Abstract
In this study, we report the insertion sequence ISPpu21 in the oprD porin gene of carbapenem-resistant Pseudomonas aeruginosa isolates from burn patients in Tehran, Iran. Antibiotic susceptibility tests for P. aeruginosa isolates were determined. Production of metallo-β-lactamases (MBLs) and carbapenemase was evaluated and the β-lactamase-encoding and aminoglycoside-modifying enzyme genes were investigated by PCR and sequencing methods. The mRNA transcription level of oprD and mex efflux pump genes were evaluated by real-time PCR. The outer membrane protein profile was determined by SDS–PAGE. The genetic relationship between the P. aeruginosa isolates was assessed by random amplified polymorphic DNA PCR. In all, 10.52% (10/95) of clinical isolates of P. aeruginosa harboured the ISPpu21 insertion element in the oprD gene. The extended-spectrum β-lactamase-encoding gene in ISPpu21-carrying isolates was blaTEM. PCR assays targeting MBL and carbapenemase-encoding genes were also negative in all ten isolates. The rmtA, aadA, aadB and armA genes were positive in all ISPpu21 harbouring isolates. The relative expression levels of the mexX, mexB, mexT and mexD genes in ten isolates ranged from 0.1- to 1.4-fold, 1.1- to 3.68-fold, 0.3- to 8.22-fold and 1.7- to 35.17-fold, respectively. The relative expression levels of the oprD in ten isolates ranged from 0.57- to 35.01-fold, which was much higher than those in the control strain P. aeruginosa PAO1. Evaluation of the outer membrane protein by SDS–PAGE suggested that oprD was produced at very low levels by all isolates. Using random amplified polymorphic DNA PCR genotyping, eight of the ten isolates containing ISPpu21 were shown to be clonally related. The present study describes a novel molecular mechanism, ISPpu21 insertion of the oprD gene, associated with carbapenem resistance in clinical P. aeruginosa isolates.
Keywords: Burn, ISPpu21, OprD porin, Pseudomonas aeruginosa, carbapenem
Introduction
Carbapenems, such as imipenem and meropenem are often used as last resort antibiotics for the treatment of multidrug-resistant (resistant to at least three different classes of antibiotics) Pseudomonas aeruginosa infections [1]. Pseudomonas aeruginosa is an opportunistic bacterial pathogen causing a variety of infections such as endocarditis, pneumonia, urinary tract infection, septicaemia, and skin, eye and ear infections. This organism is also a leading cause of morbidity and mortality among hospitalized burn patients worldwide [2]. Selecting the appropriate drug to initiate antibiotic therapy is important to control the clinical outcome. Unfortunately, treatment with the most appropriate antibiotics is complicated because of the possible resistance of this bacterium to multiple classes of antibacterial agents, such as carbapenems (imipenem and meropenem) [3]. Pseudomonas aeruginosa can use a combination of chromosomally encoded and/or plasmid-encoded mechanisms to evade carbapenem therapy. However, in non-carbapenemase-producing P. aeruginosa, resistance to doripenem and meropenem is associated with other mechanisms [4]. The main reported mechanism of resistance to carbapenems involves the loss of OprD porin from the outer membrane through deletions, mutations or insertions in the oprD gene. One important mechanism of carbapenem resistance is insertional inactivation of the oprD gene by insertion sequence (IS) elements of various sizes [5]. In this study we define a novel insertion sequence in the oprD porin gene of carbapenem-resistant P. aeruginosa isolates from burn patients in Tehran, Iran.
Materials and methods
Ethics statement
This study was approved by the Ethics Committee of Shahid Beheshti University of Medical Sciences “IR.SBMU.RETECH.REC.1396.85” and financially supported by the Infectious Diseases and Tropical Medicine Research Centre, Shahid Beheshti University of Medical Sciences, Tehran, Iran (grant No 9369).
Clinical samples and bacterial identification
A total of 95 clinical isolates of P. aeruginosa were obtained from patients hospitalized in Shahid Motahari Burn Care Centre, Tehran, Iran from March 2014 to March 2015. Wound swabs were collected from patients and immediately transported to the microbiology laboratory of the Department of Microbiology of Shahid Beheshti University of Medical Sciences, Tehran, Iran. Blood culture was also carried out in patients with a suspicion of sepsis. Strains were identified as P. aeruginosa using the standard biochemical tests, including OF, SIM and MR-VP tests, and pigment production on Mueller–Hinton agar (Merck, Darmstadt, Germany). Isolates identified as P. aeruginosa were preserved at –70°C in trypticase soy broth (Merck) supplemented with 20% glycerol until further processing [6].
