood analysis method from FastTree software (Price et al. 2010), suitable for the construction of large phylogenies, implementing the neighbor-joining method with heuristics. The Java-based Archaeopteryx software (Han and Zmasek 2009) was used to obtain a good graphical rendering of the phylogenetic trees.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Authors’ Contributions
M.V. and G.C. conceived the study. G.C. carried out FISH experiments. G.C. and G.G. analyzed the FISH results. G.C. performed the computational analysis. F.M.C. performed the phylogenetic analysis. G.C., F.M.C., M.V., and E.E.E. drafted the manuscript.
Supplementary Material
Acknowledgments
We thank T. Brown for manuscript editing and Claudia Rita Catacchio for advice and technical support. This research was supported by Futuro in Ricerca 2010 BFR103CE3 and by US National Institutes of Health (NIH) grant HG002385 to E.E.E. All authors read and approved the final manuscript. E.E.E. is an investigator of the Howard Hughes Medical Institute.
References
- Aldrup-Macdonald ME, Sullivan BA.. 2014. The past, present, and future of human centromere genomics. Genes (Basel) 5:33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov IA, Medvedev LI, Mashkova TD, Kisselev LL, Romanova LY, Yurov YB.. 1993. Definition of a new alpha satellite suprachromosomal family characterized by monomeric organization. Nucleic Acids Res. 21:2209–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen MK, Christiansen DH, Pedersen-Bjergaard J.. 2005. Centromeric breakage and highly rearranged chromosome derivatives associated with mutations of TP53 are common in therapy-related MDS and AML after therapy with alkylating agents: an M-FISH study. Genes Chromosomes Cancer 42:358–371. [DOI] [PubMed] [Google Scholar]
- Andersen MK, Pedersen-Bjergaard J.. 2000. Increased frequency of dicentric chromosomes in therapy-related MDS and AML compared to de novo disease is significantly related to previous treatment with alkylating agents and suggests a specific susceptibility to chromosome breakage at the centromere. Leukemia 14:105–111. [DOI] [PubMed] [Google Scholar]
- Avarello R, Pedicini A, Caiulo A, Zuffardi O, Fraccaro M.. 1992. Evidence for an ancestral alphoid domain on the long arm of human chromosome 2. Hum Genet. 89:247–249. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Eichler EE.. 2006. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 7:552–564. [DOI] [PubMed] [Google Scholar]
- Baldini A, Ried T, Shridhar V, Ogura K, D’iuto L, Rocchi M, Ward DC.. 1993. An alphoid DNA sequence conserved in all human and great ape chromosomes: evidence for ancient centromeric sequences at human chromosomal regions 2q21 and 9q13. Hum Genet. 90:577–583. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay R, McQuillan C, Page SL, Choo KH, Shaffer LG.. 2001. Identification and characterization of satellite III subfamilies to the acrocentric chromosomes. Chromosome Res. 9:223–233. [DOI] [PubMed] [Google Scholar]
- Berger R, Busson-Le Coniat M.. 1999. Centric and pericentric chromosome rearrangements in hematopoietic malignancies. Leukemia 13:671–678. [DOI] [PubMed] [Google Scholar]
- Cardone MF, Ballarati L, Ventura M, Rocchi M, Marozzi A, Ginelli E, Meneveri R.. 2004. Evolution of beta satellite DNA sequences: evidence for duplication-mediated repeat amplification and spreading. Mol Biol Evol. 21:1792–1799. [DOI] [PubMed] [Google Scholar]
- Catacchio CR, Ragone R, Chiatante G, Ventura M.. 2015. Organization and evolution of Gorilla centromeric DNA from old strategies to new approaches. Sci Rep. 5:14189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiatante G, Capozzi O, Svartman M, Perelman P, Centrone L, Romanenko SS, Ishida T, Valeri M, Roelke-Parker ME, Stanyon R.. 2016. Centromere repositioning explains fundamental number variability in the New World monkey genus Saimiri. Chromosoma. [DOI] [PubMed]
- Ewers E, Yoda K, Hamid AB, Weise A, Manvelyan M, Liehr T.. 2010. Centromere activity in dicentric small supernumerary marker chromosomes. Chromosome Res. 18:555–562. [DOI] [PubMed] [Google Scholar]
