Summary
The Hippo/Yki and RB/E2F pathways both regulate tissue growth by affecting cell proliferation and survival, but interactions between these parallel control systems are poorly defined. In this study, we demonstrate that interaction between Drosophila E2F1 and Sd disrupts Yki/Sd complex formation and thereby suppresses Yki target gene expression. RBF modifies these effects by reducing E2F1/Sd interaction. This regulation has significant effects on apoptosis, organ size, and progenitor cell proliferation. Using a combination of DamID-seq and RNA-seq, we identified a set of Yki targets that play a diversity of roles during development and are suppressed by E2F1. Further, we found that human E2F1 competes with YAP for TEAD1 binding, affecting YAP activity, indicating that this mode of cross-regulation is conserved. In sum, our study uncovers a previously unknown mechanism in which RBF and E2F1 modify Hippo signaling responses to modulate apoptosis, organ growth, and homeostasis.
Keywords: Hippo signaling, RBF, E2F1, Yorkie, scalloped, YAP, TEAD1, transcriptional repression, organ size control, apoptosis
Graphical Abstract
Highlights
-
•
RBF/E2F1 regulates the Hippo pathway by modulating formation of Yki/Sd complexes
-
•
E2F1 releases Yki:Sd association and suppresses a set of Yki target expression
-
•
Human E2F1 competes with YAP for TEAD1 binding and affects YAP activity
Zhang et al. uncover a mechanism whereby Drosophila RBF, via E2F1, regulates Hippo signaling by modulating the formation of Yki/Sd complexes, thereby controlling Yki transcriptional outputs and apoptosis, organ size, and gut homeostasis regulation. The RB and E2F interaction is conserved in human cells and may broadly impact tissue homeostasis.
Introduction
The balance between cell proliferation and apoptosis plays a fundamental role during development and tissue homeostasis. Perturbations in this balance often lead to cancer or degenerative diseases. The Hippo signaling pathway is an evolutionarily conserved kinase cascade that has been established as a key regulator of organ size and tissue homeostasis (Yu et al., 2015, Johnson and Halder, 2014, Pan, 2010) in Drosophila and mice. Core components of this pathway include the Hippo kinase (Hpo or MST1/2 in mammals), which phosphorylates and activates Warts (Wts or LATS1/2 in mammals), which in turn phosphorylates and inactivates the transcriptional coactivator Yorkie (Yki or YAP/TAZ in mammals) by targeting it for nuclear export and degradation (Yu et al., 2015, Johnson and Halder, 2014, Pan, 2010). Yki/YAP/TAZ trigger the transcription of target genes, the best characterized of which either promote cell proliferation or suppress apoptosis and thereby affect tissue growth (Pan, 2010). Abnormally elevated YAP/TAZ levels and nuclear enrichment of these proteins have been observed in various human cancers (Yu et al., 2015, Johnson and Halder, 2014), while hyperactivation of Hpo promotes apoptosis (Pantalacci et al., 2003, Udan et al., 2003), suggesting that proper control of Hippo signaling is crucial for tissue homeostasis. As transcriptional coactivators that lack a DNA-binding domain, Yki/YAP must interact with the DNA-binding transcription factors, namely Scalloped (Sd or TEAD1-4 in mammals), to effect target gene expression (Goulev et al., 2008, Wu et al., 2008, Zhang et al., 2008, Zhao et al., 2008). Genome-wide chromatin-binding analyses revealed that many of the effects of Yki/YAP/TAZ on transcriptional activity occur via distal enhancers (Stein et al., 2015, Zanconato et al., 2015, Oh et al., 2013), suggesting that Yki/YAP/TAZ recruit various transcription factors, chromatin modulators, or epigenetic markers to regulate expression of their targets. This idea has been confirmed in recent studies that showed Yki/YAP/TAZ can interact not only with Sd/TEADs family proteins but also with GAGA factors, SWI/SNF complex subunits, Nuclear receptor coactivator 6 (Ncoa6), and the ecdysone receptor coactivator Taiman (Tai) in various tissue contexts (Zhang et al., 2015, Zhu et al., 2015, Qing et al., 2014, Skibinski et al., 2014, Jin et al., 2013, Oh et al., 2013). It also has been shown that the Tondu-domain-containing growth inhibitor (Tgi) (VGLL4 in mammals) directly competes with Yki for Sd binding, resulting in inhibition of Yki-regulated transcription (Guo et al., 2013, Koontz et al., 2013). In mammals, VGLL4 competes with YAP for TEAD4 binding via a similar mechanism (Zhang et al., 2014). In the presence of active Hippo signaling, the lack of nuclear Yki/YAP allows Tgi/VGLL4 to complex with Sd/TEADs and promotes transcriptional repression. When Hippo signaling is silenced, Yki/YAP translocate into the nucleus and displace Tgi/VGLL4 from Sd/TEADs, leading to transcriptional activation and tissue growth (Koontz et al., 2013). These studies yielded a model in which Sd/TEADs could act as a platform for integrating different signal inputs to regulate tissue homeostasis. Since Hippo signaling has been found to cross-talk with multiple pathways in a context-dependent manner (Yu et al., 2015), we may consider Hippo signaling as a complex network rather than a single linear pathway. However, how Hippo signaling might synergize or compete with other signaling and transcription control systems that affect cell proliferation and survival remains obscure.
The RB-E2F pathway has long been known as a critical regulator of cell-cycle progression. In this system, RB binds to E2F/Dp complexes and recruits chromatin and histone regulators to E2F-regulated promoters for repression of target genes (Stevaux and Dyson, 2002), many of which promote cell proliferation. Functional inactivation of RB relieves the repression from E2F/Dp complexes, which in turn promote cell-cycle progression. RB has been established as a tumor suppressor that normally suppresses cell proliferation. In addition, its activity affects cell differentiation and rates of apoptosis (Chau and Wang, 2003). Inactivation of Rb in mouse embryo induced aberrant apoptosis that can be inhibited by E2F1 deficiency, demonstrating a function as a suppressor of E2F1-mediated apoptosis (Tsai et al., 1998). Indeed, ectopic expression of E2F1 leads to apoptosis in Drosophila, cultured cells, and transgenic mice (van den Heuvel and Dyson, 2008). Consistently, E2F1 knockout mice show higher rates of tumor development and lower rates of thymic apoptosis (Nevins, 2001). In contrast, RB can also act in a pro-apoptotic manner by functioning in a transcriptionally active RB/E2F1 complex that promotes expression of pro-apoptotic genes in highly proliferative cells (Dick and Rubin, 2013). These studies suggest that the ability of RB/E2F1 to regulate tissue homeostasis (growth versus apoptosis) may be dictated by the cellular context, although its precise mechanisms of control are less well defined.
In this study, we found that Drosophila E2F1 and RBF regulate apoptosis via interactions with Hippo signaling, and thereby affect organ size and tissue homeostasis. Analysis of this interaction revealed that E2F1 competes with Yki for binding to Sd, and that this competition affects the expression of the Yki target genes Diap1, expanded (ex), and bantam (ban). Besides these classical Yki targets, our DamID-seq and RNA-seq data define a set of Yki targets that can be negatively regulated by E2F1. Gene ontology analysis showed that these genes play diverse roles during development and tissue homeostasis, suggesting that E2F1/Yki competition is a broad phenomenon. Furthermore, our data show that RBF levels affect the output of this competition and indicate that the mechanism is conserved in human cells, where YAP, E2F1, and TEAD1 show analogous interactions. In summary, our study uncovers a previously unknown mechanism in which RBF/RB regulates Hippo signaling by modulating the balance of Yki/Sd (or YAP/TEAD1 in human cells) activator and E2F1/Sd (or hE2F1/TEAD1 in human cells) repressor complexes.
Results
Drosophila RBF Positively Regulates Yki Target Genes
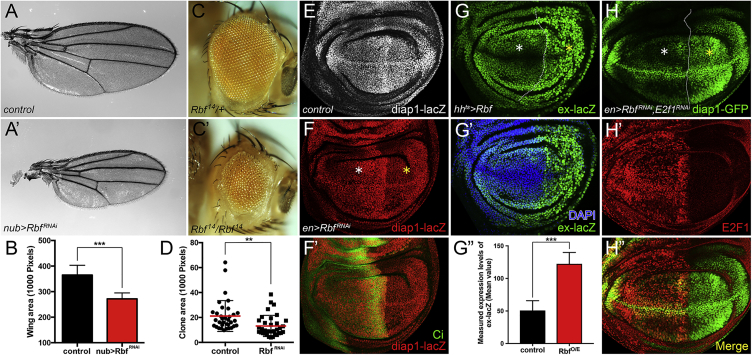
To identify new regulators for organ size control, we performed an RNAi screen in which we scored for alterations of Drosophila wing size. In the screen, we found that RNAi-mediated depletion of Rbf using nubGal4 resulted in the formation of abnormally small wings with normal wing vein patterning (Figures 1A–1B). Similarly, knockdown of Rbf using enGal4 reduced wing size specifically in the posterior compartment (Figures S1A and S1A′), and Rbf14/Rbf14 homozygous mutant eyes generated by the EGUF/hid method (Stowers and Schwarz, 1999) were significantly reduced in size (Figures 1C and 1C′). In the developing larval wing, Rbf-depleted cell clones displayed significantly reduced size (Figure 1D), as also shown in Datar et al. (2000). These observations indicate that Rbf is essential for normal cell and tissue growth. Although this is surprising when one considers RBF’s function in restraining cell proliferation (Classon and Harlow, 2002), it is consistent with previous reports from Drosophila (Tanaka-Matakatsu et al., 2009, Datar et al., 2000) that suggest that Rbf mutant cells are sub-viable.
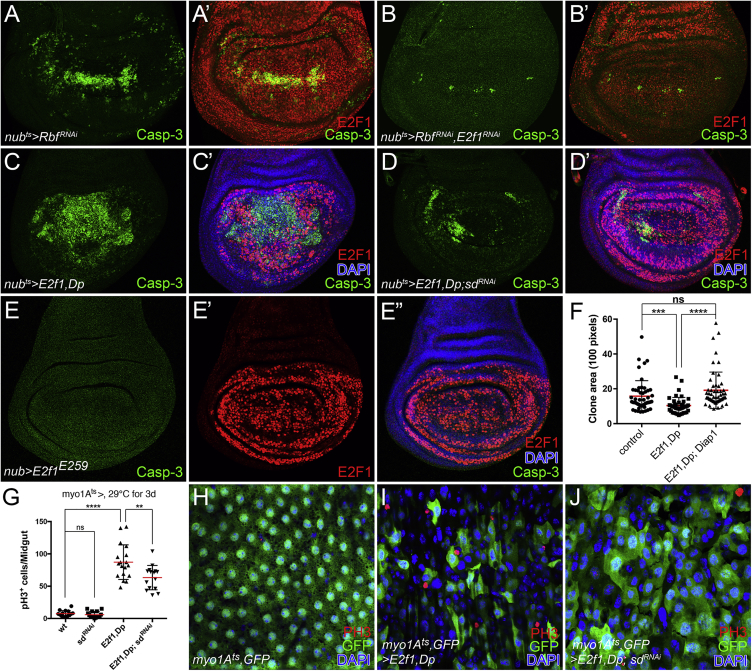
Figure 1.
Drosophila RBF Positively Regulates Yki Target Genes
(A and A′) Adult wing size: (A) nubGal4/+ control, (A′) Rbf knockdown driven by nubGal4. The pictures were taken under the same magnification.
(B) Quantification of adult wing sizes of control versus RbfRNAi (mean ± SD, n = 5, t test, ∗∗∗p < 0.001).
(C and C′) Adult eye of Rbf14/+ (C) and Rbf14/Rbf14 (C′) flies.
(D) Quantification of clone area of flip-out clones: control versus RbfRNAi (mean ± SD, n = 34, t test, ∗∗p < 0.01).
(E–F′) Diap1-lacZ expression in control (E) and Rbf knockdown (F and F′) wing discs. (F′) anterior compartment marked by Cubitus interruptus (Ci) staining.
(G–G″) Rbf overexpression in the posterior compartment was driven by hhGal4, tubGal80ts (hhts). The larvae were raised at 18°C, and then shifted to 29°C for 48 hr before dissection in third-instar stage. Nuclei were stained by DAPI. (G″) Fluorescent intensity of ex-lacZ in the anterior compartment versus the RBF-overexpressed posterior compartment was measured by ImageJ. The mean values represent the expression levels of ex-lacZ. Statistical analysis shows significant enhancement of ex-lacZ levels in the RBF-overexpressed posterior compartment (mean ± SD, t test, n = 5, ∗∗∗p < 0.001).