Susceptibility testing
Susceptibility of the P. aeruginosa isolates to imipenem (10 μg), ceftazidime (30 μg), cefotaxime (30 μg), meropenem (10 μg), doripenem (10 μg), ticarcillin (75 μg), piperacillin (100 μg), piperacillin/tazobactam (100/10 μg), ciprofloxacin (5 μg), cefepime (30 μg), aztreonam (30 μg), amikacin (30 μg) and gentamicin (10 μg) were determined using a disc diffusion method according to the CLSI [7]. The MIC values of seven selected antibiotics—amikacin, gentamicin, meropenem, imipenem, ceftazidime, ciprofloxacin and colistin—were also determined using the microbroth dilution method at a final concentration for each antimicrobial agent from 0.5 mg/L to 256 mg/L [7]. Pseudomonas aeruginosa ATCC 27853 was used as the quality control strain.
Carbapenemase and metallo-β-lactamase determination
Phenotypic determination of metallo-β-lactamase (MBL) producers and carbapenemase producers was performed by the combined disc diffusion test [8] and Modified Hodge test, respectively [1].
Conventional PCR and sequencing analysis of resistance determinants
Responsible resistance genes encoding extended spectrum β-lactamases, MBLs, carbapenemases and aminoglycoside-modifying enzymes are listed in Table 1. DNA was extracted using the DNA extraction kit (GeNet Bio Company, Daejeon, Korea; Cat. No, K-3000) according to the manufacturer's guidelines. PCR was conducted in a final volume of 25 μL with 12.5 μL of 2× Master Mix (SinaClon, Tehran, Iran; Cat. no. PR901638), including 1× PCR buffer, 0.4 mmol/L dNTPs, 3 mmol/L MgCl2 and 0.08 IU Taq DNA polymerase, 1 μL of 10 pmol of each primer and 7.5 μL of sterile distilled water. Amplification reactions were performed on a thermal cycler (Eppendorf, Mastercycler Gradient; Eppendorf, Hamburg, Germany) followed by 36 cycles of denaturation at 94°C for 45 s, annealing at 50°C to 59°C, according to the primers for each gene, for 45 s and extension at 72°C for 45 s. PCR products were electrophoresed on a 1%–1.5% agarose gel, visualized by ethidium bromide staining and photographed under UV light. Amplicons representing each studied gene were confirmed by sequencing analysis (Macrogen Korea). The obtained sequence results were examined by the NCBI BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/). Insertion sequences were characterized by the online tool ISfinder (https://www-is.biotoul.fr/).
Table 1.
DNA sequences used in PCR and real-time RT-PCR
| Primer | Sequence (5′→3′) | Expected size of amplicon (bp) | Reference |
|---|---|---|---|
| oprD for PCR | ATGAAAGTGATGAAGTGGAG CAGGATCGACAGCGGATAGT |
1329 | [13] |
| oprD for RT-PCR | CGCCGACATCAGCAACACC GGGCCGTTGAAGTCGGAGTA |
195 | [14] |
| mexB | CAAGGGCGTCGGTGACTTCCAG ACCTGGGAACCGTCGGGATTGA |
297 | [15] |
| mexD | CTCGAGCTATACGTGCCTAAC GTCCCTCTTCCCATTTCACG |
79 | [16] |
| mexT | TGTCAGTGATCCTATGCCCC ACACGATCAGCAGGTTCAGA |
165 | This study |
| mexX | GGCTTGGTGGAAGACGTG GGCTGATGATCCAGTCGC |
64 | [9] |
| rpsL | GGCGTGCGTTACCACACCGT GGACGCTTGGCGCCGTACTT |
92 | [16] |
| TEM | TCGGGGAAATGTGCGCG TGCTTAATCAGTGAGGCACC |
972 | [17] |
| SHV | TTAGCGTTGCCAGTGCTC GGTTATGCGTTATATTCGCC |
