Abstract
Background
Aicardi Goutières Syndrome (AGS) is a heritable interferonopathy associated with systemic autoinflammation causing interferon (IFN) elevation, central nervous system calcifications, leukodystrophy and severe neurologic sequelae. An infant with TREX1 mutations was recently found to have abnormal C26:0 lysophosphatidylcholine (C26:0 Lyso-PC) in a newborn screening platform for X-linked Adrenoleukodystrophy, prompting analysis of this analyte in retrospectively collected samples from individuals affected by AGS.
Methods
In this study, we explored C26:0 Lyso-PC levels and IFN signatures in newborn blood spots and post-natal blood samples in 19 children with a molecular and clinical diagnosis of AGS and in the blood spots of 22 healthy newborns. We used Nanostring nCounter™ for IFN-induced gene analysis and a high-performance liquid chromatography with tandem mass spectrometry (HPLC MS/MS) newborn screening platform for C26:0 Lyso-PC analysis.
Results
Newborn screening cards from patients across six AGS associated genes were collected, with a median disease presentation of 2 months. Thirteen out of 19 (68%) children with AGS had elevations of first tier C26:0 Lyso-PC (>0.4μM), that would have resulted in a second screen being performed in a two tier screening system for X-linked adrenoleukodystrophy (X-ALD). The median (95%CI) of first tier C26:0 Lyso-PC values in AGS individuals (0.43 μM [0.37–0.48]) was higher than that seen in controls (0.21 μM [0.21–0.21]), but lower than X-ALD individuals (0.72 μM [0.59–0.84])(p<0.001). Fourteen of 19 children had elevated expression of IFN signaling on blood cards relative to controls (Sensitivity 73.7%, 95%CI 51–88%, Specificity 95%, 95% CI 78–99%) including an individual with delayed disease presentation (36 months of age). All five AGS patients with negative IFN signature at birth had RNASEH2B mutations. Consistency of agreement between IFN signature in neonatal and post-natal samples was high (0.85).
Conclusion
This suggests that inflammatory markers in AGS can be identified in the newborn period, before symptom onset. Additionally, since C26:0 Lyso-PC screening is currently used in X-ALD newborn screening panels, clinicians should be alert to the fact that AGS infants may present as false positives during X-ALD screening.
Introduction
Aicardi-Goutières Syndrome (AGS, OMIM: 225750) is a heritable interferonopathy that mimics a congenital viral infection1; 2 and results in a devastating early onset neurologic condition. There are seven genes known to be associated with AGS: the DNA exonuclease TREX1 (TREX1)3, the three non-allelic components of RNase H2 endonuclease complex (RNASEH2A, RNASEH2B and RNASEH2C)4, the deoxynucleoside triphosphate triphosphohydrolase SAM domain and HD domain-containing protein (SAMHD1)5, the double-strand RNA editing enzyme ADAR (ADAR1)6, and the double-strand RNA cystolic sensor IFIH1/MDA5 (IFIH1)1,7. These genes encode proteins that are implicated either in intracellular nucleic acid accumulation triggering an innate immune response or directly involved in interferon signaling pathways 8; 9.
AGS typically presents as a subacute encephalopathy in the first year of life, with spasticity, dystonia, developmental delay or regression, and often rapidly progressive leukoencephalopathy with intracranial calcifications and severe atrophy on brain MRI10,11. This period is followed by relative disease stability or slow progression in later years, but brain damage incurred early in the disease results in irreversible neurologic injury often associated with very severe neurologic outcomes12. There is currently no means to screen for this disorder or to reliably identify affected individuals before the onset of neurologic devastation, despite the fact that emerging therapies offer the hope of a disease modifying intervention.