(H–H″) UAS-RbfRNAi and UAS-E2f1RNAi were coexpressed using enGal4.
White asterisks (F, G, and H) indicate the normal expression levels of each indicated reporter in anterior compartments. Yellow asterisks (F, G, and H) indicate the altered expression levels of each reporter upon different indicated genetic manipulations in posterior compartments.
In investigating RBF’s growth regulatory function, we tested interactions with Hippo/Yki signaling, a highly conserved pathway that also regulates cell proliferation and organ size. We first investigated whether RBF is required for the expression of Yki target genes. In the wing disc, RNAi depletion of Rbf led to a significant reduction in the expression of Diap1-lacZ (Figures 1E–1F′), a well-characterized Yki target. Knockdown of Rbf using mirrGal4 in the dorsal part of the eye disc also reduced Diap1-GFP expression (Figure S1B). Consistently, overexpression of CycD/Cdk4, whose kinase activity can inactivate RBF (Connell-Crowley et al., 1997), also suppressed the expression of Diap1-lacZ (Figure S1C). Conversely, overexpression of Rbf significantly increased the expression of Diap1-GFP (Figures S1D and S1D′) and another Yki target, ex-lacZ (Figures 1G–1G″). Thus, RBF regulates the expression of Yki target genes.
E2F1/Dp Negatively Regulates Yki Target Genes
Given that a major function of RBF is to act as a corepressor for the E2F1 and E2F2 transcription factors, we examined whether RBF regulated Hippo signaling in an E2F-dependent manner. We found that the reduction of Diap1-GFP induced by the depletion of Rbf could be reversed by E2f1 knockdown (Figures 1H–1H″). This suggests that RBF could affect organ size through E2F1 and Yki. Further tests showed that overexpression of E2f1 significantly decreased levels of Diap1-GFP (Figures 2A–2B′). However, overexpression of E2f2, a non-essential paralog of E2f1 that acts as a repressor, showed no effect on Diap1-GFP expression (Figure S1E), suggesting a specific role for E2F1. We then investigated the suppressive effect of E2F1PIP3A, a stabilized but active form of E2F1 (Zielke et al., 2011), on Yki activity. Overexpressed E2f1PIP3A strongly reduced the expression of Diap1 as indicated by Diap1-lacZ and Diap1 staining (Figures S1F–S1F″).
Figure 2.
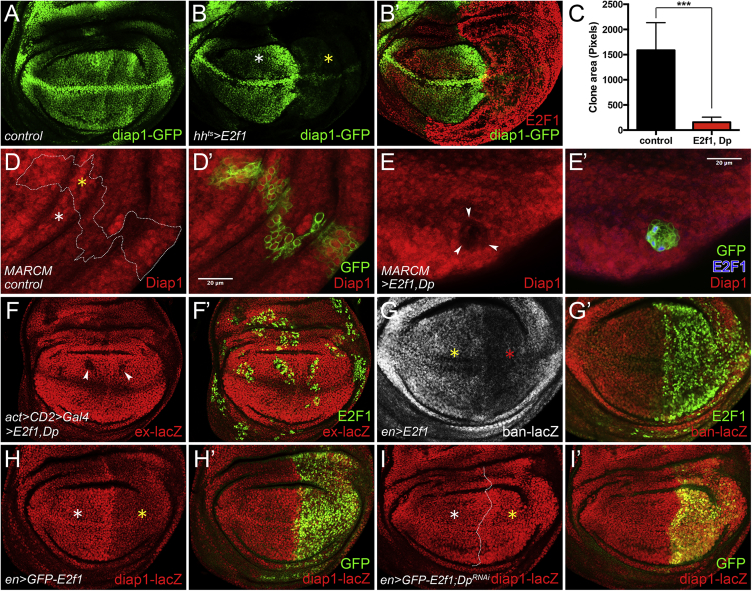
E2F1/Dp Negatively Regulates Yki Target Genes
(A) Diap1-GFP expression in control wing disc.
(B and B′) The larvae were raised at 18°C and then shifted to 29°C for 72 hr before dissection in third-instar stage. Overexpression of E2f1 in posterior part (indicated by yellow asterisk in B) of wing discs using hhts. Anterior compartment labelled by a white asterisk.
(C–E′) UAS-E2f1+Dp-overexpressing clones were generated using the MARCM system. (C) Quantification of clone area of MARCM clones: control versus E2f1+Dp (1,587 ± 182.0 versus 158.1 ± 49.12, mean ± SD, t test, ∗∗∗p < 0.001). (D–E′) Diap1 staining in control (D and D′) and E2f1+Dp overexpression (E and E′) wing discs. Yellow asterisk (D) indicates Diap1 in a control MARCM clone, while the white asterisk (D) indicates Diap1 expression outside of the control MARCM clone. Arrowheads in (E) indicate reduced Diap1 levels in a E2f1+Dp-overexpressing clone.
(F and F′) ex-lacZ levels were detected in flip-out clones overexpressing E2f1+Dp. Arrowheads in (F) showed the reduction of ex-lacZ.
(G and G′) E2f1 overexpression was driven by enGal4. Red asterisk in (G) indicated the decreasing of ban-lacZ. White asterisk in (G) indicates the normal levels of ban-lacZ in the anterior compartment.
(H and H′) GFP-E2f1 was overexpressed using enGal4.
(I and I′) UAS-GFP-E2f1 and UAS-DpRNAi were coexpressed using enGal4. White asterisks (H and I) label the normal expression of diap1-lacZ in anterior compartments, while the yellow asterisks indicate expression of diap1-lacZ upon the indicated genetic manipulations in posterior compartments.
Scale bars, 20 μm.
Next, we performed a MARCM clone assay to investigate whether E2f1 overexpression might inhibit tissue growth in the wing disc. Cell clones overexpressing E2f1 and Dp grew significantly less than wild-type controls (Figures 2C–2E′), and the expression of Diap1 in these clones was reduced, suggesting that the transcriptional activity of Yki was suppressed (Figure 2E). Consistent with this, clones overexpressing E2f1+Dp showed less ex-lacZ expression (Figures 2F and 2F′). Overexpression of E2f1 also led to a significant reduction in the expression of ban-lacZ (Figures 2G and 2G′), another established target of Yki (Nolo et al., 2006, Thompson and Cohen, 2006). Since E2F1 requires complex formation with Dp to function, we asked whether the E2F1-mediated suppression of Yki targets was Dp-dependent. As expected, knockdown of Dp blocked the inhibitory effect of E2F1 on the expression of Diap1-lacZ (Figures 2H–2I′). These results demonstrate that E2F1 suppresses Yki activity in a Dp-dependent manner.
Yki-E2F1 Epistasis
We next investigated the epistatic relationships between E2F1 and Hippo pathway components. We found that depletion of hpo using RNAi did not affect the reduction in Diap1-GFP expression caused by E2f1+Dp overexpression (Figures 3A and 3A′). This suggests that the suppression of Yki target gene expression by E2F1 occurs downstream of Hpo. We checked whether the transcription of the Yki dimerization partner, scalloped (sd) might be regulated by E2F1. However, levels of sd-lacZ were not affected by ectopic E2f1+Dp (Figures 3B and 3B′), suggesting that E2F1 does not suppress Yki activity by controlling sd expression. Interestingly, in cells overexpressing E2f1+Dp, protein levels of Yki were reduced (Figures 3C–3D′), despite a lack of change in levels of yki mRNA (Figure 3E). Knockdown of E2f1, however, caused no detectable change in the protein levels of Yki (Figures 3F and 3F′). This observation suggested that overexpression of yki might rescue the E2f1 overexpression phenotype. Indeed, overexpression of a constitutively active mutant version of Yki, YkiS168A, reversed the loss of ex-lacZ expression caused by high levels of E2f1 (Figures 3G–3H′). Taken together, these data indicate that E2F1 acts in parallel or upstream of Yki to regulate the expression of Hippo pathway targets (e.g., Diap1 and ex).
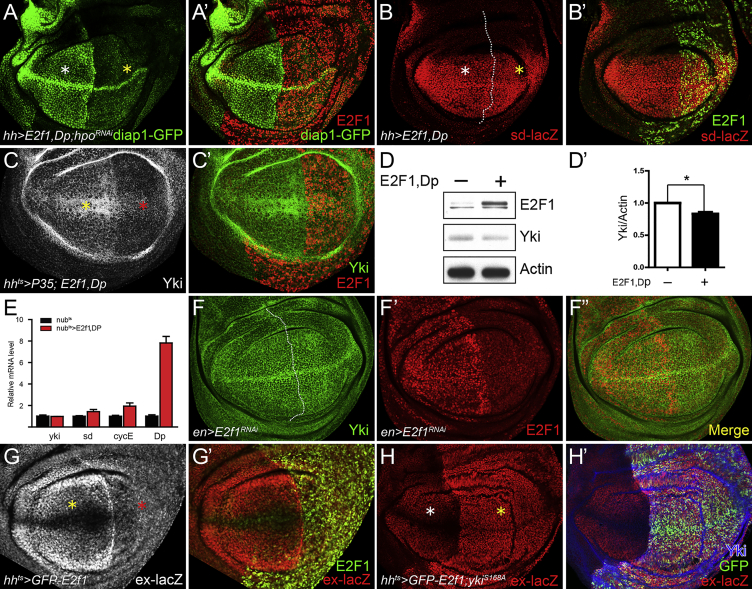
Figure 3.
Yki-E2F1 Epistasis
(A–B′) The larvae were raised at 18°C. (A and A′) UAS-E2f1+Dp and UAS-hpoRNAi were coexpressed using hhGal4. Yellow asterisk in (A) indicates the decrease of Diap1-GFP expression. White asterisk (A) indicates normal expression levels of diap1-GFP in the anterior compartment.
(B and B′) UAS-E2f1+Dp was overexpressed using hhGal4. Transcription levels of sd were indicated by sd-lacZ reporter. White asterisk in (B) indicates the normal expression levels of sd-lacZ in the anterior compartment.
(C and C′) Larvae were raised at 18°C and then shifted to 29°C for 48 hr before dissection in third-instar stage. UAS-P35 and UAS-E2f1+Dp were coexpressed using hhts. P35 was coexpressed to block the potential side effect that apoptosis induces universal reduction of total protein levels. The reduction of Yki protein levels is indicated by a red asterisk in (C). The yellow asterisk indicates the normal expression of Yki in the anterior compartment.
(D and E) Larvae were raised at 18°C and then shifted to 29°C for 48 hr before dissection in third-instar stage. The control and E2f1+Dp overexpression (driven by nubGal4, tubGal80ts, henceforth referred to as nubts) wing discs were dissected for western blot and qRT-PCR. (D′) The relative levels of Yki as shown in (D) were quantified after normalization against actin. Values represent mean ± SEM (n = 3, t test, ∗p < 0.05).
(F–F″) UAS-E2f1RNAi was overexpressed using enGal4. Anterior/posterior boundary in (F) is shown by the dotted line.
(G and G′) Larvae were raised at 18°C, and then shifted to 29°C for 48 hr before dissection in third-instar stage. (G and G′) GFP-E2f1 overexpression was driven by hhts. The decrease of ex-lacZ is indicated by the red asterisk. Yellow asterisk in (G) indicates normal expression of ex-lacZ in the anterior compartment.
(H and H′) UAS-GFP-E2f1 and UAS-ykiS168A were coexpressed using hhts. The E2f1-induced ex-lacZ reduction (red asterisk in G) was reversed by ykiS168A overexpression (yellow asterisk in H). White asterisk in (H) indicates normal expression of ex-lacZ in the anterior compartment.
We next investigated the effects of loss of E2F1 function on the expression of Yki targets. In E2f1-depleted cells, the expression of either Diap1-GFP (Figures S2A and S2A′) or ex-lacZ (Figures S2C and S2C′) was increased. Similarly, knockdown of Dp also significantly increased the expression of Diap1-GFP (Figures S2B and S2B′). Further, the upregulation of ex caused by E2f1-RNAi could be reversed by depletion of yki (Figures S2D and S2D′). Moreover, the upregulation of Diap1 caused by hpo knockdown (Figures S2E and S2E′) or yki overexpression (Figures S2G and S2G′) could be strengthened by coexpression of E2f1-RNAi (Figures S2F and S2F′ and Figures S2H and S2H′, respectively). Overall these results are consistent with the conclusion that E2F1 acts as a repressor of Yki target gene expression.
