Abstract
Aim
Identify solutions to the most important policy barriers to the clinical adoption of next-generation sequencing.
Materials & methods
Four-round modified policy Delphi with a multistakeholder panel of 48 experts. The panel deliberated policy solutions to (previously reported) challenges deemed most important to address.
Results
The group advocated using consensus panels to promote consistency in payer policies and to standardize test reporting, and favored making genomic data-sharing a condition of regulatory clearance, certification, or accreditation processes. They were split on the role of US FDA.
Conclusion
Panelists found common ground on solutions for health plan coverage policy consistency, data-sharing, and standardizing reporting, but were sharply divided on the role of the FDA in mitigating risks to patients.
Keywords: clinical data reporting, clinical genomics, coverage and reimbursement, intellectual property, next-generation sequencing, personalized medicine, policy, regulation
Significant investment is being made to leverage new genomic technologies to enable more precise ways of preventing and treating disease – the hallmark of what is often called ‘precision’ or ‘personalized’ medicine [1]. The advent of high-throughput genomic-sequencing techniques [2] makes it feasible to sequence large numbers of genes at a price point increasingly competitive with other forms of genetic testing, potentially laying the foundation for clinical integration of genomic data. These ‘next-generation sequencing’ (NGS) techniques facilitate testing of whole genomes and exomes as well as more targeted testing of large panels of genes. Yet multiple challenges hamper the integration of NGS into clinical practice, including the complexity of genomic information [3], the inadequacy of many clinical studies [4], the growing complexity of bio-informatics [5], and the economics of personalized medicine [6]. From the perspective of public and private payers in the USA, enthusiasm has outpaced clinical evidence for NGS-based clinical testing. These payer organizations separately assess and write policies for coverage and reimbursement of new clinical tests, and have been slow to embrace new NGS-based clinical testing. More rapid evidence development will depend on many factors, including the status and implementation of proposals for regulatory oversight of laboratory-developed tests (LDTs) and NGS-based testing, intellectual property law, and policies for data-sharing. There are thus significant policy issues that must be addressed, particularly related to coverage and reimbursement, intellectual property and data sharing, and regulatory oversight of genomic tests [7–12]. To respond effectively, policymakers need to tackle the most important and tractable challenges first, taking account of the perspectives of multiple stakeholders.
To assess the relative importance and tractability of challenges arising in the clinical integration of NGS, we carried out a four-round Delphi study focusing on policy questions. We used a multistakeholder panel of experts to identify and rank the most important policy challenges and deliberate about solutions to the top four challenges. The first two rounds of this study (reported elsewhere) established the group’s collective judgement about which barriers were most important to address and most tractable [13]. Here we report the results of the second two rounds, which explored a series of potential policy solutions to the challenges previously identified as most important to address for effective clinical adoption of NGS.
Materials & methods
We conducted a modified policy Delphi, an iterative structured communication technique conducted in rounds with a select group of experts to assess a specific policy question. The policy Delphi approach uses a heterogeneous group of experts to explore differences of opinion and produce a range of policy options [14–16]. We used a ‘modified’ Delphi approach, which includes facilitated direct interaction among Delphi panelists [17].
For purposes of this study, we defined ‘expert’ as a person with professional expertise or significant lay knowledge in the field of genetics, government, health policy, patient advocacy, or law. A history of NGS-related publications, presentations, or service on task forces or professional or public committees was evidence of expertise. Experts were contacted by email and offered an honorarium of $100 per round of the completed Delphi. Out of 93 invitations sent, 48 experts in the USA agreed to participate and 27/28 of these completed rounds three and four, respectively. Table 1 shows the stakeholder composition of the experts from this panel who participated in rounds three and four of the Delphi process. Delphi surveys are not a form of statistical population sampling. The group participating in this study was purposively sampled [18] according to our criteria for expertise in specified stakeholder categories. Our claim is not that this group is necessarily representative of a larger population of experts, but that their perspectives are inherently valuable and warrant serious consideration. Note that for Table 1 we asked the experts to identify their ‘primary professional role’. However, many of these participants have multiple professional roles. For example, three ‘genomics researchers’ were also physicians and one was also the director of a molecular diagnostics laboratory. By the same token, one of the ‘product developers’ was a laboratory director, another was a software designer. The table notes and labels convey supplemental information about these categories.
Table 1.
Expert stakeholder composition of Delphi Panel, rounds three and four.
| Primary profession† | Round three (n) | Round four (n) |
|---|---|---|
| Genomic researcher‡ | 2 | 3 |
| Lawyer or legal scholar | 3 | 4 |
| Payer (included public and commercial payers)§ | 3 | 2 |
| Research funder | 0 | 0 |
| Regulator or policy maker | 1 | 3 |
| Clinician or healthcare provider (as primary role) | 1 | 1 |
| Industry funder (venture capitalist) | 0 | 0 |
| Health economist | 2 | 2 |
| Product developer (includes laboratories) | 1 | 3 |
| Patient advocate | 3 | 4 |
| Social scientist | 1 | 1 |
| Informatician | 1 | 0 |
| Other¶ | 9 | 5 |
| Total | 27 | 28 |
As self-identified by participants when asked for ‘primary professional roles’. Participating experts typically have more than one relevant role. Additional detail provided in notes and on table.
Includes oncologist/physicians, genomics laboratory director, and representative of the National Institutes of Health, which is also a research funder.
One ‘payer’ is also a molecular diagnostics laboratory director
Other self-identified roles provided by participants: ethicist, administrator/educator, public health, laboratory director, software developer, ethics and policy analysis, physician performing translational research, policy advocate, policy analyst.
The first two rounds of this study were conducted by survey and established the group’s collective judgement about which policy challenges in the clinical integration of NGS were most important to address (i.e., if unaddressed, the challenge would pose a barrier to patient and provider’s access to accurate, valid clinical information generated using NGS technology) and most tractable (i.e., overall, the challenge can be addressed effectively). As reported elsewhere [13], a total of 29 prospective policy challenges were presented to the panelists for evaluation, encompassing: 19 challenges across the domains of coverage and reimbursement, intellectual property, and regulation framed by the project team based on preliminary research and the content expertise of project team members, and ten additional challenges suggested by the panelists in round one without restriction as to domain. The four challenges identified in the first two rounds of the Delphi as most important were:
Payer variation in evidence standards: different payers have different evidentiary standards for assessing clinical utility, leading to inconsistent policies on coverage and reimbursement for NGS-based testing.
Proprietary databases: diagnostic companies are able to maintain proprietary databases on the substantial variety of clinically meaningful mutations found in patients. Data-sharing promotes more rapid learning on the clinical significance of discovered variants, leading to more rapid demonstration of clinical utility and adoption of clinically useful NGS tests.
Role of regulation in reducing risks: there is a lack of consensus about what mechanisms should be used to address the clinical risks to patients from NGS-based LDTs. For example, there is continued debate as to whether US FDA oversight should be used to address these risks.
Reporting genomic test results: there is a lack of standardization for reporting NGS test results (e.g., determining which results to report, how to effectively communicate findings and to whom those findings should be communicated).