Individuals with AGS have elevated interferon-α levels in cerebrospinal fluid13, CSF pleocytosis, elevated CSF neopterin14, and an interferon (IFN) signature characterized by increased expression of genes induced by the IFN pathway15. These inflammatory markers are thought to be present very early in the disease course16; 17, and may be present in pre-symptomatic patients. A recently implemented newborn screening program for X-linked adrenoleukodystrophy (X-ALD) in the state of New York (USA) unintentionally identified a single individual with AGS when the child’s first tier C26:0 lysophosphatidylcholine (C26:0 Lyso-PC) blood levels tested above the program’s threshold of 0.4μM (unpublished data). As a phosphatidylcholine, C26:0 Lyso-PC might reasonably be expected to be increased as an acute phase reactant and we hypothesized that AGS individuals might have elevation on newborn screening, due to the severe autoinflammatory disease seen in these individuals. We evaluated C26:0 Lyso-PC and Interferon Stimulatory Gene expression (IFN signature) in an additional 18 retrospectively collected newborn screening cards from children with confirmed AGS. Our objective was to explore the potential for unintended detection of AGS under some X-ALD newborn screening protocols.
Methods
Human subjects participation
Individuals were recruited as part of a leukodystrophy associated registry, the Myelin Disorders Bioregistry at Children’s Hospital of Philadelphia (USA), and as part of bioregistry efforts at the Royal Children’s Hospital of Melbourne (Australia) and the C. Besta Neurological Institute of Milano (Italy), and the National Institutes of Health Undiagnosed Diseases Program (USA) according to IRB approved protocols. When available, both neonatal blood spots and post-natal blood samples (taken in PAX gene tubes) were collected. Clinical information was abstracted for year of birth (to establish length of time blood cards were stored), age at symptom onset, age at diagnosis, and functional disease severity as measured by previously used signatures in AGS (Gross Motor Functional Classification System)18; 19.
Interferon (IFN) signature
An IFN signature was calculated to assess the presence of autoinflammation in affected individuals. RNA was extracted from dried blood spots using Norgen Total RNA Purification Micro Kit or post-natal blood samples, which was collected in PAXgene Blood RNA tubes (PreAnalytiX), using PAXgene Blood RNA kit (Qiagen), obtained from 19 AGS individuals and 22 healthy controls during the newborn period and from post-natal blood samples from 11/19 of the studied AGS individuals and from 51 controls. Extracted RNA was quantified on Qubit 2.0 fluorometer and RNA integrity number determined using Agilent 2100 Bioanalyzer. Copy number of mRNA transcripts of the 6 previously reported type I IFN-inducible genes with increased levels in individuals with AGS (IFI27, IFI44L, IFIT1, ISG15, RSAD2, and SIGLEC1)15 and 4 housekeeping genes (ALAS1, HPRT1, TBP, and TUBB) was quantified using Nanostring nCounter™ Digital Analyzer as described (Supplemental methods). The raw copy number of mRNA transcripts of each type I IFN-inducible gene was standardized (stdGene) using the geometric mean of the 4 housekeeping genes for each individual. Then, a Z score for each IFN inducible gene was calculated using the mean and the standard deviation (SD) in the normal control group according to the type of sample (see formula below and supplemental material):
The 6 gene IFN signature in each individual was calculated using the median of the Z scores of the 6 genes. The IFN signature as considered positive (IFN high) if their value was ≥1.96 (>98centile) (one tail analysis was used since only positive IFN signatures in AGS patients were expected).
Lyso-PC- MS/MS analysis of neonatal blood spots
Lyso-PC tandem mass spectrometry (MS/MS) analysis was performed as per previously completed studies20 using residual banked newborn screening cards (supplemental methods) in 19 individuals with mutation confirmed AGS. At least one 3mm punch of the newborn screening blood spot of consented individuals was used for analysis. High-purity grade high performance liquid chromatography (HPLC) solvents were used (J.T. Baker Co or Burdick & Jackson, Inc.) as well as standards D4-26:0-lyso-PC (internal standard) and 26:0-lyso-PC from Avanti Polar Lipids, Inc. Quantitation of 26:0 Lyso-PC was performed by analyzing the signal of the target analyte compared with the signal of the internal standard. Data from the calibration curves for 26:0 Lyso-PC show a linear response (R2 = 0.99) in dynamic range of 0.1–20 pmoles per 3mm newborn screening spot21. All prepared samples with first tier C26:0 Lyso-PC concentrations that were greater than or equal to 0.4 μM were verified by reflex analysis by use of HPLC-MS/MS as described previously20 to minimize false-positive results (second tier 26:0 Lyso-PC). Those samples with results from 0.24 to 0.39 μM on second tier analysis were considered borderline (in routine newborn screening a repeat specimen would be requested) and those samples with HPLC-MS/MS results ≥0.4 μM were considered positive screens for X-ALD22. The values of C26:0 Lyso-PC determined in blood spots of the 19 AGS individuals were compared with those determined in blood spots of controls (670,524 controls in the first tier and 11,353 for the second tier) and 24 ALD individuals, from the X-ALD screen program in the state of New York determined by the same procedures.