- Fan Y, Newman T, Linardopoulou E, Trask BJ.. 2002. Gene content and function of the ancestral chromosome fusion site in human chromosome 2q13-2q14.1 and paralogous regions. Genome Res. 12:1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez MD, Forster DW, Jones EP, Johannesdottir F, Gabriel SI, Panithanarak T, Scascitelli M, Merico V, Garagna S, Searle JB, et al. 2017. A half-century of studies on a chromosomal hybrid zone of the house mouse. J Hered. 108:25–35. [DOI] [PubMed] [Google Scholar]
- Glusman G, Lancet D.. 2000. GESTALT: a workbench for automatic integration and visualization of large-scale genomic sequence analyses. Bioinformatics 16:482–483. [DOI] [PubMed] [Google Scholar]
- Golfier G, Chibon F, Aurias A, Chen XN, Korenberg J, Rossier J, Potier MC.. 2003. The 200-kb segmental duplication on human chromosome 21 originates from a pericentromeric dissemination involving human chromosomes 2, 18 and 13. Gene 312:51–59. [DOI] [PubMed] [Google Scholar]
- Han MV, Zmasek CM.. 2009. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics 10:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann N, Scherthan H.. 2004. Characterization of ancestral chromosome fusion points in the Indian muntjac deer. Chromosoma 112:213–220. [DOI] [PubMed] [Google Scholar]
- Heerema NA, Maben KD, Bernstein J, Breitfeld PP, Neiman RS, Vance GH.. 1996. Dicentric (9;20)(p11;q11) identified by fluorescence in situ hybridization in four pediatric acute lymphoblastic leukemia patients. Cancer Genet Cytogenet. 92:111–115. [DOI] [PubMed] [Google Scholar]
- Henikoff JG, Thakur J, Kasinathan S, Henikoff S.. 2015. A unique chromatin complex occupies young alpha-satellite arrays of human centromeres. Sci Adv. 1(1): pii: e1400234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmuz M, Glotzbach CD, Bailey KA, Bandyopadhyay R, Shaffer LG.. 2007. The evolution of satellite III DNA subfamilies among primates. Am J Hum Genet. 80:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen AL, Bostock CJ, Bak AL.. 1986. Chromosome-specific subfamilies within human alphoid repetitive DNA. J Mol Biol. 187:185–196. [DOI] [PubMed] [Google Scholar]
- IJdo JW, Baldini A, Ward DC, Reeders ST, Wells RA.. 1991. Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proc Natl Acad Sci U S A. 88:9051–9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer KM, Brock JA, Bloom K, Moore JK, Haber JE.. 1994. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol Cell Biol. 14:1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA.. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune J, Dutrillaux B, Rethore MO, Prieur M.. 1973. Comparison of the structure of chromatids of Homo sapiens and Pan troglodytes (author’s transl). Chromosoma 43:423–444. [DOI] [PubMed] [Google Scholar]
- Mackinnon RN, Campbell LJ.. 2011. The role of dicentric chromosome formation and secondary centromere deletion in the evolution of myeloid malignancy. Genet Res Int. 2011:643628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet T, Kidd JM, Ventura M, Graves TA, Cheng Z, Hillier LW, Jiang Z, Baker C, Malfavon-Borja R, Fulton LA, et al. 2009. A burst of segmental duplications in the genome of the African great ape ancestor. Nature 457:877–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga KH. 2016. Chromosome-specific centromere sequences provide an estimate of the ancestral chromosome 2 fusion event in hominin genomes. J Hered. 108(1):45–52. [DOI] [PubMed]
- Page SL, Shaffer LG.. 1998. Chromosome stability is maintained by short intercentromeric distance in functionally dicentric human Robertsonian translocations. Chromosome Res. 6:115–122. [DOI] [PubMed] [Google Scholar]
- Page SL, Shin JC, Han JY, Choo KH, Shaffer LG.. 1996. Breakpoint diversity illustrates distinct mechanisms for Robertsonian translocation formation. Hum Mol Genet. 5:1279–1288. [DOI] [PubMed] [Google Scholar]
- Patsouris C, Michael PM, Campbell LJ.. 2002. A new nonrandom unbalanced t(17;20) in myeloid malignancies. Cancer Genet Cytogenet. 138:32–37. [DOI] [PubMed] [Google Scholar]
- Pennaneach V, Kolodner RD.. 2009. Stabilization of dicentric translocations through secondary rearrangements mediated by multiple mechanisms in S. cerevisiae. PLoS One 4:e6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera H, Ayala-Madrigal LM, Gutierrez-Angulo M, Vasquez AI, Ramos AL.. 2003. Isodicentric Y chromosomes and secondary microchromosomes. Genet Couns. 14:227–231. [PubMed] [Google Scholar]