This paper focuses on rounds three and four of the study: solutions to the challenges identified in the first two rounds.
Round three consisted of group discussions by telephone in March 2015 to explore the specific nature of each of the four top ranking challenges and potential solutions. Participants were given the option of remaining blinded, but all chose to voluntarily un-blind their identities to one another for the purpose of the telephone conversations. The participating Delphi panelists were divided into four groups, each of which discussed two challenges. Each group included between five and nine panelists; a total of 27 panelists participated in round three (see Table 1 for composition). We sought to maintain a balance of stakeholders in each discussion group; we also invited panelists to elect participation in the groups of most interest to them. The discussions were facilitated by a member of the research team and were 1 h in length, with approximately 30 min allocated to each challenge. Proposed solutions to each challenge taken from a previous group survey (done in February 2015) were presented to the panelists in advance. Round three discussions were audio-recorded, with permission, transcribed, and analyzed by members of the research team, who developed an initial list of key discussion points, which were then reviewed and refined by all members of the project team.
In round four, we surveyed the entire panel in June 2015, asking them to assess the round three discussion in three different ways:
By assessing their agreement with each key discussion point individually using four-point Likert scales (from strongly agree to strongly disagree, with a ‘don’t know’ option but not a neutral point on the scale; list of discussion points is provided in Table 2).
By assessing each proposed solution according to the perceived effectiveness and feasibility to implement using the same four-point Likert scale.
By selecting the single most effective solution to each challenge from a list.
Table 2.
Challenges with potential solutions in rank order (based on percentage of panelists who selected the solution as ‘best to pursue’ in round four).
| Challenges with potential solutions | Best to pursue (%) |
|---|---|
| Challenge A: Different payers have different evidentiary standards for assessing clinical utility, which leads to inconsistent policies on coverage and reimbursement for NGS-based tests | |
| Multistakeholder consensus panels that include payers and patients should be convened to set evidentiary standards | 37 |
| Expert panels (e.g., ACMG, NAM, EGAPP, etc.) should develop recommendations for evidentiary standards for all payers to use | 33 |
| Fund research on methods of establishing standards, and pilot different approaches to determine the best one | 15 |
| All payers, public and private, should cooperate to develop consistent standards | 7 |
| No policy action needed. Accept variability in evidence standards and coverage inconsistency among payers and focus on other challenges | 7 |
| There should be a government or legislative mandate for payers to cover all NGS tests | 0 |
| There should be a legislative mandate for payers to cover FDA-approved or cleared tests only | 0 |
| Challenge B: Diagnostic companies are able to maintain proprietary databases on a substantial variety of clinically meaningful mutations found in patients. Refusal to share this type of information could impede the development of clinically useful NGS tests. | |
| Make data sharing and the possibility of independent verification a condition of approval/clearance, certification or accreditation (specifically, a requirement set forth by FDA, CMS/CLIA and/or CAP) | 54 |
| Use positive-economic incentives so that payers reimburse more for tests from laboratories that share data | 18 |
| Use negative incentives. For example, payers refuse to pay, or healthcare providers refuse to order tests, if there is no data sharing | 14 |
| Make data sharing and sufficient transparency for independent verification a condition of funding (specifically, a requirement for NIH funding) | 14 |
| Public shaming of companies that do not share | 0 |
| Do nothing. You cannot or should not compel data disclosure in this space | 0 |
| Challenge C: There is a lack of consensus about what mechanisms should be used to address the clinical risks to patients from NGS-based laboratory-developed tests. For example, there is continued debate as to whether FDA oversight should be used to address these risks | |
| Use FDA oversight as a mechanism to address the risks to patients from NGS-based LDTs | 41 |
| Use laboratory accreditation and proficiency testing by professional societies as a mechanism to address the risks to patients from NGS-based LDTs | 1 |
| Use payers to address certain risks to patients from NGS-based LDTs, specifically, restrict payment to accredited laboratories | 7 |
| Use payers to address certain risk to patients from NGS-based LDTS, specifically, condition coverage on consultation with a genetic counselor | 7 |
| Rely on state regulation to address the risks to patients from NGS-based LDTs | 4 |
| Use payers to address certain risks to patients from NGS-based LDTs, specifically, restrict reimbursement to FDA-approved or cleared tests | 0 |
| Rely on self-regulation to address the clinical risks to patients from NGS-based LDTs | 0 |
| Challenge D: There is a lack of standardization for reporting NGS test results (e.g., determining which results to report, how to effectively communicate findings, and to whom those findings should be communicated | |
| A consensus panel could designate a general framework and nomenclature for how to present information (formatting reports) | 82 |
| No action at this point. It is premature to come out with any guidelines on formatting reports | 14 |
| Standardize communication to patients for providers and genetic counselors | 4 |
ACMG: American College of Medical Genetics; CAP: College of American Pathologists; CLIA: Clinical Laboratory Improvement Amendment; CMS: Centers for Medicare and Medicaid Services; EGAPP: Evaluation of Genomic Applications in Practice and Prevention; NAM: National Academies of Medicine; LDT: Laboratory-developed test; NGS: Next-generation sequencing.
28 panelists completed the survey in round four (see Table 1 for composition). Note that while the Institutional Review Board gave us permission to allow group members to un-blind themselves to each other, the Institutional Review Board still requires that we maintain the anonymity of the group in publications.
For purposes of reporting results of the evaluations of effectiveness/feasibility of individual potential solutions to challenges (Figures 2–5), we created scores weighted and normalized such that: if all respondents selected ‘strongly agree’, the score would be 100; if all respondents selected ‘agree’, the score would be 50; if all respondents selected ‘disagree’, the score would be −50; and if all respondents selected ‘strongly disagree’, the score would be −100. Selections of ‘don’t know’ are not included in the normalized scores. They are instead separately represented on the graphs with lines (% not known is the percentage of respondents who selected ‘don’t know’ when prompted to evaluate the effectiveness and/or feasibility of a given potential solution).
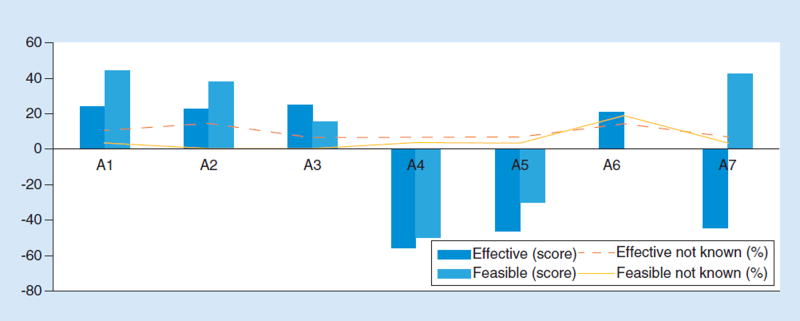
Figure 2. Effectiveness and feasibility scores for solutions to inconsistent payer evidence standards.