Post-natal testing of plasma very long chain fatty acids, lyso-PC and associated peroxisomal function testing
Very long chain fatty acids (VLCFA) and C26:0 Lyso-PC were measured in a total of 56 plasma samples from 36 AGS individuals, including 8 individuals from the 19 AGS affected individuals studied by newborn screening. VLCFA were measured according to standard clinical testing approaches at the Kennedy Krieger Institute 23. Post-natal plasma C26:0 Lyso-PC measurements were also performed according to previously published methods 24. Descriptive analysis of these results was performed.
Peroxisomal functional testing on fibroblasts in AGS individuals
Standard clinical laboratory assays of peroxisomal function were performed on cultured skin fibroblasts obtained from six AGS individuals unrelated to the individuals tested by newborn blood spots. These assays include very long chain fatty acid content25, peroxisomal plasmalogen synthesis enzyme activity 26, branched chain fatty acid (phytanic and pristanic) enzyme activity27; 28, and catalase distribution 29.
Statistical analysis
Descriptive data as age or IFN signatures and C26:0 Lyso-PC levels are presented by their median and interquartile range (IQR). Factors influencing the value of IFN signature in blood spots, post-natal blood samples, and the value of C26:0 Lyso-PC in blood spots were assessed by multivariate linear regression models, including all variables with p>0.1 in univariable linear regression (data not shown), and approached by forward stepwise procedure; only the variables that remained significant were considered as independent predictors. Linear regression coefficients and 95% confidence interval (CI) were used to measure the effect of predictors. Study of consistency of agreement between neonatal and post-natal samples for IFN signature was done using the interclass correlation coefficient (ICC) in which ICC<0.4 bad, 0.75 good, >0,75 very good correlation)30. 95%CI for sensitivity and specificity of IFN signature for the diagnosis of AGS was assessed with Wilson procedure. Comparison of values of C26:0 Lyso-PC between groups (AGS, controls and ALD individuals) was done by Kruskall Wallis test as Barlett’s test showed non homogenous variances between groups. Statistical software (STATA version 13.1; StataCorp, Texas, USA) was used for all the analyses.
Results
Genotype and phenotypic characteristics of AGS individuals
Nineteen individuals with mutations across six of the seven AGS associated genes were identified for this study, with an equal distribution across genders and disease presentation in the first three years of life (Table 1 and supplemental Table 1). There was broader variation of age at diagnosis, in part because some individuals were identified before genes associated with AGS had been identified (Table 1 and supplemental Table 1). AGS was suspected in 17 patients because of clinical symptoms, in 1 patient because a sibling was affected, and in 1 patient based on newborn screening for X-ALD. Molecular diagnosis was confirmed by Sanger sequencing (11/19), Next generation Sequencing (7/19), or MLPA (1/19). Cerebrospinal fluid analysis for interferon-α and neopterin/tetrahydrobiopterin was performed in 5 and 9 individuals respectively (12/19 in total) and were abnormal in all but one individual with RNASEH2B mutations in whom it was performed at 19 years of age, more than 18 years after symptom onset (data not shown).
Table 1.