861 | [17] |
| CTX-M | TTTGCGATGTGCAGTACCAGTAA CGCTATCGTTGGTGGTGCCATA |
544 | [17] |
| VEB | CGACTTCCATTTCCCGATGC GGACTCTGCAACAAATACGC |
643 | [18] |
| GES | TTGCAATGTGCTCAACGTTC TATCACAACCAATATTGTCG |
351 | This study |
| KPC | CGTCTAGTTCTGCTGTCTTG CTTGTCATCCTTGTTAGGCG |
798 | [19] |
| IMP | GAAGGCGTTTATGTTCATAC GTAAGTTTCAAGAGTGATGC |
587 | [19] |
| VIM | GATGGTGTTTGGTCGCATA CGAATGCGCAGCACCAG |
390 | [20] |
| NDM | GGTTTGGCGATCTGGTTTTC CGGAATGGCTCATCACGATC |
621 | [20] |
| GIM | TCGACACACCTTGGTCTGAA AACTTCCAACTTTGCCATGC |
477 | [20] |
| SPM | AAAATCTGGGTACGCAAACG ACATTATCCGCTGGAACAGG |
271 | [20] |
| rmtA | CTAGCGTCCATCCTTTCCTC TTTGCTTCCATGCCCTTGCC |
635 | [21] |
| aadA | ATGAGGGAAGCGGTGATCG TTATTTGCCGACTACCTTGGTG |
320 | [21] |
| aadB | ATGGACACAACGCAGGTCGC TTAGGCCGCATATCGCGACC |
120 | [21] |
| armA | TGGGAAGTTAAAGACGACGA CCATTCCCTTCTCCTTTCCA |
212 | [21] |
Real-time RT-PCR
The expression levels of oprD, mexB, mexD, mexT and mexX genes were assessed by RT-PCR. After an overnight culture of bacteria on Luria–Bertani broth [9], RNAs were extracted using the RNX-Plus kit (Cat. No., RN7713C; SinaClon) according to the manufacturer's instructions. The contaminating DNA was removed by DNase I (Fermentas, Waltham, MA, USA). The total RNA concentration was determined using the Nanodrop (DS-11 Spectrophotometer, USA). DNase-treated RNA was reverse-transcribed into cDNA using the Takara kit (Shiga, Japan). The primers used for real-time RT-PCR are shown in Table 1. Real-time RT-PCR assay was performed using the Power SYBR Green PCR Master Mix (Bioneer, Daejeon, Korea) on a Corbett Rotor-Gene 6000 Real-Time rotary analyser (Corbett Life Science, Sydney, Australia). Samples were run in triplicate and contained 2 μL of cDNA per reaction. Controls run without reverse transcriptase confirmed the absence of contaminating cDNA in any of the samples. The relative expression of the investigated genes was normalized against the rpsL housekeeping gene, and was calculated based on a 2-ΔΔCT method. Results were obtained as the relative expression of the mRNA compared with that of P. aeruginosa PAO1. MexAB-OprM, MexCD-OprJ, MexEF-OprN and MexXY-oprM efflux systems and OprD porin were considered as overexpressed when mexB, mexC, mexE, mexX and oprD transcriptional levels were at least 3-, 100-, 100-, 4- and 2-fold higher than those in P. aeruginosa PAO1 strain, respectively [10], [11], [12].
Outer membrane protein analysis
Bacterial outer membrane proteins were examined using previously described methods [9]. Following sonication, membranes were collected by ultracentrifugation at 100 000 g for 45 min. Outer membrane proteins were separated by SDS–PAGE, and gels were stained with Coomassie blue.
DNA fingerprinting analysis
Random amplified polymorphic DNA (RAPD)-PCR was performed as described previously [6]. The RAPD fingerprints were analysed and genotypes were assigned on the basis of weight and number of band differences.
Nucleotide sequence accession numbers
The DNA sequences of ISPpu21/oprD and ISPpu21 have been deposited in GenBank under accession numbers KT736319 and KT728193, respectively.