(A1) Expert panels (e.g., ACMG, Institute of Medicine or CDC/Evaluation of Genomic Applications in Practice and Prevention) should develop recommendations for evidentiary standards for all payers. (A2) Fund research on methods of establishing standards, and pilot different approaches to determine the best one. (A3) Multistakeholder consensus panels that include payers and patients should be convened to set evidentiary standards. (A4) There should be a government or legislative mandates for payers to cover tests. (A5) There should be a legislative mandate for payers to cover FDA-approved or cleared tests only. (A6) All payers, public and private, should cooperate to develop consistent standards across all payers. (A7) No policy action needed. Accept variability in evidence standards and coverage inconsistency among payers and focus on other challenges. ACMG: American College of Medical Genetics; CDC: Centers for Disease Control and Prevention.
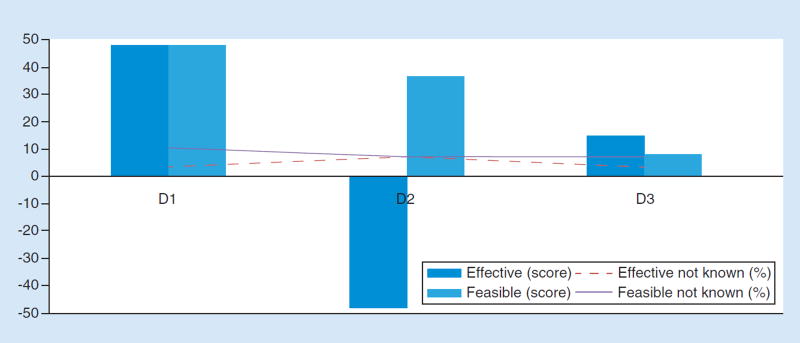
Figure 5. Effectiveness and feasibility scores by solutino for challenge D.
(D1) A consensus panel could designate a general framework and nomenclature for how to present information (formatting reports). (D2) No action at this point. It is premature to come out with any guidelines on how to format reports. (D3) Standardize communication to patients for providers and genetic counselors.
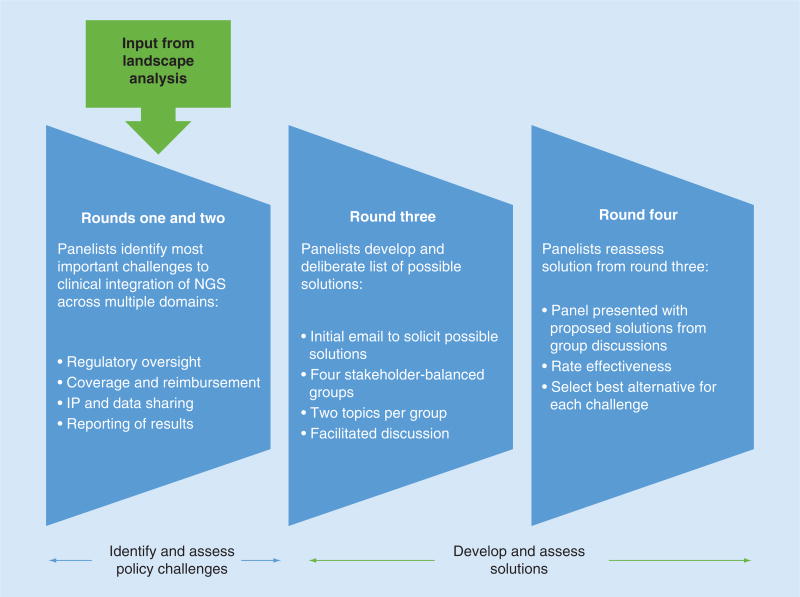
The entire Delphi process underlying this report is summarized in Figure 1. All components of the study were approved by an institutional review board at Baylor College of Medicine.
Figure 1. Delphi process.
NGS: Next-generation sequencing; IP: Intellectual property.
Results
As shown in Table 1, 27 of the experts participated in the round three discussion groups and 28 responded to the round four survey. Table 2 summarizes the results of the ranking exercise for potential solutions for each challenge, showing the percentage of respondents who selected a potential solution as ‘best to pursue’ to address the relevant challenge. Figures 2–5 illustrate the effectiveness and feasibility scores obtained for potential solutions to each of the four individual challenges (one figure per challenge). Table 3 exhibits our reassessment in round four of the talking points arising from discussion groups conducted in the previous round (round three). What follows is a report of the results by individual challenge, starting with the perceived variation in payer evidence standards. For each challenge we summarize key points from the discussions of possible solutions to that challenge (round three) and then report on the formal reassessment of those key talking points (round four).
Table 3.
Number of panelists agreeing with key discussion points for proposed solutions to challenges.
| Key discussion points | Disagree, n (%) | Agree, n (%) |
|---|---|---|
| Challenge A – aligning payer evidence standards | ||
| A1: Expert panels to develop recommendations | ||
| No single body can be viewed as authoritative | 6 (21) | 21 (75) |
| Recommendations go out of date quickly | 14 (50) | 14 (50) |
| Even with shared evidentiary standards, each payer will make individual choices on utility and coverage | 3 (11) | 24 (89) |
| Even if overall coverage determinations were the same across payers, there could still be substantive differences in eligibility requirements, reimbursement levels, preferred care pathways, etc. | 2 (7) | 26 (90) |
| Standards cannot be imposed on payers from the outside; standard-setting must include payer involvement | 6 (21) | 22 (76) |
| A3: Fund research and pilot different approaches | ||
| Payers do not come to the table; they have no incentive to discuss or disclose | 16 (53) | 12 (40) |
| It is difficult for any one group to develop a consensus statement that would be authoritative (e.g., NAM) | 9 (30) | 21 (70) |
| A4: Mandate coverage (tests) | ||
| This solution is not practical given our Congress | 8 (29) | 19 (68) |
| This is a technical problem, not a political one (i.e., keep out of the political process) | 7 (25) | 19 (68) |
| A5: Mandate coverage (FDA-approved or -cleared tests only) | ||
| Since very few LDTs have FDA approval or clearance, such a mandate would cause too many to apply for FDA approval, and FDA does not have the capacity | 11 ( 39 ) | 15 (54) |
| Alignment of FDA approval with payer standards is not feasible because FDA and payers have differing information needs for their respective decisions | 7 (24) | 20 (69) |
| This solution may be more feasible 15–20 years in the future | 13 (46) | 8 (29) |
| A6: All payers cooperate to develop consistent standards | ||
| This solution will not work because payers are prohibited by antitrust law from colluding (i.e., although payers can talk to each other about evidence, they must set policies for coverage independently) | 14 (48) | 12 (41) |
| This is a technical problem, not a political one (i.e., keep out of political process) | 11 (38) | 12 (41) |
| Challenge B (Proprietary Data) | ||
| B1: Require data sharing and transparency for approval/certification/accreditation | ||
| This solution might require legislative action, and if so, may prove politically infeasible | 8 (29) | 17 (61) |
| Keep FDA out at all costs | 21 (78) | 5 (19) |
| B2: Require data-sharing and transparency for funding | ||
| Depositing data are already required with NIH-funded research. The major challenge for this solution is when NIH-funded research is commercialized and when the data are not contributing to a publication | 3 (11) | 23 (82) |
| B3: Use positive-economic incentives/reimburse more for data sharing | ||
| Negative-economic incentives are also needed | 10 (36) | 13 (46) |
| B4: Use negative-economic incentives/exclude nonsharers from payment, ordering | ||
| Positive-economic incentives are generally more welcomed than negative ones | 2 (7) | 23 (82) |
| There are legal repercussions (i.e., legal challenges likely if payers punish labs for keeping proprietary data; also antitrust implications if payers collude on not paying). These are more troublesome than the solution of withholding payment | 7 (26) | 14 (52) |
| B5: Publicly shame companies that do not share | ||
| Some companies do not feel shame | 5 (18) | 19 (75) |
| Challenge C (mitigating risks to patients from NGS-based LDTs) | ||
| C1: Use FDA oversight to address risks | ||
