Abstract
Glucose regulated protein 94 (Grp94) is the ER resident isoform of the 90 kDa heat shock protein family (Hsp90) and represents a promising therapeutic target for the treatment of many diseases. Modification of the cis-amide bioisostere to alter the angle between the resorcinol ring and the benzyl side chain via cis-amide replacements produced compounds with improved Grp94 affinity and selectivity. Structure-activity relationship studies led to the discovery of 30, which exhibits 540 nM affinity and 73-fold selectivity towards Grp94. Grp94 is responsible for the maturation and trafficking of proteins associated with cell signaling and motility, including select integrins. The Grp94-selective inhibitor 30 was shown to exhibit potent anti-migratory effects against multiple aggressive and metastatic cancers.
Keywords: Hsp90, Grp94, metastatic cancer, medicinal chemistry, chaperones
Graphical Abstract

Stop right there! Modifications to the cis-amide bioisostere imidazole of the first generation Grp94-selective inhibitors were pursued to develop more potent and selective Grp94 inhibitors. Reduction of the angle between the resorcinol moiety and the benzyl side chain of BnIm led to 30 which exhibited improved Grp94 affinity and selectivity. KUNG65 exhibited nanomolar anti-migratory activity against aggressive and metastatic cancers.
Introduction
The 90 kDa heat shock protein (Hsp90) family is responsible for the maturation of nascent polypeptides and the rematuration of denatured proteins. Hsp90 has gained considerable interest as a therapeutic target because Hsp90-dependent proteins are directly associated with all ten hallmarks of cancer.[1] Therefore, inhibition of Hsp90 results in simultaneous disruption of multiple oncogenic pathways that are essential to cancer progression via a single molecular target. Seventeen small molecule Hsp90 inhibitors have progressed into clinical trials for the treatment of various forms of cancer.[2] Unfortunately, these inhibitors have produced various toxicities that have dampened enthusiasm for Hsp90 as a therapeutic target.[3] All of these clinical candidates are pan-Hsp90 inhibitors that target all four Hsp90 isoforms with similar affinity which has been suggested to be the cause of some on-target toxicities.[4] Alternative approaches toward Hsp90 inhibition are needed to overcome these potential liabilities associated with pan-Hsp90 inhibition.
One approach that has gained considerable interest in recent years is the development of Hsp90 isoform-selective inhibitors.[5] The Hsp90 protein family is composed of four isoforms: Hsp90α and Hsp90β reside in the cytosol, Trap1 is localized to the mitochondria, and Grp94 is found in the endoplasmic reticulum. Grp94 is responsible for the maturation and trafficking of proteins associated with cell signaling and motility.[6] Some Grp94-dependent proteins have been elucidated and include the Toll-like receptors, integrins, insulin-like growth factors I and II, LRP6, and mutant myocilin.[6–7] Consequently, Grp94 has been implicated for the treatment of disease states that include metastatic cancer (integrins), glaucoma (mutant myocilin), multiple myeloma (LRP6), and hepatocellular carcinoma. While Grp94 is essential during embryonic development, it is non-essential in developed organisms.[8] Therefore, Grp94 inhibition represents a non-toxic approach toward the treatment of some diseases (e.g. glaucoma). However, some disease states rely more heavily upon a functional ER chaperone system, such as multiple myeloma and hepatocellular carcinoma, wherein Grp94 knockouts decrease the viability of these cancers, highlighting Grp94 as a potential target for these cancers. The development of Hsp90 isoform-selective inhibitors is hindered by >85% identity within the N-terminal ATP-binding site of all four isoforms which poses a significant challenge to the rational design of Hsp90 isoform-selective inhibitors.[9]
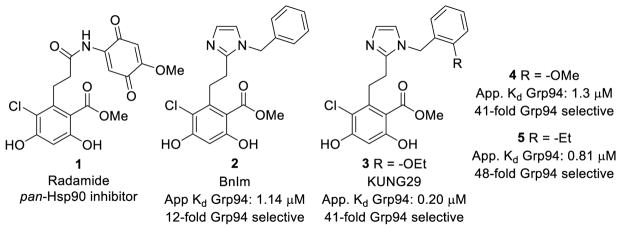
The nucleotide binding site of Grp94 is the most unique among the four isoforms, due to a five amino acid insertion into its primary sequence, which results in secondary binding pockets that can be exploited for selective inhibition. The first Grp94-selective inhibitor was developed via incorporation of a cis-amide bioisostere into the radamide scaffold (1) to predispose the side chain into the Grp94 exclusive pocket, ultimately leading to the development of BnIm (2), which manifested ~12-fold selectivity via a fluorescence polarization assay.[10] Subsequent structure-activity relationship studies on the benzyl side chain of 2 resulted in the development of KUNG29 (3), which exhibited improved affinity and selectivity for Grp94 as compared to 2 (Figure 1).[11] Interestingly, substitutions at the 2-position of the benzyl side chain (3–5) manifested the greatest Grp94-selectivity (Figure 1), and suggested that projection of the side chain into this region could improve selectivity. Analysis of the co-crystal structure of KUNG38 bound to Grp94 suggested that modification of the cis-amide bioisostere (imidazole) could orient the side chain to mimic substitutions present in 3–5 and increase both selectivity and affinity.
