Abstract
Carbapenem resistant Klebsiella pneumoniae (CR-KP) infections are an ever-increasing global issue, especially in the Indian subcontinent. Here we report genetic insight into a blaOXA-181 harbouring Klebsiella pneumoniae, belonging to the pandemic lineage ST147, that represents an intermediate stage towards pan-drug resistance. The CR-KP isolate DA48896 was isolated from a patient from Pakistan and was susceptible only to tigecycline and colistin. It harboured blaOXA-181 and was assigned to sequence type ST147. Analysis from whole genome sequencing revealed a very high sequence similarity to the previously sequenced pan-resistant K. pneumoniae isolate MS6671 from the United Arab Emirates. The two isolates are very closely related with only 46 chromosomal nucleotide differences, 14 indels and differences in plasmid content. Both carry a substantial number of plasmid-borne and chromosomally encoded resistance determinants. Interestingly, the two differences in susceptibility between the isolates could be attributed to DA48896 lacking an insertion of blaOXA-181 into the mgrB gene that results in colistin resistance in MS6671 and SNPs affecting AcrAB efflux pump expression likely to result in tigecycline resistance. These differences between the otherwise very similar isolates indicate that strong selection has occurred for resistance towards these last-resort drugs and illustrates the trajectory of resistance evolution of OXA-181-producing versions of the ST147 international risk clone.
Introduction
Over the last two decades carbapenems have emerged as last line of effective therapy for treating the worldwide disseminated extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. Various studies from Pakistan have reported prevalence of ESBL-producing Klebsiella pneumoniae ranging from 26% to 59% during 2002 to 2013 [1–4]. Hence there is a significantly increasing dependence upon carbapenems for treating such MDR K. pneumoniae. Although carbapenemases in Enterobacteriaceae were relatively rare a decade ago, their emergence and rapid dissemination is now raising concern in health communities all over the globe [5]. Until 1990’s, carbapenemases were considered species-specific and a problem of clonal spread, but in the past decade the interspecies dissemination of carbapenemases has made it a global issue [6]. Hence it is of great significance to understand the genetic context of carbapenemases and how carbapenemase-producing strains further evolve resistance to the last remaining active antibiotics such as colistin. Carbapenemases including VIM, IMP, OXA-48, KPC and NDM-1 have been reported from Pakistan recently [7–9]. However, the knowledge about the variants of these acquired carbapenemases and their genetic contexts still remain scarce.
OXA-181, differing by 4 amino acid changes from OXA-48 while sharing the same hydrolytic properties against carbapenems, has been identified in enterobacterial isolates from India and from patients with a link with the Indian subcontinent [10, 11]. Since the identification of OXA-181 in India in 2007, OXA-181-producing Enterobacteriaceae have been reported from several other countries i.e. Bangladesh, Sri Lanka, Nepal, Oman, South Africa, Canada, USA, Australia, France and the United Kingdom [12–21]. Certain K. pneumoniae sequence types are particularly important in the spread of carbapenemases. Recently, K. pneumoniae ST147 has been associated with the carriage of OXA-181 in isolates from UAE, Abu Dhabi and USA [22–24]. Here we report and genetically characterize the first carbapenem-resistant ST147 K. pneumoniae clinical isolate harbouring blaOXA-181 from Pakistan.
Materials and methods
Bacterial isolation and identification
Strain DA48896 was received at the microbiology laboratory of the tertiary hospital Pakistan Institute of Medical Sciences, Islamabad as part of a larger study of carbapenemase producing K. pneumoniae at the hospital. It was identified as Klebsiella pneumoniae using Gram stain and standard biochemical tests including Oxidase test, SIM (Sulfur, Indole, Motility) test, TSI (Tripple sugar iron) test and Simmon citrate agar test.
Antibiotic susceptibility profiling
The Etest method (Biomerieux, France) was used to determine MIC against routinely used antibiotics according to the manufacturer’s instructions. For colistin the broth dilution method was used according to the protocol of CLSI. The susceptibility results were interpreted according to EUCAST (the European Committee on Antimicrobial Susceptibility Testing) guidelines (www.eucast.org).