| FDA does not have the bandwidth to cover all possible risks | 8 (31) | 16 (62) |
| FDA’s domain for oversight includes risks associated with clinical validity only | 19 (66) | 6 (21) |
| Because there are different risks, different approaches are needed (i.e., it cannot be just FDA) | 10 (36) | 14 (50) |
| C2: Use laboratory accreditation and proficiency testing to address risks | ||
| This is the approach appropriate to assure preanalytic/ analytic validity only | 7 (24) | 19 (66) |
| C3a: Restrict reimbursement to FDA-approved or cleared tests | ||
| Very few LDTs have FDA approval or clearance. | 13 (45) | 14 (48) |
| C3c: Condition coverage on consultation with a genetic counselor | ||
| There is a national shortage of genetic counselors. This requirement would be unreasonably restrictive | 10 (34) | 12 (48) |
| C5: Rely on state regulation to address risks | ||
| There are too many different standards (e.g., CLIA, New York, and CAP all have different standards) | 3 (11) | 23 (82) |
| There is no common language for analytic standards | 3 (11) | 17 (61) |
| Challenge D (standardizing genomic testing results) | ||
| D1: Consensus panel to designate general framework and nomenclature | ||
| While achieving consensus for formatting through expert panels is useful, ultimately laboratories will choose to create reports as they wish (i.e., expert panels have no ‘teeth’ to enforce formats) | 8 (29) | 18 (64) |
| Any guideline created should be high-level and general (as opposed to fine-grained) | 6 (22) | 19 (70) |
| D3: Attempt to standardize communication | ||
| It is hard to standardize verbal communication and explanations | 5 (19) | 22 (81) |
| Any attempt at standardization should be in broad strokes | 6 (21) | 21 (75) |
CAP: College of American Pathologists; CLIA: Clinical Laboratory Improvement Amendment; NAM: National Academies of Medicine; LDT: Laboratory-developed test.
Payer variation in evidence standards
Round three
The panelists discussing this topic grappled with the true nature of the underlying challenge. Is it that the evidence standards (including definitions of clinical utility) actually vary between payers? Or are the standards similar but there is variation in how they are applied? On the one hand, differing standards can still lead to the same or very similar coverage decisions. On the other hand, even if payers agree a given test should be covered, many differences arise in the conditions and criteria for determining who qualifies for testing and what elements of the resulting care pathway will be covered. Ultimately, while recognizing these complexities, the group’s discussion focused on between-payer consistency and transparency of evidence standards for genetic and genomic testing.
Both of the groups assigned to this challenge gave consideration to, and then rejected, suggestions that payers collaborate to agree on evidence standards, or that legislation be used to compel greater consistency. On the former suggestion, it was noted that payers avoid any appearance of collusion on coverage decision-making, which is prohibited by antitrust law. The latter suggestion was dismissed due to a lack of confidence in the US Congress to function effectively on the issue. With these options set aside, both discussion groups gravitated toward the use of expert panels, studies, and consensus groups. A disadvantage of this approach is that no single set of recommendations is typically viewed as definitive. Agreement nevertheless coalesced around the notion that large multistakeholder group deliberations could be successful in achieving greater consistency of standards. Technical expert panels were also viewed favorably, but substantive inclusion of payers, patients, and other stakeholders was seen as advantageous (although some panelists argued payers lack incentives to ‘come to the table’).
Round four
In the round four assessment of the single best solution to the payer standards challenge, the top selection (with 37% of respondents making the choice) was ‘multistakeholder consensus panels that include payers’ followed by ‘expert panels’ (33%; Table 2). Runner-up options were: research studies on methods to align standards (15%); payers cooperating to develop standards (7.4%); and ‘no policy action needed’ (7.4%; Table 2). Some members of the full panel expressed skepticism about the real extent of antitrust liability exposure connected with payer collaboration to develop standards (Table 3). Nevertheless, payer collaboration was not ultimately seen as a highly feasible alternative (Figure 2). In addition, new statutory law was seen as undesirable and impractical (Table 3) and ultimately likely to prove ineffective (Figure 2). Indeed, zero panelists selected legislative mandates for payers to cover tests, or to only cover FDA-approved or cleared tests, as the best policy option to pursue ( Table 2 ).
Proprietary databases
Round three
One of the discussion groups considering the challenge of proprietary databases focused on how to promote sharing of new variant information by laboratories, while the other group focused on the need for a sustainable, long-term approach to maintaining trustworthy, reliable shared resources. In reviewing specific suggested solutions, several panelists spoke up in favor of a combination of approaches, with some favoring a variety of positive incentives and others endorsing the view that combining a ‘carrot’ and a ‘stick’ would be optimal to accelerate progress.
Of the possible approaches, legislative or regulatory mandates for data sharing as a condition of market access or laboratory accreditation were seen as potentially the most effective (Figure 3). A few panelists expressed an aversion to FDA involvement, with one voicing a desire to ‘keep FDA out of it at all costs’, but others disagreed. Economic incentives were seen as likely to be more effective than ‘public shaming’ of companies that refuse to share data because, as one panelist noted, some companies are ‘incapable of [feeling shame]’. The groups also discussed making data sharing a condition of research funding. NIH policy already requires NIH-funded researchers to share data, although some panelists noted that NIH research results can be taken up by a commercial testing enterprise that does not share.
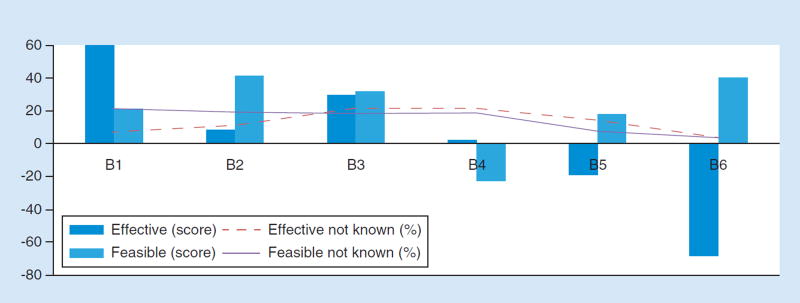
Figure 3. Effectiveness and feasibility scores for solutions to proprietary databases.