Results
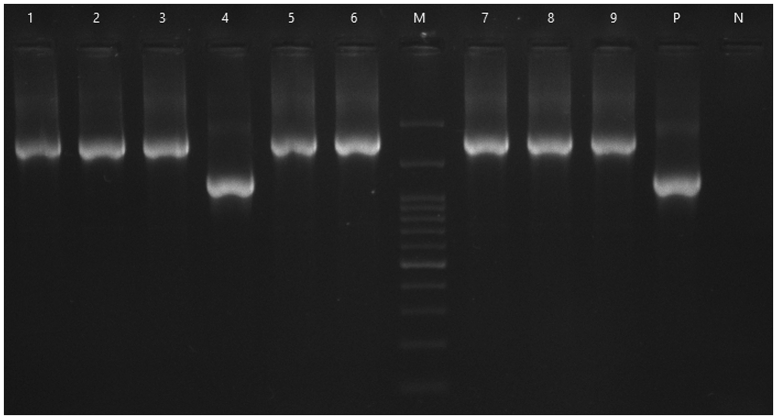
In this study, 95 P. aeruginosa isolates were identified as imipenem-resistant with MICs ranging from 4 to 256 mg/L. Antibiotic susceptibility tests in this study showed that P. aeruginosa isolates were resistant to all currently used antibiotics, including β-lactams, fluoroquinolones and aminoglycosides, but remained susceptible to colistin. In all, 10.52% (10/95) clinical isolates of P. aeruginosa harboured an insertion element in the oprD gene (Fig. 1).
Fig. 1.
PCR amplification of oprD gene from carbapenem-resistant Pseudomonas aeruginosa. M, DNA Ladder 100 bp; N, negative control; P, P. aeruginosa PAO1 strain; lane 4, oprD gene; lanes 2,3,5–9, ISPpu21/oprD combination.
Phenotypic and genotypic determination of carbapenemase and MBLs in 10.52% (10/95) of P. aeruginosa isolates revealed absence of any carbapenemase activity and production of MBL in isolates. In contrast, these isolates were positive for the ESBL gene (blaTEM). In this study, the rmtA, aadA, aadB and armA genes were positive in all ISPpu21-harbouring isolates. The relative expression levels of the mexX, mexB, mexT and mexD genes in ten isolates ranged from 0.1-fold to 1.4 fold, 1.1-fold to 3.68-fold, 0.3-fold to 8.22-fold and 1.7-fold to 35.17-fold, respectively and relative expression levels of the oprD in ten isolates ranged from 0.57-fold to 35.01-fold, which was higher than those in the control strain P. aeruginosa PAO1 (Table 2).
Table 2.
Antibiotic susceptibility, PCR results for genes and gene expression data for ISPpu21-carrying Pseudomonas aeruginosa isolates
| Isolate | MIC (μg/mL) |
ESBL genes | MBL genes | Aminoglycoside-Modifying Enzyme Genes | Relative expression levels |
Inserted into oprD | ISPpu21/oprD | RAPD-PCR | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CO | GEN | AK | IMP | MEM | CAZ | CIP | mexX | mexB | mexD | mexT | oprD | |||||||
| PA11 | ≤2 | 256 | 64 | 8 | 64 | 128 | 16 | blaTEM | − | aadA- aadB- rmtA-armA | 1.2 | 1.1 | 21.11 | ND | 12.22 | + | + | A |
| PA17 | ≤2 | 128 | 32 | 4 | 128 | 128 | 32 | blaTEM | − | aadA- aadB-rmtA | 2.1 | 1.2 | 35.17 | 0.97 | 10.25 | + | + | A |
| PA22 | ≤2 | 32 | 32 | 256 | 64 | 128 | 16 | blaTEM | − | aadA- aadB- rmtA-armA | 1.3 | 1.1 | 1.7 | 0.89 | 0.57 | + | + | A |
| PA31 | ≤2 | 32 | 32 | 256 | 128 | 128 | 128 | blaTEM | − | aadA- aadB-armA | 1.2 | 1.3 | 4.56 | 0.35 | 1.37 | + | + | A |
| PA42 | ≤2 | 256 | 256 | 256 | 128 | 64 | 32 | blaTEM | − | aadA- aadB- rmtA-armA | 1.2 | 1.1 | 19.2 | 8.22 | 30.36 | + | + | A |
| PA43 | ≤2 | 256 | 256 | 256 | 128 | 128 | 32 | blaTEM | − | aadA- aadB- rmtA-armA | 1.3 | 1.1 | 4.22 | 2.07 | 16.14 | + | + | A |
| PA46 | ≤2 | 256 | 256 | 256 | 8 | 64 | 4 | blaTEM | − | aadA- aadB- rmtA-armA | 0.18 | 1.2 | 15.24 | 2.31 | 25.56 | + | + | C |
| PA48 | ≤2 | 128 | 256 | 4 | 8 | 256 | 8 | blaTEM | − | aadA- aadB- rmtA-armA | 0.53 | 1.2 | 23.57 | 1.06 | 9.01 | + | + | A |
| PA49 | ≤2 | 32 | 256 | 4 | 8 | 16 | 16 | blaTEM | − | aadA- rmtA | 1.4 | ND | ND | ND | 4.07 | + | + | A |
| Psa6 | ≤2 | 256 | 256 | 4 | 4 | 4 | 16 | blaTEM | − | aadA- rmtA- | 1.10 | 3.68 | 2.21 | 2.36 | 35.01 | + | + | B |
Abbreviations: GEN, gentamicin; AK, amikacin; IMP, imipenem; MEM, meropenem; CAZ, cefotaxime; CIP, ciprofloxacin; ESBL, extended-spectrum β-lactamases; MBL, metallo-β-lactamases; RAPD-PCR, random amplified polymorphic DNA polymerase chain reaction.