Figure 1.
First generation Grp94-selective inhibitors.
Replacement of the first generation imidazole with a phenyl ring was hypothesized to serve multiple purposes: 1) The larger 6-membered ring would reduce the angle between the benzyl side chain and the resorcinol moiety (Figure 2a) and mimic the binding mode of 3–5 to improve selectivity; 2) Removal of the polar nitrogen atoms within the imidazole ring could improve interactions with the more hydrophobic binding site present in Grp94 (Figure 2b);[12] and 3) The phenyl ring could provide a synthetic handle to identify substituents that interact with polar residues at the solvent exposed region without reconstituting the extensive hydrogen bond network present in Hsp90 (Figure 2c). The ortho-substituted phenyl derivative, 6, reduced the angle between the resorcinol moiety and the benzyl side chain (~72°→~60°), which can mimic 2-substitutions on the benzyl side chain of the first generation Grp94 inhibitors and potentially improve Grp94 selectivity. Alternatively, the meta-substituted phenyl derivative, 7 increased the angle between these two substituents (from ~72°to ~120°) which should reduce Grp94 affinity and selectivity due to steric limitations that result from the backbone carbonyl of Asn162. Herein, the synthesis and biological evaluation of Grp94-selective inhibitors with a modified cis-amide bioisostere linker is reported.
Figure 2.
Rational design for second generation Grp94-selective inhibitors. (a) Modification to the cis-amide linker to decrease (6) and increase (7) the angle between the benzylic side chain and the resorcinol moiety. (b) The water mediated hydrogen bonding network in Hsp90 (right, PDBID: 2FXS) is much more extensive than that of Grp94 (left, PDBID: 2GFD, trans-RDA omitted for clarity). (c) Polar residues are present in the region which the linker is proposed to bind can be utilized to increase affinity for Grp94.
Results and Discussion
Synthesis of the designed analogues, 6 and 7, was envisioned to occur via a Heck coupling between aryl bromide 8 and 2- or 3-benzylstyrene (9a and 9b, respectively), followed by hydrogenolysis of the benzyl ethers and concomitant reduction of the alkene. This synthetic route allowed for late-stage diversification and provided access to various analogues in a succinct manner (Scheme 1). Aryl bromide 8 was synthesized from 1-bromo-3,5-dimethoxybenzene (10) in 6 steps, 28% overall yield, and required only one chromatographic separation. 10 was demethylated via pyridinium hydrochloride, which provided the free phenols after an acidic workup. The phenols were subsequently converted to the benzyl ethers (11) upon treatment with benzyl bromide and potassium carbonate. Silver-promoted formylation of 11 and Pinnick oxidation yielded benzoic acid 12.[13] Conversion of 12 to the corresponding methyl ester was achieved upon exposure to dimethyl sulfate. Regioselective chlorination was achieved upon reaction with sulfuryl chloride at −30°C, which provided the desired resorcinol, 8 (Scheme 1).[14] Synthesis of 9a and 9b commenced via a palladium-catalyzed cross coupling reaction between commercially available 2- and 3-formylphenyl boronic acid and benzyl bromide to provide the corresponding 2- and 3-benzylbenzaldehydes (13a and 13b, respectively). Subsequent Wittig olefination afforded the 2- and 3-benzylstyrenes (9a and 9b, respectively), which were then subjected to Heck olefination conditions with aryl bromide 8.[15] Hydrogenolysis of the benzyl ethers and reduction of the olefin under a hydrogen atmosphere provided the desired analogues 6 and 7 (Scheme 1).
Scheme 1.
Synthesis of 6 and 7.
Compounds 6 and 7 were evaluated via a fluorescence polarization assay to determine Grp94 affinity and selectivity.[16] As seen in Table 1, 6 manifested an apparent Kd of 0.63 μM along with 32-fold selectivity for Grp94 over Hsp90α, which represented ~2-fold improvement over 2. Alternatively, 7 exhibited a significant reduction in both affinity and selectivity (apparent Kd = 10.4 μM and >10-fold selective) for Grp94. These data suggested that reduction of the angle between the resorcinol moiety and the benzyl side chain was beneficial for Grp94 affinity and selectivity. Therefore, structure-activity relationship studies were performed on the phenyl linker of 6 to probe interactions between the phenyl ring and polar residues surrounding this linker.
Table 1.
Evaluation of 6 and 7 via a fluorescence polarization assay against Grp94 and Hsp90α
| Compound | Apparent Kd Grp94 (μM) | Apparent Kd Hsp90α (μM) | Fold Selective for Grp94 |
|---|---|---|---|
| 6 | 0.63 ± 0.03 | 20.7 ± 0.5 | 32 |
| 7 | 10.4 ± 0.2 | >100a | >10 |
100 μM was the highest concentration tested. Apparent Kd values are the average of two independent experiments ± SEM.