(B1) Make data sharing and the possibility of independent verification a condition of approval/clearance, certification, or accreditation (specifically, a requirement set forth by FDA, CMS/Clinical Laboratory Improvement Amendments, and/or College of American Pathologists). (B2) Make data sharing and sufficient transparency for independent verification a condition of funding (specifically, a requirement for NIH funding. (B3) Use positive-economic incentives so that payers reimburse more for tests from laboratories that share data. (B4) Use negative-economic incentives. For example, payers refuse to pay, or healthcare providers refuse to order tests, if there is no data sharing. (B5) Public shaming of companies that do not share. (B6) Do nothing. You cannot or should not compel data disclosure in this space.
CMS: Centers for Medicare and Medicaid Services.
Round four
In the round four evaluation of key discussion points, panelists were skeptical that any solution depending on Congressional action or regulatory change was feasible. Overall, the group favored positive-economic incentives (e.g., higher reimbursement rates for laboratories that share data) as more effective than negative ones (e.g., no reimbursement if data are not shared; Figure 3). Ultimately, however, despite skepticism about the feasibility of solutions involving regulatory change, when asked to select the single best solution to the challenge of proprietary databases, 54% selected making data-sharing a condition of approval/clearance, certification, or accreditation. 18% selected positive-economic incentives; 14% selected negative-economic incentives; 14% selected making data-sharing a condition of research funding; and 0% selected public shaming ( Table 2 ).
Role of regulation to reduce risk to patients
Round three
Both round three discussion groups identified similar sets of risks to patients from NGS-based LDTs. These were: flawed or inconsistent detection of the presence or absence of genomic variants (analytic validity); flawed variant calling and interpretation; incorrect association of detected variants with a phenotype of interest (clinical validity); and errors or misunderstandings in communicating results (related to clinical utility). Some of the panelists thought that, if the FDA is going to exert oversight of NGS-based LDTs, it should focus only on clinical validity, but this view was not shared by all discussants. Regarding non-FDA approaches to managing risk, the groups discussed three mechanisms by which payers could control risks to patients: restricting reimbursement only to FDA-cleared or approved tests; restricting payment to accredited laboratories; and conditioning coverage on consultation with a genetic counselor.
Round four
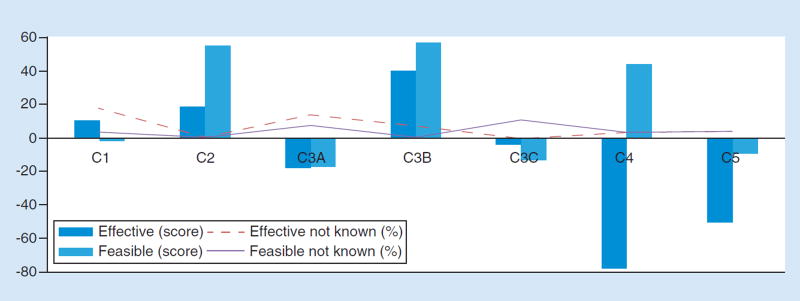
Although using the FDA to manage risks to patients from LDTs received only weak support when assessed individually for effectiveness and feasibility (Figure 4), in the selection of the single best alternative, the panelists were split between FDA oversight (41%) and laboratory accreditation and proficiency testing by professional societies (41%; Table 2 ). The evaluations of key discussion points suggest a way of understanding these findings: a majority agreed that laboratory accreditation and proficiency testing are appropriate tools to assure analytic validity and also saw some role for FDA in oversight of LDTs, but there were significant concerns about the FDA’s resources (‘bandwidth’) to regulate all aspects of risk (Table 3). The remaining options received little support (7% for restricting payment to accredited laboratories; 7% for condition coverage on consultation with a genetic counselor; 4% for relying on state regulation; 0% for conditioning coverage on FDA clearance; 0% on self-regulation of laboratories; Table 2). Also of note, in the evaluation of potential solutions along dimensions of effectiveness and feasibility, payers restricting coverage to accredited laboratories scored highly on both dimensions (Figure 4), suggesting that this approach is viewed by many as promising, even if few regard it as the single best strategy for addressing risks to patients. In survey questions addressing making coverage contingent on genetic counseling, 48% of the participating panelists agreed (vs 34% who disagreed) that this condition would be too restrictive given a lack of genetic counselors outside of major medical centers (Table 3). Self-regulation of laboratories and reliance on state regulation were also rejected as ineffective options (Figure 4). Finally, the panel overwhelmingly agreed that too many different sets of standards are currently in force for clinical laboratory accreditation (State of New York, Clinical Laboratory Improvement Amendments (CLIA) regulations, State of California, etc.); these should be aligned for more consistent, less cumbersome oversight of LDTs (Table 3).
Figure 4. Effectiveness and feasibility scores for solutions to mitigating risk to patients from NGS-based LDTs.
(C1) Use FDA oversight as a mechanism to address the risks to patients from next-generation sequencing (NGS)-based laboratory-developed tests (LDTs). (C2) Use laboratory accreditation and proficiency testing by professional societies as a mechanism to address the risks to patients from NGS-based LDTs. (C3A) Use payers to address certain risks to patients from NGS-based LDTs, specifically, restrict reimbursement. (C3B) Use payers to address certain risks to patients from NGS-based LDTs, specifically, to restrict payment to accredited laboratories. (C3C) Use payers to address certain risks to patients from NGS-based LDTs, specifically, condition coverage on consultation with a genetic counselor. (C4) Rely on self-regulation to address the clinical risks to patients from NGS-based LDTs. (C5) Rely on state regulation to address the risks to patients from NGS-based tests.
LDT: Laboratory-developed test; NGS: Next-generation sequencing.
Reporting genomic test results
Round three
One of the round three discussion groups sought to clarify specifically what kind of reporting represented the most important challenge to address. Discussants agreed that there are three aspects of reporting that represent challenges. The first is the question of which genomic information should be reported (in particular, handling of variants of uncertain significance). This first question was set aside as already being addressed by professional societies and other groups. The second question is how the information should be presented in a clinical report. The third question is how the reported information should be communicated verbally to patients. While the question of verbal communication is important, the group felt that standards could not readily be created for this aspect of reporting, especially since, with recent amendments to the Health Insurance Portability and Accountability Act and CLIA, 17 patients now have the option of side-stepping a clinician or genetic counselor and receiving reports directly from the laboratory. The group therefore chose to focus on the second question and the challenge of developing standards for consistent formatting and nomenclature in genomic reports, and this question also received significant attention from the other discussion group. Panelists in both groups believed that an expert consensus panel could designate a format and nomenclature.
Round four
Responses to the round four survey confirmed the preference for use of a consensus panel to address reporting of results of NGS-based tests. In the final ranking exercise, 82% of respondents selected this option as the best approach ( Table 2), in line with favorable evaluations of effectiveness and feasibility (Figure 5). They largely rejected the idea that no action is needed (only 14% selected this option), or that standards should be created for communications of providers to patients (only 4% selected this option; Table 2). At the same time, the majority recognized the limits of the consensus panel approach, agreeing that expert panels have no ‘teeth’ to enforce standards against nonconforming laboratories, and believing that guidelines emerging from such a process should be kept high-level and general (Table 3).