In the course of these experiments we also discovered that the expression of an E2f1 transcriptional reporter, e2f1-lacZ (Figure S2I), E2F1 protein (Figure S2J), and the E2F1 target gene PCNA-GFP (Figures S2K and S2K′) were all upregulated by yki overexpression. These results confirm previous observations suggesting that E2f1 is a downstream target of Hippo signaling (Goulev et al., 2008, Nicolay and Frolov, 2008). In the context of our other results, this relationship suggests that Yki might negatively regulate its own activity by stimulating E2F1 expression. This kind of regulation, wherein a transcriptional activator indirectly suppresses its own targets, is not infrequent in nature and has been termed “incoherent” regulation.
E2F1 DNA Binding Is Required for Interaction with Yki/Sd
We next examined the biochemical basis underlying E2F1’s suppression of Yki activity. We found that both E2F1 and Dp could be coimmunoprecipitated (coIPed) with Yki’s transcriptional coactivator, Sd from S2 or HEK293 cells (Figures 4A and S3A). However, no direct interaction was detected between E2F1 and Yki (Figures 4B and S3B). Interestingly, overexpression of E2F1 reduced the amount of Yki pulled down by Sd (Figure 4C), and conversely, overexpression of Yki diminished the amount of E2F1 pulled down by Sd (Figure 4D). Moreover, increasing concentrations of E2F1 decreased the amount of Yki bound to Sd (Figure S3C). These results indicate that E2F1 can compete with Yki for Sd interaction. This competition mechanism is similar to that reported for Tgi, which was demonstrated to play a role in Sd-mediated default repression (Guo et al., 2013, Koontz et al., 2013). Hence, we reasoned that, if E2F1 uses a similar mechanism, knockdown of sd should relieve the suppression of Yki targets by E2F1. Confirming this prediction, knockdown of sd partially reversed the reduction of Diap1-lacZ caused by overexpressed E2f1 (Figures S3D–S3E′). These results suggest that E2F1 competes with Yki for Sd interaction and is required for Sd-mediated default repression of Diap1 in the wing disc. Furthermore, overexpression of Rbf diminished the amount of E2F1 pulled down by Sd (Figure S4F), suggesting that RBF reflects the competition of E2F1 and Yki for Sd interaction.
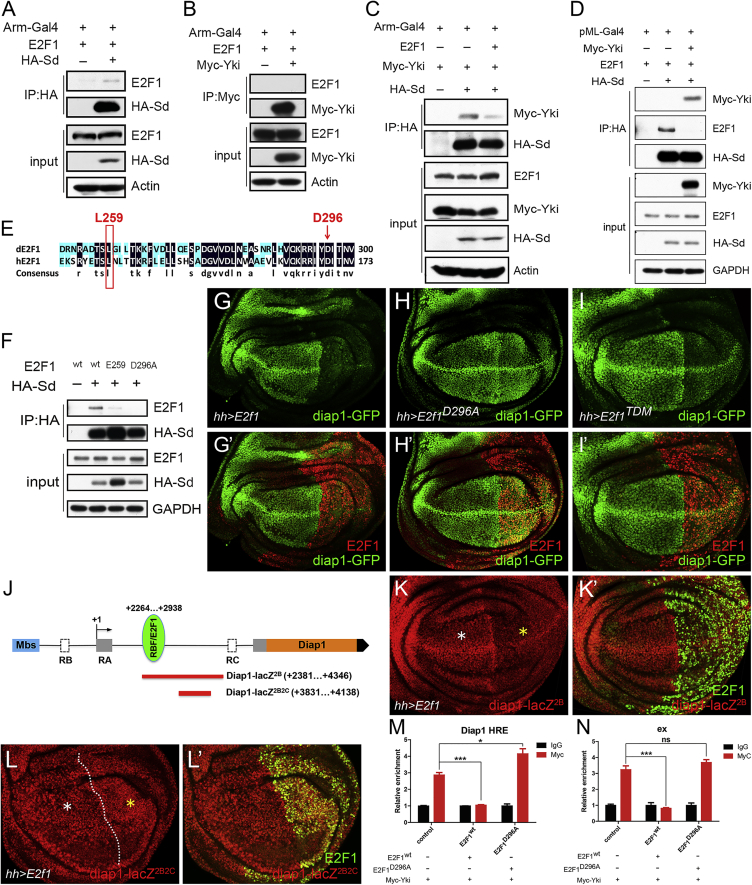
Figure 4.
E2F1 DNA Binding Is Required for Interaction with Yki/Sd
(A–C) The indicated plasmids were transfected into Drosophila S2 cells (A–C) and HEK293T cells (D and F), respectively, for coIP assays. E2F1 coIPed with HA-Sd (A) rather than with My-Yki (B). (C) Overexpression of E2F1 diminished the amount of Myc-Yki pulled down by HA-Sd.
(D) Overexpression of Myc-Yki reduced the interaction between E2F1 and HA-Sd.
(E) Sequence alignment of the DNA-binding domains of Drosophila E2F1 (dE2F1) and human E2F1 (hE2F1) with DNAMAN software, followed by manual refinement.
(F) To create the E2F1E259 mutant, the DNA sequence at Drosophila E2F1 codons L259 and G260 (CTGGGA) were replaced with an EcoRI restriction site sequence (GAATTC). E2F1D296A was generated using the site-directed mutagenesis system. Neither E2F1E259 nor E2F1D296A was pulled down by HA-Sd.
(G–I′) Compared with wild-type E2F1 (G and G′), neither E2F1E259 (H and H′) nor E2F1D296A (I and I′) showed suppression on the expression of Diap1-GFP.
(J) Schematic representation of the Diap1 loci and a putative binding site of RBF/E2F1. The binding of RBF/E2F1 on Diap1 was analyzed based on published data (Oh et al., 2013, Korenjak et al., 2012). Diap1-lacZ2B, but not Diap1-lacZ2B2C, contained this binding site.
(K–L′) Overexpression of E2f1 was driven by hhGal4, respectively. The levels of Diap1-lacZ2B (K and K′), rather than Diap1-lacZ2B2C (L and L′), was decreased by E2F1 overexpression. White asterisks (K and L) indicate the normal expression of each indicated reporter in anterior compartments, whereas yellow asterisks (K and L) indicate the altered expression of each reporter upon E2f1 overexpression in posterior compartments. Dotted line in (L) indicates the anterior/posterior compartment boundary.
(M and N) ChIP assays were performed in Drosophila S2 cells transfected with indicated plasmids. Chromatins were precipitated by control IgG or Myc antibody. The enrichment of ChIP products on the intronic Hippo-responsive element (HRE) of diap1 and ex promoter were measured by real-time PCR (mean ± SEM, n = 3, t test, ∗p < 0.05, ∗∗∗p < 0.001, ns, not significant).
We next tested whether the DNA-binding activity of E2F1 is required for its repression of Yki activity. In previous studies in mammalian cells, the L132 site of human E2F1 was reported to be essential for DNA binding and activation (Cress et al., 1993). By sequence alignment analysis, amino acid hE2F1L132 was found to be conserved as E2F1L259 in Drosophila (Figure 4E). The DNA sequence at Drosophila E2F1 codons L259 and G260 was replaced with an EcoRI restriction site sequence (here designated as E2F1E259). In addition, E2F1D296, which is conserved with hE2F1D169, is required for DNA binding and transcriptional activation in Drosophila (Royzman et al., 1999). A D296 to A296 point mutation of E2F1 (E2F1D296A) was generated. In coIP assays, neither E2F1E259 nor E2F1D296A were pulled down efficiently with HA-Sd (Figure 4F). Furthermore, overexpression of either E2f1D296A or E2f1E259 did not suppress Diap1-GFP expression (Figures 4G–4H′, S3H, and S3H′). By chromatin immunoprecipitation (ChIP) assays, we found that overexpression of wild-type E2f1 significantly reduced the Yki binding on the intronic Hippo-responsive element (HRE) of diap1 (Figure 4M) and the promoter region of ex (Figure 4N). However, E2f1D296A lost the repression of Yki binding on Diap1 and ex (Figures 4M and 4N). These results demonstrate E2F1’s DNA-binding domain is required for it to interact with Sd and suppress Yki target genes.
Further, we undertook to map the interacting region on E2F1 and Sd. CoIP data showed that both HA-SdN, which contains the DNA-binding domain of Sd, and HA-SdC, which lacks the DNA-binding domain, could interact with E2F1 (Figure S3G). This surprising result indicates that the DNA-binding domain of Sd is not required for its interaction with E2F1. Moreover, through analyzing published E2F1 and RBF ChIP data from third-instar larvae (Korenjak et al., 2012), we found a potential binding site of RBF/E2F1, which is close to the intronic HRE of Diap1 (Figure 4J). Hence, we tested whether a reporter gene that lacks this RBF/E2F1 binding site but still has Sd binding sites could be regulated by E2F1. Wu et al. (2008) generated such constructs. The longer form of their reporter, Diap1-lacZ2B, contains the potential RBF/E2F1 binding site, while the shorter form of the reporter, Diap1-lacZ2B2C, lacks this site (Figure 4J). Consistent with our expectation, we found that the expression of Diap1-lacZ2B but not Diap1-lacZ2B2C could be suppressed by E2f1 overexpression (Figures 4K–4L′). To determine if the competition between E2F1 and Yki depends on the transactivating function of E2F1, we generated a truncation mutant of the transactivation domain of E2f1, E2f1TDM, in which the Q525 was mutated to a stop codon (Royzman et al., 1999). We found that E2f1TDM could interact with HA-Sd by coIP (Figure S3I), and that overexpressed E2f1TDM could suppress Diap1-GFP (Figures 4I and 4I′). By further in vitro binding assay, we found that E2F1 physically binds to Sd, and Yki could diminish this physical interaction (Figure S3J). These results indicate that the competition effect and the binding of E2F1 to Sd do not require E2F1’s transactivation domain.
To further investigate the potential interactions between Yki and E2F1 on target gene promoters, published E2F1 ChIP-chip data from third-instar larvae (Korenjak et al., 2012) and Yki ChIP-seq data from third-instar wing discs (Oh et al., 2013) were analyzed. The nearest promoter for each binding peak was determined according to FlyBase gene annotation. Compared with randomized peaks distributed in the Drosophila genome, both Yki binding peaks and E2F1 binding peaks were enriched in promoter regions (Figure S4A). Focusing on genes targeted by both Yki and E2F1 (i.e., both Yki and E2F1 peaks detected within a ±1 kb region surrounding their transcription start sites), Yki and E2F1 peaks were significantly overlapped according to a Jaccard index calculation (Figure S4B). Overall, these data indicate that E2F and Yki/Sd bind adjacent sites in the vicinity of target gene promoters. This genomic colocalization may facilitate the competition between E2F1/Dp and Yki/Sd that affects target gene expression.
E2F1/Sd Interactions Modulate Apoptosis
Since loss of Rbf function reduced wing and eye sizes, RBF may play a role in cell survival during organ development. We found that knockdown of Rbf induced obvious apoptosis in wing discs, as indicated by cleaved caspase-3 staining (Figures 5A and 5A′). This apoptosis could be suppressed by simultaneous E2f1 knockdown (Figures 5B and 5B′), suggesting that Rbf loss promotes apoptosis by potentiating E2F1. Consistent with this and a previous report (Neufeld et al., 1998), overexpression of E2f1+Dp caused significant cell death in wing discs (Figures 5C and 5C′). Interestingly, this phenotype was significantly diminished by sd knockdown, suggesting that E2F1 may induce apoptosis by interacting with Sd (Figures 5D and 5D′). This led us to ask whether E2F1 triggers cell death via its DNA-binding activity. Indeed, the E2F1E259 DNA-binding mutant had a greatly reduced ability to induce apoptosis (Figures 5E–5E″), indicating that DNA binding of E2F1 is required. Further, overexpression of diap1 successfully restored the reduced clone size caused by E2f1+Dp overexpression (Figure 5F), indicating that the reduction in clone size was likely due to increased apoptosis. Taken together, our data are consistent with a model in which RBF/E2F1-regulated apoptosis depends on the balance of pro-apoptotic E2F1/Sd complexes and anti-apoptotic Yki/Sd complexes.
