Abstract
Pseudomonas aeruginosa is a ubiquitous, Gram-negative opportunistic pathogen that can cause disease in various sites within the human body. This bacterium is a major source of nosocomial infections that are often difficult to treat due to high intrinsic antibiotic resistance and coordinated virulence factor production. P. aeruginosa utilizes three cell-to-cell signaling systems to regulate numerous genes in response to cell density. One of these systems utilizes the small molecule 2-heptyl-3-hydroxy-4-quinolone (Pseudomonas quinolone signal [PQS]) as a signal that acts as a co-inducer for the transcriptional regulator PqsR. Quinolone signaling is required for virulence in multiple infection models, and PQS is produced during human infections, making this system an attractive target for potential drug development. In this study we have examined the role of a TetR-type transcriptional regulator, PsrA, in the regulation of PQS production by P. aeruginosa. Previous studies showed that PsrA regulates genes of the fatty acid β-oxidation pathway, including PA0506, which encodes a FadE homolog. In this report, we show that deletion of psrA resulted in a large decrease in PQS production and that co-deletion of PA0506 allowed PQS production to be restored to a wild type level. We also found that PQS production could be restored to the psrA mutant by the addition of oleic or octanoic acid. Taken together, our data suggest that psrA positively affects PQS production by repressing the transcription of PA0506, which leads to a decrease in the conversion of acyl-CoA compounds to enoyl-CoA compounds, thereby allowing some octanoyl-CoA to escape the ß-oxidation pathway and serve as a PQS precursor.
Introduction
Pseudomonas aeruginosa is a Gram-negative, ubiquitous bacterium and an important opportunistic human pathogen that causes chronic infections in both immunocompromised and cystic fibrosis (CF) patients [1, 2]. These infections have high morbidity and mortality rates due to intrinsic antibiotic resistance and the well-coordinated expression of a wide array of virulence factors [2, 3]. P. aeruginosa is very adaptable to diverse environments and is capable of surviving on many different carbon sources while generating a multitude of secondary metabolites [4], which in some cases affect virulence [5, 6]. This opportunistic pathogen also utilizes cell-to-cell signaling systems to regulate numerous genes, many of which are important for virulence [7, 8]. The LasRI and RhlRI quorum sensing systems respond to the acyl-homoserine lactone signals N-(3-oxododecanoyl)-L-homoserine lactone and N-butyryl-L-homoserine lactone, respectively [9, 10], while the quinolone signaling system functions through 2-heptyl-3-hydroxy-4-quinolone (Pseudomonas quinolone signal [PQS]) [11]. PQS serves as a coinducer for PqsR (also known as MvfR), which then transcriptionally activates the pqsABCDE operon to create a positive feedback loop that results in more PQS synthesis [12]. This feedback loop also leads to an increase in PqsE expression and this protein is required to mediate virulence through RhlR-C4-HSL via an unknown mechanism [13]. The first step in PQS synthesis is carried out by PqsA, which charges anthranilate with CoA to form athraniloyl-CoA [14]. Malonyl-CoA is then added via the action of PqsD, after which PqsB and PqsC catalyze the ligation of octanoyl-CoA to the newly formed 2-aminobenzoylacetate to create 2-heptyl-4-quinolone (HHQ) [15, 16]. The monooxygenase PqsH then catalyzes the formation of 2-heptyl-3-hydroxy-4quinolone (PQS) from HHQ [17]. An alternative product of the PQS synthesis pathway is 4-hydroxy-2-heptylquinoline N-oxide (HQNO) which is produced by the probable FAD-dependent monooxygenase, PqsL [14].
Previously, a study on factors that control the autolytic phenotype of P. aeruginosa demonstrated that deletion of pqsL caused an overproduction of PQS and colonies of this mutant grown on agar plates exhibited an autolytic phenotype [18]. Increased PQS levels in the pqsL mutant were attributed to a shift in the quinolone synthesis pathway because HQNO production stopped and more precursors for PQS synthesis were available. The study also identified ten other genes that affected PQS production [18]. One of these mutants harbored a disruption of gene PA3006 (psrA), which is a transcriptional regulator belonging to the TetR family of regulators. PsrA directly activates the stationary phase sigma factor RpoS and also regulates genes involved in fatty acid metabolism in P. aeruginosa, including the fadBA5 operon and PA0506 [19–21]. Specifically, a prior microarray study suggested that psrA had a negative effect on the transcription of PA0506, a FadE homolog which is one of many predicted acyl-CoA dehydrogenases in strain PAO1 [21]. This enzyme is important for the conversion of acyl-CoA moieties (a PQS precursor) into enoyl-CoA during the first step of fatty acid β-oxidation. P. aeruginosa appears to have multiple sets of enzymes responsible for the breakdown of long chain fatty acids, including three fadBA operons which putatively encode a 3-hydroxyl-CoA dehydrogenase (FadB) and a 3-ketoacyl-CoA thiolase (FadA), which respectively catalyze the last two steps of fatty acid β-oxidation [22]. PsrA has been shown to bind to and repress the fadBA5 and PA0506 promoters at the C/GAAAC N4 GTTTG/C palindromic motif, and repression is relieved by long chain fatty acids [19, 21]. The fact that the loss of PsrA had a negative effect on PQS production suggested that the fatty acid β-oxidation pathway was important for PQS synthesis. In this study, we explore the link between PsrA and PQS production in P. aeruginosa and present data which suggest that negative regulation asserted by PsrA over the fatty acid β-oxidation pathway is required to allow acyl-CoA fatty acids to escape the β-oxidation pathway and be shuttled into quinolone production.