Discussion
This multidisciplinary expert panel identified and deliberated potential solutions to four policy challenges that, if unaddressed, were felt to pose substantial barriers to clinical integration of NGS technologies. As noted, to make payer evidence standards (and, by extension, coverage decisions) more consistent, participants in our Delphi panel favored the use of expert panels and especially multistakeholder consensus efforts that include payers. In the round three discussions, panel members repeatedly mentioned the Institute of Medicine (IOM, now the National Academies of Medicine, or NAM) as an example of the type of organization that could be employed for this purpose. In fact, the IOM hosted a consensus study group called Policy Issues in the Clinical Development and Use of Biomarkers for Molecularly Targeted Therapies. In its consensus report [19], the group recommended that the Secretary of Health and Human Services ‘should facilitate the development of common clinical utility evidentiary standards that are applied for initial and ongoing coordinated regulatory, coverage, and reimbursement decisions for biomarker tests for molecularly targeted therapies.’ One mechanism for doing so, said the report, was through ‘convening one or more independent, public-private, multistakeholder bodies’ ( [19] recommendation 1).
Other groups were also mentioned by panelists as potential models for consensus development: the US Centers for Disease Control and Prevention, the US Preventive Services Task Force, the Evaluation of Genomic Applications in Practice and Prevention Working Group, and the Green Park Collaborative (GPC). Notably, the GPC has hosted a series of multistakeholder discussions that included major national payers in an effort to reach consensus on clinical utility and coverage for NGS. Building on previously issued guidelines representing the payer perspective on clinical utility evidence needed for covering molecular diagnostic tests [20], this additional effort focused on how large gene panels and large-scale sequencing differ from traditional diagnostics for purposes of clinical utility evidence and coverage decision-making. Although not binding, the GPC’s report made initial recommendations for coverage that could have the effect of making payer policies for genomics more consistent [21].
Both the IOM and GPC efforts recognize that coverage policy is only one among several interrelated challenges to be addressed simultaneously for patients to gain access to any benefits of clinical genomics. Due to space constraints, we bracket complex questions concerning broader social goals, such as the development of substantive ethical and economic criteria for prioritizing expenditures on genomic and other health services and integration of cost-effectiveness studies early in the innovation cycle, while acknowledging their importance. See, for example [22,23]. Consistent curation of variants is required to assure the validity of variant-disease associations. Organizations like ClinGen are using expert panels very effectively for this purpose. However, even if these associations were well established and payers had broadly consistent requirements for demonstrating clinical utility, sparse empirical evidence of clinical utility continues to be a key barrier to coverage, as well as to the more effective use of genomics in clinical medicine [24]
This is why proprietary databases were identified as a highly important challenge to clinical adoption of NGS. Aggregation of clinical and genomic data across large repositories is essential for making sense of the genome for healthcare – to establish the clinical utility of genomic variants in patient care. This is the reasoning behind the US Precision Medicine Initiative (now called the All of Us research program) plan to build a million-person cohort to follow longitudinally [25]. Yet even a repository of this size may lack power to tease out genotype–phenotype relationships for certain subpopulations. The key will be to share resources from existing repositories and new ones as they are built, and to connect many kinds of data globally [26]. This is an important enough priority that 53% of our Delphi panel favored a top-down approach (which they generally eschewed in other contexts) to compel data sharing by making it a condition of FDA clearance or approval, CLIA certification, or accreditation by the College of American Pathologists.
The group did not consider another way to compel data-sharing: making it a condition of acceptance of manuscripts for journal publication. The International Committee of Medical Journal Editors has proposed a requirement that authors agree to share deidentified individual-patient data within 6 months after publication as a condition of consideration by a member journal [27]. However, the proposal is limited to interventional clinical trials and has not yet been adopted. Further, to be truly effective, a data-sharing requirement would have to be embraced by other major journals, and even then it would only influence organizations that prize peer-reviewed publication. This may be why the group overlooked discussion of this possibility in favor of more comprehensive top-down approaches. The group also overlooked microattribution (recognition of contributors to databases) to encourage submissions to databases [28,29]. While this approach has been shown to incentivize submissions to variant databases, it would not be likely to overcome for-profit, proprietary data-holding. Many in the group acknowledged a legitimate role for commercial enterprises to collect, store, and use proprietary data for their own purposes.
Additional challenges exist for data-sharing. In different ways, the continuing IOM and GPC efforts noted above acknowledge that for data-pooling to be effective, common core data elements must be collected across repositories, and these elements must be gathered using common standards and data definitions that permit direct comparisons and aggregation. This is another challenge that may be solved by expert and multistakeholder consensus-building and has international as well as US dimensions. Recently, GPC has teamed with the Medical Evidence Development Consortium, the NIH Cancer Genome Atlas, the American Association for Cancer Research GENIE program, the American Society of Clinical Oncology, and other major cancer genome repositories to agree on essential clinical data elements that should always be collected for these data-bases. A multistakeholder workshop for this purpose was hosted by GPC in April 2016 and a consensus set of recommendations is forthcoming. Other promising new initiatives include BloodPAC, a US-based public-private consortium of liquid biopsy companies, clinicians, data experts, patient advocates, and others to share analytic and clinical validation data on blood based tumor DNA profiling, with the goal of creating a regulatory-grade data platform for more rapid liquid biopsy test development [30]. At the global level, the Global Alliance for Genomics and Health has a number of working groups that are addressing various technical and ethical and regulatory aspects of data sharing [31].
For optimal comprehension in clinical use, relevant information from NGS must be reported with consistent and well-defined format and nomenclature – a framework which the Delphi group overwhelmingly said could be developed through the work of a consensus panel. The literature now includes guidance from the American College of Medical Genetics and other professional societies on standardizing terminology related to interpretation of sequence variants [32] and templates for genome sequencing reports [33,34], as well as studies of patient preferences related to reporting [35,36]. Relatedly, groups in the USA and Europe have been working to achieve consensus on reporting for incidental findings [37,38]. These are promising signs that this set of challenges can be resolved through these mechanisms in a relatively timely fashion.
Finally, development of a regulatory framework to address clinical risks to patients from NGS-based LDTs has now been suspended by the FDA [39]. Many of our panelists were at least somewhat supportive of FDA involvement in this space (Table 2 & Figure 4). The New York State Department of Health recently published a proposed policy for risk-based evaluation of LDTs [40], which in principle is consistent with the approach advocated by the FDA. However, our panelists were nearly unanimous in their agreement that too many different sets of standards are currently in force for oversight of laboratory testing; harmonization of existing and emerging standards should be a top policy priority in the regulatory domain. The majority believed that other complementary approaches are needed to protect against risks to patients (Table 3). Specifically, the majority supported development and implementation of consistent standards for accreditation of laboratories and proficiency testing to address analytic validity, and limiting payment to accredited laboratories as potentially effective and highly feasible strategies to help mitigate or manage risks (Figure 4). Notably, the College of American Pathologists (CAP) has developed an accreditation and proficiency testing program for NGS. Participation in this program could be used by payers as a criterion for payment [41,42] (GPC recommendations call for this type of provision [21]). However, the program will only assure the quality of laboratory procedures and validation; it will not address what our panel perceived as shifting requirements by payers for evidence of clinical utility, or a lack of clinical outcomes evidence as perceived by payers.