Figure 5.
E2F1/Sd Interactions Modulate Apoptosis and Intestinal Stem Cell Proliferation
(A–B′) Larvae were raised at 18°C and then shifted to 29°C for 48 hr before dissection in third-instar stage. Rbf knockdown or Rbf/E2f1 double knockdown was driven by nubts. Apoptosis was detected by caspase-3 staining.
(C–D′) Larvae were raised at 18°C and then shifted to 29°C for 36 hr before dissection in third-instar stage. E2f1+Dp overexpression or E2f1+Dp overexpression in conjunction with sd knockdown was driven by nubts, respectively. Apoptosis was detected by caspase-3 staining.
(E–E″) E2F1E259 lost the ability to induce apoptosis.
(F) Quantification of clone area of flip-out clones expressing different transgenes as indicated. Values represent mean ± SD (t test, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ns, not significant).
(G–J) Adult females were shifted to 29°C after eclosion for 3 days and midguts were stained with anti-GFP and anti-PH3 antibodies and DAPI. The enterocyte (EC)-specific driver, myo1A-Gal4, tubGal80ts, UAS-GFP (myo1Ats,GFP), was used to drive the expression of each of the transgenes shown in (G). ECs were labeled by GFP. Intestinal stem cell (ISC) mitosis was scored and quantified by PH3+ cells. Nuclei in (H, I, and J) were stained by DAPI. Quantification data shown in (G) represents the mean ± SD (t test, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, ns, not significant).
E2F1/Sd Interactions Modulate Intestinal Stem Cell Proliferation
Given that Hippo signaling plays critical roles in regulating intestinal stem cell (ISC) division during normal midgut homeostasis and regeneration (Karpowicz et al., 2010, Ren et al., 2010, Shaw et al., 2010, Staley and Irvine, 2010, Morris et al., 2006), we addressed the relevance of E2F1 to Hippo signaling during this process. As widely documented in previous studies (Jiang et al., 2011, Jiang et al., 2009), the apoptosis of midgut enterocytes (ECs) stimulates cytokine (Upd2, 3) and growth factor (Vn, Spi, Krn) production that non-cell autonomously triggers ICS mitoses and accelerates midgut turnover. Since E2f1 overexpression can induce apoptosis in many cell types, we investigated whether E2F1 non-cell autonomously regulates ISC mitosis by modifying Hippo signaling. Overexpression of E2f1+Dp in ECs, which are post-mitotic, robustly triggered the division of ISCs (Figure 5G). In addition, the number of mature GFP+ ECs was significantly reduced compared with wild-type control (Figures 5H and 5I). These observations are consistent with the induction of EC apoptosis by E2f1+Dp overexpression. Although RNAi-mediated depletion of sd in ECs had no detectable effect on ISC proliferation (Figure 5G), depletion of sd in ECs overexpressing E2f1+Dp significantly suppressed the non-cell autonomous stimulation of ISC proliferation induced by E2f1+Dp (Figures 5G and 5J). These results are consistent with those we obtained in wing discs and further support a model in which E2F1/Sd interactions promote apoptosis, whereas Yki/Sd interactions suppress apoptosis, and E2F1 and Yki compete for the interaction with Sd.
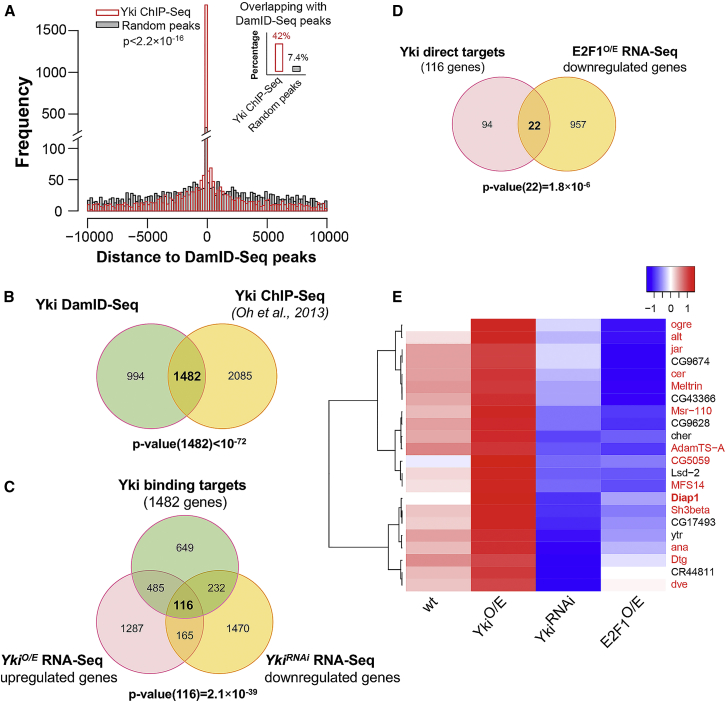
Genome-wide Interactions between Yki and E2F1
To investigate whether E2F1/Yki competition is restricted to a few targets, such as Diap1 and ex, or is a broader phenomenon, we used a combination of DamID-seq (Papagiannouli et al., 2014, Choksi et al., 2006, van Steensel et al., 2001) and RNA-seq to map Yki targets genome-wide in wing discs and assess how these are affected by E2F1. Using DamID-seq, we identified 4,986 binding sites with strong Dam-Yki binding (log2 fold change >3, false discovery rate [FDR] <0.001) when compared with a Dam-only control (Table S1). The gene set associated with these binding sites had a very significant overlap (p < 10−72, hypergeometric test) with a previous Yki ChIP study also performed on wing discs (Oh et al., 2013) (Figures 6A and 6B). Using RNA-seq, we identified 281 genes that could be upregulated by yki overexpression and downregulated by depletion of yki using RNAi (Figure 6C; Tables S2 and S3, FDR <0.05). As a way to identify potentially direct transcriptional targets of Yki, the RNA-seq datasets were cross-compared with our Yki DamID data and the Yki ChIP data of Oh et al. Among 281 genes transcriptionally regulated by Yki, 116 had Yki binding sites as assayed by both DamID and ChIP (Figure 6C and Table S4). This set of 116 genes defines likely direct transcriptional targets of Yki. Gene ontology analysis suggested that the above-mentioned 116 genes mediate a broad range of functions, including imaginal disc development and growth, regulation of programmed cell death, tissue homeostasis, stem cell fate commitment, etc (Table S5).
Figure 6.
Genome-wide Interactions between Yki and E2F1
(A) Histogram showing the distribution of Yki ChIP-seq peaks (Oh et al., 2013) to Yki DamID-seq peaks (red) and also the distance distribution of random peaks (by shuffling the ChIP-seq peaks in the genome) to Yki DamID-seq peaks as background (gray). Compared with the random peaks, the ChIP-seq peaks were significantly colocalized with DamID-seq peaks (p < 2.2×10−16, Kolmogorov-Smirnov test). The MACS software was used to identify the ChIP binding peaks of Yki. The p value cutoff was set to 10−5. If more than half the length of one peak region overlapped with an annotated gene, the peak was assigned to that gene.
(B) Venn diagram showing the overlap between Yki DamID-seq targets and Yki ChIP-seq targets in wing discs. The significance of overlap (p < 10−72) was assessed by hypergeometric test.
(C) Venn diagram showing the overlap of genes upregulated by yki overexpression, downregulated by ykiRNAi and associated with significant Yki binding. The significance of overlap (p = 2.1 × 10−39) was assessed by hypergeometric test. The overlapping 116 genes were categorized as direct Yki targets.
(D) Venn diagram showing the overlap between the 116 genes identified in (C) and genes downregulated by E2f1 overexpression (p = 1.8 × 10−6, hypergeometric test).
(E) Heatmap displaying alterations in mRNA expression of 22 genes shown in (D) after altering yki or E2f1 expression. Each row represents one gene. Normalized gene expression level is colored blue to red to indicate low to high. Genes marked in red have been shown to be differentially regulated during implantation and wound-induced wing disc regeneration process (Blanco et al., 2010).
In order to determine which Yki targets could be modulated by E2F1, we also generated RNA-seq data following overexpression of E2F1. This experiment identified 3,740 genes that were either activated or repressed by overexpressed E2F1 (Tables S6 and S7, FDR <0.05). By comparing these 116 direct Yki targets with data from RNA-seq profiling following E2F1 overexpression (Table S7), we identified 22 genes, including Diap1, that were transcriptionally downregulated by E2f1 overexpression (Figures 6D and 6E, p = 1.8 × 10−6, hypergeometric test). Interestingly, 14 of these 22 genes (red labels, Figure 6E) have been shown to be differentially regulated during implantation and wound-induced wing disc regeneration (Blanco et al., 2010). This suggests that E2F1-regulated Yki activity might play a role in damage-induced regeneration. In addition, jaguar (jar), a recently identified Hippo pathway regulator (Kwon et al., 2013), was found in this 22-gene list, suggesting another mechanism of feedback regulation in Hpo signaling. Altogether, our DamID-seq and RNA-seq analyses confirm that E2F1-Yki interactions affect many target genes and are likely to have diverse biological effects.
Human E2F1 Suppresses YAP Activity by Interacting with TEAD1
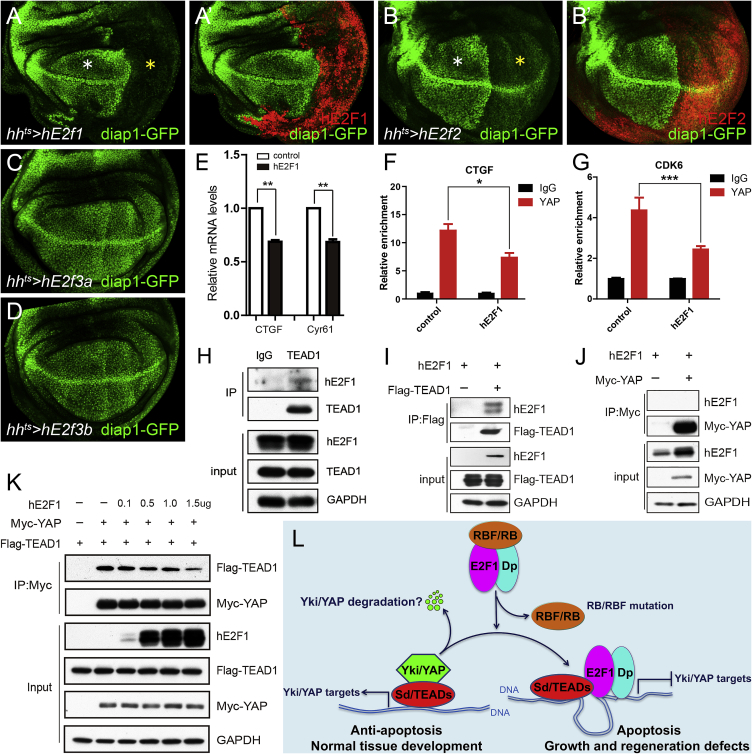
We next examined whether the Yki/E2F1 interaction we describe in Drosophila is conserved in mammalian cells. Drosophila E2f1 is an ortholog of human E2f1, E2f2, and E2f3. First, we generated transgenic flies carrying these human genes and tested their roles in regulating the Hippo pathway in Drosophila wing discs. We found that overexpression of either hE2f1 or hE2f2 dramatically reduced Diap1-GFP levels (Figures 7A–7B′). Overexpression of hE2f3a and hE2f3b, however, showed no repression of Yki activity (Figures 7C and 7D). These data suggest that human E2f1 and E2f2 are functionally conserved orthologs of Drosophila E2f1 with respect to the ability to regulate Yki activity.
Figure 7.
Human E2F1 Suppresses YAP Activity by Interacting with TEAD1
(A–D) UAS-hE2f1, 2, 3a, 3b were overexpressed driven by hhts, respectively. The larvae were raised at 18°C, and then shifted to 29°C for 24 hr before dissection in third-instar stage. White asterisks (A and B) indicate normal expression of diap1-GFP in anterior compartments, and yellow asterisks (A and B) indicate altered expression of diap1-GFP upon human E2f1 (or human E2f2) overexpression in posterior compartments.
(E) MCF7 cells were infected by hE2F1 adenovirus. The qPCR assay showed that hE2F1 represses the mRNA levels of CTGF and Cyr61. Values represent means ± SEM (n = 3, t test, ∗∗p < 0.01).