Materials and methods
Bacterial strains, plasmids, and culture conditions
All bacterial strains and plasmids are listed in Table 1. E. coli and P. aeruginosa strains were maintained in 10% glycerol and 10% Difco skim milk (Becton, Dickinson, and Co.), respectively, at -80°C. All strains were freshly plated to start each experiment and then grown at 37°C in Difco lysogeny broth (LB; Becton, Dickinson, and Co.). Growth was monitored spectrophotometrically based on the optical density at 660 nm (OD660) or 600 nm (OD600) for P. aeruginosa or E. coli strains, respectively. Strains were grown in the presence of either 200 μg/ml or 100 μg/ml carbenicillin (Research Products, Inc.) as needed to maintain plasmids for P. aeruginosa or E. coli strains, respectively. When needed, L-arabinose (0.5% final concentration) (Sigma-Aldrich) was added to cultures to induce genes controlled by the pBad promoter.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or Plasmid | Relevant genotype or phenotype | Reference or source |
|---|---|---|
| E. coli DH5α | λ- φ80dlacZΔM15 Δ(lacZYA-argF) U196 recA1 endA1 hsdR17 (rK- mK-) supE44 thi-1 gyrA relA1 | [23, 24] |
| P. aeruginosa strains | ||
| PAO1 | Wild type | [25] |
| PGW-ΔpsrA | psrA deletion in PAO1 | This Study |
| PGW-ΔPA0506 | PA0506 deletion PAO1 | This Study |
| PGW-ΔpsrAΔPA0506 | PA0506 deletion in PGW-ΔpsrA | This Study |
| PGW-ΔPA0507 | PA0507 deletion in PAO1 | This Study |
| PGW-ΔpsrAΔPA0507 | PA0507 deletion in PGW-ΔpsrA | This Study |
| PGW-ΔPA0508 | PA0508 deletion in PAO1 | This Study |
| PGW-ΔpsrAΔPA0508 | PA0508 deletion in PGW-ΔpsrA | This Study |
| Plasmids | ||
| pHERD20T | E. coli/P. aeruginosa shuttle expression vector | [26] |
| pEX18Ap | Suicide vector for P. aeruginosa | [27] |
| pEX1.8 | Expression plasmid | [28] |
| pMW105 | rpoS in pEX1.8 | [29] |
| pPQSsynOE | PBAD-pqsABCD on pHERD20T | [30] |
| pGW-1 | psrA deletion in pEX18Ap | This Study |
| pGW-2 | PBAD-psrA on pHERD20T | This Study |
| pGW-14 | PsrA-6HIS in pHERD20T | This Study |
| pGW-15 | PA0506 deletion in pEX18Ap | This Study |
| pGW-16 | PA0507 deletion in pEX18Ap | This Study |
| pGW-18 | PBAD-PA0507 on pHERD20T | This Study |
| pGW-19 | PA0508 deletion in pEX18Ap | This Study |
| pGW-20 | PBAD-PA0508 on pHERD20T | This Study |
| pGW-21 | PBAD-PA0506 on pHERD20T | This Study |
| pGW-22 | PBAD-fadAB5 on pHERD20T | This Study |
Generation of mutant strains and plasmids
All oligonucleotide primers are listed in S1 Table. Mutant P. aeruginosa strains were derived by a modified version of a previously published protocol [31]. The mutant alleles were generated by using a splicing-by-overlapping PCR extension of primers [32]. The alleles were constructed to harbor in-frame deletions in the DNA coding sequence corresponding to amino acids 18 to 226 for psrA (89% of protein sequence), 19 to 585 for PA0506 (94% of protein sequence), 36 to 553 for PA0507 (86% of protein sequence), and 38 to 535 for PA0508 (86% of protein sequence). Oligonucleotide primers utilized for this procedure were designed to generate at least 1000 bp of DNA upstream and downstream from the junction. Strain PAO1 chromosomal DNA was utilized as template for the PCR. The final PCR product was cloned into pEX18Ap using PstI-HF (psrA) and HindIII-HF (PA0506-08) (New England Biolabs) restriction enzymes, respectively.
Plasmids harboring ΔpsrA (pGW-1), ΔPA0506 (pGW-15), ΔPA0507 (pGW-16), ΔPA0508 (pGW-19) were sequenced to confirm in-frame deletions and transformed into strain PAO1 by electroporation [33, 34]. Mutants were selected as described by Hoang et al. [27]. Potential mutant colonies containing a psrA deletion in strain PAO1 (PGW-ΔpsrA) were screened by PCR utilizing appropriate flanking primers and confirmed by DNA sequencing of the PCR product. Plasmids pGW-15, pGW-16, and pGW-19 were transformed into strain PAO1 to generate single mutants, or strain PGW-ΔpsrA to generate double mutants.
To generate expression plasmids for complementation experiments, genes psrA (702 bp), PA0506 (1821 bp), PA0507 (1796 bp), PA0508 (1778 bp), and fadAB5 (3354 bp) were amplified by PCR from strain PAO1 chromosomal DNA. The oligonucleotide primers with engineered restriction enzyme sites were utilized to harbor both the start codon and the stop codon for all genes. After digesting with restriction enzymes, the PCR fragment was ligated into the digested pHERD-20T which contains an araC pBAD (PBAD) promoter to control gene expression. These clonings resulted in plasmids pGW-2, pGW-21, pGW-18, pGW-20, and pGW-22, respectively.
Measuring PQS production
Washed cells from an overnight culture were used to inoculate fresh 10 ml cultures to an OD660 of 0.05. After 24 hours of growth at 37°C with vigorous shaking, 300 μl of each culture was extracted with 900 μl of acidified ethyl acetate as previously described [35]. One-half of the organic phase was completely evaporated at 37°C, and 50 μl of 1:1 acidified ethyl acetate-acetonitrile was used to reconstitute the extract. Samples were analyzed by thin-layer chromatography (TLC), visualized by long-wave UV light, photographed, and quantified by densitometric analysis [35].
Measuring pyocyanin production
Pyocyanin measurements were completed using a modified protocol from Essar et al. [36]. Washed cells from an overnight culture were used to inoculate 10 ml of LB media to an OD660 of 0.05, which were incubated at 37°C with vigorous shaking. After 24 hours of growth, cultures were centrifuged to remove bacterial cells and 500 μl of culture supernatant was extracted with 300 μl of chloroform. The organic phase was extracted with 100 μl of 0.2 N HCL, which produced a pink solution containing pyocyanin. The absorbance of this solution at 520 nm was measured using a NanoDrop ND-1000 spectrophotometer. Data are presented in absorbance as the mean ± standard deviation [σ(n-1)] of three independent experiments.
Monitoring fatty acid effects of PQS
Octanoic acid or oleic acid (Sigma-Aldrich) was added to a final concentration of 10 mM to 10 ml of LB media and the pH was adjusted to 7.0 with sodium hydroxide. Flasks were inoculated with strain PAO1, PGW-ΔpsrA, PGW-ΔPA0506, PGW-ΔPA0507, PGW-ΔPA0508, or PGW-ΔpsrA, 0506 and incubated at 37°C with vigorous shaking overnight. Washed cells from these cultures were utilized to inoculate fresh 10 ml cultures containing the same fatty acid to an OD660 of 0.05 and incubated at 37°C with vigorous shaking. After 24 hours of growth, PQS was extracted and quantified as described above.
Purification of PsrA
Overnight cultures of E. coli strain DH5α harboring the control plasmid (pHERD20T) or the expression vector (pGW-14) that contains a 6-His-tag-PsrA fusion protein were subcultured with a starting OD600 of 0.05. Cultures were incubated at 37°C with vigorous shaking for 2.5 hours to an OD600 of approximately 0.5, and L-arabinose was added to a final concentration of 0.5% to induce expression of PsrA. The cultures were then incubated for 3 hours and cells were harvested by centrifugation at 6,000 × g for 10 min at 4°C. Bacterial cell pellets were resuspended in 1 ml of STE buffer (pH 7.5; 10 mM Tris-HCl, 1 mM EDTA, 100 mM NaCl), and this suspension was passed through a French pressure cell at 16,000 lb/in2 to yield a whole-cell lysate. The 6-His-tag-PsrA protein was purified via Ni-NTA agarose (Qiagen) utilizing the native protein methods recommended by Qiagen. Protein concentration for DNA mobility shift assays was determined using a Bradford assay (Bio-Rad).
DNA mobility shift assay
PCR was used to generate DNA fragments containing the kynA (228 bp), psrA (298 bp), PA0506 (349-bp), PA0507 (233-bp), or PA0508 (342-bp) promoter region. DNA probes were labeled with 32P using [γ-32P] ATP (Perkin-Elmer) and T4 polynucleotide kinase (New England Biolabs). Binding reactions were carried out in buffer containing 20 mM HEPES (pH 7.6), 1mM EDTA, 10 mM ammonium sulfate, 150 mM potassium chloride, 1 mM DTT, and 5% glycerol. Each reaction mixture contained 0.3 μg of salmon sperm DNA, 3000 cpm of radiolabeled probe, and 0 to 50 ng of purified 6-His-tagged PsrA protein. Reaction mixtures were incubated at room temperature for 20 min and separated by electrophoresis on a native 6% polyacrylamide gel in 0.5X Tris-borate-EDTA buffer at 4°C. Gels were then exposed to X-ray film to visualize radiolabeled bands.