As seen in Figure 2c, the linker was proposed to bind adjacent to polar residues (Glu158 and Asn162) and at the edge of the binding pocket. Therefore, hydrogen bond donors and acceptors were incorporated onto the linker to evaluate Grp94 affinity. In addition, a pyridine ring (in lieu of the phenyl linker) was included to investigate the electronic nature of the linker. Both 9c and 9d were synthesized using similar conditions to 9a and 9b outlined in Scheme 1. As shown in Scheme 2, commercially available aryl bromides, 14a–d and 15a–c, were subjected to a palladium-cross coupling reaction with potassium benzyltrifluoroborate, RuPhos, and palladium (II) acetate to provide benzaldehydes 13e–k. Conversion of benzaldehydes 13e–k to the corresponding styrenes, 9c–k, was achieved via a Wittig reaction with methyltriphenylphosphonium bromide and n-butyllithium. The corresponding alkenes were then subjected to a Heck reaction with 8, followed by simultaneous cleavage of the benzyl ethers and reduction of the alkene to provide analogues 16–24. Under hydrogenation conditions, the nitro group was also reduced to the corresponding aniline (19 and 20) and hydrogenolysis of the benzyl ethers produced the free phenols, 22–24.
Scheme 2. Synthesis of 16–24.
Conditions: (a) Pd(PPh3)4, K2CO3; (b) MePh3P+Br−, nBuLi; (c) Potassium benzyltrifluoroborate, Pd(OAc)2, RuPhos, K3PO4; (d) BnBr, K2CO3; (e) 9c–9k, Pd(OAc)2, triethanolamine; (f) H2, Pd/C
Data obtained via fluorescence polarization with these compounds (Table 2) suggested that incorporation of hydrogen bond acceptors (16–18) did not significantly improve affinity (apparent Kd’s = 0.45–0.73 μM). However, hydrogen bond donors (19, 20, 22–24) did improve Grp94 affinity and resulted in compounds that manifested an apparent Kd value between 0.24–0.54 μM. However, both hydrogen bond acceptors and donors produced a significant loss in Grp94 selectivity. Of these analogues, 18 manifested the highest selectivity (19-fold), which was considerably less than analogue 6 which was 32-fold selective. Replacement of the phenyl ring with a pyridine linker (21) resulted in a substantial increase in affinity (apparent Kd = 0.18 μM) as compared to 6, however this substitution also produced high affinity for Hsp90α (0.16 μM) and resulted in no selectivity between these two isoforms.
Table 2.
Evaluation of 16–24 via a fluorescence polarization assay against Grp94 and Hsp90α
| Compound | Apparent Kd Grp94 (μM) | Apparent Kd Hsp90α (μM) | Fold Selective for Grp94 |
|---|---|---|---|
| 16 | 0.45 ± 0.04 | 2.2 ± 0.2 | 5 |
| 17 | 0.68 ± 0.04 | 2.6 ± 0.6 | 4 |
| 18 | 0.73 ± 0.1 | 13.7 ± 1.4 | 19 |
| 19 | 0.28 ± 0.04 | 1.0 ± 0.1 | 4 |
| 20 | 0.24 ± 0.04 | 0.34 ± 0.07 | n/a |
| 21 | 0.18 ± 0.01 | 0.16 ± 0.02 | n/a |
| 22 | 0.30 ± 0.03 | 2.1 ± 0.1 | 7 |
| 23 | 0.33 ± 0.01 | 0.92 ± 0.03 | 3 |
| 24 | 0.54 ± 0.02 | 2.0 ± 0.2 | 4 |
Apparent Kd values are the average of two independent experiments ± SEM.
Due to the low selectivity produced by 16–24, additional structure-activity relationship studies were sought to improve both Grp94 selectivity and affinity. 16–24 were synthesized to contain the benzyl side chain of 2 to rapidly evaluate modifications about the linker, however, the side chain of 2 was previously optimized to achieve >40-fold Grp94-selectivity (see Figure 1). Incorporation of the Grp94-selective 2-ethoxy- and 2-methoxybenzyl side chains (3 and 4) onto the phenyl-linked analogue was proposed to further increase Grp94 selectivity. Synthesis of these analogues was achieved through a route similar to that utilized to construct 6 (Scheme 3). Upon their preparation, 25 and 26 were evaluated and shown to manifest a significant loss in affinity (apparent Kd = 6.1 μM and 7.8 μM, respectively, Table 3). The loss in affinity exhibited by 25 (compared to 3) suggests the phenyl linker does not bind in an overlapping manner similar to the original imidazole-linked inhibitors. Therefore, a chloride scan was performed to identify an optimal side chain for the phenyl-linked analogues. Synthesis of 27–29 was performed in a manner similar to that used for 25 (Scheme 3). Once prepared, these analogues were evaluated for Grp94 affinity and selectivity via fluorescence polarization and as shown in Table 3, 27 and 29 manifested >18- and >24-fold selectivity for Grp94. However, this gain in selectivity was accompanied by ~10-fold reduction in Grp94 affinity as compared to 6, which suggests a potential steric clash between the chloride-substituted side chains and the hydrophobic pocket of Grp94. Due to the size difference between the hydrogen of 6 and the chloride of 29, a fluoride was incorporated into the 4-position of the side chain to reduce detrimental steric interactions. Gratifyingly, 30 produced an apparent Kd of 0.54 μM and was 73-fold selective for Grp94. The smaller fluoride substitution (compared to the chloride of 29) appears to minimize steric interactions with the hydrophobic pocket and produce increased Grp94 affinity as compared to 6.