–, Negative; +, Positive; ND, not determined.
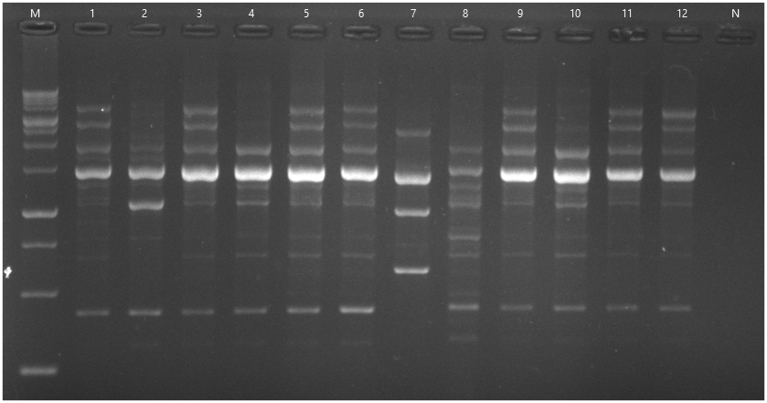
Outer membrane profiles obtained by SDS–PAGE indicated that except for one isolate, nine remaining isolates had a corresponding band for oprD in comparison to that of PAO1. Using RAPD-PCR genotyping, eight of the ten isolates containing ISPpu21 were shown to be clonally related (Fig. 2, Table 2).
Fig. 2.
Random amplified polymorphic DNA-PCR fingerprints of Pseudomonas aeruginosa isolates from burn patients using primer 272. M, DNA molecular weight marker (1 kb); N, negative control; 1–12, clinical samples.
Discussion
Carbapenems, such as imipenem and meropenem have been widely used as active antimicrobials to treat infections associated with P. aeruginosa. Producing carbapenemases has been reported as a major mechanism of carbapenem resistance in Iran [22]. Loss of OprD has also contributed to carbapenem resistance, especially imipenem resistance, in Iran [10]. The present study describes isolates of P. aeruginosa that exhibit reduced susceptibility to carbapenems other than carbapenemase production [1]. Antibiotic susceptibility tests in this study showed that P. aeruginosa isolates were resistant to all antibiotics, but remained susceptible to colistin. PCR assays targeting MBL and carbapenemase-encoding genes were also negative in all ten isolates. It seems that imipenem resistance in such bacteria is not mediated by carbapenemase enzymes. It has been shown that one of the most important mechanisms for imipenem resistance in P. aeruginosa is the absence of OprD production due to insertions, mutations and/or deletions in the oprD gene, as well as the down-regulation of oprD transcription [23]. In the present study, PCR assay using the oprD-specific primers demonstrated that 10.52% (10/95) of imipenem-resistant P. aeruginosa isolates harbored an IS element in the oprD gene. PCR of oprD gene using the corresponding primer is expected to obtain an amplicon with size 1323 bp or 1329 bp, while a fragment with a size about 2511 bp was produced. After purification and sequencing, it was found that the oprD gene was disrupted at nucleotide position 728 by the insertion of a 1179-bp sequence (GenBank accession no. KT736319) containing features of an ISPpu21 element. This insertional inactivation of the oprD gene mediated by ISPpu21 was found to be associated with ten of 95 isolates. These findings are in agreement with Fowler and Hanson, in which insertion of an IS element has resulted in a reduction of carbapenem susceptibility and also loss of OprD production [4]. Different IS elements have been identified worldwide that may inactivate the oprD gene and confer resistance to imipenem in clinical isolates of P. aeruginosa. To date, the presence of IS elements that disrupt the oprD gene has been reported in South Africa (ISPa26), Croatia (ISRP10) [1], Iran (ISPa1328) [2], Spain (ISPa133), China (ISPa1328, ISPre2), the USA (ISPa8, ISPa1635 and ISPa1328) and France (ISPa46 and ISPa1328) [1]. Mobile elements, such as IS can help Gram-negative bacteria to promote their survival and adaptation to altered environmental niches through the interruption of genes and genomic modifications. This type of adaptation has been well reported in clinical isolates of P. aeruginosa that are resistant to β-lactams [4]. ISPpu21, identified in the present study, is a 1179-bp insertion sequence with a couple of 6-bp inverted repeats (TCTGAA) at its extremes. Interestingly, ISPpu21 