Detection of carbapenemase encoding genes
252 carbapenem resistant K. pneumoniae isolated during April- September 2015 were screened for the presence of acquired carbapenemase encoding genes including blaIMP, blaVIM, blaNDM, blaSPM, blaAIM, blaDIM, blaGIM, blaSIM, blaKPC, blaBIC, blaOXA-48 according to a protocol described previously [25]. Reaction I included primers for blaIMP, blaVIM and blaSPM. Reaction II included primers for blaNDM, blaKPC and blaBIC. Reaction III included primers for blaAIM, blaDIM, blaGIM and blaSIM. The PCR program consisted of following conditions in T3000 Thermocycler 48 (Biomerta, Germany), initial denaturation at 94°C for 10min, followed by 36 cycles of DNA denaturation at 94°C for 30sec, primer annealing at 52°C for 40sec and primer extension at 72°C for 50sec. The final extension was performed at 72°C for 5min.
Whole-genome sequencing and bioinformatics
Genomic DNA for whole genome sequencing was prepared using MasterPure™ DNA Purification Kit (Epicentre Technologies, Madison, Wisconsin). Pacific Biosciences sequencing was performed at the Science for Life laboratories sequencing platform at Uppsala University using Pacific Biosciences II technology and Illumina MiSeq was performed in-house using the NexteraXT technology. CLC Genomic Workbench v 10 (CLC Bio/Qiagen) with Microbial Genomics Module and Microbial Genome Finishing Tools was used for de novo assembly of reads and of reference assembly and variance analysis to K. pneumoniae LN824133. The sequence data including raw sequence reads and assembled contigs of the chromosome and plasmids have been deposited at NCBI with accession numbers CP024429-CP024436 under BioProject PRJNA348457.
The contigs were submitted to the ResFinder and PlasmidFinder databases at the Centre of Genomic Epidemiology (www.genomicepidemiology.org) to identify antibiotic resistance genes and plasmid replicons [26–28]. MLST was performed by submitting contigs at Institut Pasteur MLST Databases (http://www.pasteur.fr/mlst/).
Results
In June 2015, a carbapenem resistant K. pneumoniae was isolated from the tracheal secretion and an endotracheal tube of a 32-year-old female patient at the intensive care unit at the tertiary hospital, Pakistan Institute of Medical sciences, Islamabad. Isolate DA48896 was found to be multi-drug resistant and sensitive only to Tigecycline and Colistin (Table 1). The isolate carried a blaOXA-48-like carbapenemase encoding gene 100% identical to blaOXA-181. This was the only OXA-181 producing isolate identified out of 252 screened K. pneumoniae at the hospital during the study period.
Table 1. MICs against routinely used antibiotics.
| Antimicrobial Class | Antimicrobial agent | DA48896 MIC (mg/L) |
EUCAST interpretation | MS6671 MIC (mg/L) |
|---|---|---|---|---|
| Carbapenems | Meropenem | 8 | R | 8 |
| Ertapenem | >32 | R | >32 | |
| Doripenem | 6 | R | 4 | |
| Imipenem | 24 | R | 4 | |
| Penicillin | Ampicillin | >256 | R | >256 |
| Cephalosporins | Cefuroxime | >256 | R | >256 |
| Cefaclor | >256 | R | - | |
| Cefpriome | >256 | R | - | |
| Cefepime | >256 | R | 32 | |
| Ceftazidime | >256 | R | - | |
| Penicillin and beta-lactamase inhibitor | Amoxycillin/clavulanate | >256 | R | >256 |
| Piperacillin/Tazobactam | >32 | R | >256 | |
| Fuoroquinolones | Ciprofloxacin | 4 | R | >32 |
| Levofloxacin | 4 | R | - | |
| Tetracyclines | Tetracycline | >256 | R | 32 |
| Doxycycline | 16 | R | 32 | |
| Tigecycline | 0.25 | S | 4 | |