A limitation of the study is that, given the mixed expertise of the panel members, the results are conditioned by which experts agreed to participate, and also by the differing composition of the expert groups actually participating in each round of the Delphi. There is no way to predict whether or how these variables affected the outcomes of the project. While some might see this as a weakness of the method, it is designed to be informative as a structured exercise in expert opinion-making, not to produce systematic, generalizable knowledge. Further, because the panelists actually participating in each round were drawn from across important stakeholder groups (representatives of key groups did participate in every round, except venture capitalists, who were represented in rounds one and two, but not in three and four), we believe there is considerable value in our findings to highlight areas of agreement and disagreement. Additionally, our use of multiple question formats allowed us to provide insight into at least some of the factors shaping participants’ policy preferences.
As of this writing, Congress passed the 21st Century Cures Act (in late 2016) with bipartisan support. The legislation will significantly augment research funding for personalized medicine, but can only do so if bipartisan support for authorization is sustained in the face of Trump administration proposals for deep cuts to the National Institutes of Health. Under the Trump administration, less regulatory intervention at the Federal level may be expected than in recent years. Hence, while the 21st Century Cures Act contains a number of provisions intended to foster data sharing, any further initiatives seem likely to come through the types of voluntary efforts already underway. The FDA’s suspension of efforts to regulate LDTs make accreditation and proficiency testing for genomic laboratories by professional societies even more crucial. The lack of FDA oversight in this area could serve to widen the coverage gaps for personalized medicine testing (or at least not help to close it), since FDA processes of oversight allow payers somewhat better transparency than the proficiency testing programs of professional societies. Nevertheless, we expect that in the current political environment, collaborative and voluntary programs, with engagement of industry, health plans, patients, and all key stakeholders working together, will be essential for continued progress in precision medicine.
Conclusion
The Delphi panel identified a series of policy challenges to be addressed to clear the way for more rapid clinical adoption of genomic sequencing and deliberated a series of solutions to these challenges. On the challenges of inconsistency in payer evidence requirements and uniform clinical data reporting standards, the panel repeatedly considered, and in most cases rejected, Congressional and other top-down approaches to problem-solving as either impractical or undesirable. Instead they favored voluntary, multistakeholder collaborative efforts, even while recognizing that recommendations from such efforts are not binding. At the same time, we found sharp division – a nearly 50–50 split – on the best way to mitigate clinical risks to patients. Defining a role for the FDA remains problematic. The group found the holding of proprietary data to be among the highest challenges for the growth of personalized medicine. While divided on this issue, the group was more inclined towards top-down measures to compel desired behaviors on this (e.g., to compel data-sharing) than on any other of the challenges.
Summary points.
Through a policy Delphi method, a multistakeholder expert panel identified the following challenges as highly important barriers to clinical adoption of next-generation sequencing: inconsistency of health plans in defining clinical utility and thus in coverage policies for genomic sequencing; ability of commercial laboratories to maintain proprietary mutation databases; uncertainty on oversight needed to protect patients from clinical risks of next-generation sequencing and advanced sequencing techniques; and lack of standardization in the way genomic data are reported out to clinicians and patients.
To make payer evidence standards and policies for coverage more consistent, panelists rejected the possibility of between-payer collaboration, concluding that a more effective approach would be convening expert workgroups, especially ones having multistakeholder representation to include payers.
A slim majority of the group favored compelling data-sharing by making it a condition of regulatory clearance, certification or accreditation processes.
The panel was split on how to manage risks to patients from laboratory developed tests, with almost half favoring FDA regulation as the most effective tool, and almost half of the panel favoring reliance on laboratory accreditation and proficiency testing; there was agreement on the need to harmonize existing and emerging standards.
A strong consensus was ultimately achieved that inconsistencies in reporting could be addressed through the action of expert workgroups.
In sum, we found modest-to-strong consensus among expert stakeholders on solutions to three of four major policy challenges related to clinical adoption of next-generation sequencing, with accompanying concerns (and guidance) noted for policy makers and others engaged in efforts aligned with favored solutions, but no majority opinion on the role of the FDA in mitigating clinical risks to patients.
Acknowledgments
Financial
This project was supported by grant R01HG006460 from the US National Human Genome Research Institute (National Institutes of Health). D Messner is employed by the nonprofit Center for Medical Technology Policy (CMTP) and runs the Green Park Collaborative (GPC) program noted in the Discussion, but receives no direct remuneration from the GPC program.