- Rivera H, Dominguez MG, Vasquez AI, Ramos AL, Fragoso R.. 1993. Centromeric association of a microchromosome in a Turner syndrome patient with a pseudodicentric Y. Hum Genet. 92:522–524. [DOI] [PubMed] [Google Scholar]
- Roberto R, Misceo D, D’Addabbo P, Archidiacono N, Rocchi M.. 2008. Refinement of macaque synteny arrangement with respect to the official rheMac2 macaque sequence assembly. Chromosome Res. 16:977–985. [DOI] [PubMed] [Google Scholar]
- Slijepcevic P. 1998. Telomeres and mechanisms of Robertsonian fusion. Chromosoma 107:136–140. [DOI] [PubMed] [Google Scholar]
- Stanyon R, Rocchi M, Capozzi O, Roberto R, Misceo D, Ventura M, Cardone MF, Bigoni F, Archidiacono N.. 2008. Primate chromosome evolution: ancestral karyotypes, marker order and neocentromeres. Chromosome Res. 16:17–39. [DOI] [PubMed] [Google Scholar]
- Stimpson KM, Matheny JE, Sullivan BA.. 2012. Dicentric chromosomes: unique models to study centromere function and inactivation. Chromosome Res. 20:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson KM, Song IY, Jauch A, Holtgreve-Grez H, Hayden KE, Bridger JM, Sullivan BA.. 2010. Telomere disruption results in non-random formation of de novo dicentric chromosomes involving acrocentric human chromosomes. PLoS Genet. 6(8): pii: e1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Schwartz S.. 1995. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum Mol Genet. 4:2189–2197. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Willard HF.. 1998. Stable dicentric X chromosomes with two functional centromeres. Nat Genet. 20:227–228. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Wolff DJ, Schwartz S.. 1994. Analysis of centromeric activity in Robertsonian translocations: implications for a functional acrocentric hierarchy. Chromosoma 103:459–467. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Sylvester JE, Gonzalez IL, Costanzi CC, Gillespie D.. 1989. Definition of a second dimeric subfamily of human alpha satellite DNA. Nucleic Acids Res. 17:2769–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler-Smith C, Oakey RJ, Larin Z, Fisher RB, Crocker M, Affara NA, Ferguson-Smith MA, Muenke M, Zuffardi O, Jobling MA.. 1993. Localization of DNA sequences required for human centromere function through an analysis of rearranged Y chromosomes. Nat Genet. 5:368–375. [DOI] [PubMed] [Google Scholar]
- Ventura M, Catacchio CR, Sajjadian S, Vives L, Sudmant PH, Marques-Bonet T, Graves TA, Wilson RK, Eichler EE.. 2012. The evolution of African great ape subtelomeric heterochromatin and the fusion of human chromosome 2. Genome Res. 22:1036–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Mudge JM, Palumbo V, Burn S, Blennow E, Pierluigi M, Giorda R, Zuffardi O, Archidiacono N, Jackson MS, et al. 2003. Neocentromeres in 15q24-26 map to duplicons which flanked an ancestral centromere in 15q25. Genome Res. 13:2059–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Weigl S, Carbone L, Cardone MF, Misceo D, Teti M, D’Addabbo P, Wandall A, Bjorck E, de Jong PJ, et al. 2004. Recurrent sites for new centromere seeding. Genome Res. 14:1696–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandall A, Tranebjaerg L, Tommerup N.. 1998. A neocentromere on human chromosome 3 without detectable alpha-satellite DNA forms morphologically normal kinetochores. Chromosoma 107:359–365. [DOI] [PubMed] [Google Scholar]
- Wang P, Spielberger RT, Thangavelu M, Zhao N, Davis EM, Iannantuoni K, Larson RA, Le Beau MM.. 1997. dic(5;17): a recurring abnormality in malignant myeloid disorders associated with mutations of TP53. Genes Chromosomes Cancer 20:282–291. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ.. 2009. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson N, Dunlop L, Robson L, Sharma P, Smith A.. 2000. 17p- syndrome arising from a novel dicentric translocation in a patient with acute myeloid leukemia. Cancer Genet Cytogenet. 118:159–162. [DOI] [PubMed] [Google Scholar]
- Willard HF, Waye JS.. 1987. Chromosome-specific subsets of human alpha satellite DNA: analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J Mol Evol. 25:207–214. [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Prakash O.. 1982. The origin of man: a chromosomal pictorial legacy. Science 215:1525–1530. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.