RNA isolation
Overnight cultures of strains PAO1 and PGW-ΔpsrA were washed in LB media and used to inoculate 10 ml of LB media to an OD660 of 0.05. Cultures were incubated at 37°C for 3 hours (or 6 hours for S2 Fig) or to an OD660 of approximately 1.5 prior to centrifugation at 4°C to harvest bacterial cells. Total cellular RNA was isolated from P. aeruginosa cells using the RNeasy Midiprep kit according to the manufacturer’s protocol (Qiagen). Contaminating DNA was removed by treatment of RNA samples with RQ1 DNase as per the manufacturer’s protocol (Promega). RNA was then extracted with 1:1 phenol-chloroform and then precipitated with ethanol. Purified RNA was resuspended in nuclease-free water. RNA concentration was determined with a NanoDrop ND-1000 spectrophotometer.
cDNA synthesis for quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from strains PAO1 and PGW-ΔpsrA, and purified as described above. cDNA was synthesized in a 21 μl reaction volume from 5 μg of total RNA using a 1:1 mixture of GC-rich hexamers (Gene Link) and random hexamers (Invitrogen) for priming with 40 μM deoxynucleoside triphosphates (dNTPs) (USB). Reactions were then heated at 65°C for 5 min, followed by cooling to 4°C for 1 min. Following this step, 200 U of SuperScript III reverse transcriptase in First Strand buffer (Invitrogen) with 0.1 M dithiothreitol and RNase Out RNase inhibitor (Invitrogen) were added to each reaction mixture, yielding a final volume of 30 μl. Reaction mixtures were then heated to 25°C for 5 min, followed by 50°C for 1 hour and then 75°C for 15 min.
qRT-PCR
cDNA and total RNA to be used as the template and negative control, respectively, were diluted 1:200 in nuclease-free water. Oligonucleotide primer pairs for qRT-PCR were generated by the Primer-BLAST program available at www.ncbi.nlm.nih.gov/tools/primer-blast/. Primers were designed to amplify a 200 bp fragment of clpX (control), a 132 bp fragment of lasR, a 150 bp fragment of pqsA, a 139 bp fragment of PA0506, a 124 bp fragment of PA0507, and a 117 bp fragment of PA0508 as target genes. Quantitative RT-PCR was performed using FastStart SYBR green master mix (Roche Diagnostics) with Bio-Rad CFX96. The following cycle was utilized to amplify and quantify fragments: 95°C for 10 min and then 95°C for 15 s, 56°C for 15 s, and 72°C for 20 s, repeated 40 times. Melt curve data were collected to ensure amplification of one fragment by heating samples from 65°C to 95°C in 0.5°C increments. Data were generated from three separate RNA and cDNA preparations and at least two technical replicates for each primer set. Relative expression of each gene was determined by comparing target genes with the control gene (clpX) using the Pfaffl method [37].
Reverse transcription polymerase chain reaction
DNase treated RNA (400 ng) from wild type strain PAO1 was used as a template for reverse transcriptase PCR (RT-PCR) performed utilizing the Promega AccessQuick RT-PCR system by following the manufacturer’s protocol. Primer pairs were designed to cover the intergenic regions of all genes of interest. All cDNA synthesis and PCR amplification steps were performed in an Eppendorf Mastercycler with the following parameters: cDNA synthesis at 45°C for 30 min; 95°C for 2 min; 30 cycles of 95°C for 45 s, 55°C for 45 s, and 72°C for 40 s. The final cycle was for 5 min at 72°C. Positive controls were performed using genomic DNA for strain PAO1 (100 ng), and negative controls were performed without adding reverse transcriptase. Reaction products were analyzed by agarose gel electrophoresis.
Results
PsrA positively controls PQS production in P. aeruginosa
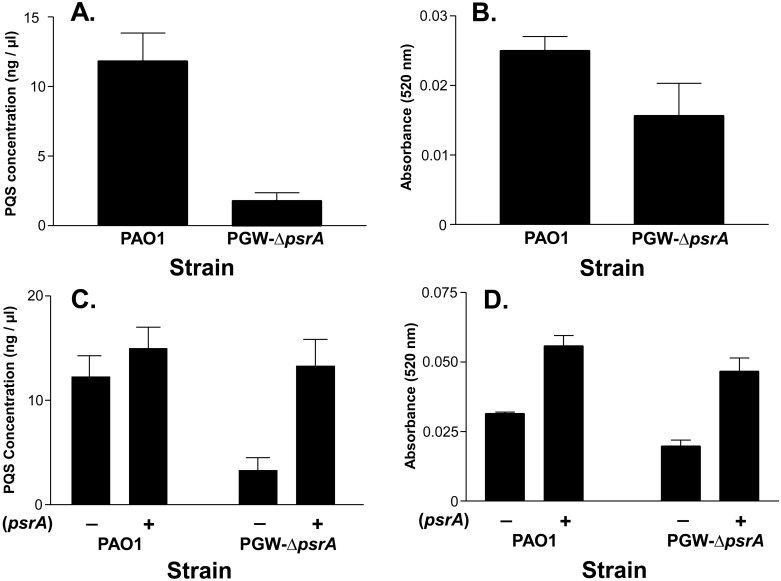
Our previous studies have shown that a pqsL mutant was autolytic and overexpressed PQS [18]. When this mutant was randomly mutagenized, a transposon insertion in psrA (PA3006) caused both the suppression of the autolytic phenotype and a decrease in PQS production [18]. To expand on this result, we constructed an isogenic psrA mutant (PGW-ΔpsrA) and examined its phenotype. As expected, strain PGW-ΔpsrA produced less PQS than the parent strain PAO1 (Fig 1A), and it also produced less of the PQS-controlled virulence factor pyocyanin (Fig 1B). PQS and pyocyanin production were both restored to at least the wild type level in strain PGW-ΔpsrA containing a psrA expression plasmid (Fig 1C and 1D), indicating that mutation of psrA did not have polar effects on adjacent genes. Taken together, these data indicate that psrA positively controls PQS production.
Fig 1. PsrA positively controls PQS production in P. aeruginosa.
Strains PAO1 and PGW-ΔpsrA were grown for 24 h in LB medium, then (A and C) PQS or (B and D) pyocyanin was extracted and quantified as described in Materials and Methods. The presence of psrA expression plasmid (pGW-2) or pHERD20T (control plasmid) is indicated by a plus or minus, respectively. For C and D, LB media was supplemented with 0.5% L-arabinose to induce psrA. All data are presented as the average ± SD of three independent experiments.