| Folate-pathway inhibitors | Trimethoprim/Sulfamethoxazole | >256 | R | 8 |
| Aminoglycosides | Gentamicin | >256 | R | >256 |
| Netilmicin | >256 | R | >256 | |
| Phosphonic acids | Fosfomycin | >1024 | R | 64 |
| Polymyxins | Colistin | 0.125 | S | 128 |
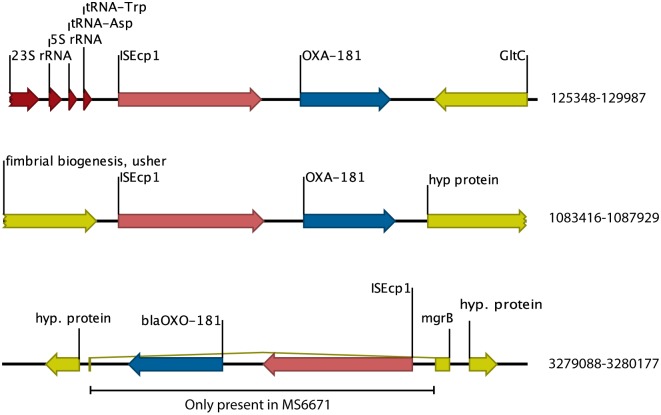
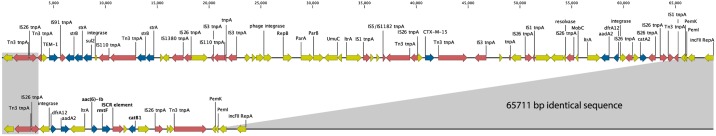
Whole genome sequencing was performed with both Pacific Biosciences and Illumina MiSeq techniques to generate a complete sequence of the genome and associated plasmids. The DA48896 strain was assigned to sequence type 147 and capsular serotype K64 and the chromosomal sequence had a very high sequence similarity to the previously sequenced K. pneumoniae isolate MS6671 (accession no. LN824133). MS6671 was isolated from a patient in the United Arab Emirates and resistant to all tested antibiotics including tigecycline and colistin, to which DA48896 was sensitive [22]. The chromosomes of the two strains were very closely related with only 46 individual nucleotide differences (S1 Table), 16 of which were clustered frame shifts restoring putatively frame-shifted reading frames compared to MS6671. These 16 differences occurred within a stretch of 2500 bp indicating either a recombinational event or previous sequence problems in this region. Furthermore, the chromosomes differed by 14 structural variations (insertions/deletions) the majority found in mobile genetic elements and intergenic regions (S2 Table). This high degree of sequence identity indicates a very recent common origin of the two isolates. The blaOXA-181 was found downstream of ISEcp1 as reported previously. DA48896 has two copies of blaOXA-181 present on the chromosome at positions 127900 and 1086004, compared to 3 copies in MS6671 (Fig 1). Interestingly, the IS insertion disrupting the mgrB gene in MS6671 was missing in DA48896 explaining the difference in colistin susceptibility between the two strains. One copy of blaCTX-M-15 coupled to ISEcp1 was present on the chromosome in the identical place as in MS6671 and a second copy of blaCTX-M-15 was carried on p48896_1, which also carried blaTEM-1B, sul1, strA and strB in a >70 kbp insertion compared to the otherwise very similar MS6671 plasmid E (LN824138) (Fig 2 and Table 2). This region was recently shown to vary extensively among other closely related OXA-181 producing ST147 isolates (see Discussion below) [23, 24]. DA48896 carried variants of 3 of the 5 plasmids of K. pneumoniae MS6671 (missing MS6671 plasmids A (LN824134) and C (LN824136)) but in addition three plasmids not found in MS6671 (Table 2). None of these extra plasmids contained any resistance genes. However, some plasmids contained larger replacements or rearrangements that affected the presence or locations of resistance genes (S3 Table).
Fig 1. Comparison of blaOXA-181 insertions between DA48896 and MS6671.
The OXA-181 reading frame is depicted in blue, the associated ISEcp1 is depicted in red, surrounding proteins coding genes are depicted in yellow and tRNA and rRNA genes in dark red. Positions indicated to the right are from the DA48896 genome.
Fig 2. Comparison of resistance cassettes between pDA48896_1 and pMS6671_E.
Resistance genes are depicted in blue, mobile genetic elements are depicted in red, surrounding proteins coding genes are depicted in yellow. The shaded areas depict identical sequences flanking the variable region. The plasmids contain a backbone of 69 kbp of highly similar sequence only shown for pMS6671 and not drawn to scale.