Footnotes
competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest; •• of considerable interest
- 1.Collins FS, Varmus H. A new initiative on precision medicine. N. Engl. J. Med. 2015;372(9):793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kulkarni S, Pfeifer J, editors. Clinical Genomics. Elsevier; MA, USA: 2014. [Google Scholar]
- 3.Arnedos M, Vielh P, Soria J-C, Andre F. The genetic complexity of common cancers and the promise of personalized medicine: is there any hope?: personalized treatment in cancer. J. Pathol. 2014;232(2):274–282. doi: 10.1002/path.4276. [DOI] [PubMed] [Google Scholar]
- 4.Holmes MV, Shah T, Vickery C, Smeeth L, Hingorani AD, Casas JP. Fulfilling the promise of personalized medicine? systematic review and field synopsis of pharmacogenetic studies. PLoS ONE. 2009;4(12):e7960. doi: 10.1371/journal.pone.0007960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernald GH, Capriotti E, Daneshjou R, Karczewski KJ, Altman RB. Bioinformatics challenges for personalized medicine. Bioinformatics. 2011;27(13):1741–1748. doi: 10.1093/bioinformatics/btr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis JC, Furstenthal L, Desai AA, et al. The microeconomics of personalized medicine: today’s challenge and tomorrow’s promise. Nat. Rev. Drug Discov. 2009;8(4):279–286. doi: 10.1038/nrd2825. [DOI] [PubMed] [Google Scholar]
- 7.Chan IS, Ginsburg GS. Personalized medicine: progress and promise. Annu. Rev. Genomics Hum. Genet. 2011;12(1):217–244. doi: 10.1146/annurev-genom-082410-101446. [DOI] [PubMed] [Google Scholar]
- 8•.Kaufman D, Curnutte M, McGuire AL. Clinical integration of next-generation sequencing: a policy analysis. J. Law Med. Ethics. 2014;42(Suppl. 1):5–8. doi: 10.1111/jlme.12158. An introduction to a series of articles describing and analyzing critical policy issues in personalized medicine and adoption of clinical high-throughput DNA sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Javitt GH, Carner KS. Regulation of next-generation sequencing. J. Law Med. Ethics. 2014;42(Suppl. 1):9–21. doi: 10.1111/jlme.12159. Environmental review of the coverage and reimbursement landscape, discussing the need for clinical utility evidence to satisfy third-party payers. [DOI] [PubMed] [Google Scholar]
- 10.Deverka PA, Dreyfus JC. Clinical integration of next-generation sequencing: coverage and reimbursement challenges. J. Law Med. Ethics. 2014;42(Suppl. 1):22–41. doi: 10.1111/jlme.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Cook-Deegan R, Chankrasekharan S. Patents and genome-wide DNA sequence analysis: is it safe to go into the human genome? J. Law Med. Ethics. 2014;42(Suppl. 1):42–50. doi: 10.1111/jlme.12161. Examination of case law related to the patenting of human genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Evans BJ. Economic regulation of next-generation sequencing. J. Law Med. Ethics. 2014;42(Suppl. 1):51–66. doi: 10.1111/jlme.12162. Discussion of the ways in which the next-generation sequencing industry is evolving and new business models are emerging, with implications for regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Messner DA, Al Naber J, Koay P, et al. Barriers to clinical adoption of next generation sequencing: perspectives of a policy Delphi panel. Appl. Transl. Genomic. 2016;10:19–24. doi: 10.1016/j.atg.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linstone HA, Turoff M, editors. The Delphi Method: Techniques and Applications. Addison-Wesley Publishing; MA, USA: 1975. [Google Scholar]
- 15.Turoff M. The design of a policy Delphi. Technol. Forecast Soc. Change. 1970;2(2):149–171. [Google Scholar]
- 16.Adler M, Ziglio E, editors. Gazing into the Oracle: The Delphi Method and its Application to Social Policy and Public Health. Jessica Kingsley Publishers; PA, USA: 1996. [Google Scholar]
- 17.Fitch K, Bernstein SJ, Aguilar MD, et al. RAND/UCLA appropriateness method user’s manual. 2001 www.rand.org/content/dam/rand/pubs/monograph.
- 18.Bryman A. Social Research Methods. 4. Oxford University Press; Oxford; NY, USA: 2012. [Google Scholar]
- 19•.National Academies of Sciences Engineering, Medicine. Biomarker tests for molecularly targeted therapies: key to unlocking precision medicine. www.nap.edu/catalog/21860/biomarker-tests Report and recommendations to address barriers to using molecularly targeted testing and therapies, including recommendations to address inconsistencies across regulatory and payer definitions of clinical utility.
- 20.Deverka PA, Messner DA, Dutta T. Evaluation of clinical validity and clinical utility of actionable molecular diagnostic tests in adult oncology. Center for Medical Technology Policy. www.cmtpnet.org/docs/resources/MDX_EGD.pdf.
- 21•.Center for Medical Technology Policy. Initial Medical Policy and Model Coverage Guidelines for Clinical next-generation sequencing in Oncology, Report and Recommendations. 2015 www.cmtpnet.org/docs/resources/Full_Release_Version Green Park Collaborative report and recommendations from a multistakeholder dialog to address US payer policies on next-generation sequencing.
- 22.Van Nimwegen KJ. Feasibility of the headroom analysis in early economic evaluation of innovative diagnostic technologies with no immediate treatment implications. Value Health. 2014;17(7):A550. doi: 10.1016/j.jval.2014.08.1792. [DOI] [PubMed] [Google Scholar]
- 23.Rogowski WH, Grosse SD, Schmidtke J, Marckmann G. Criteria for fairly allocating scarce health-care resources to genetic tests: which matter most? Eur. J. Hum. Genet. 2014;22(1):25–31. doi: 10.1038/ejhg.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hresko A, Haga S. Insurance coverage policies for personalized medicine. J. Pers. Med. 2012;2(4):201–216. doi: 10.3390/jpm2040201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fact sheet: President Obama’s precision medicine initiative | The White House. www.whitehouse.gov/the-press-office/2015/01/30/fact-sheet.
- 26.Majumder MA, Cook-Deegan R, McGuire AL. Beyond our borders? Public resistance to global genomic data sharing. PLOS Biol. 2016;14(11):e2000206. doi: 10.1371/journal.pbio.2000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taichman DB, Backus J, Baethge C, et al. Sharing clinical trial data: a proposal from the international committee of medical journal editors. Ann. Intern. Med. 2016;164(7):505. doi: 10.7326/M15-2928. [DOI] [PubMed] [Google Scholar]
- 28.Giardine B, Borg J, Higgs DR, et al. Systematic documentation and analysis of human genetic variation in hemoglobinopathies using the microattribution approach. Nat. Genet. 2011;43(4):295–301. doi: 10.1038/ng.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sosnay PR, Siklosi KR, Van Goor F, et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat. Genet. 2013;45(10):1160–1167. doi: 10.1038/ng.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.BloodPAC. www.bloodpac.org.
- 31.Global Alliance for Genomics and Health. http://genomicsandhealth.org.
- 32.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17(5):405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dorschner MO, Amendola LM, Shirts BH, et al. Refining the structure and content of clinical genomic reports. Am. J. Med. Genet. C Semin. Med. Genet. 2014;166(1):85–92. doi: 10.1002/ajmg.c.31395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vassy JL, McLaughlin HL, MacRae CA, et al. A one-page summary report of genome sequencing for the healthy adult. Public Health Genomics. 2015;18(2):123–129. doi: 10.1159/000370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haga SB, Mills R, Pollak KI, et al. Developing patient-friendly genetic and genomic test reports: formats to promote patient engagement and understanding. Genome Med. 2014;6(7):58. doi: 10.1186/s13073-014-0058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stuckey H, Williams JL, Fan AL, et al. Enhancing genomic laboratory reports from the patients’ view: a qualitative analysis. Am. J. Med. Genet. A. 2015;167(10):2238–2243. doi: 10.1002/ajmg.a.37174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 2013;15(7):565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hehir-Kwa JY, Claustres M, Hastings RJ, et al. Towards a European consensus for reporting incidental findings during clinical NGS testing. Eur. J. Hum. Genet. 2015;23(12):1601–1606. doi: 10.1038/ejhg.2015.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.US FDA. Discussion paper on laboratory developed tests (LDTs) www.fda.gov/downloads/medicaldevices.
- 40.NYSDOH proposed policy for risk-based evaluation of laboratory developed tests (LDT) NY State Dep. Health Wadsworth Cent; 2016. www.wadsworth.org/news/nysdoh-proposed-policy-for-risk. [Google Scholar]
- 41.Aziz N, Zhao Q, Bry L, et al. College of American pathologists’ laboratory standards for next-generation sequencing clinical tests. Arch. Pathol. Lab. Med. 2015;139(4):481–493. doi: 10.5858/arpa.2014-0250-CP. [DOI] [PubMed] [Google Scholar]
- 42.Laboratory accreditation program – college of american pathologists. www.cap.org/web/home/lab/accreditation.