Scheme 3. Synthesis of 25–31.

Conditions: (a) Pd(PPh3)4, K2CO3, benzyl bromides; (b) MePh3P+Br−, nBuLi; (c) Pd(dppf)Cl2, K3PO4, 4-fluorobenzylboronic acid pinacol ester; (d) Pd(OAc)2, 9l–9r, triethanolamine; (e) H2, Pd/C
Table 3.
Evaluation of 25–31 via a fluorescence polarization assay against Grp94 and Hsp90α
| Compound | Apparent Kd Grp94 (μM) | Apparent Kd Hsp90α (μM) | Fold Selective for Grp94 |
|---|---|---|---|
| 25 | 6.1 ± 0.3 | >100a | >16 |
| 26 | 7.8 ± 0.4 | >100a | >13 |
| 27 | 5.5 ± 0.1 | >100a | >18 |
| 28 | 11.9 ± 1.8 | 44.6 ± 2.0 | 4 |
| 29 | 4.2 ± 0.7 | >100a | 24 |
| 30 | 0.54 ± 0.05 | 39.2 ± 2.7 | 73 |
| 31 | 0.36 ± 0.02 | 0.46 ± 0.03 | n/a |
100 μM was the highest concentration tested. Apparent Kd values are the average of two independent experiments ± SEM.
Due to the high selectivity of the 4-fluorobenzyl side chain, it too was incorporated into the highest affinity linker in an effort to combine the high affinity and selectivity of 21 with 30 in the form of 31. Synthesis of 31 was accomplished via a palladium-catalyzed cross coupling reaction between 14d and 4-fluorobenzylboronic acid pinacol ester (32) to provide benzaldehyde 13r which was then converted to the corresponding styrene 9r. A Heck coupling reaction between 9r and 8 followed by hydrogenolysis of the benzyl ethers and reduction of the alkene provided 31 (Scheme 3). Evaluation of 31 against Grp94 and Hsp90α via fluorescence polarization gave apparent Kd values of 0.36 μM and 0.46 μM, respectively. Ultimately, the 4-fluoro side chain produced ~2-fold loss in Grp94 affinity (compared to 21) and a complete loss of selectivity. These data suggest the pyridine linker ring oriented the side chain towards the solvent exposed region of the binding pocket in order to avoid repulsive interactions between the pyridine nitrogen and the hydrophobic pocket of Grp94.
Crystal structures of the Grp94 N-terminal domain in complex with 6 and 30 revealed binding modes that can explain differences in their selectivity and affinity profiles (Crystallographic statistics SI Table 1) as well as the binding properties for similar compounds within this series (Figure 3b and 3f). The resorcinol ring anchored within the N-terminal ATP-binding site through direct interactions with Asp149 and Thr245 and a hydrogen bonding network with water molecules (Figrue 3c and 3g).[11–12] Interestingly, both inhibitors were best modeled via two partially occupied binding modes. The first, termed “benzyl in”, aligns with the previously observed binding mode of the first generation Grp94-selective inhibitor, KUNG38, where the benzylic side chain is oriented towards the Grp94-exclusive binding pocket.[11] When crystallized with 6, Grp94 loop residues 167–170, which make up a portion of the aforementioned unique binding region, appeared disordered, suggesting that this interaction was not stabilizing this loop (Figure 3a). Alternatively, when crystallized with 30, the main chain of these loop residues could be built into visible electron density, suggesting that the hydrophobic interactions mediated by the fluoride substituent stabilized these residues, leading to slightly improved Grp94 binding affinity (Figure 3e). The second binding mode, termed “benzyl out”, positioned the benzyl side chain in the solvent-exposed region of the binding pocket. The phenyl linker of 6 is rotated ~180° in this conformation compared to the “benzyl in” conformation. This orients the terminal benzene ring in the vicinity of the ε-NH2 of Lys114, suggesting a stabilized cation-π interaction (Figure 3c and 3d); direct confirmation of such an interaction is limited by the partial occupancy of 6 in this orientation, but appears likely. Alternatively, the phenyl linker of 30 only rotated ~90° and did not appear to orient the terminal benzene ring in a position to form this interaction (Figure 3g and 3h). Since Lys114 is conserved amongst Hsp90 isoforms (Lys44 cytosolic Hsp90s), interactions with this residue are likely to affect selectivity. In sum, the improved selectivity of 30 appears to result from decreased affinity for Hsp90, rather than a significant improvement in affinity for Grp94. Similarly, non-selective inhibitors, like 16–24, may favor the binding mode in which Lys114/Lys44 forms a cation-π interaction. Such information will be useful to guide the design of next generation inhibitors.
Figure 3.