Expression of pqsABCD restores PQS activity in a psrA mutant
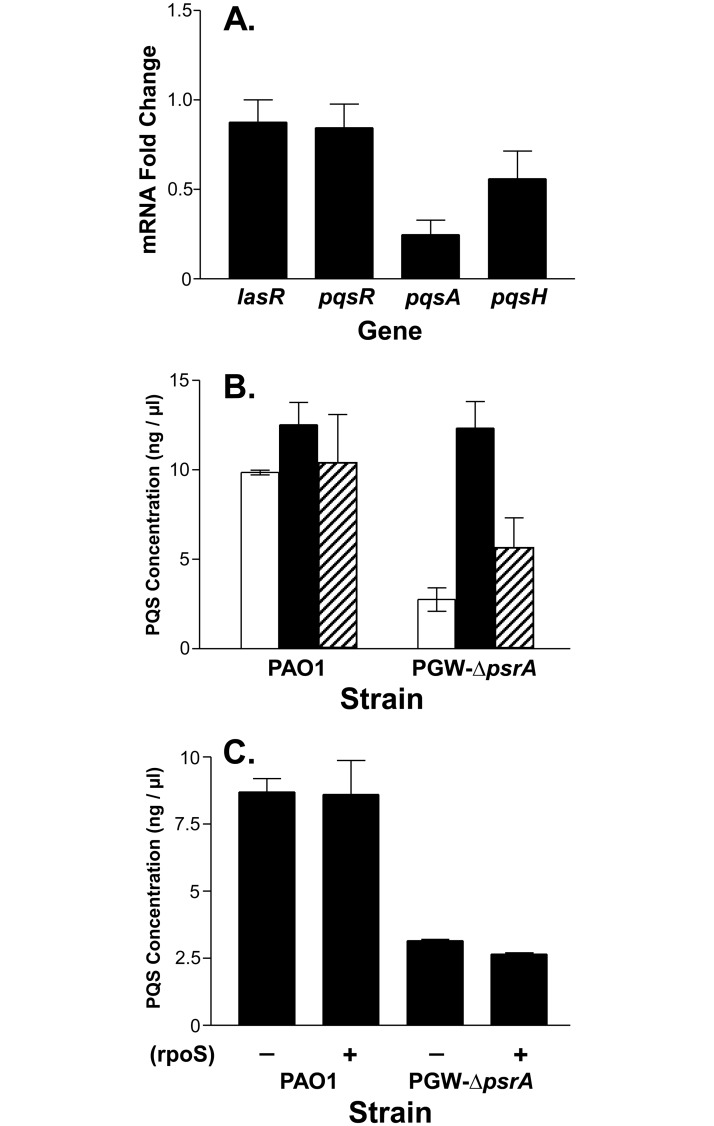
Since PsrA has been shown to act as a transcriptional regulator that influences the expression of a wide range of genes, we decided to use qRT-PCR to assess the relative expression of genes required for PQS synthesis in a psrA mutant. LasR directly activates pqsR and pqsH, and PqsR directly activates pqsA, and the loss of any of these genes causes a drastic reduction in PQS production [12, 38, 39]. Our data showed that the expression of neither lasR nor pqsR was altered in the psrA mutant, indicating that the effect of psrA on PQS was not mediated through either of these two PQS-controlling regulators (Fig 2A). The expression of pqsA and pqsH decreased to 0.23 and 0.57, respectively, when compared to the parent strain PAO1 set to a value of 1 (Fig 2A). Both of these genes are directly involved in PQS synthesis and the pqsABCDE operon is positively controlled by PQS, so a decrease in the expression of pqsA would be expected if less PQS were present. This was confirmed by results which showed that expressing pqsABCD from an inducible promoter on a plasmid caused PQS production to return to a wild type level in the psrA mutant (Fig 2B). We also found that adding PQS to the psrA mutant containing a pqsA-lacZ reporter fusion caused pqsA expression to return to a wild type level (data not shown), indicating again that the observed effect of psrA was caused by lower PQS production in the psrA mutant. In addition, the overexpression of pqsH caused a small increase in PQS production in the psrA mutant to approximately half that of the wild type strain, suggesting that pqsH is not the mediator of the psrA effect on PQS production (Fig 2B).
Fig 2. PsrA positively controls pqsA.
(A) qRT-PCR was performed on RNA from strains PAO1 and PGW-ΔpsrA to analyze the relative expression of lasR, pqsH, pqsR and pqsA. Data are presented as average fold change ± SD in the psrA mutant compared to the wild type strain (set to a value of 1). The gene clpX was used as a reference gene to normalize expression and each experiment was completely repeated three times. (B and C) PQS production by strains PAO1 or PGW-ΔpsrA expressing (B) PQS synthetic genes or (C) RpoS. Cultures were grown for 24 h in LB medium supplemented with 0.5% L-arabinose to induce genes. PQS was then extracted and quantified as described in Materials and Methods. Data are presented as the average ± SD of three independent experiments. (B) Plasmids contained by strains are: open bars, control vector; solid bars, pqsABCD expression vector; and hatched bars, pqsH expression vector. (C) Presence of the rpoS expression vector is indicated by a plus (+) symbol.
We also noted from previous studies that PsrA was shown to have a positive effect on rpoS transcription, and that RpoS negatively regulates quorum sensing [19, 20, 29]. This led us to question whether the effect of PsrA on PQS production was mediated by RpoS. To test this, we examined PQS production in strain PGW-ΔpsrA containing plasmid pMW105, which harbors an inducible rpoS. These data showed that RpoS had no effect on PQS production in the psrA mutant (Fig 2C), and eliminated this avenue of regulation as an explanation for the effect of psrA on PQS.