elements introduced at insertion site 1 and insertion site 2 generate a pair of 7-bp and a pair of 4-bp target site duplication direct repeats, respectively. Just a single open reading frame was discovered in ISPpu21, which encodes a protein with 326 amino acids. The ISPpu21 nucleotide sequence is 100% identical to a region of O antigen biosynthesis gene cluster (GenBank accession no. AF498406) of P. aeruginosa serotype O15-ATCC with the open reading frame in this ISPpu21 orthologue transcribed in the opposite direction from the P. aeruginosa chromosomal gene cluster [24]. The putative product of the open reading frame in ISPpu21 has a homology (identities from 61% to 93%) to transposase proteins belonging to the IS5 family of transposases [24]. A previous study has demonstrated that resistance to carbapenems, such as meropenem and doripenem in P. aeruginosa isolates that do not produce metallo-β-lactamase, could be developed through both down-regulation and loss of OprD in the outer membrane and up-regulation of MexAB-OprM in these strains [4]. In this study, the relative expression levels of the mexX, mexB, mexT and, mexD genes in the isolates ranged from 0.1-fold to 1.4-fold, 1.1-fold to 3.68-fold, 0.3-fold to 8.22-fold, and 1.7-fold to 35.17-fold, respectively, and the oprD expression level in ISPpu21-carrying isolates ranged from 0.57-fold to 35.01-fold, which was much higher than in the control strain P. aeruginosa PAO1 [4]. However, the susceptibility analysis results for nine isolates were contrary to others, so imipenem resistance could not be attributed to the increased expression of the oprD gene. These results were similar to those by Sun and Dennis, in which the relative expression levels of the oprD gene in IS-positive isolates were higher than the control strain P. aeruginosa ATCC 27853 [24]. In addition, we found one isolate that showed decreased expression of the oprD gene. These results were also similar to those of Wolter et al., who reported how an IS element with 97% similarity to ISPa16 inserted in the promoter of the oprD gene in two isolates, P. aeruginosa PA323 and P. aeruginosa PA415, reduced the oprD expression and increased imipenem MIC value [25]. Outer membrane profiles obtained by SDS–PAGE indicated that, with the exception of one isolate, the nine remaining isolates had a corresponding band for oprD in comparison to that of PAO1. Although these results signify the presence of a functional OprD in our clinical isolates, they strongly indicate that the OprD is absent or produced at very low levels. These results were similar to those of Shen et al., who reported a reduction in the size of OprD through the insertion of an IS element in the oprD gene of six isolates. In contrast, both imipenem-susceptible strains and the PAO1 control strain showed the expected protein band for OprD at 46 000 MW [26]. In the other study, Fournier et al., showed that an IS insertion in the oprD gene of one isolate may be associated with down-regulated OprD production [27]. RAPD-PCR was performed for typing all the isolates harbouring ISPpu21 in order to survey whether the IS-carrying strains collected in this study were associated with nosocomial infections. On observing DNA fingerprinting results, three patterns of these isolates were obtained. In the other word, eight of the ten isolates were clonally related. The results showed that the high prevalence of ISPpu21-carrying isolates was related to nosocomial infections.