Table 2. Plasmids found in DA48896.
| Name | Size (bp) | Resistance genes | Plasmid replicons | Best match in MS6671 | Best match in Genbank |
|---|---|---|---|---|---|
| p48896_1 (CP024430) |
131,243 | aadA2, strAB, blaCTX-M-15, blaTEM-1, sul2, dfrA12, catA2 | IncFII, IncR | LN824138 (62% coverage/99% identity) | KT725788 (63% coverage/99% identity) |
| p48896_2 (CP024431) |
114,815 | rmtf, aacA4, aac(6’)Ib-cr | IncFII | LN824135 (87% coverage/99% identity) | CP021758 (100% coverage/99% identity) |
| p48896_3 (CP024432) |
45,291 | ND | ND | ND | CP011864 (91% coverage/98% identity) |
| p48896_4 (CP024433) |
55,118 | ND | ND | LN824139 (88% coverage/99% identity) | CP017988 (94% coverage/99% identity) |
| p48896_5 (CP024434) |
4,644 | ND | ND | LN824137 (100% coverage/100% identity) | LN824137 (100% coverage/100% identity) |
| p48896_6 (CP024435) |
4,167 | ND | ND | ND | EU932690 (100% coverage/100% identity) |
| p48896_7 (CP024436) |
2,054 | ND | ColpVC | ND | KU302803 (99% coverage/95% identity) |
ND: Not detected.
Like MS6671, DA48896 was multi-resistant and carried an extensive number of resistance markers: blaOXA-181, blaCTX-M-15, blaTEM-1, blaSHV-11, rmtf, aadA2, strAB, aacA4, oqxAB, aac(6’)Ib-cr, fosA, ARR-2, catA2, sul2 and dfrA12. DA48896 and MS6671 also differed by four SNPs in chromosomal genes previously known to affect resistance through efflux pumps, AcrR R18L, MarA V26D, S50G and RcsC L60M. BLASTp analysis showed that the mutation in AcrR (repressor of the AcrAB-TolC efflux pump) is unique to MS6671 as is the V26D mutation in MarA (multiple antibiotic resistance protein) while the MarA S50G and the RcsC (two-component sensor histidine kinase) L60M mutations were unique to DA48896. Examination of porin genes showed that both isolates carry an IS insertion associated with blaCTX-M-15 in the ompk35 porin gene and an insertion of two additional amino acids in Ompk36. Also, a single non-synonymous mutation (Arg3Ser) was found in RamR, a transcriptional repressor of ramA. Inactivation of RamR has previously been shown to downregulate porin-expression and increase efflux resulting in antibiotic resistance.
Discussion
DA48896 is a MDR K. pneumoniae belonging to the internationally spread clone ST147, and is very closely related to the pan-drug resistant K. pneumoniae MS6671 isolated in the UAE in 2014 [22]. Although they differed in several respects with regard to plasmid content, both isolates had almost the same antibiotic susceptibility profile except that DA48896 was sensitive to tigecycline and colistin. Colistin susceptibility could be explained by the absence of the IS insertion disrupting the mgrB gene in MS6671 [22]. It is intriguing that addition of more gene copies of the OXA-181 gene by replicative transposition, that likely is an adaptation to increased resistance to carbapenems, also has led to resistance to colistin, the last resort antibiotic for carbapenemases-producing multi-resistant K. pneumoniae. Furthermore, inactivation of RamR, a negative regulator of the ramA gene, leads to increased expression of acrAB which has been linked to tigecycline resistance [29]. MS6671 contain a non-synonymous mutation (Arg3Ser) in ramR, which was previously suggested to have caused tigecycline resistance in MS6671 [22]. However, DA48896 has the same non-synonymous mutation (Arg3Ser) in RamR but is susceptible to tigecycline. Instead, MS6671 contains a unique R187L mutation in AcrR, the repressor of acrAB, which could potentially give increased efflux through this pathway if it impedes the AcrR function. The otherwise very close relatedness between isolates DA48896 and MS6671 with very few SNPs suggests that strong selection for increased resistance to both these last resort antibiotics has occurred for MS6671 and indicates the direction of further resistance evolution in this internationally recognized pathogenic lineage. The genetic makeup and resistance profiles of the isolates indicate that the isolate found from Pakistan is an intermediate stage towards pan-resistance evolution of this particular clone. Mutations in ompK36 present in both the isolates lead to generation of a previously described variant ompk36v with reduced influx of ertapenem [30]. OmpK35 was also inactivated by IS insertion. The combination of multiple copies of blaOXA-181 and reduced permeability of both general porins may explain the high MIC of Ertapenem in these isolates.