PsrA regulates PQS production via the fatty acid β-oxidation cycle
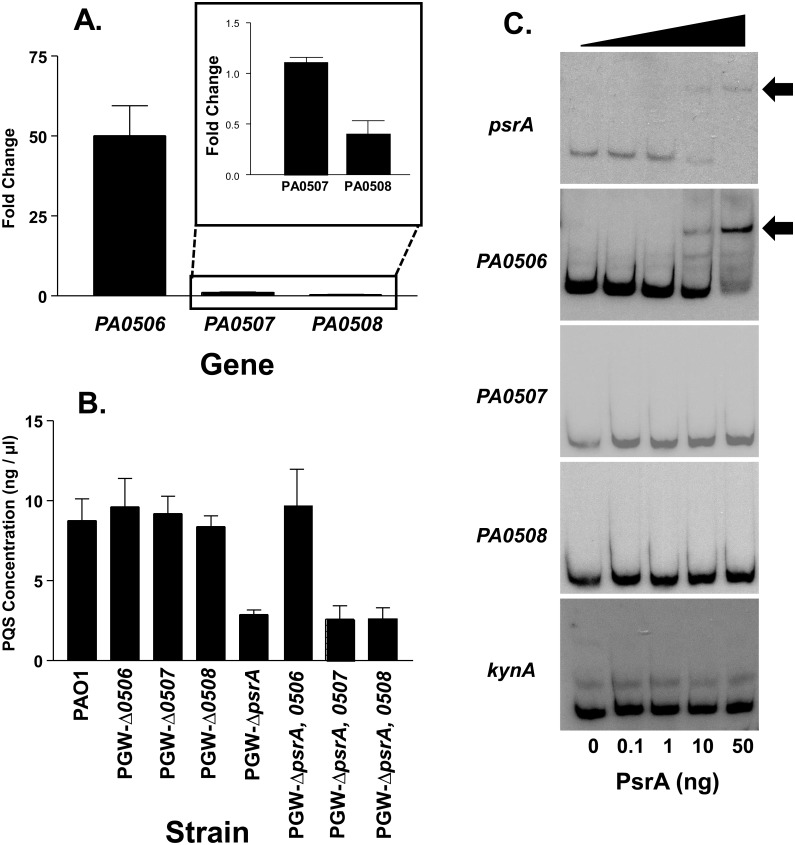
Several fatty acid β-oxidation enzymes have been identified in P. aeruginosa and there seems to be overlapping pathways and complex regulation designed to ensure that various fatty acids can be broken down when needed. For example, this organism has three putative fadE genes (PA0506, PA0507, and PA0508) which are adjacent to one another and encode acyl-CoA dehydrogenase homologs that would catalyze the first step of fatty acid β-oxidation. PsrA appears to be one of the major regulators of some fatty acid degradation enzymes as it has been shown to bind and repress both PA0506 and the fadBA5 operon, which is important for the breakdown of fatty acids with chain lengths of 12 or greater [19, 21, 40]. The repression of fadBA5 by PsrA was relieved in the presence of fatty acids, with the repression decreasing as fatty acid chain length increased [21]. In addition, a prior microarray study suggested that psrA had a negative effect on PA0506 and PA0507 [21]. These reports, along with the fact that PQS production requires an octanoyl-CoA which can be derived from β-oxidation of long chain fatty acids, led us to test whether PQS concentrations were affected by one of the PsrA-regulated FadE homologs (PA0506, PA0507, and PA0508). We first attempted to confirm the prior microarray studies by using quantitative real time PCR to monitor the expression of PA0506, PA0507, and PA0508 in a psrA mutant and its parent strain. This experiment showed that in the psrA mutant, PA0506 expression was increased 50 fold as expected, PA0507 expression was unchanged, and PA0508 expression decreased to half that of the wild type strain (Fig 3A). The potential for differential regulation of these three genes was confirmed by reverse transcription-PCR, which showed that each gene is transcribed in a separate operon (S1 Fig). Since FadE catalyzes the first step in the fatty acid β-oxidation cycle, and PQS synthesis requires an octanoyl-CoA, we hypothesized that the overexpression of the FadE homolog PA0506 in the psrA mutant was probably causing a decrease in octanoyl-CoA available for PQS synthesis. To begin to test this, we created isogenic in-frame mutants of all three putative FadE homolog genes, PA0506, PA0507, and PA0508. While our results above suggested that a PA0506 mutant might produce more PQS, we saw that all three mutants produced PQS similar to the wild type strain (Fig 3B). In retrospect, this was not really a surprise since the likely overlapping function of the three adjacent homologs probably negates any effects of a single mutant. To try to further establish the link between psrA, a FadE homolog, and PQS production, we also created and tested double mutants which had deletions of psrA and either PA0506, PA0507, or PA0508 (Fig 3B). Analysis of PQS production by these three strains showed that the psrA, PA0506 double mutant produced a wild type amount of PQS. This indicated that in order for psrA to control PQS production, PA0506 had to be present. Removal of PA0507 or PA0508 in the psrA mutant had no effect on PQS production (Fig 3B). Furthermore, EMSA analysis confirmed that PsrA only binds to the PA0506 promoter region (Fig 3C). Taken together, our data suggest that PQS production is controlled by PsrA via its negative regulation of PA0506, which prevents or slows down the fatty acid β-oxidation cycle of P. aeruginosa to allow the escape of the octanoyl-CoA needed for PQS synthesis.
Fig 3. PsrA directly regulates PA0506 to control PQS production.
(A) qRT-PCR was performed on RNA from strains PAO1 and PGW-ΔpsrA. Data are presented as average fold change ± SD of expression in the psrA mutant as compared to expression in the wild type strain (set to a value of 1). The gene clpX was used as a reference for normalization and experiments were repeated three separate times. Targeted genes are listed below the columns. (B) The indicated single and double mutants were assessed for PQS production after 24 h growth. (C) EMSA for promoter regions of psrA (positive control) [41], PA0506, PA0507, PA0508, and kynA (negative control) was performed using 0, 0.1, 1.0, 10, and 50 ng of PsrA-his tag protein. Binding is indicated by an arrow and data are representative of three independent experiments.
Long chain fatty acids induce PQS production
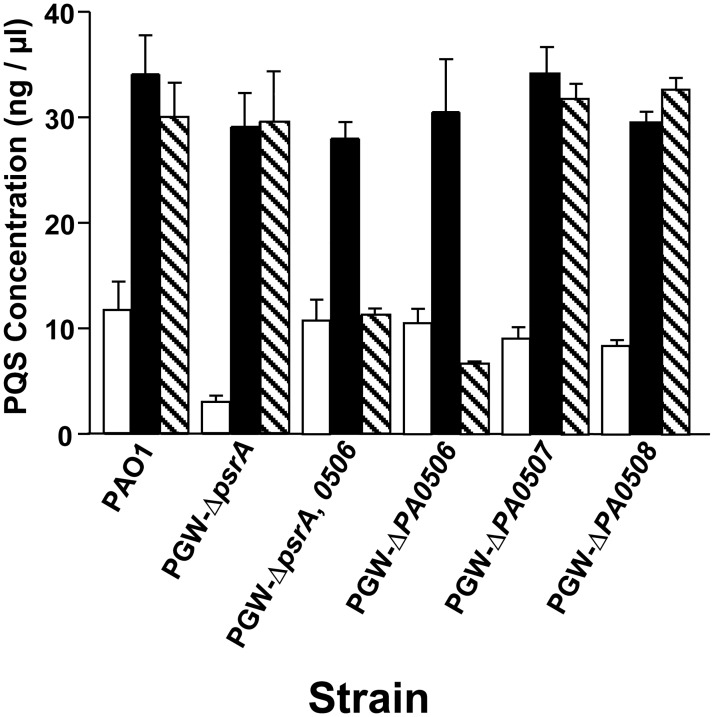
PsrA has been shown to bind and repress the fadBA5 operon, which is important for the breakdown of fatty acids with chain lengths of 12 or greater [21]. This repression by PsrA was also shown to be relieved in the presence of fatty acids, with medium and long chain fatty acids being more efficient at relieving repression than short chain fatty acids [21]. In addition, PsrA has also been shown to bind and repress PA0506 [19]. This led us to hypothesize that long chain fatty acids would relieve the effect that PsrA had on PQS production. To test this, we grew various P. aeruginosa strains in the presence of octanoic acid or oleic acid. We found that both fatty acids caused an increase in PQS production in the wild type strain PAO1 and also fully restored PQS production in the psrA mutant (Fig 4). Most interestingly, PQS production in the PA0506 mutant did not increase in response to oleic acid, while the PA0507 and PA0508 mutants produced PQS levels that were similar to those seen from the wild type strain grown with oleic acid (Fig 4). These data imply that of the three adjacently encoded FadE homologs, PA0506, PA0507, and PA0508, only PA0506 is important for breaking down long chain fatty acids that will be used for PQS synthesis.
Fig 4. PA0506 is required for C18 fatty acid to serve as a PQS precursor.
Cultures of the indicated strains were grown for 24 h in LB medium alone (open bars) or supplemented with either octanoic acid (black bars) or oleic acid (hatched bars). PQS was extracted and quantified as described in Materials and Methods. Data are presented as the average ± SD of three independent experiments.