Conclusion
Carbapenem resistance mediated by OprD deficiency in P. aeruginosa clinical isolates may be more frequent in Iranian hospitals, which could in turn be derived from the insertional inactivation. In this study, we report the emergence of ISPpu21 in carbapenem-resistant P. aeruginosa strains isolated from burn patients in Iran.
Transparency declaration
The authors declare that they have no competing interests.
Acknowledgements
The present study was financially supported by the Infectious Diseases and Tropical Medicine Research Centre, Shahid Beheshti University of Medical Sciences, Tehran, Iran (grant No 9369).
References
- 1.Al-Bayssari C., Valentini C., Gomez C., Reynaud-Gaubert M., Rolain J.-M. First detection of insertion sequence element ISPa1328 in the oprD porin gene of an imipenem-resistant Pseudomonas aeruginosa isolate from an idiopathic pulmonary fibrosis patient in Marseille, France. New Microb New Infect. 2015;7:26–27. doi: 10.1016/j.nmni.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadredinamin M., Hashemi A., Goudarzi H., Tarashi S., Nojookambari N.Y., Erfanimanesh S. Detection of ISPa1328 and ISPpu21, two novel insertion sequences in the OprD porin and bla IMP-1 gene among metallo-β-lactamase-producing Pseudomonas aeruginosa isolated from burn patients. Arch Trauma Res. 2016;6(1) [Google Scholar]
- 3.Lister P.D., Wolter D.J., Hanson N.D. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fowler R.C., Hanson N.D. Emergence of carbapenem resistance due to the novel insertion sequence IS Pa 8 in Pseudomonas aeruginosa. PloS One. 2014;9 doi: 10.1371/journal.pone.0091299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diene S.M., L’homme T., Bellulo S., Stremler N., Dubus J.-C., Mely L. ISPa46, a novel insertion sequence in the oprD porin gene of an imipenem-resistant Pseudomonas aeruginosa isolate from a cystic fibrosis patient in Marseille, France. Int J Antimicrob Agents. 2013;42:268–271. doi: 10.1016/j.ijantimicag.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Nanvazadeh F., Khosravi A.D., Zolfaghari M.R., Parhizgari N. Genotyping of Pseudomonas aeruginosa strains isolated from burn patients by RAPD-PCR. Burns. 2013;39:1409–1413. doi: 10.1016/j.burns.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute . CLSI; Wayne, PA: Jan 2015. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. CLSI document M100–S25. [Google Scholar]
- 8.Hakemi Vala M., Hallajzadeh M., Hashemi A., Goudarzi H., Tarhani M., Sattarzadeh Tabrizi M. Detection of Ambler class A, B and D ß-lactamases among Pseudomonas aeruginosa and Acinetobacter baumannii clinical isolates from burn patients. Ann Burns Fire Disasters. 2014;27:8–13. [PMC free article] [PubMed] [Google Scholar]
- 9.Quale J., Bratu S., Gupta J., Landman D. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother. 2006;50:1633–1641. doi: 10.1128/AAC.50.5.1633-1641.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campana E.H., Xavier D.E., Petrolini F.V.-B., Cordeiro-Moura J.R., de Araujo M.R.E., Gales A.C. Carbapenem-resistant and cephalosporin-susceptible: a worrisome phenotype among Pseudomonas aeruginosa clinical isolates in Brazil. Braz J Infect Dis. 2017;21(1):57–62. doi: 10.1016/j.bjid.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabot G., Ocampo-Sosa A.A., Tubau F., Macia M.D., Rodríguez C., Moya B. Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa isolates from bloodstream infections: prevalence and impact on resistance in a Spanish multicenter study. Antimicrob Agents Chemother. 2011;55:1906–1911. doi: 10.1128/AAC.01645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ocampo-Sosa A.A., Cabot G., Rodríguez C., Roman E., Tubau F., Macia M.D. Alterations of OprD in carbapenem-intermediate and-susceptible strains of Pseudomonas aeruginosa isolated from patients with bacteremia in a Spanish multicenter study. Antimicrob Agents Chemother. 