Crystal structures of 6 (a–d, PDBID: 6AOM) and 30 (e–h, PDBID: 6AOL) bound to Grp94. (a, e) Surface representation of nucleotide binding pocket of Grp94 occupied by 6 (a) and 30 (e) which are shown as ball and stick. Alternate conformations of inhibitors indicated by off-shade coloration. Arrow in (e) indicates loop stabilized at the mouth of the binding pocket through hydrophobic interactions with fluorine substituent of 30. (b, f) Cartoon view of Grp94 active site highlighting compound occupancy with direct protein-compound interactions featured. Final 2Fo−Fc electron density (blue mesh), contoured at 1.3 σ for amino acid residues and 1.0 σ for ligands, is superimposed with initial Fo−Fc density (green mesh) contoured at 2.5 σ after molecular replacement. See Experimental Section for details. Compounds shown as ball and stick, waters indicated as small red spheres. (c, g) Hydrogen-bond-mediated protein-inhibitor interactions (dashes) in nucleotide binding pocket between Grp94 and 6 (c) or 30 (g). (d, h) 2-D model of protein-compound interaction network, with hydrogen bonding interactions indicated by dashes and hydrophobic interactions represented by dashed crescents. Spheres: red, oxygen; blue, nitrogen; cyan, water molecules. “Benzyl out” conformation and conformation-specific interactions indicated by purple coloration.
Based on the structure-activity relationship studies performed with the second generation Grp94-selective inhibitors, 30 was selected for further evaluation in a cellular model of metastatic cancer. Grp94 overexpression has been proposed as a biomarker for aggressive and metastatic cancer, due to its upregulation in these cells compared to less aggressive cancers.[16–17] Previously, Grp94-selective inhibition (pharmacological and genetic) has demonstrated a reduction in the migratory capabilities of aggressive breast and prostate cancer cell lines.[11, 18] Integrins are dependent upon Grp94 for their maturation and trafficking to the cell surface and interact with the extracellular matrix to provide a mechanism for oncogenic cell migration away from the primary tumor site to form metastatic lesions.[19]
Evaluation of 30 in a wound healing scratch assay was used to determine the potential of Grp94 inhibitors against different cancers. As previously reported, Grp94-selective inhibition was effective at reducing the migratory capabilities of metastatic breast (MDA-MB-231) and prostate (PC3-MM2) cell lines. Against these cell lines, treatment with 30 produced 38–40% migration (~60% inhibition) at 500 nM after 24 h and exhibited good effects at lower concentrations (Figure 4 and SI Figure S1a–S1b). This anti-migratory effect was observed at concentrations much lower than the GI50 manifested by 30 against these cell lines (SI Table 2), which together demonstrate that Grp94-selective inhibition results in an anti-migratory effect that is not due to decreased proliferation. Based on these results, 30 was evaluated in a melanoma cell line to determine whether Grp94-selective inhibition could provide therapeutic value in reducing the migration of other aggressive cancers. Metastatic melanoma (SK-MEL-28) cells demonstrated a susceptibility to Grp94 inhibition after 24 h (Figure 4, SI Figure 1c), however, the anti-migratory effect was less than that observed in breast and prostate cancers, suggesting Grp94-selective inhibition may provide anti-migratory effects against certain cancers.
Figure 4.
Representative images of a wound healing scratch assay in different cancer cell lines after 24 h treatment with 30 or vehicle (0.25% final concentration of DMSO). Scale bar = 100 μm.
Increased integrin expression has been positively correlated with increased metastatic aggressiveness for lung cancer and therefore Grp94 inhibition could reduce the metastatic potential of this cancer.[20] Grp94-selective inhibition with 30 was evaluated against a non-metastatic lung cancer cell line (A549) and demonstrated an anti-migratory effect at 500 nM, however this effect was reduced when compared to the highly metastatic breast, prostate, and melanoma cancers above due to the non-aggressive nature of the A549 cell line (Figure 4, SI Figure 1d). The limited efficacy of Grp94 inhibition in non-aggressive cancers and the overexpression of Grp94 in aggressive cancers suggests that the efficacy of Grp94-selective inhibition correlates directly with the aggressiveness of a type of cancer.
The integrin α2 subunit is dependent upon Grp94 for its maturation and trafficking to the cell surface.[18b] Integrin α2 is found as a heterodimer with the β1 integrin subunit and is responsible for binding collagen in the extracellular matrix, which promotes cancer metastasis and invasion.[21] Increased expression of integrin α2β1 has been reported, in both primary and metastatic tissue samples of melanoma from patients, suggesting this cancer utilizes Grp94-dependent integrins to facilitate migration.[22] As shown in Figure 6, treatment with 30 resulted in degradation of the integrin α2 subunit at 2.5 μM in the metastatic cancer cell lines (MDA-MB-231, PC3-MM2, SK-MEL-28, Figure 5). When treated with 30, the non-metastatic cancer cell line (A549) exhibited the degradation of integrin α2, however, this effect was not observed until 5 μM, which correlated with the diminished anti-migratory activity exhibited against this cell line. Treatment with 30 did not induce the degradation of Akt, a cytosolic Hsp90-dependent protein, nor did it result in the induction of Hsp70. This is in contrast to the natural product and pan-Hsp90 inhibitor geldanamycin (G, Figure 5), which induced the degradation of Akt and increased the levels of Hsp70 in all cell lines. These data demonstrate that 30 selectively targets Grp94 and induces the degradation of Grp94-specific client proteins without decreasing the levels of proteins that depend upon the cytosolic Hsp90 isoforms.
Figure 5.