Overexpression of PA0506 negatively affects PQS production
The data presented so far caused us to hypothesize that negative regulation of PA0506 by psrA allows normal levels of PQS to be produced and that the decrease in PQS production seen in the psrA mutant is due to the increased expression of PA0506, which should pull more long chain fatty acids into the β-oxidation cycle, and away from PQS synthesis. To test this idea, we overexpressed PA0506 in strains PAO1, PGW-ΔpsrA, PGW-ΔPA0506, and PGW-ΔpsrA, ΔPA0506. As expected, PQS production was decreased approximately 50 percent when PA0506 was overexpressed in the wild type strain and PA0506 mutants (Fig 5). We must note here that the psrA mutant containing the PA0506 expression plasmid had an increase of PA0506 expression of approximately 8 fold (see S2 Fig), which is less than that seen without a plasmid present (Fig 3A). The reason for this is not clear and we can only speculate that the plasmid and/or the antibiotic selection either increased the expression of our qRT-PCR reference gene or decreased (compared to the strain without a plasmid) expression of PA0506. Nevertheless, the data of Fig 5 confirm that the expression of PA0506 will cause PQS production to decrease. We also determined that when overexpressed from a plasmid, PA0507 and PA0508 had a small negative effect on PQS production (S3 Fig). This suggests that these two FadE homologs are capable of changing the supply of octanoyl-CoA, and more importantly, that PA0506 is the FadE homolog that acts on the precursor needed for PQS to be synthesized.
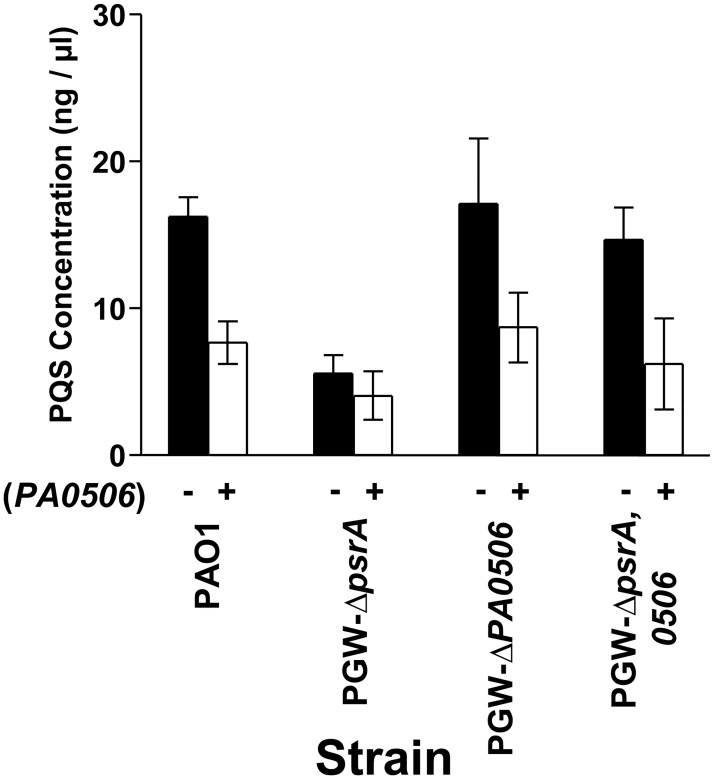
Fig 5. Increased expression of PA0506 alters PQS production.
The indicated strains were grown for 24 h in LB medium harboring pHERD20T (control plasmid) or pGW-21 (PA0506 expression plasmid) indicated by a minus or plus symbol, respectively. Cultures were supplemented with 0.5% L-arabinose to induce PA0506 expression, and PQS was then extracted and quantified. Data are presented as the average ± SD of three independent experiments.
Discussion
PsrA is a member of the TetR family of transcriptional regulators that has been shown to regulate numerous genes, including rpoS, PA0506, and fadBA5 [19, 41]. It was also linked to PQS production [18] and the data reported here extends these prior studies by showing how PsrA indirectly controls PQS production. PQS production was decreased approximately 85% in a psrA mutant (Fig 1), and since lasR, pqsR, pqsH, and pqsA were previously identified as genes that controlled the pqsABCDE operon [12, 42, 43], we looked at expression of these genes in a psrA mutant. Neither lasR, pqsR, nor pqsH, was affected at a level that would produce the observed decrease in PQS production (Fig 2), and these genes, along with rpoS were ruled out as potential targets for psrA-mediated regulation of PQS. This led us to search for an indirect connection between PsrA and PQS synthesis and the most likely connector seemed to be the fatty acid octanoyl-CoA, which serves as a PQS precursor.
In the fatty acid β-oxidation pathway, FadE catalyzes the conversion of an acyl-CoA to an enoyl-CoA [22]. There are several probable fadE genes within the P. aeruginosa genome, including PA0506, PA0507, and PA0508, and psrA has been shown to directly repress PA0506 [19, 40, 44]. We analyzed the expression of PA0506, PA0507, and PA0508 in the psrA mutant and found that they were differentially expressed (Fig 3), with PA0506 expression increasing as seen before [21, 40]. The genetic organization of the genes suggested that they may be in an operon, but their differential expression indicated that they were independently transcribed, which we confirmed (S1 Fig). At this point, we surmised that the overexpression of PA0506 in the psrA mutant could be causing the rapid conversion of acyl-CoA molecules into enoyl-CoA molecules, leaving little precursor for the synthesis of PQS. This idea was strengthened by our data which showed that mutating PA0506 in the psrA mutant led to a restoration of PQS production to a wild type level (Fig 3). Additionally, PQS production was decreased approximately 50% in strain PAO1 expressing PA0506 from an inducible promoter on a plasmid (Fig 5), which further confirms our hypothesis that PA0506 repression by PsrA maintains acyl-CoA precursors for PQS synthesis. Mutating PA0507 or PA0508 in the psrA mutant had no effect on PQS production, and PsrA did not interact with the promoter for either of these genes (Fig 3). Taken together, these data suggested that acyl-CoA molecules that serve as PQS precursors were most likely being processed by the β-oxidation pathway that utilized PA0506 as the enzyme which converts them into enoyl-CoA molecules. Although previous data showed that PsrA also represses the fadBA5 operon [21], these enzymes act at a later stage in the β-oxidation cycle, after PA0506 exerts its activity. We found that expressing fadBA5 from a plasmid had no effect on PQS production (S4 Fig). The reason for this is not obvious but we can speculate that fadBA5 expression had no effect because an enzyme earlier in the β-oxidation pathway catalyzes the rate-limiting step.
Since a C8 fatty acid serves as a direct PQS precursor, and PsrA is known to regulate fatty acid degradation [16, 19, 21], we investigated whether the addition of medium or long chain fatty acids would restore PQS production in our psrA mutant. The addition of octanoic acid to any strain tested caused a major increase in PQS production, even in the psrA mutant, indicating that this precursor is greatly limited in this strain (Fig 4). At this point, all of our observations seemed to be pointing to the overexpression of PA0506 and subsequent loss of acyl-CoA molecules to serve as PQS precursors, as the reason for a decrease in PQS production in the psrA mutant. This is interesting because it suggests that PA0506 is important for PQS synthesis from the breakdown of C18 fatty acids but that too much PA0506 is detrimental to the process.