Capsular serotype K64 has been reported as a major serotype associated with carbapenem resistance within the internationally widespread clone ST147 that is linked with a multitude of different virulence factors [31]. Presence of the Yersinia high-pathogenicity island and ABC transporter kfu in MS6671 and DA48896 makes these isolates likely to be hypervirulent [32]. The ST147 K. pneumoniae has previously been linked to the spread of ESBLs (especially CTX-M-15), VIM, OXA-48, KPC and recently also to NDM-1 in different clinical settings. The emergence of OXA-181 among this globally spread high-risk clone is worrying, leaving few therapeutic options available and contributing to a great impact in infection control measures [33, 34]. We screened 252 K. pneumoniae isolates from the same hospital over a period of 6 months and no further isolates with same blaOXA-181 genotype were recovered. In case of MS6671, no further isolates with the same genotype were found in the index patient’s hospital for 6 months [22]. Recently, closely related OXA-181 producing ST147 pan-drug resistant isolates have been described from 3 patients at a hospital in Abu Dhabi and a patient from USA [23, 24]. Complete whole genome sequences were not generated from these isolates but WGS plasmid data showed that they carried an IncFII plasmid with >66 kbp of identical backbone sequence as pMSS6671_E and pDA48896_1. However, the resistance regions of these plasmids also included blaNDM-5 and further varied extensively between the isolates illustrating the highly dynamic nature of resistance plasmids in this lineage. Finding such closely related ST147 isolates from multiple different geographical locations is of particular concern. OXA-181 is believed to have originated from the Indian sub-continent and bacteria harbouring this gene have been reported frequently from this region [10, 13, 15].
Conclusions
In conclusion, to the best of our knowledge, we report the first OXA-181 producing ST147 K. pneumoniae from Pakistan. Its close relatedness to MS6671 from UAE gives an insight into the role of ST147 in OXA-181 dissemination, the rapid evolution of resistance towards the two last resort antibiotics, colistin and tigecycline, and global transfer of this international risk clone.
Supporting information
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
The authors would like to acknowledge support of the National Genomics Infrastructure (NGI) / Uppsala Genome Center and UPPMAX for providing assistance in massive parallel sequencing and computational infrastructure.
Data Availability
The sequence data including raw sequence reads and assembled contigs of the chromosome and plasmids have been deposited at NCBI with accession numbers CP024429-CP024436 under BioProject PRJNA348457.
Funding Statement
This work was supported by the Swedish Research Council—Medicine and Health (Grant no K2013-99X-22208-01-5) to LS and by Higher Education Commission (Pakistan) under the Indigenous PhD Fellowship Program (5000 Fellowships, 112-25656-2Bm1-034) and the International Research Support Initiative Program (29BMS46) to FN. Work performed at NGI / Uppsala Genome Center has been funded by RFI / VR and Science for Life Laboratory, Sweden. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Khan E, Ejaz M, Zafar A, Jabeen K, Shakoor S, Inayat R, et al. Increased isolation of ESBL producing Klebsiella pneumoniae with emergence of carbapenem resistant isolates in Pakistan: report from a tertiary care hospital. JPMA The Journal of the Pakistan Medical Association. 2010;60(3):186–90. . [PubMed] [Google Scholar]
- 2.Ullah F, Malik SA, Ahmed J. Antimicrobial susceptibility and ESBL prevalence in Pseudomonas aeruginosa isolated from burn patients in the North West of Pakistan. Burns: journal of the International Society for Burn Injuries. 2009;35(7):1020–5. doi: 10.1016/j.burns.2009.01.005 . [DOI] [PubMed] [Google Scholar]
- 3.Riaz S, Faisal M, Hasnain S. Prevalence and comparison of Beta-lactamase producing Escherichia coli and Klebsiella spp from clinical and environmental sources in Lahore, Pakistan. African Journal of Microbiology Research 2012;6(2):6. [Google Scholar]