Characteristics of AGS affected individuals
| Age of onset (median, range) | 2 months (0–36 months) | |
|
| ||
| Age at diagnosis (median, rang) | 9 months (0–236 months) | |
|
| ||
| Gender | male 9/19 and female (10/19) | |
|
| ||
| Genotype | TREX1 (4/19) | 21% |
| RNASEH2A (1/19) | 5.3% | |
| RNASEH2B (9/19) | 47.3% | |
| RNASEH2C (1/19) | 5.3% | |
| SAMHD1 (3/19) | 15.7% | |
| IFIH1 (1/19) | 5.3% | |
|
| ||
| Presenting symptom | Developmental delay (7/19) or regression (4/19) | 57.8% |
| Seizures including infantile spasms (hypsarrythmia in 1 individual) (3/19) | 15.8% | |
| Irritability (5/19) | 26.3% | |
|
| ||
| Additional disease features present in subsets of patients | Feeding impairment (7/19) | 36.8% |
| Sleep abnormalities (2/19) | 10.5% | |
| Microcephaly (7/19) | 36.8% | |
| Tone abnormalities (7/19) | 36.8% | |
| Sterile pyrexia (3/19) | 15.8% | |
| Hepatomegaly, hematochezia, excessive startle reactions, gait anomalies and easy fatigability were described in one individual each | 5.3% each | |
Individuals presented with developmental abnormalities, irritability or seizures (Table 1). Individuals were very neurologically impaired, as typically seen in AGS, based on retrospective assignment of Gross Motor Function Classification System (based on examinations on subjects at average age of 81.7 months, range 5 – 276 months). Most had severe motor impairment (level V in 16/19, 84.2%), with only a few with some preservation of motor function (level IV in 2/19, 10.5% and level III in 1/19, 5.3%).
The infant identified as part of newborn screening (individual 4 Table 2) for X-ALD with elevated C26:0 Lyso-PC (index case) was symptomatic, suggestive of a non X-ALD etiology. The infant subsequently had negative Sanger sequencing analysis for mutations in ABCD1 and genes associated with peroxisomal disorders, thus excluding X-ALD, and had molecular testing confirming AGS.
Table 2.
Newborn screening results in AGS patients
| Sample ID | Individual | Gene | Years blood spot stored | C26:0 Lyso-PC | Interferon signature | |||
|---|---|---|---|---|---|---|---|---|
| First tier result C26:0 LPC(μM) | Second tier result C26:0 LPC(μM) | Natal | Post-natal | Age at post-natal sample | ||||
| LD_0838.0 | 1 | TREX1 | 2012 | 0.45 | 0.17 | 30.5 | NA | NA |
| LD_0993.0 | 2 | TREX1 | 2014 | 0.45 | 0.22 | 25.1 | NA | NA |
| LD_1126.0 | 3 | TREX1 | 2014 | 0.42 | 0.21 | 13.5 | NA | NA |
| LD_1179.0 | 4 | TREX1 | 2014 | 0.50 | 0.35 | 11.2 | NA | NA |
| LD_0945.0 | 5 | RNASEH2A | 2014 | 0.53 | 0.30 | 27.2 | 11.4 | 23 mo |
| LD_0184.0A | 6 | RNASEH2B | 1992 | 0.43 | 0.14 | 0.3 | 7.9 | 23 y |
| LD_0184.0B | 7 | RNASEH2B | 1995 | 0.71 | 0.34 | 18.6 | 16.1 | 17 y |
| LD_0195.0 | 8 | RNASEH2B | 2006 | 0.48 | 0.13 | 0.4 | 4.7 | 6 y 6 mo |
| LD_0201.0 | 9 | RNASEH2B | 2004 | 0.37 | 0.10 | 2.8 | 0.5 | 8 y 2 mo |
| LD_0655.0A also UDP_5356 | 10 | RNASEH2B | 1993 | 0.53 | 0.14 | 0.2 | 7.0 | 19 y |
| LD_0655.0B also UDP_5367 | 11 | RNASEH2B | 1998 | 0.58 | 0.20 | 0.4 | 8.6 | 14 y |
| LD_0990.0 | 12 | RNASEH2B | 2014 | 0.29 | 0.08 | −0.1 | −0.9 | 12 mo |
| LD_1124.0 | 13 | RNASEH2B | 2015 | 0.41 | 0.12 | 12.6 | 5.6 | NA |
| LD_1127.0 | 14 | RNASEH2B | 2012 | 0.35 | 0.11 | 8.6 | NA | NA |
| LD_0981.0 | 15 | RNASEH2C | 2014 | 0.19 | 0.08 | 14.6 | 9.4 | 11 mo |
| LD_0803.0 | 16 | SAMHD1 | 2012 | 0.35 | 0.18 | 13.7 | 16.1 | 3 y 8 mo |
| LD_1125.0 | 17 | SAMHD1 | 2011 | 0.42 | 0.07 | 13.9 | NA | NA |
| LD_1169.0 | 18 | SAMHD1 | 2011 | 0.41 | 0.08 | 12.2 | NA | NA |
| LD_0943.0 also UDP_8031 | 19 | IFIH1 | 2004 | 0.21 | 0.13 | 29.4 | 43.6 | 22 y |
Abbreviations: C26:0 Lyso-PC: C26:0-lysophosphatidylcholine; AGS: Aicardi Goutiere Syndrome; NA: non available. Grey shading levels over the cutt-off.