Taken together, our data suggest that long chain fatty acids that serve as PQS precursors undergo β-oxidation via a pathway that utilizes PA0506, the gene for which is strongly repressed by PsrA. To illustrate this, we present a model (Fig 6) of how the regulation of fatty acid β-oxidation controls PQS production in P. aeruginosa. It should also be noted here that P. aeruginosa synthesizes many different quinolones that do not act as intercellular signals but have other physiological functions, and that their synthesis would also be affected by the PsrA-controlled availability of various acyl-CoA molecules. Furthermore, it could be speculated that PsrA-dependent control of the β-oxidation cycle could additionally affect malonyl-CoA availability (another PQS precursor) via altered acetyl-CoA production and this further demonstrates the importance of PsrA regulatory activity during PQS synthesis.
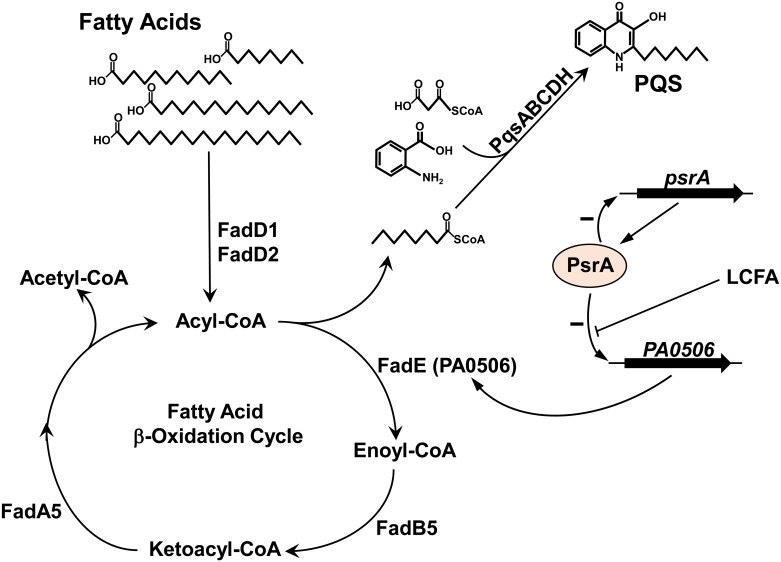
Fig 6. Proposed model of PsrA regulation of PQS production in P. aeruginosa.
In this model, PsrA represses PA0506 (fadE) which allows acyl-CoA to be available for the PQS biosynthesis pathway. Long chain fatty acids (LCFA) relieve this repression. When PsrA is mutated, the expression of PA0506 is greatly elevated and excess PA0506 will rapidly convert acyl-CoA to enoyl-CoA. This apparently causes a shortage of the octanoyl-CoA needed for PQS synthesis and therefore PQS production greatly decreases.
An interesting aspect of this study is that it links PQS production, and thus virulence factor control via cell to cell signaling, to fatty acid degradation via the β-oxidation pathways. These pathways were shown to be induced in P. aeruginosa growing within the lungs of cystic fibrosis patients where the long chain fatty acids of phosphatidylcholine can serve as a carbon source [5]. P. aeruginosa β-oxidation pathway mutants were also less virulent in a mouse lung infection model, indicating the importance of fatty acid degradation for virulence in vivo [45]. We speculate that this decrease in virulence is at least partly caused by the dysregulation of quinolone signaling that would occur when the cells can no longer convert long chain fatty acids into the appropriate precursor needed for PQS production.
Supporting information
Primer locations are indicated by the letters on the gene map and can be matched to the reactions electrophoresed on the agarose gels. For each primer set, lane 1 contains an RT-PCR reaction in which no reverse transcriptase was added (negative control); lane 2 contains a reaction in which chromosomal DNA was added (positive control); and lane 3 contains the experimental reaction.
(TIF)
qRT-PCR was performed using RNA from strains PAO1 and PGW-ΔpsrA containing either a control plasmid or one with an inducible promoter that controls PA0506 (pHERD20T and pGW-21, respectively). All cultures were supplemented with 0.5% L-arabinose except for a control culture of strain PAO1 (pHERD20T), which served as a reference for which the fold change value was set at 1. Data from three independent repeats are presented as average fold change ± SD of expression of PA0506 as compared to the reference culture.
(TIF)
Strain PAO1 harboring pHERD20T (control plasmid), pGW-21, pGW-18 or pGW-20 (PA0506, PA0507, or PA0508 expression plasmids, respectively) were grown for 24 h in LB medium supplemented with 0.5% L-arabinose. PQS was then extracted and quantified as described in Materials and Methods. Data are presented as the average ± SD of three independent experiments.
(TIF)
The indicated strains harboring pGW-22 (fadBA5 expression plasmid) were grown for 24 h in LB medium with or without 0.5% L-arabinose as indicated by a plus or minus symbol, respectively. PQS was then extracted and quantified as described in Materials and Methods. Data are presented as the average ± SD of three independent experiments.
(TIF)
(PDF)
Acknowledgments
We thank J. Farrow, S. Lotlikar, and J. Coleman for thoughtful insight and proofreading of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Funding provided by National Institute of Allergy and Infectious Disease grant number R01-AI076272.
References
- 1.Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerceo E, Deitelzweig SB, Sherman BM, Amin AN. Multidrug-Resistant Gram-Negative Bacterial Infections in the Hospital Setting: Overview, Implications for Clinical Practice, and Emerging Treatment Options. Microb Drug Resist. 2016;22:412–431. doi: 10.1089/mdr.2015.0220 [DOI] [PubMed] [Google Scholar]
- 3.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu Rev Microbiol. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635 [DOI] [PubMed] [Google Scholar]
- 4.Leisinger T, Margraff R. Secondary metabolites of the fluorescent pseudomonads. Microbiological Reviews. 1979;43:422–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Son MS, Matthews WJ Jr., Kang Y, Nguyen DT, Hoang TT. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect Immun. 2007;75:5313–5324. doi: 10.1128/IAI.01807-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weir TL, Stull VJ, Badri D, Trunck LA, Schweizer HP, Vivanco J. Global gene expression profiles suggest an important role for nutrient acquisition in early pathogenesis in a plant model of Pseudomonas aeruginosa infection. Appl Environ Microbiol. 2008;74:5784–5791. doi: 10.1128/AEM.00860-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1995;92:1490–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearson JP, Gray KM, Passador L, Tucker KD, Eberhard A, Iglewski BH, et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci U S A. 1994;91:197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pesci EC, Milbank JB, Pearson JP, McKnight S, Kende AS, Greenberg EP, et al. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1999;96:11229–11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wade DS, Calfee MW, Rocha ER, Ling EA, Engstrom E, Coleman JP, et al. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J Bacteriol. 2005;187:4372–4380. doi: 10.1128/JB.187.13.4372-4380.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farrow JM 3rd, Sund ZM, Ellison ML, Wade DS, Coleman JP, Pesci EC. PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J Bacteriol. 2008;190:7043–7051. doi: 10.1128/JB.00753-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman JP, Hudson LL, McKnight SL, Farrow JM 3rd, Calfee MW, Lindsey CA, et al. Pseudomonas aeruginosa PqsA is an anthranilate-coenzyme A ligase. J Bacteriol. 2008;190:1247–1255. doi: 10.1128/JB.01140-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bredenbruch F, Nimtz M, Wray V, Morr M, Muller R, Haussler S. Biosynthetic pathway of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines. J Bacteriol. 2005;187:3630–3635. doi: 10.1128/JB.187.11.3630-3635.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dulcey CE, Dekimpe V, Fauvelle DA, Milot S, Groleau MC, Doucet N, et al. The end of an old hypothesis: the Pseudomonas signaling molecules 4-hydroxy-2-alkylquinolines derive from fatty acids, not 3-ketofatty acids. Chem Biol. 2013;20:1481–1491. doi: 10.1016/j.chembiol.2013.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schertzer JW, Brown SA, Whiteley M. Oxygen levels rapidly modulate Pseudomonas aeruginosa social behaviours via substrate limitation of PqsH. Mol Microbiol. 2010;77:1527–1538. doi: 10.1111/j.1365-2958.2010.07303.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D′Argenio DA, Calfee MW, Rainey PB, Pesci EC. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol. 2002;184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kojic M, Jovcic B, Vindigni A, Odreman F, Venturi V. Novel target genes of PsrA transcriptional regulator of Pseudomonas aeruginosa. FEMS Microbiol Lett. 2005;246:175–181. doi: 10.1016/j.femsle.2005.04.003 [DOI] [PubMed] [Google Scholar]