(F and G) ChIP assays were performed in HeLa cells transfected with indicated plasmids. Chromatin was precipitated by control IgG or endogenous YAP antibody. The enrichment of ChIP products on CTGF and CDK6 promoters were measured by real-time PCR (mean ± SEM, n = 3, t test, ∗p < 0.05, ∗∗∗p < 0.001).
(H) The endogenous coIP of hE2F1 and TEAD1 was conducted in HeLa cells.
(I and J) HEK293T cells were transfected with indicated plasmids and subjected to immunoprecipitation. hE2F1 coIPed with Flag-TEAD1 (I) but not Myc-YAP (J).
(K) Increasing concentrations (dose, 0.1–1.5 μg) of hE2F1 reduced the amount of Flag-TEAD1 pulled down by Myc-Yki.
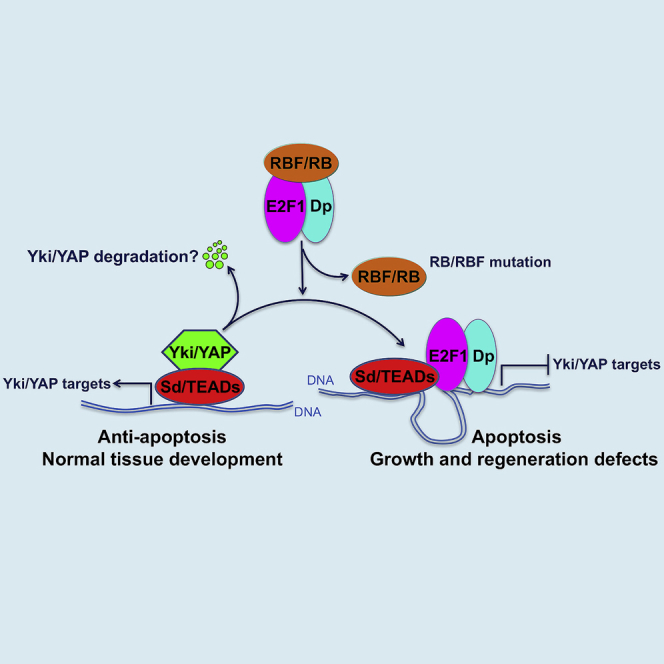
(L) The model shows that the interaction in which E2F1 suppresses Yki/YAP target gene expression by interfering with Yki/YAP binding to Sd/TEAD is functionally conserved between humans and Drosophila. This regulation has significant effects on apoptosis, organ growth, and homeostasis. A question mark indicates uncertainty or yet-to-be-established finding.
Next, we examined the ability of hE2f1 to inhibit the human ortholog of Yki, YAP, in mammalian cells. Overexpression of hE2f1 in MCF7 cells significantly inhibited the expression of CTGF and Cyr61, which are endogenous downstream targets of YAP (Figure 7E). Further, ChIP assays showed that overexpression of hE2f1 in HeLa cells significantly reduced the binding of YAP on its characterized targets, CTGF and CDK6 (Figures 7F and 7G). Consistently, coIP assays showed that the hE2F1 protein interacts with the Sd homolog TEAD1 (Figures 7H and 7I) but not YAP (Figures 7J), and that hE2F1 interferes with YAP binding to TEAD1 in a dose-dependent manner (Figure 7K), just as observed for the Drosophila orthologs of these genes. Taken together, these results indicate that the interaction in which E2F1 suppresses Yki/YAP target gene expression by interfering with Yki/YAP binding to Sd/TEAD is functionally conserved between humans and Drosophila (Figure 7L).
Discussion
The Hippo and RB-E2F signaling pathways are both highly conserved, and both regulate cell proliferation and survival in diverse scenarios. The overlapping functions of these two pathways suggest that their activities should be coordinated to regulate cell fates. However, few interactions between these pathways have been reported. In this study, we uncover a previously unknown mechanism in which RBF/RB, via E2F1, regulates Hippo signaling by modulating the formation of Yki/Sd (YAP/TEAD1) activator complexes. We show that Drosophila E2F1 counteracts the binding of Yki, a coactivator, to its DNA-binding partner, Sd. Yki/Sd complexes simulate transcription of target genes (Diap1, ex, and ban, etc) that promote cell growth and survival, whereas E2F1 releases Yki:Sd association and suppresses Yki target gene expression, thereby promoting apoptosis and other developmental defects. RBF modified these effects by reducing the activity of the E2F1/Sd repressor complexes, potentially also by a competitive mechanism. This competition mechanism is similar to that reported for Tgi, which was demonstrated to play a role in Sd-mediated default repression (Guo et al., 2013, Koontz et al., 2013). To determine how specific E2F1’s effects on Hippo signaling are, we examined whether E2F1 regulates the activity of four other signaling pathways: Notch (N), Wingless (Wg), Hedgehog (Hh), and Decapentaplegic (Dpp). Like Hpo signaling, each of these pathways is important for wing development. Tests in wing discs using reporter genes for these pathways did not reveal any modifying effects of E2F1 (Figures S5A–S5D″). These data suggest that E2F1’s ability to repress Yki activity is a pathway-specific mechanism, rather than a general transcriptional effect.
The Drosophila genome contains two E2f genes (E2f1 and E2f2). E2F1 (a functionally conserved homolog of mammalian E2F1-3) acts primarily as a transcriptional activator, whereas E2F2 (a functionally conserved homolog of mammalian E2F4-5) represses transcription (Gordon and Du, 2011). While either RB loss of function or E2F1 gain of function may sensitize cells to apoptosis, the mechanisms underlying these effects are not well understood. In the Drosophila wing, dE2F1-induced apoptosis is sensitive to the levels of the anti-apoptotic factor Diap1 (Morris et al., 2006). Since Diap1 is by many accounts a downstream target of Hippo signaling, this raises the possibility that RB/E2F1 might regulate apoptosis by controlling this branch of the Hippo pathway. Indeed, our clonal analysis showed that overexpression of Diap1 successfully restored the reduced clone size caused by E2f1 overexpression, strongly indicating that RBF/E2F1-regulated apoptosis depends on the balance of pro-apoptotic E2F1/Sd complexes and anti-apoptotic Yki/Sd complexes. A previous report showed that the induction of apoptosis in Drosophila by RBF depends on E2F2 (Clavier et al., 2014). However, we observed no suppression of Diap1 expression by E2F2, suggesting a specific role for RB-E2F1 signaling. This is consistent with previous findings showing that E2F-induced apoptosis is associated with some E2F family members (human and Drosophila E2F1) but not with others (Lazzerini Denchi and Helin, 2005, Moon et al., 2005). Further, our data confirmed previous observations suggesting that E2f1 is a downstream target of Hippo signaling (Goulev et al., 2008, Nicolay and Frolov, 2008) (Table S2). This suggests that Yki, via E2F1, might be able to suppress its own activity. Overall the results we present are consistent with a model in which E2F1 acts as a repressor of Yki target gene expression. While a previous study reported that Yki/Sd and E2F1 can activate a set of common targets (e.g., Dachs, PCNA, and Dp) in eye discs (Nicolay et al., 2011), our analysis did not find that these three genes were activated by both Yki/Sd and E2F1 in wing discs (Figures S6A–S6H′, Tables S2 and S6). These discrepancies are likely due to differences in cell type, which can have large effects on transcriptional outputs.
Our in vivo and in vitro data show that competitive interactions between E2F and Yki/Sd require E2F1’s DNA-binding activity. This raises the possibility that E2F1-mediated repression of Yki targets might be mediated through the transcriptional regulation of other unknown repressor(s), which thereby repress Yki activity via an indirect mechanism. To address this, we generated a transactivation domain-defective mutant form of E2f1, E2f1TDM. This mutant still showed strong inhibition on Diap1 expression, suggesting that the inhibition of Yki targets by E2F1 cannot be attributed to effects via E2F1’s transcriptional target(s). To further test this, we performed in vitro binding assays. These experiments showed that purified E2F1 and Sd physically interact quite strongly in vitro. Moreover, this in vitro interaction could be diminished by the presence of Yki (Figure S3J). Taken together, these data are consistent with the idea that E2F1 and Yki compete for Sd binding through direct protein-protein interaction. The fact that the E2F DNA-binding domain is required may indicate the involvement of a DNA bridging interaction or may suggest that E2F1 needs to bind to DNA to take on an active conformation that interacts with Sd. Through analyzing published E2F1 ChIP-chip data (Korenjak et al., 2012) and Yki ChIP-seq data (Oh et al., 2013), we found that E2F1 and Yki typically bind in close proximity to each other on common target loci (Figures S4A and S4B). These data suggest that the adjacent binding of E2F1 and Yki/Sd complex on certain regions of the genome may facilitate the negative effect of E2F1 on Yki. However, given our current data, we still cannot rule out the possibility that E2F1 recruits transcriptional repressor(s) to Yki targets, thereby repressing their expression.
To better understand whether the E2F1/Yki interaction we discovered is restricted to a few genes or is a broader phenomenon, we used a combination of RNA-seq and DamID-seq to investigate how Yki targets genome-wide could be affected by E2F1 activity. Our DamID-seq and RNA-seq data define a set of Yki targets that can be negatively regulated by E2F1. Many of these genes have been shown to be differentially regulated during implantation and wound-induced wing disc regeneration, suggesting that E2F1 might play an important role in regulating YAP-controlled regeneration process. Importantly, our data demonstrate that the E2F1/Yki competition is an evolutionarily conserved mechanism. First, overexpression of hE2f1 or hE2f2 in Drosophila wing discs dramatically suppressed expression of the Yki target Diap1. In addition, hE2F1 competed with YAP for binding to the Sd ortholog TEAD1, and effectively suppressed YAP target gene expression in human cells. While it has been previously reported that ectopic expression of hE2f1, hE2f2, and hE2f3 induce apoptosis (Classon and Harlow, 2002), our experiments suggest that only hE2F1 and hE2F2 retain the ability to repress YAP target genes. Overall, our study describes a previously unappreciated mechanism in which RB/E2F1 activity modifies Hippo signaling by modulating the formation of Yki/Sd activator and E2F1/Sd repressor complexes and thereby influences organ size control and tissue homeostasis.