For C26:0 Lyso-PC the cutt-off of the first tier is ≥0.4 and for the 2nd tier is >0.24 for reanalysis and ≥0.4 for postive score. For the IFN score the cut-off for a positive signature is ≥1.96.
IFN signaling gene expression in blood spots and correlation with post-natal blood and blood spots samples in Aicardi Goutières Syndrome
Extracted RNA from neonatal blood spots obtained during the newborn period was available in 19 AGS individuals and 22 controls. Twelve AGS individuals had follow up blood post-natal samples (median age at the sample collection was 110 months old, IQR 26–202). Median age at which post-natal blood samples were obtained in the 51 control individuals was 213 months old (IQR 58–523).
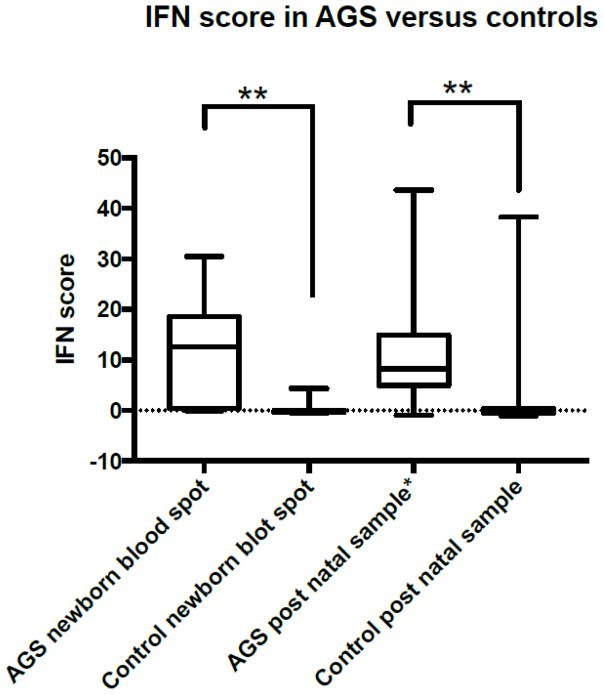
Overall 17/19 (94%) AGS individuals and 6/73 (8%) control individuals had at least one positive IFN signature in one of the samples, p<0.001 (Table 1 and Figure 1). In neonatal blood spots samples, the IFN signature was positive in 14/19 patients and 1/22 controls (Sensitivity 73.7%, 95%CI 51–88%, Specificity 95%, 95% CI 78–99%). The median of the IFN signature value in neonatal blood spots samples was 12.6 (IQR 0.4–18.6) in AGS individuals and 0.28 (IQR −0.36 – 0.01) in controls, p<0.001 (Figure 1).
Figure 1. IFN signaling genes in newborn blood spots and post natal samples in AGS versus controls.
Interferon signaling gene score (IFN signature), calculated in each individual using the median of the Z scores of the 6 genes in previously published scores (see methods). The IFN signature was considered positive (IFN high) if their value was ≥1.96 (>98centile). IFN signature was elevated in a majority of neonatal samples and correlated with post-natal signatures where available. * post-natal signatures were used only for those patients for whom neonatal signatures were available and do not represent the full range of post-natal signatures in the AGS population.