- 4.Ejaz H, Ul-Haq I, Mahmood S, Zafar A, Mohsin Javed M. Detection of extended-spectrum beta-lactamases in Klebsiella pneumoniae: comparison of phenotypic characterization methods. Pakistan journal of medical sciences. 2013;29(3):768–72. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canton R, Akova M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2012;18(5):413–31. doi: 10.1111/j.1469-0691.2012.03821.x . [DOI] [PubMed] [Google Scholar]
- 6.Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clinical microbiology reviews. 2007;20(3):440–58, table of contents. doi: 10.1128/CMR.00001-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nahid F, Khan AA, Rehman S, Zahra R. Prevalence of metallo-beta-lactamase NDM-1-producing multi-drug resistant bacteria at two Pakistani hospitals and implications for public health. Journal of infection and public health. 2013;6(6):487–93. doi: 10.1016/j.jiph.2013.06.006 . [DOI] [PubMed] [Google Scholar]
- 8.Hasan B, Perveen K, Olsen B, Zahra R. Emergence of carbapenem-resistant Acinetobacter baumannii in hospitals in Pakistan. Journal of medical microbiology. 2014;63(Pt 1):50–5. doi: 10.1099/jmm.0.063925-0 . [DOI] [PubMed] [Google Scholar]
- 9.Sattar H, Toleman M, Nahid F, Zahra R. Co-existence of blaNDM-1 and blaKPC-2 in clinical isolates of Klebsiella pneumoniae from Pakistan. Journal of chemotherapy. 2016;28(4):346–9. doi: 10.1179/1973947814Y.0000000223 . [DOI] [PubMed] [Google Scholar]
- 10.Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrobial agents and chemotherapy. 2011;55(3):1274–8. doi: 10.1128/AAC.01497-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalpoe JS, Al Naiemi N, Poirel L, Nordmann P. Detection of an Ambler class D OXA-48-type beta-lactamase in a Klebsiella pneumoniae strain in The Netherlands. Journal of medical microbiology. 2011;60(Pt 5):677–8. doi: 10.1099/jmm.0.028308-0 . [DOI] [PubMed] [Google Scholar]
- 12.Dimou V, Dhanji H, Pike R, Livermore DM, Woodford N. Characterization of Enterobacteriaceae producing OXA-48-like carbapenemases in the UK. The Journal of antimicrobial chemotherapy. 2012;67(7):1660–5. doi: 10.1093/jac/dks124 . [DOI] [PubMed] [Google Scholar]
- 13.Balm MN, La MV, Krishnan P, Jureen R, Lin RT, Teo JW. Emergence of Klebsiella pneumoniae co-producing NDM-type and OXA-181 carbapenemases. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2013;19(9):E421–3. doi: 10.1111/1469-0691.12247 . [DOI] [PubMed] [Google Scholar]
- 14.Hall JM, Corea E, Sanjeewani HD, Inglis TJ. Molecular mechanisms of beta-lactam resistance in carbapenemase-producing Klebsiella pneumoniae from Sri Lanka. Journal of medical microbiology. 2014;63(Pt 8):1087–92. doi: 10.1099/jmm.0.076760-0 . [DOI] [PubMed] [Google Scholar]
- 15.Williamson DA, Heffernan H, Sidjabat H, Roberts SA, Paterson DL, Smith M, et al. Intercontinental transfer of OXA-181-producing Klebsiella pneumoniae into New Zealand. The Journal of antimicrobial chemotherapy. 2011;66(12):2888–90. doi: 10.1093/jac/dkr396 . [DOI] [PubMed] [Google Scholar]
- 16.Potron A, Nordmann P, Lafeuille E, Al Maskari Z, Al Rashdi F, Poirel L. Characterization of OXA-181, a carbapenem-hydrolyzing class D beta-lactamase from Klebsiella pneumoniae. Antimicrobial agents and chemotherapy. 2011;55(10):4896–9. doi: 10.1128/AAC.00481-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brink AJ, Coetzee J, Corcoran C, Clay CG, Hari-Makkan D, Jacobson RK, et al. Emergence of OXA-48 and OXA-181 carbapenemases among Enterobacteriaceae in South Africa and evidence of in vivo selection of colistin resistance as a consequence of selective decontamination of the gastrointestinal tract. Journal of clinical microbiology. 2013;51(1):369–72. doi: 10.1128/JCM.02234-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mataseje LF, Boyd DA, Hoang L, Imperial M, Lefebvre B, Miller M, et al. Carbapenem-hydrolyzing oxacillinase-48 and oxacillinase-181 in Canada, 2011. Emerging infectious diseases. 2013;19(1):157–60. doi: 10.3201/eid1901.120706 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathers AJ, Hazen KC, Carroll J, Yeh AJ, Cox HL, Bonomo RA, et al. First clinical cases of OXA-48-producing carbapenem-resistant Klebsiella pneumoniae in the United States: the "menace" arrives in the new world. Journal of clinical microbiology. 2013;51(2):680–3. doi: 10.1128/JCM.02580-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sidjabat HE, Kennedy K, Silvey A, Collignon P, Paterson DL. Emergence of bla(OXA-181)-carrying ColE plasmid in Klebsiella pneumoniae in Australia. International journal of antimicrobial agents. 2013;41(3):294–6. doi: 10.1016/j.ijantimicag.2012.11.003 . [DOI] [PubMed] [Google Scholar]