Since cancerous malignancies are caused by multiple mutations (Loeb et al., 2003), a major challenge in the study of tumorigenesis is to determine the relative contribution of individual molecular lesions. While Rb is often mutated or inactivated in cancers, the singular loss of Rb function in mice increases apoptosis in some tissues, rather than leading to tumorigenesis (Classon and Harlow, 2002). Our discovery that E2F1 can suppress YAP activity provides one possible explanation for this conundrum. A previous study showed that low Wnt activity in Rb-deficient cells was due to E2F1-mediated abrogation of β-catenin target gene expression and induction of β-catenin degradation (Morris et al., 2008). The authors concluded that repression of β-catenin by E2F1 may contribute to E2F1-induced apoptosis. In conjunction with the findings reported here, we suggest that a block to apoptosis may be necessary for Rb mutations to contribute to tumorigenesis and that increased YAP activity may be one way to achieve this. Emerging evidence shows that the Hippo pathway is deregulated in many human cancers (Yu et al., 2015). However, it remains unknown whether the deregulation of the Hippo pathway contributes to the malignancy of Rb-deficient tumors by suppressing apoptosis. Although elevated levels and nuclear localization of YAP/TAZ occur in various human cancers, mutations in Hippo pathway genes are rare (Yu et al., 2015, Johnson and Halder, 2014). This indicates that the Hippo pathway may be regulated by many other pathways influencing oncogenesis. However, it is still unknown whether Yki activity is reduced to relatively low levels in cancer cells with defective RB function, or whether other oncogenic pathways bypass Hippo signaling to block RB mutation-induced apoptosis. This will be an interesting avenue for future study.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse α-β-galactosidase | Promega | Cat#Z3781; RRID: AB_430877 |
| Rat α-Ci | DHSB | 2A1; RRID: AB_2109711 |
| Guinea pig α-E2F1 | Xiaolin Bi (DMU) | N/A |
| Rabbit α-E2F1 | Xiaolin Bi (DMU) | N/A |
| Mouse α-En | DHSB | 4D9; RRID: AB_528224 |
| Mouse α-Diap1 | Bruce Hay (Caltech) | N/A |
| Rabbit α-Yki | Lei Zhang (SIBCB) | N/A |
| Rabbit α-hE2F1 | CST | Cat#3742; RRID: AB_2096936 |
| Rabbit α-hE2F2 | Santa Cruz Biotechnology | Cat#Sc-632; RRID: AB_2277708 |
| Rabbit α-Cleaved Caspase-3 | CST | Cat#9661; RRID: AB_2341188 |
| Chicken α-GFP | Thermo Fisher Scientific | Cat#A10262; RRID: AB_2534023 |
| Rabbit α-PH3 | Millipore | Cat#06-570; RRID: AB_310177 |
| Mouse α-Wg | DHSB | 4D4; RRID: AB_528512 |
| Guinea pig α-Sens | (Zhang et al., 2011) | N/A |
| Mouse α-Dll | Xinhua Lin (CCHMC) | N/A |
| Mouse α-Ptc | DHSB | Apa 1.3; RRID: AB_528441 |
| Mouse α-Smo | DHSB | 20C6; RRID: AB_528472 |
| Rabbit α-Sal | (Zhang et al., 2011) | N/A |
| Rat α-Dachs | David Strutt (University of Sheffield) | N/A |
| Mouse α-Dp | Nicholas Dyson (MGH) | N/A |
| Mouse α-V5 | Thermo Fisher Scientific | Cat#MA5-15253; RRID: AB_10977225 |
| EZview™ Red α-HA Affinity Gel | Sigma-Aldrich | Cat#E6779; RRID: AB_10109562 |
| EZview™ Red α-Flag Affinity Gel | Sigma-Aldrich | Cat#F2426; RRID: AB_2616449 |
| Mouse α-Myc | MBL International | Cat#M047-3; RRID: AB_591112 |
| Rabbit α-YAP | ABclonal | Cat#A1002 |
| Mouse α-Flag | Sigma-Aldrich | Cat#F4042; RRID: AB_439686 |
| Mouse α-HA | Santa Cruz Biotechnology | Cat# sc-7392; RRID: AB_627809 |
| Mouse α-TEF-1(TEAD1) | BD Biosciences | Cat#610923; RRID:AB_398238 |
| Mouse α-GAPDH | CWBiotech | Cat#CW0100M |
| Mouse α-β-actin | Proteintech Group | Cat#60008-1-Ig; RRID: AB_2289225 |
| Bacterial and Virus Strains | ||
| TOP10 | CWBiotech | Cat#CW0807 |
| BL21(DE3) | CWBiotech | Cat#CW0809 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DAPI | Thermo Fisher Scientific | Cat#D1306; RRID:AB_2629482 |
| Proteinase K | Roche | Cat#03115828001 |
| RNase A | QIAGEN | Cat#19101 |
| T4 DNA ligase | NEB | Cat#M0202L |
| DpnI | NEB | Cat#R0176L |
| DpnII | NEB | Cat#R0543L |
| Sau3AI | NEB | Cat# R0169L |
| Critical Commercial Assays | ||
| QIAquick PCR purification kit | QIAGEN | Cat#28106 |
| Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific | Cat#Q32851 |
| TruSeq DNA PCR-Free Library Preparation Kit | Illumina | Cat#FC-121-3001 and FC-121-3002 |
| Experimental Models: Cell Lines | ||
| Drosophila S2 cells | Thermo Fisher Scientific | Cat#R69007 |
| HEK293T | ATCC | Cat#CRL-3216 |
| HeLa | ATCC | Cat#CCL-2 |
| MCF7 | ATCC | Cat#HTB-22 |
| Experimental Models: Organisms/Strains | ||
| Drosophila: UAS-RbfRNAi | BDSC | 36744 |
| Drosophila: UAS-Rbf | BDSC | 50747 |
| Drosophila: UAS-E2f1RNAi | BDSC | 27564 |
| Drosophila: UAS-DpRNAi | BDSC | 33372 |
| Drosophila: UAS-E2f1+Dp (II) | BDSC | 4774 |
| Drosophila: UAS-E2f1+Dp (III) | BDSC | 4770 |
| Drosophila: UAS-P35 | BDSC | 5072 |
| Drosophila: ban-LacZ | BDSC | 10154 |
| Drosophila: UAS-hpoRNAi | BDSC | 33614 |
| Drosophila: UAS-ykiRNAi | BDSC | 31965 |
| Drosophila: UAS-sdRNAi | BDSC | 29352 |
| Drosophila: UAS-E2f2 | FlyORF | F000069 |
| Drosophila: Rbf14FRT19A | (Buttitta et al., 2010) | N/A |
| Drosophila: UAS-GFP-E2f1 | (Buttitta et al., 2010) | N/A |
| Drosophila: UAS-GFP-E2f1PIP3A | (Buttitta et al., 2010) | N/A |
| Drosophila: UAS-cycD, cdk4 | (Buttitta et al., 2010) | N/A |
| Drosophila: PCNA-GFP | (Buttitta et al., 2010) | N/A |
| Drosophila: MARCM82B | Tao Wang (NIBS) | N/A |
| Drosophila: EGUF19A | Tao Wang (NIBS) | N/A |
| Drosophila: UAS-wtsRNAi | VDRC | 106174 |
| Drosophila: sd-lacZ | Georg Halder (KU Leuven) | N/A |
| Drosophila: Diap1-GFP3.5 | (Zhang et al., 2008) | N/A |
| Drosophila: Diap1-lacZ | (Zhang et al., 2008) | N/A |
| Drosophila: act>CD2>Gal4 | (Zhang et al., 2008) | N/A |
| Drosophila: ex-lacZ | (Zhang et al., 2008) | N/A |
| Drosophila: UAS-yki | (Zhang et al., 2008) | N/A |
| Drosophila: Diap1-lacZ2B | (Wu et al., 2008) | N/A |
| Drosophila: Diap1-lacZ2B2C | (Wu et al., 2008) | N/A |
| Drosophila: UAS-E2f1 | This paper | N/A |
| Drosophila: UAS-E2f1E259 | This paper | N/A |
| Drosophila: UAS-E2f1D296A | This paper | N/A |
| Drosophila: UAS-V5-E2f1TDM | This paper | N/A |
| Drosophila: UAS-hE2f1 | This paper | N/A |
| Drosophila: UAS-hE2f2 | This paper | N/A |
| Drosophila: UAS-hE2f3a | This paper | N/A |
| Drosophila: UAS-hE2f3b | This paper | N/A |
| Drosophila: UAS-Dam-yki | This paper | N/A |
| Drosophila: UAS-Dam | Andrea Brand (The Gurdon Institute) | N/A |
| Drosophila: hhGal4, tubGal80ts | This paper | N/A |
| Drosophila: nubGal4, tubGal80ts | This paper | N/A |
| Drosophila: Myo1AGal4, tubGal80ts | This paper | N/A |
| Drosophila: apGal4, tubGal80ts | This paper | N/A |
| Oligonucleotides | ||
| E2f1 mutagenesis primers | This paper, Table S8 | N/A |
| qRT-PCR primers | This paper, Table S8 | N/A |
| ChIP-PCR primers | (Qing et al., 2014, Yimlamai et al., 2014, Xie et al., 2013), Table S8 | N/A |
| Oligos used for DamID-Seq | (Vogel et al., 2007), Table S8 | N/A |
| Recombinant DNA | ||
| Plasmid: pUASTattB-E2f1 | This paper | N/A |
| Plasmid: pUASTattB-E2f1E259 | This paper | N/A |
| Plasmid: pUASTattB-E2f1D296A | This paper | N/A |
| Plasmid: pUASTattB-V5-E2f1TDM | This paper | N/A |
| Plasmid: pUASTattB-hE2f1 | This paper | N/A |
| Plasmid: pUASTattB-hE2f2 | This paper | N/A |
| Plasmid: pUASTattB-hE2f3a | This paper | N/A |
| Plasmid: pUASTattB-hE2f3b | This paper | N/A |
| Plasmid: pUASTattB-V5-Dp | This paper | N/A |
| Plasmid: pUASTattB-V5-Rbf | This paper | N/A |
| Plasmid: pET-28a-His-Yki | This paper | N/A |
| Plasmid: pGEX-4T-1-GST-Sd | This paper | N/A |
| Plasmid: pcDNA6A-Myc-His-E2f1 | This paper | N/A |
| Plasmid: pCMV-Flag- hE2f1 | This paper | N/A |
| Plasmid: pUASTattB-LT3-NDam | Andrea Brand (The Gurdon Institute) | N/A |
| Plasmid: pUASTattB-LT3-Dam-yki | This paper | N/A |
| Plasmid: pCMV-Flag-TEAD1 | This paper | N/A |
| Plasmid: pCMV-Flag-yki | This paper | N/A |
| Plasmid: pCMV-Myc-Yap | This paper | N/A |
| Plasmid: pUAST-Myc-yki | Lei Zhang (SIBCB) | N/A |
| Plasmid: pUAST-HA-Sd | Lei Zhang (SIBCB) | N/A |
| Plasmid: pUAST-HA-Sd-N | Lei Zhang (SIBCB) | N/A |
| Plasmid: pUAST-HA-Sd-C | Lei Zhang (SIBCB) | N/A |
| Plasmid: arm-Gal4 | Xinhua Lin (CCHMC) | N/A |
| Plasmid: pML-Gal4 | Xinhua Lin (CCHMC) | N/A |
| Software and Algorithms | ||
| Bowtie 2 | (Langmead and Salzberg, 2012) | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Damidseq_pipeline | (Marshall and Brand, 2015) | https://owenjm.github.io/damidseq_pipeline/ |
| TopHat2 version 2.0.9 | (Bolger et al., 2014, Kim et al., 2013) | N/A |
| HTSeq python package | http://htseq.readthedocs.io | N/A |
| R packages | https://www.r-project.org | N/A |
| DNAMAN version 6 | Lynnon Biosoft | http://www.lynnon.com |
| Prism 7 | GraphPad Software | RRID:SCR_002798 |
| ImageJ | https://fiji.sc | N/A |
Contact for Reagent and Resource Sharing
Requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Bruce A. Edgar (bruce.edgar@hci.utah.edu).
Experimental Model and Subject Details
All fly strains were kept on standard fly medium at 25°C. For temperature shift experiments, larvae were raised at 18°C prior to shifting to the temperature conditions described in the corresponding figure legends and Method Details. Imaginal discs were dissected from 3rd instar larvae of both sexes. Midguts were dissected from adult females.
Method Details
E2f1 Mutagenesis
Different E2f1 mutations (E2f1E259, E2f1D296A and E2f1TDM) were generated using Site-Directed mutagenesis kit (Thermo Fisher Scientific). Primers used for E2f1 mutagenesis are shown in Table S8.
Cell Culture
HeLa, HEK293T and MCF7 cell lines were cultured in DMEM supplemented with 10% FBS (Gibco) in 5% CO2 atmosphere at 37°C. Plasmid transfection was conducted with Lipofectamine (Thermo Fisher Scientific). Drosophila S2 cells were maintained at 25°C in Schneider’s medium (Gibco) supplemented with 10% FBS (Gibco).
Immunostaining
After dissection, samples were fixed in PBS with 4% paraformaldehyde (20 min for wing discs or 30 min for midguts), washed in PBS with 0.1% Triton X-100, and blocked in PBS with 0.1% Triton X-100 and 10% NGS for at least 30 min at room temperature. All samples were then stained with primary antibodies at 4°C overnight with the following dilutions: mouse α-β-gal (Promega, 1:500), rat α-Ci (DHSB, 1:20), guinea pig α-E2F1 (X. Bi, 1:200), mouse α-En (DHSB, 1:20), mouse α-Diap1 (B. Hay, 1:200), rabbit α-Yki (L. Zhang, 1:50), rabbit α-hE2F1 (CST, 1:100), rabbit α-hE2F2 (Santa Cruz Biotechnology, 1:100), rabbit α-Cleaved Caspase-3 (CST, 1:100), chicken α-GFP (Thermo Fisher Scientific, 1:1000), rabbit α-PH3 (Millipore, 1:1000), mouse α-Wg (DHSB, 1:5), guinea pig α-Sens (Zhang et al., 2011), mouse α-Dll (X. Lin, 1:200), mouse α-Ptc (DHSB, 1:50), mouse α-Smo (DHSB, 1:100), rabbit α-Sal (Zhang et al., 2011), rat α-Dachs (D. Strutt, 1:500) and mouse α-Dp (N. Dyson, 1:5). DAPI (Thermo Fisher Scientific, 1:1000) was used to label nuclei. Staining was detected by Alexa Fluor 488, 568, or 633 conjugated species appropriate secondary antibodies (Thermo Fisher Scientific, 1:1000).