- 20.Kojic M, Venturi V. Regulation of rpoS gene expression in Pseudomonas: involvement of a TetR family regulator. J Bacteriol. 2001;183:3712–3720. doi: 10.1128/JB.183.12.3712-3720.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang Y, Nguyen DT, Son MS, Hoang TT. The Pseudomonas aeruginosa PsrA responds to long-chain fatty acid signals to regulate the fadBA5 beta-oxidation operon. Microbiology. 2008;154:1584–1598. [DOI] [PubMed] [Google Scholar]
- 22.Fujita Y, Matsuoka H, Hirooka K. Regulation of fatty acid metabolism in bacteria. Mol Microbiol. 2007;66:829–839. doi: 10.1111/j.1365-2958.2007.05947.x [DOI] [PubMed] [Google Scholar]
- 23.Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, et al. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989;17:3469–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, et al. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Research. 1989;17:3469–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holloway BW, Krishnapillai V, Morgan AF. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl Environ Microbiol. 2008;74:7422–7426. doi: 10.1128/AEM.01369-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. [DOI] [PubMed] [Google Scholar]
- 28.Pearson JP, Pesci EC, Iglewski BH. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whiteley M, Parsek MR, Greenberg EP. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J Bacteriol. 2000;182:4356–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tipton KA, Coleman JP, Pesci EC. QapR (PA5506) represses an operon that negatively affects the Pseudomonas quinolone signal in Pseudomonas aeruginosa. J Bacteriol. 2013;195:3433–3441. doi: 10.1128/JB.00448-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knoten CA, Hudson LL, Coleman JP, Farrow JM 3rd, Pesci EC. KynR, a Lrp/AsnC-type transcriptional regulator, directly controls the kynurenine pathway in Pseudomonas aeruginosa. J Bacteriol. 2011;193:6567–6575. doi: 10.1128/JB.05803-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warrens AN, Jones MD, Lechler RI. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene. 1997;186:29–35. [DOI] [PubMed] [Google Scholar]
- 33.Enderle PJ, Farwell MA. Electroporation of freshly plated Escherichia coli and Pseudomonas aeruginosa cells. Biotechniques. 1998;25:954–956. [DOI] [PubMed] [Google Scholar]
- 34.Choi KH, Kumar A, Schweizer HP. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods. 2006;64:391–397. doi: 10.1016/j.mimet.2005.06.001 [DOI] [PubMed] [Google Scholar]
- 35.Collier DN, Anderson L, McKnight SL, Noah TL, Knowles M, Boucher R, et al. A bacterial cell to cell signal in the lungs of cystic fibrosis patients. FEMS Microbiol Lett. 2002;215:41–46. [DOI] [PubMed] [Google Scholar]
- 36.Essar DW, Eberly L, Hadero A, Crawford IP. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172:884–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farrow JM 3rd, Pesci EC. Distal and proximal promoters co-regulate pqsR expression in Pseudomonas aeruginosa. Mol Microbiol. 2017;104:78–91. doi: 10.1111/mmi.13611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilbert KB, Kim TH, Gupta R, Greenberg EP, Schuster M. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol Microbiol. 2009;73:1072–1085. doi: 10.1111/j.1365-2958.2009.06832.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock RE. Induction by cationic antimicrobial peptides and involvement in intrinsic polymyxin and antimicrobial peptide resistance, biofilm formation, and swarming motility of PsrA in Pseudomonas aeruginosa. J Bacteriol. 2008;190:5624–5634. doi: 10.1128/JB.00594-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang Y, Lunin VV, Skarina T, Savchenko A, Schurr MJ, Hoang TT. The long-chain fatty acid sensor, PsrA, modulates the expression of rpoS and the type III secretion exsCEBA operon in Pseudomonas aeruginosa. Mol Microbiol. 2009;73:120–136. doi: 10.1111/j.1365-2958.2009.06757.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pesci EC, Pearson JP, Seed PC, Iglewski BH. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGrath S, Wade DS, Pesci EC. Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol Lett. 2004;230:27–34. [DOI] [PubMed] [Google Scholar]
- 44.Miller RM, Tomaras AP, Barker AP, Voelker DR, Chan ED, Vasil AI, et al. Pseudomonas aeruginosa twitching motility-mediated chemotaxis towards phospholipids and fatty acids: specificity and metabolic requirements. J Bacteriol. 2008;190:4038–4049. doi: 10.1128/JB.00129-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang Y, Zarzycki-Siek J, Walton CB, Norris MH, Hoang TT. Multiple FadD acyl-CoA synthetases contribute to differential fatty acid degradation and virulence in Pseudomonas aeruginosa. PLoS One. 2010;5:e13557 doi: 10.1371/journal.pone.0013557 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer locations are indicated by the letters on the gene map and can be matched to the reactions electrophoresed on the agarose gels. For each primer set, lane 1 contains an RT-PCR reaction in which no reverse transcriptase was added (negative control); lane 2 contains a reaction in which chromosomal DNA was added (positive control); and lane 3 contains the experimental reaction.
(TIF)
qRT-PCR was performed using RNA from strains PAO1 and PGW-ΔpsrA containing either a control plasmid or one with an inducible promoter that controls PA0506 (pHERD20T and pGW-21, respectively). All cultures were supplemented with 0.5% L-arabinose except for a control culture of strain PAO1 (pHERD20T), which served as a reference for which the fold change value was set at 1. Data from three independent repeats are presented as average fold change ± SD of expression of PA0506 as compared to the reference culture.
(TIF)
Strain PAO1 harboring pHERD20T (control plasmid), pGW-21, pGW-18 or pGW-20 (PA0506, PA0507, or PA0508 expression plasmids, respectively) were grown for 24 h in LB medium supplemented with 0.5% L-arabinose. PQS was then extracted and quantified as described in Materials and Methods. Data are presented as the average ± SD of three independent experiments.
(TIF)
The indicated strains harboring pGW-22 (fadBA5 expression plasmid) were grown for 24 h in LB medium with or without 0.5% L-arabinose as indicated by a plus or minus symbol, respectively. PQS was then extracted and quantified as described in Materials and Methods. Data are presented as the average ± SD of three independent experiments.
(TIF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.