Western blot analysis for the Grp94-dependent client protein Integrin α2, the cytosolic Hsp90-dependent client protein Akt, Hsp70, and loading control actin after treatment with 30 at indicated concentrations for 24 h (0.1% DMSO final concentration). D = DMSO, G = geldanamycin, a natural product, pan-Hsp90 inhibitor (0.5 μM).
Conclusion
Incorporation of a phenyl linker in lieu of the imidazole ring, reduced the angle between the resorcinol moiety and the benzyl side chain and led to the identification of 6, which manifests 0.64 μM Grp94 affinity and 32-fold Grp94 selectivity. Substitutions about the phenyl linker provided compounds with improved affinity, however, in all cases this was accompanied by a loss in selectivity. Subsequent modification to the side chain revealed alternative binding modes between the first and second generation of Grp94-selective inhibitors. Structure-activity relationship studies on the side chain of 6 ultimately led to the discovery of 30, which manifests 0.54 μM affinity and 73-fold selectivity for Grp94 versus other Hsp90 isoforms. Evaluation of 30 in a wound healing scratch assay against multiple aggressive and metastatic cancers revealed that Grp94-selective inhibition results in decreased migratory capabilities at 500 nM and induced degradation of the Grp94-specific protein, integrin α2. When combined, these data suggest that Grp94 inhibition may provide a suitable therapeutic option to reduce the migration/metastasis of many aggressive cancers.
Experimental Section
Chemistry General
1H NMR were recorded at 400 (Bruker AVIIIHD 400 MHz NMR with a broadband X-channel detect gradient probe) or 500 MHz (Avance AVIII 500 MHz spectrometer with a dual carbon/proton cryoprobe), 19F NMR were recorded at 376 MHz (Bruker AVIIIHD 400 MHz NMR equipped with a multinuclear broadband fluorine observe probe), and 13C NMR were recorded at 125 MHz (Bruker AVIII spectrometer equipped with a cryogenically cooled carbon observe probe); chemical shifts are reported in δ (ppm) relative to the internal standard (CDCl3,7.26 ppm for 1H and 77.2 for 13C). HRMS spectra were recorded with a LCT Premier with ESI ionization. All biologically tested compounds were determined to be >95% pure. TLC analysis was performed on glass backed silica gel plates and visualized by UV light. All solvents were reagent grade and used without further purification. Synthesis and characterization of compounds is provided in the supplemental information.
Fluorescence Polarization
The assay was performed in 96-well black, flat-bottom plates with a final volume of 100 μL. 25 μL of assay buffer (20 mM HEPES, pH 7.3, 50 mM KCl, 5 mM MgCl2, 20 mM Na2MoO4, 2 mM DTT, 0.1 mg/mL BGG, and 0.01% NP-40) were added, followed by 25 μL of assay buffer containing 6 nM FITC-GDA (fluorescent tracer, stock in DMSO, diluted in assay buffer) and 50 μL of assay buffer containing 10 nM of either Grp94 or Hsp90α were added to each well. For each plate, wells containing buffer only (background), tracer in buffer only (low polarization control) and protein, tracer, and 1% DMSO (final concentration, high polarization control) were included. Compounds were then added with a final concentration of DMSO = 1%. Plates were incubated at 4°C with rocking for 24 h. Polarization values (in mP units) were measured at 37°C with an excitation filter at 485 nm and an emission filter at 528 nm. Polarization values were correlated to % tracer bound and compound concentrations. The concentration at which the tracer was 50% displaced by compound of interested were calculated and reported as apparent Kd’s.
Docking Studies
The Surlfex-Docking module in Sybyl v8.0 was used for docking studies using the crystal structure of KUNG38 bound to Grp94 (PDBID: 5IN9). Pymol was used for visualization of the results.
Structure Determination of Grp94 in Complex with Compounds 6 and 30
The N-terminal domain construct of Grp94, NΔ41 (Canis lupus familiaris origin, amino acid residues 69–337, with the segment 287–327 replaced by a GGGG stretch as in Dollins et al.[23], cloned into a custom codon-optimized vector by ATUM, Menlo Park, CA was expressed as a tobacco etch virus (TEV) protease-cleavable maltose binding protein (MBP) fusion and purified as previously reported.[11] Purified NΔ41 protein was exchanged into 100 mM bicine buffer pH 7.8 and concentrated to 30 mg/mL; protein concentration was determined by Bradford assay (Amresco), with BSA used as a calibration standard. Apo NΔ41 crystals were grown using the sitting drop vapor diffusion method. Crystallization drops containing a mixture of 2 μL protein and 2 μL mother liquor were equilibrated against 500 μL of mother liquor reservoir solutions composed of 35–39% PEG400, 7.5–10% glycerol, 100 mM bicine at pH 7.8, and 75 mM MgCl2. Typically, crystals grew within a one-week period. Protein crystals were harvested and transferred to wells containing fresh mother liquor mixed with 2 mM compound (20% v/v DMSO overall), and were allowed to soak for 4 days. After soaking, 100% glycerol was layered atop the soak well solution to a total concentration of ~25%, and crystals were rapidly pulled through this cryoprotecting glycerol layer and immediately flash-cooled in liquid nitrogen.