- 21.Ruppe E, Armand-Lefevre L, Estellat C, El-Mniai A, Boussadia Y, Consigny PH, et al. Acquisition of carbapenemase-producing Enterobacteriaceae by healthy travellers to India, France, February 2012 to March 2013. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2014;19(14). . [DOI] [PubMed] [Google Scholar]
- 22.Zowawi HM, Forde BM, Alfaresi M, Alzarouni A, Farahat Y, Chong TM, et al. Stepwise evolution of pandrug-resistance in Klebsiella pneumoniae. Scientific reports. 2015;5:15082 doi: 10.1038/srep15082 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonnevend A, Ghazawi A, Hashmey R, Haidermota A, Girgis S, Alfaresi M, et al. Multihospital Occurrence of Pan-Resistant Klebsiella pneumoniae Sequence Type 147 with an ISEcp1-Directed blaOXA-181 Insertion in the mgrB Gene in the United Arab Emirates. Antimicrobial agents and chemotherapy. 2017;61(7). doi: 10.1128/AAC.00418-17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rojas LJ, Hujer AM, Rudin SD, Wright MS, Domitrovic TN, Marshall SH, et al. NDM-5 and OXA-181 Beta-Lactamases, a Significant Threat Continues To Spread in the Americas. Antimicrobial agents and chemotherapy. 2017;61(7). doi: 10.1128/AAC.00454-17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagnostic microbiology and infectious disease. 2011;70(1):119–23. doi: 10.1016/j.diagmicrobio.2010.12.002 . [DOI] [PubMed] [Google Scholar]
- 26.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. The Journal of antimicrobial chemotherapy. 2012;67(11):2640–4. doi: 10.1093/jac/dks261 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrobial agents and chemotherapy. 2014;58(7):3895–903. doi: 10.1128/AAC.02412-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cosentino S, Voldby Larsen M, Moller Aarestrup F, Lund O. PathogenFinder—distinguishing friend from foe using bacterial whole genome sequence data. PloS one. 2013;8(10):e77302 doi: 10.1371/journal.pone.0077302 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruzin A, Immermann FW, Bradford PA. Real-time PCR and statistical analyses of acrAB and ramA expression in clinical isolates of Klebsiella pneumoniae. Antimicrobial agents and chemotherapy. 2008;52(9):3430–2. doi: 10.1128/AAC.00591-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Fernandez A, Miriagou V, Papagiannitsis CC, Giordano A, Venditti M, Mancini C, et al. An ertapenem-resistant extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae clone carries a novel OmpK36 porin variant. Antimicrobial agents and chemotherapy. 2010;54(10):4178–84. doi: 10.1128/AAC.01301-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan YJ, Lin TL, Lin YT, Su PA, Chen CT, Hsieh PF, et al. Identification of capsular types in carbapenem-resistant Klebsiella pneumoniae strains by wzc sequencing and implications for capsule depolymerase treatment. Antimicrobial agents and chemotherapy. 2015;59(2):1038–47. doi: 10.1128/AAC.03560-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–18. doi: 10.4161/viru.22718 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuelsen O, Toleman MA, Hasseltvedt V, Fuursted K, Leegaard TM, Walsh TR, et al. Molecular characterization of VIM-producing Klebsiella pneumoniae from Scandinavia reveals genetic relatedness with international clonal complexes encoding transferable multidrug resistance. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2011;17(12):1811–6. doi: 10.1111/j.1469-0691.2011.03532.x . [DOI] [PubMed] [Google Scholar]
- 34.Damjanova I, Toth A, Paszti J, Hajbel-Vekony G, Jakab M, Berta J, et al. Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type beta-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005—the new ‘MRSAs’? The Journal of antimicrobial chemotherapy. 2008;62(5):978–85. doi: 10.1093/jac/dkn287 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The sequence data including raw sequence reads and assembled contigs of the chromosome and plasmids have been deposited at NCBI with accession numbers CP024429-CP024436 under BioProject PRJNA348457.