2012;56:1703–1713. doi: 10.1128/AAC.05451-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pirnay J.-P., Bilocq F., Pot B., Cornelis P., Zizi M., Van Eldere J. Pseudomonas aeruginosa population structure revisited. PLoS One. 2009;4(11) doi: 10.1371/journal.pone.0007740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomás M., Doumith M., Warner M., Turton J.F., Beceiro A., Bou G. Efflux pumps, OprD porin, AmpC β-lactamase, and multiresistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob Agents Chemother. 2010;54:2219–2224. doi: 10.1128/AAC.00816-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh H., Stenhoff J., Jalal S., Wretlind B. Role of efflux pumps and mutations in genes for topoisomerases II and IV in fluoroquinolone-resistant Pseudomonas aeruginosa strains. Microb Drug Resist. 2003;9:323–328. doi: 10.1089/107662903322762743. [DOI] [PubMed] [Google Scholar]
- 16.Purssell A., Poole K. Functional characterization of the NfxB repressor of the mexCD–oprJ multidrug efflux operon of Pseudomonas aeruginosa. Microbiology. 2013;159:2058–2073. doi: 10.1099/mic.0.069286-0. [DOI] [PubMed] [Google Scholar]
- 17.Goudarzi H., Aghamohammad S., Hashemi A., Nikmanesh B., Noori M. Distribution of bla TEM, bla SHV and bla CTX-M genes among Escherichia coli isolates causing urinary tract infection in children. Arch Clin Infect Dis. 2013;8(3) [Google Scholar]
- 18.Shahcheraghi F., Nikbin V.-S., Feizabadi M.M. Prevalence of ESBLs genes among multidrug-resistant isolates of Pseudomonas aeruginosa isolated from patients in Tehran. Microb Drug Resist. 2009;15:37–39. doi: 10.1089/mdr.2009.0880. [DOI] [PubMed] [Google Scholar]
- 19.Gholipourmalekabadi M., Bandehpour M., Mozafari M., Hashemi A., Ghanbarian H., Sameni M. Decellularized human amniotic membrane: more is needed for an efficient dressing for protection of burns against antibiotic-resistant bacteria isolated from burn patients. Burns. 2015;41:1488–1497. doi: 10.1016/j.burns.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 20.Poirel L., Walsh T.R., Cuvillier V., Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Kashfi M., Hashemi A., Eslami G., Amin M.S., Tarashi S., Taki E. The prevalence of aminoglycoside-modifying enzyme genes amongst Pseudomonas aeruginosa strains isolated from burn patients. Arch Clin Infect Dis. 2017;12(1) [Google Scholar]
- 22.Roodsari M.R., Fallah F., Taherpour A., Vala M.H., Hashemi A. Carbapenem-resistant bacteria and laboratory detection methods. Arch Pediatr Infect Dis. 2014;2:188–191. [Google Scholar]
- 23.Sanbongi Y., Shimizu A., Suzuki T., Nagaso H., Ida T., Maebashi K. Classification of OprD sequence and correlation with antimicrobial activity of carbapenem agents in Pseudomonas aeruginosa clinical isolates collected in Japan. Microbiol Immunol. 2009;53:361–367. doi: 10.1111/j.1348-0421.2009.00137.x. [DOI] [PubMed] [Google Scholar]
- 24.Sun X., Dennis J.J. A novel insertion sequence derepresses efflux pump expression and preadapts Pseudomonas putida S12 for extreme solvent stress. J Bacteriol. 2009;191:6773–6777. doi: 10.1128/JB.00832-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolter D.J., Acquazzino D., Goering R.V., Sammut P., Khalaf N., Hanson N.D. Emergence of carbapenem resistance in Pseudomonas aeruginosa isolates from a patient with cystic fibrosis in the absence of carbapenem therapy. Clin Infect Dis. 2008;46:e137–e141. doi: 10.1086/588484. [DOI] [PubMed] [Google Scholar]
- 26.Shen J., Pan Y., Fang Y. Role of the outer membrane protein OprD 2 in carbapenem-resistance mechanisms of Pseudomonas aeruginosa. PloS One. 2015;10(10) doi: 10.1371/journal.pone.0139995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fournier D., Richardot C., Müller E., Robert-Nicoud M., Llanes C., Plésiat P. Complexity of resistance mechanisms to imipenem in intensive care unit strains of Pseudomonas aeruginosa. J Antimicrob Chemother. 2013;68:1772–1780. doi: 10.1093/jac/dkt098. [DOI] [PubMed] [Google Scholar]