Co-immunoprecipitation and Western Blot
For co-immunoprecipitation, cell lysates were prepared in the lysis buffer (50mM Tris-HCl, pH7.5; 150mM NaCl, 1mM EDTA, 1mM EGTA, 5mM Na4P2O7, 25mM NaF, 1% Triton X-100) with protease inhibitors (cocktail) on ice for 30min. After clarification by centrifugation, the lysates were incubated for 2-6h at 4 °C with antibodies pre-bound to protein A/G agarose beads. Beads were washed four times with washing buffer (50mM Tris-HCl, pH7.5; 150mM NaCl, 1mM EDTA, 1mM EGTA, 5mM Na4P2O7, 25mM NaF, 0.5% Triton X-100), and eluted in 1x SDS loading buffer. Eluted samples were analyzed by western blotting. Antibodies used for immunoprecipitation were: mouse α-Myc (MBL International), mouse α-V5 (Thermo Fisher Scientific), mouse α-TEAD1 (BD Biosciences), EZview™ Red Anti-HA Affinity Gel (Sigma-Aldrich) and EZview™ Red Anti-Flag Affinity Gel (Sigma-Aldrich). Primary antibodies used for western blot were: rabbit α-E2F1 (X. Bi, 1:2000), mouse α-Flag (Sigma-Aldrich, 1:10000), rabbit α-Yki (L. Zhang, 1:5000), mouse α-TEAD1 (BD Biosciences, 1:500), mouse α-HA (Santa Cruz Biotechnology, 1:1000), mouse α-V5 (Thermo Fisher Scientific, 1:20000), mouse α-Myc (MBL International, 1:2000), rabbit α-hE2F1 (CST, 1:1000), mouse α-GAPDH (CWBiotech, 1:1000) and mouse α-β-actin (Proteintech Group, 1:5000).
qRT-PCR
Quantitative real-time PCR was performed using 2×SYBR Green PCR master mix (CWBiotech) in an Agilent Mx3005P qRT-PCR system. The mRNA levels were normalized to rp49 (for wing disc) or GAPDH (for cell line) expression levels. Primers used for qRT-PCR are listed Table S8.
ChIP Assays
Cells were cultured in 15cm dishes (about 90% density), cross-linked with 1% formaldehyde for 15min in room temperature and then sonicated to an average fragment size of ∼600 bp. Control IgG or specific antibodies, including mouse anti-Myc (MBL), rabbit anti-YAP (Abclonal) were used in each ChIP assay. The immunoprecipitated DNA was quantified using real-time PCR. All values were normalized to the input. Primers for analyzing the ChIP DNA are shown in Table S8 (Qing et al., 2014, Yimlamai et al., 2014, Xie et al., 2013).
DamID-Seq and Data Analysis
The yki cDNA was cloned into pUASTattB-LT3-NDam (A. Brand) and transgenic flies were generated. Flies carrying the UAS-Dam alone transgene (A. Brand) were used as a control for nonspecific Dam activity. The expression of all transgenes used in DamID-Seq was driven by the nubts system. Larvae were raised at 18°C, and then shifted to 29°C for 24 hours before dissection of third instar wing discs in ice-cold PBS. Around 100 wing discs were pooled in 200 μl of lysis (TENS) buffer. 2 μl Proteinase K (20 mg/ml, Roche) was added to each sample before homogenization followed by incubation for 7 hours at 65°C. Samples were incubated for 30 min at 37°C with 0.5 mg/ml RNase A (QIAGEN) before phenol/chloroform extraction of genomic DNA (gDNA). Genomic DNA from each genotype (∼2.5 μg) was digested with DpnI (NEB) for 16 hr at 37°C in a total volume of 10 μl. 5 μl of DpnI-digested gDNA was ligated to 40 pmol of a double-stranded unphosphorylated adaptor using T4 DNA ligase (NEB) in a total volume of 20 μl. The adaptor was generated by mixing equal volumes of oligo AdRt (100 μM) (5’-CTAATACGACTCACTATAGGGCAGCGTGGTCGCGGCCGAGGA-3’) and oligo AdRb (100 μM) (5’-TCCTCGGCCG-3’) (Vogel et al., 2007), at 100°C for 1 min, followed by cooling to RT. To prevent amplification of DNA fragments containing unmethylated GATCs, the adaptor-ligated DNA was cut with DpnII (NEB) for 2 hr at 37°C in a total volume of 80 μl. 20 μl of DpnII-cut DNA was amplified by PCR in a total volume of 80 μl using the primer (5’-GGTCGCGGCCGAGGATC-3’) designed to fit to the adaptor (Vogel et al., 2007). 3-3.5 μg of PCR product was diluted to 45 μl total volume and sonicated to produce 300-400 bp fragments. The sonicated DNA was then digested by Sau3AI at 37°C overnight to remove the adaptors, as Sau3AI can cut the both methylated and non-methylated GATC sites. The Sau3AI-digested product was purified using a QIAquick PCR purification kit (QIAGEN). Purified product was quantified by Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific). The sequencing library was prepared using a TruSeq DNA PCR-Free Sample Preparation kit (Illumina).
DamID-Seq raw reads were mapped to the Drosophila genome (dm6, genome.ucsc.edu) using Bowtie 2 (Langmead and Salzberg, 2012) using default settings. Unmapped reads and reads with a mapping quality score <10 were excluded for downstream analysis. The retained reads were extended 300 base pair toward the 3’-end, and the resulting bam files were subjected to the damidseq_pipeline analysis (Marshall and Brand, 2015) with parameters --bamfiles --bins=75 --max_norm_value=100 --min_norm_value=-100 --norm_method=kde --qscore1max=1 --qscore1min=0.1 --qscore2max=1. In brief, the method determined the best normalization factor between Dam-fusion and Dam-only samples based on Gaussian kernel density estimation, and determined the optimal number of pseudo-counts added to both samples in order to reduce background noise. The pipeline yielded log2-transformed read count ratios between Dam-fusion and Dam-only experiments in continuous non-overlapping 75-bp windows, which were then used for peak calling. The methods of peak calling and the false discovery rate (FDR) estimation were adapted from the previous publication (Wolfram et al., 2012). In brief, all 75-bp windows with binding intensity (that is log2 read count ratios) above certain thresholds (here, quantiles from 0.95 to 0.99 with a 0.01 step) were listed and merged. To assess the FDR of these candidate peaks, the intensity values of all windows were randomly shuffled for 100 times, and the mean frequency of difference size of consecutive windows with intensity score above each threshold was calculated. In the shuffled dataset, the relationship between the number of consecutive windows and the frequency of observation such windows was log linear, and could thus be effectively modeled for any number of windows with linear regression. The FDR was calculated as the ratio of expected over observed peaks for different number of consecutive windows above a given intensity threshold. Annotation of significant peaks (FDR<0.01) was based on overlaps between the peaks and genes annotated in the FlyBase or Ensembl databases.
RNA-Seq and Data Analysis
The expression of each transgenes used in RNA-Seq was driven by the nubts system. Larvae were raised at 18°C, and then shifted to 29°C for 48 hours before dissection. Third instar wing discs of each samples were dissected for RNA-Seq according to standard protocols. Trimmomatic-processed raw RNA-Seq reads were mapped to the FlyBase Drosophila genome version 6.13 using TopHat2 version 2.0.9 (Bolger et al., 2014, Kim et al., 2013). Mapped reads were counted using HTSeq python package (http://htseq.readthedocs.io). Differentially expressed genes between experimental and control samples were determined using the MARS method (MA-plot-based method with random sampling model) from the R-package DEGseq, filtering for FDR<0.05 and absolute change bigger than 50 reads as previously described (Jiang et al., 2013).
Statistical Analysis
Statistical analyses were performed using the Graphpad Prism 7 software package. Statistical significance (P values) of all results were calculated by unpaired two-tailed Student's t-test. Statistical significance was denoted as follows: ∗ P<0.05, ∗∗ P<0.01, ∗∗∗ P<0.001 and ∗∗∗∗ P<0.0001.
Author Contributions
Conceptualization, P.Z.; Methodology, P.Z. and C.P.; Investigation, P.Z. and C.P.; Formal Analysis, X.W., J.X., B.-F.S., M.M., Y.C., and X.Q.; Writing – Original Draft, P.Z.; Writing – Review & Editing, P.Z. and B.A.E; Funding Acquisition, P.Z., B.A.E., and Z.Y.; Resources, J.-W.X. and Y.-P.S.; Supervision, B.A.E. and Z.Y.
Acknowledgments
We thank X. Bi, L. Zhang, G. Halder, X. Lin, T. Wang, B. Hay, D. Strutt, N. Dyson, A. Brand, and D. J. Pan for fly stocks, plasmids, and antibodies; S. Pronovost and T. Roy provided critical comments on the manuscript. This work was supported by grants from the National Natural Science Foundation of China (81200993 to P.Z., 81125010 and 81030025 to Z.Y.), National Basic Research Program of China (973-2012CB910701 and 2013DFA31990 to Z.Y.), the European Research Council (ERC AdG 268515 to B.A.E.), and the Huntsman Cancer Foundation (to B.A.E).
Published: December 4, 2017
Footnotes
Supplemental Information includes six figures and eight tables and can be found with this article online at https://doi.org/10.1016/j.devcel.2017.10.033.
Contributor Information
Bruce A. Edgar, Email: bruce.edgar@hci.utah.edu.
Zengqiang Yuan, Email: zyuan620@yahoo.com.
Supplemental Information
This table shows Yki binding peaks from Yki DamID-seq in third-instar wing disc cells overexpressing Dam-Yki fusion protein for 24 hr (cutoff: Log2 fold change of Dam-Yki/Dam-only >3, FDR <0.001).
This table includes a list of genes that are significantly upregulated (FDR <0.05) in third-instar wing disc cells overexpressing yki for 48 hr.
This table includes a list of genes that are significantly downregulated (FDR <0.05) in third-instar wing disc cells overexpressing ykiRNAi for 48 hr.
This list includes 116 genes that are differentially regulated both upon yki overexpression and yki knockdown and are also associated with one or more Yki binding peak. These genes are scored as direct transcriptional targets of Yki.
Gene ontology (GO) enrichment analysis was executed in R (version 3.1.1) using custom scripts with R packages GO.db, AnnotationDbi, and org.Dm.e.g.db. A number of 14,782 genes expressed in any conditions were taken as the background set, and the 116 genes were tested for enrichment in biological process (BP) ontology terms. p values were calculated with hypergeometric tests and thereafter adjusted with the Benjamini-Hochberg procedure (adjusted p values).
This table includes a list of genes that are significantly upregulated (FDR <0.05) in third-instar wing disc cells overexpressing E2f1 for 48 hr.
This table includes a list of genes that are significantly downregulated (FDR <0.05) in third-instar wing disc cells overexpressing E2f1 for 48 hr.
This table includes all of the primers and oligos used for E2f1 mutagenesis, qRT-PCR, ChIP-PCR, and DamID-seq in this study.