Diffraction data for inhibitor-soaked NΔ41 crystals were collected at the Advanced Photon Source, Argonne National Laboratories Beamline Southeast Region Collaborative Access Team (SER-CAT) 22-BM. Data pertaining to the structure of NΔ41 with 6 were processed with HKL-3000[24]. Co-crystal structure data with compound 30 were initially processed with XDS[25]; the data were subsequently assigned a space group, merged and scaled with Aimless within the CCP4 suite[26]. Both structures were solved by molecular replacement with Phaser[27] using as a search model the polypeptide chain of the Grp94-RDA structure (PDBID: 2GFD). Models were iteratively built and refined with Coot[28] and Phenix refine[29], respectively. Models of and additional restraints for compounds 6 and 30, as well as for PEG400 and glycerol, were prepared using eLBOW in Phenix refine[29] using as input SMILES strings generated in ChemDraw (version 15.1.0.144, Perkin Elmer). The structures have been deposited in the Protein Data Bank with the PDB accession codes: 6AOM (6) and 6AOL (30).
Cell Culture
MDA-MB-231, PC3-MM2, SK-MEL-28, and A549 cells were grown in a water jacketed incubator at 37°C with 5% CO2 in DMEM (MDA-MB-231 and PC3-MM2), EMEM (SK-MEL-28) or F-12K (A549) media supplemented with 10% FBS and 1% penicillin/streptomycin.
Anti-proliferation. cells were counted via Trypan blue exclusion and seeded in 96-well plates at 2000 cells/0.1 mL/well then returned to the incubator for 24 h. Compounds or vehicle were administered in DMSO (1% DMSO final concentration) and the plates were placed back in the incubator. After 72 h, the % viable cells were determined using the MTS/PMS cell proliferation kit (Promega) per the manufacturer’s instructions. Cells treated with vehicle were normalized to 100% viable and compound treated wells were adjusted accordingly. GI50 values were calculated via GraphPad Prism and reported as the average of 2 or more independent experiments ± SEM.
Wound Healing Scratch Assay
Cells were counted via Trypan blue exclusion and seeded in 12-well plates at 200,000 cells/mL/well and returned to the incubator for 24 h. Scratches were made with a 0.1–10 μL pipette tip, then cells were washed with PBS and fresh media was added. Compound or DMSO control were then added (0.25% DMSO final concentration) and 0 h pictures taken with a camera-mounted Olympus IX-71 microscope (10X objective). Plates were returned to the incubator until 24 h pictures were taken. Images were processed and % migration determined via ImageJ. All experiments were performed in quadruplicate.
Western Blot Analysis
Cells were counted via Trypan blue exclusion and were seeded at 100,000 cells/mL in 10 cm dishes and placed back in the incubator for 24 h. Compounds or vehicle were dosed (0.25% DMSO final concentration) and incubated together for 24 h. Cells were harvested in cold PBS and lysed using MPER (Thermo Scientific) supplemented with protease inhibitors (Roche) according the manufacturer’s instructions. Cell lysates were obtained by centrifugation at 15,000 rpm for 10 min at 4°C. Protein concentrations were determined using the Pierce BCA assay kit following the manufacturer’s instructions. Equal amounts of protein were separated via gel electrophoresis under reducing conditions (10% acrylamide gels) then transferred to PVDF membranes and immunoblotted with the corresponding primary antibodies. Membranes were then incubated with the correct HRP-labeled secondary antibody, developed with a chemiluminescent substrate, and visualized.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (EY024232 to BSJB, EY021205 to RLL). VMC is supported by the NCI (F99CA212467). Support for NMR instrumentation was provided by NIH Shared Instrumentation Grants (S10OD016360, S10RR024664) and NSF Major Research Instrumentation Grant (0320648). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
References
- 1.a Hanahan D, Weinberg RA. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]; b Miyata Y, Nakamoto H, Neckers L. Curr Pharm Des. 2013;19:347–365. doi: 10.2174/138161213804143725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a Travers J, Sharp S, Workman P. Drug Discov Today. 2012;17:242–252. doi: 10.1016/j.drudis.2011.12.021. [DOI] [PubMed] [Google Scholar]; b Khandelwal A, Crowley VM, Blagg BS. Med Res Rev. 2016;36:92–118. doi: 10.1002/med.21351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a Neckers L, Workman P. Clin Cancer Res. 2012;18:64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Taipale M, Jarosz DF, Lindquist S. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 4.Ernst JT, Liu M, Zuccola H, Neubert T, Beaumont K, Turnbull A, Kallel A, Vought B, Stamos D. Bioorg Med Chem Lett. 2014;24:204–208. doi: 10.1016/j.bmcl.2013.11.036. [DOI] [PubMed] [Google Scholar]
- 5.Gewirth DT. Curr Top Med Chem. 2016;16:2779–2791. doi: 10.2174/1568026616666160413141154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marzec M, Eletto D, Argon Y. Biochim Biophys Acta. 2012;1823:774–787. doi: 10.1016/j.bbamcr.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a Hua Y, White-Gilbertson S, Kellner J, Rachidi S, Zusmani S, Chiosis G, Depinho R, Li Z, Liu B. Clin Cancer Res. 2013;19:6242–6251. doi: 10.1158/1078-0432.CCR-13-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Stothert AR, Suntharalingam A, Huard DJ, Fontaine SN, Crowley VM, Mishra S, Blagg BS, Lieberman RL, Dickey CA. Hum Mol Genet. 2014;23:6470–6480. doi: 10.1093/hmg/ddu367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Randow F, Seed B. Nat Cell Biol. 2001;3:891–896. doi: 10.1038/ncb1001-891. [DOI] [PubMed] [Google Scholar]
- 9.a Chen B, Piel WH, Gui L, Bruford E, Monteiro A. Genomics. 2005;86:627–637. doi: 10.1016/j.ygeno.2005.08.012. [DOI] [PubMed] [Google Scholar]; b Taldone T, Patel PD, Patel M, Patel HJ, Evans CE, Rodina A, Ochiana S, Shah SK, Uddin M, Gewirth D, Chiosis G. J Med Chem. 2013;56:6803–6818. doi: 10.1021/jm400619b. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Song HY, Dunbar JD, Zhang YX, Guo D, Donner DB. J Biol Chem. 1995;270:3574–3581. [PubMed] [Google Scholar]
- 10.Duerfeldt AS, Peterson LB, Maynard JC, Ng CL, Eletto D, Ostrovsky O, Shinogle HE, Moore DS, Argon Y, Nicchitta CV, Blagg BS. J Am Chem Soc. 2012;134:9796–9804. doi: 10.1021/ja303477g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowley VM, Khandelwal A, Mishra S, Stothert AR, Huard DJ, Zhao J, Muth A, Duerfeldt AS, Kizziah JL, Lieberman RL, Dickey CA, Blagg BS. J Med Chem. 2016;59:3471–3488. doi: 10.1021/acs.jmedchem.6b00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Immormino RM, Metzger LEt, Reardon PN, Dollins DE, Blagg BS, Gewirth DT. J Mol Biol. 2009;388:1033–1042. doi: 10.1016/j.jmb.2009.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohsawa K, Yoshida M, Doi T. J Org Chem. 2013;78:3438–3444. doi: 10.1021/jo400056k. [DOI] [PubMed] [Google Scholar]
- 14.Dutton BL, Kitson RR, Parry-Morris S, Roe SM, Prodromou C, Moody CJ. Organic & biomolecular chemistry. 2014;12:1328–1340. doi: 10.1039/c3ob42211a. [DOI] [PubMed] [Google Scholar]
- 15.Li HJWL. Eur J Org Chem. 2006;2006:5099–5102. [Google Scholar]
- 16.Hu T, Xie N, Qin C, Wang J, You Y. Tumour Biol. 2015;36:9357–9364. doi: 10.1007/s13277-015-3635-4. [DOI] [PubMed] [Google Scholar]
- 17.Dejeans N, Glorieux C, Guenin S, Beck R, Sid B, Rousseau R, Bisig B, Delvenne P, Buc Calderon P, Verrax J. Free Radic Biol Med. 2012;52:993–1002. doi: 10.1016/j.freeradbiomed.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 18.a Muth A, Crowley V, Khandelwal A, Mishra S, Zhao J, Hall J, Blagg BS. Bioorg Med Chem. 2014;22:4083–4098. doi: 10.1016/j.bmc.2014.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Ghosh S, Shinogle HE, Galeva NA, Dobrowsky RT, Blagg BS. J Biol Chem. 2016;291:8309–8323. doi: 10.1074/jbc.M115.688374. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Mishra SJ, Ghosh S, Stothert AR, Dickey CA, Blagg BS. ACS Chem Biol. 2017;12:244–253. doi: 10.1021/acschembio.6b00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.aB Luo H, Carman CV, Springer TA. Annu Rev Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wu S, Hong F, Gewirth D, Guo B, Liu B, Li Z. J Biol Chem. 2012;287:6735–6742. doi: 10.1074/jbc.M111.309526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.a Guo L, Zhang F, Cai Y, Liu T. Pathol Res Pract. 2009;205:847–853. doi: 10.1016/j.prp.2009.07.005. [DOI] [PubMed] [Google Scholar]; b Gogali A, Charalabopoulos K, Constantopoulos S. Exp Oncol. 2004;26:106–110. [PubMed] [Google Scholar]
- 21.a Naci D, Vuori K, Aoudjit F. Semin Cancer Biol. 2015;35:145–153. doi: 10.1016/j.semcancer.2015.08.004. [DOI] [PubMed] [Google Scholar]; b Haidari M, Zhang W, Caivano A, Chen Z, Ganjehei L, Mortazavi A, Stroud C, Woodside DG, Willerson JT, Dixon RA. J Biol Chem. 2012;287:32981–32992. doi: 10.1074/jbc.M112.395905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schadendorf D, Gawlik C, Haney U, Ostmeier H, Suter L, Czarnetzki BM. J Pathol. 1993;170:429–434. doi: 10.1002/path.1711700405. [DOI] [PubMed] [Google Scholar]
- 23.Dollins DE, Warren JJ, Immormino RM, Gewirth DT. Mol Cell. 2007;28:41–56. doi: 10.1016/j.molcel.2007.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. Acta Crystallogr D. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch W. Acta Crystallogr D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans PR, Murshudov GN. Acta Crystallogr D. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Lohkamp B, Scott WG, Cowtan K. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.