References
- Blanco E., Ruiz-Romero M., Beltran S., Bosch M., Punset A., Serras F., Corominas M. Gene expression following induction of regeneration in Drosophila wing imaginal discs. Expression profile of regenerating wing discs. BMC Dev. Biol. 2010;10:94. doi: 10.1186/1471-213X-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttitta L.A., Katzaroff A.J., Edgar B.A. A robust cell cycle control mechanism limits E2F-induced proliferation of terminally differentiated cells in vivo. J. Cell Biol. 2010;189:981–996. doi: 10.1083/jcb.200910006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau B.N., Wang J.Y. Coordinated regulation of life and death by RB. Nat. Rev. Cancer. 2003;3:130–138. doi: 10.1038/nrc993. [DOI] [PubMed] [Google Scholar]
- Choksi S.P., Southall T.D., Bossing T., Edoff K., de Wit E., Fischer B.E., van Steensel B., Micklem G., Brand A.H. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Classon M., Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- Clavier A., Baillet A., Rincheval-Arnold A., Coleno-Costes A., Lasbleiz C., Mignotte B., Guenal I. The pro-apoptotic activity of Drosophila Rbf1 involves dE2F2-dependent downregulation of diap1 and buffy mRNA. Cell Death Dis. 2014;5:e1405. doi: 10.1038/cddis.2014.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell-Crowley L., Harper J.W., Goodrich D.W. Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol. Biol. Cell. 1997;8:287–301. doi: 10.1091/mbc.8.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress W.D., Johnson D.G., Nevins J.R. A genetic analysis of the E2F1 gene distinguishes regulation by Rb, p107, and adenovirus E4. Mol. Cell. Biol. 1993;13:6314–6325. doi: 10.1128/mcb.13.10.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datar S.A., Jacobs H.W., de la Cruz A.F., Lehner C.F., Edgar B.A. The Drosophila cyclin D-Cdk4 complex promotes cellular growth. EMBO J. 2000;19:4543–4554. doi: 10.1093/emboj/19.17.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick F.A., Rubin S.M. Molecular mechanisms underlying RB protein function. Nat. Rev. Mol. Cell Biol. 2013;14:297–306. doi: 10.1038/nrm3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon G.M., Du W. Conserved RB functions in development and tumor suppression. Protein Cell. 2011;2:864–878. doi: 10.1007/s13238-011-1117-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulev Y., Fauny J.D., Gonzalez-Marti B., Flagiello D., Silber J., Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Guo T., Lu Y., Li P., Yin M.X., Lv D., Zhang W., Wang H., Zhou Z., Ji H., Zhao Y. A novel partner of Scalloped regulates Hippo signaling via antagonizing Scalloped-Yorkie activity. Cell Res. 2013;23:1201–1214. doi: 10.1038/cr.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Grenley M.O., Bravo M.J., Blumhagen R.Z., Edgar B.A. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell. 2011;8:84–95. doi: 10.1016/j.stem.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Patel P.H., Kohlmaier A., Grenley M.O., McEwen D.G., Edgar B.A. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Zhang J., Wang J.J., Wang L., Zhang L., Li G., Yang X., Ma X., Sun X., Cai J. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell. 2013;153:773–784. doi: 10.1016/j.cell.2013.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Xu J., Yin M.X., Lu Y., Hu L., Li P., Zhang P., Yuan Z., Ho M.S., Ji H. Brahma is essential for Drosophila intestinal stem cell proliferation and regulated by Hippo signaling. Elife. 2013;2:e00999. doi: 10.7554/eLife.00999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R., Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat. Rev. Drug Discov. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz P., Perez J., Perrimon N. The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development. 2010;137:4135–4145. doi: 10.1242/dev.060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koontz L.M., Liu-Chittenden Y., Yin F., Zheng Y., Yu J., Huang B., Chen Q., Wu S., Pan D. The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev. Cell. 2013;25:388–401. doi: 10.1016/j.devcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenjak M., Anderssen E., Ramaswamy S., Whetstine J.R., Dyson N.J. RBF binding to both canonical E2F targets and noncanonical targets depends on functional dE2F/dDP complexes. Mol. Cell. Biol. 2012;32:4375–4387. doi: 10.1128/MCB.00536-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y., Vinayagam A., Sun X., Dephoure N., Gygi S.P., Hong P., Perrimon N. The Hippo signaling pathway interactome. Science. 2013;342:737–740. doi: 10.1126/science.1243971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzerini Denchi E., Helin K. E2F1 is crucial for E2F-dependent apoptosis. EMBO Rep. 2005;6:661–668. doi: 10.1038/sj.embor.7400452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb L.A., Loeb K.R., Anderson J.P. Multiple mutations and cancer. Proc. Natl. Acad. Sci. USA. 2003;100:776–781. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall O.J., Brand A.H. damidseq_pipeline: an automated pipeline for processing DamID sequencing datasets. Bioinformatics. 2015;31:3371–3373. doi: 10.1093/bioinformatics/btv386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon N.S., Frolov M.V., Kwon E.J., Di Stefano L., Dimova D.K., Morris E.J., Taylor-Harding B., White K., Dyson N.J. Drosophila E2F1 has context-specific pro- and antiapoptotic properties during development. Dev. Cell. 2005;9:463–475. doi: 10.1016/j.devcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Morris E.J., Ji J.Y., Yang F., Di Stefano L., Herr A., Moon N.S., Kwon E.J., Haigis K.M., Naar A.M., Dyson N.J. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris E.J., Michaud W.A., Ji J.Y., Moon N.S., Rocco J.W., Dyson N.J. Functional identification of Api5 as a suppressor of E2F-dependent apoptosis in vivo. PLoS Genet. 2006;2:e196. doi: 10.1371/journal.pgen.0020196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld T.P., de la Cruz A.F., Johnston L.A., Edgar B.A. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- Nevins J.R. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- Nicolay B.N., Bayarmagnai B., Islam A.B., Lopez-Bigas N., Frolov M.V. Cooperation between dE2F1 and Yki/Sd defines a distinct transcriptional program necessary to bypass cell cycle exit. Genes Dev. 2011;25:323–335. doi: 10.1101/gad.1999211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay B.N., Frolov M.V. Context-dependent requirement for dE2F during oncogenic proliferation. PLoS Genet. 2008;4:e1000205. doi: 10.1371/journal.pgen.1000205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo R., Morrison C.M., Tao C., Zhang X., Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr. Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- Oh H., Slattery M., Ma L., Crofts A., White K.P., Mann R.S., Irvine K.D. Genome-wide association of Yorkie with chromatin and chromatin-remodeling complexes. Cell Rep. 2013;3:309–318. doi: 10.1016/j.celrep.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev. Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantalacci S., Tapon N., Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- Papagiannouli F., Schardt L., Grajcarek J., Ha N., Lohmann I. The Hox gene Abd-B controls stem cell niche function in the Drosophila testis. Dev. Cell. 2014;28:189–202. doi: 10.1016/j.devcel.2013.12.016. [DOI] [PubMed] [Google Scholar]
- Qing Y., Yin F., Wang W., Zheng Y., Guo P., Schozer F., Deng H., Pan D. The Hippo effector Yorkie activates transcription by interacting with a histone methyltransferase complex through Ncoa6. Elife. 2014;3 doi: 10.7554/eLife.02564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F., Wang B., Yue T., Yun E.Y., Ip Y.T., Jiang J. Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc. Natl. Acad. Sci. USA. 2010;107:21064–21069. doi: 10.1073/pnas.1012759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royzman I., Austin R.J., Bosco G., Bell S.P., Orr-Weaver T.L. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes Dev. 1999;13:827–840. doi: 10.1101/gad.13.7.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw R.L., Kohlmaier A., Polesello C., Veelken C., Edgar B.A., Tapon N. The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development. 2010;137:4147–4158. doi: 10.1242/dev.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibinski A., Breindel J.L., Prat A., Galvan P., Smith E., Rolfs A., Gupta P.B., Labaer J., Kuperwasser C. The Hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell Rep. 2014;6:1059–1072. doi: 10.1016/j.celrep.2014.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley B.K., Irvine K.D. Warts and Yorkie mediate intestinal regeneration by influencing stem cell proliferation. Curr. Biol. 2010;20:1580–1587. doi: 10.1016/j.cub.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein C., Bardet A.F., Roma G., Bergling S., Clay I., Ruchti A., Agarinis C., Schmelzle T., Bouwmeester T., Schubeler D. YAP1 exerts its transcriptional control via TEAD-mediated activation of enhancers. PLoS Genet. 2015;11:e1005465. doi: 10.1371/journal.pgen.1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevaux O., Dyson N.J. A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 2002;14:684–691. doi: 10.1016/s0955-0674(02)00388-5. [DOI] [PubMed] [Google Scholar]
- Stowers R.S., Schwarz T.L. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M., Xu J., Cheng L., Du W. Regulation of apoptosis of rbf mutant cells during Drosophila development. Dev. Biol. 2009;326:347–356. doi: 10.1016/j.ydbio.2008.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B.J., Cohen S.M. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Tsai K.Y., Hu Y., Macleod K.F., Crowley D., Yamasaki L., Jacks T. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol. Cell. 1998;2:293–304. doi: 10.1016/s1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- Udan R.S., Kango-Singh M., Nolo R., Tao C., Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- van den Heuvel S., Dyson N.J. Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 2008;9:713–724. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Delrow J., Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat. Genet. 2001;27:304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- Vogel M.J., Peric-Hupkes D., van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat. Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- Wolfram V., Southall T.D., Brand A.H., Baines R.A. The LIM-homeodomain protein islet dictates motor neuron electrical properties by regulating K(+) channel expression. Neuron. 2012;75:663–674. doi: 10.1016/j.neuron.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Liu Y., Zheng Y., Dong J., Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Xie Q., Chen J., Feng H., Peng S., Adams U., Bai Y., Huang L., Li J., Huang J., Meng S. YAP/TEAD-mediated transcription controls cellular senescence. Cancer Res. 2013;73:3615–3624. doi: 10.1158/0008-5472.CAN-12-3793. [DOI] [PubMed] [Google Scholar]
- Yimlamai D., Christodoulou C., Galli G.G., Yanger K., Pepe-Mooney B., Gurung B., Shrestha K., Cahan P., Stanger B.Z., Camargo F.D. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.X., Zhao B., Guan K.L. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F., Forcato M., Battilana G., Azzolin L., Quaranta E., Bodega B., Rosato A., Bicciato S., Cordenonsi M., Piccolo S. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 2015;17:1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Robinson B.S., Xu W., Yang L., Yao B., Zhao H., Byun P.K., Jin P., Veraksa A., Moberg K.H. The ecdysone receptor coactivator Taiman links Yorkie to transcriptional control of germline stem cell factors in somatic tissue. Dev. Cell. 2015;34:168–180. doi: 10.1016/j.devcel.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ren F., Zhang Q., Chen Y., Wang B., Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Wu Y., Belenkaya T.Y., Lin X. SNX3 controls Wingless/Wnt secretion through regulating retromer-dependent recycling of Wntless. Cell Res. 2011;21:1677–1690. doi: 10.1038/cr.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Gao Y., Li P., Shi Z., Guo T., Li F., Han X., Feng Y., Zheng C., Wang Z. VGLL4 functions as a new tumor suppressor in lung cancer by negatively regulating the YAP-TEAD transcriptional complex. Cell Res. 2014;24:331–343. doi: 10.1038/cr.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J.D., Wang C.Y., Chinnaiyan A.M. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Li D., Wang Y., Pei C., Liu S., Zhang L., Yuan Z., Zhang P. Brahma regulates the Hippo pathway activity through forming complex with Yki-Sd and regulating the transcription of Crumbs. Cell Signal. 2015;27:606–613. doi: 10.1016/j.cellsig.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Zielke N., Kim K.J., Tran V., Shibutani S.T., Bravo M.J., Nagarajan S., van Straaten M., Woods B., von Dassow G., Rottig C. Control of Drosophila endocycles by E2F and CRL4(CDT2) Nature. 2011;480:123–127. doi: 10.1038/nature10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This table shows Yki binding peaks from Yki DamID-seq in third-instar wing disc cells overexpressing Dam-Yki fusion protein for 24 hr (cutoff: Log2 fold change of Dam-Yki/Dam-only >3, FDR <0.001).
This table includes a list of genes that are significantly upregulated (FDR <0.05) in third-instar wing disc cells overexpressing yki for 48 hr.
This table includes a list of genes that are significantly downregulated (FDR <0.05) in third-instar wing disc cells overexpressing ykiRNAi for 48 hr.
This list includes 116 genes that are differentially regulated both upon yki overexpression and yki knockdown and are also associated with one or more Yki binding peak. These genes are scored as direct transcriptional targets of Yki.
Gene ontology (GO) enrichment analysis was executed in R (version 3.1.1) using custom scripts with R packages GO.db, AnnotationDbi, and org.Dm.e.g.db. A number of 14,782 genes expressed in any conditions were taken as the background set, and the 116 genes were tested for enrichment in biological process (BP) ontology terms. p values were calculated with hypergeometric tests and thereafter adjusted with the Benjamini-Hochberg procedure (adjusted p values).
This table includes a list of genes that are significantly upregulated (FDR <0.05) in third-instar wing disc cells overexpressing E2f1 for 48 hr.
This table includes a list of genes that are significantly downregulated (FDR <0.05) in third-instar wing disc cells overexpressing E2f1 for 48 hr.
This table includes all of the primers and oligos used for E2f1 mutagenesis, qRT-PCR, ChIP-PCR, and DamID-seq in this study.