In post-natal blood samples obtained from PAX gene tubes, the IFN signature was positive in 10/12 AGS individuals, and in 6/51 controls (Sensitivity 83%, 95%CI 55–95%, Specificity 88%, 95%CI 76–94%). The median IFN signature value in post-natal blood samples was 8.24 (IQR 5.1–13.7) in AGS individuals and −0.33 (−0.53 – 0.36) in controls, p<0.001 (Figure 1).
The 6 individuals in whom at least 1 sample showed a negative IFN signature all had RNASEH2B mutations. Three individuals had blood spot negative but post-natal positive IFN signature, 1 individual had positive blood spot but negative IFN signature in a post-natal sample, and 2 individuals had negative IFN signature in both blood spot and post-natal samples. The value of the IFN signature was lower in individuals with RNASEH2B mutation (p<0.001). Overall, we did not find significant differences in the value of the IFN signature when comparing neonatal blood spots with post-natal blood samples (mean 12.4 vs 10.4, p=0.627). Consistency of agreement between IFN signature in samples obtained during neonatal period (blood spots) and post-natal age (blood samples) was high (interclass correlation coefficient=0.85, 95%CI 0.48–0.96, p=0.002).
In multivariate linear regression analysis, only the absence of an RNASEH2B mutation (coef 15.1 95%CI 7.8–22.5) was an independent factor influencing the value of IFN signature in blood spots. We found that sex, age at onset, and storage time of the newborn blood spot had no effect on IFN signatures. The value of the IFN signature in post-natal blood sample was influenced positively by the absence of a RNASEH2B mutation (coef 10.4, 95%CI 1.9–19.0) and an older age at the onset of symptoms (coef 0.8, 95%CI 0.4–1.2).
Neonatal blood spot C26:0 Lyso-PC, VLCFA, and peroxisomal studies in AGS
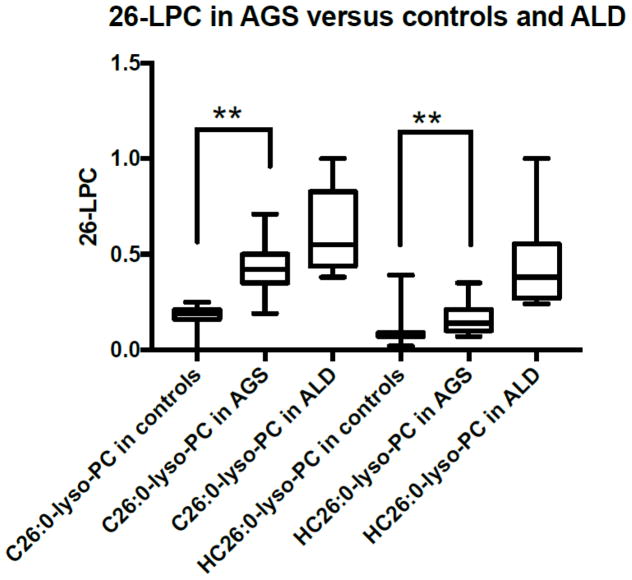
In the first tier analysis, 13 of out 19 (68%) individuals with AGS had elevations of first tier C26:0 Lyso-PC (>0.4μM) that would have resulted in determination of second tier 26:0 Lyso-PC via a second screen being performed in a two tier screening system for X-ALD (Table 2). In the first tier, the mean of C26:0 Lyso-PC values in AGS individuals was 0.43 μM (IC95% 0.37–0.48), which was higher than controls (mean 0.21 μM, IC95% 0.21–0.21), but lower than ALD individuals (mean 0.72 μM, IC95% 0.59–0.84) (p<0.001) (Figure 2).
Figure 2. C26:0 lysophosphatidylcholine (LPC) levels (Tier 1 C26:0 LPC and Tier 2 C26:0 LPC) in newborn blood spots from control, AGS affected and ALD affected infants.
AGS affected infants have C26:0 LPC levels in the range of requirement for a repeat analysis (p<0.001) in a majority of cases. Second tier analysis is more reliable in discriminating AGS from ALD individuals, but second tier 26:0 LPC in AGS is still above the typical range of controls (p<0.001).
In the second tier, only 3 AGS individuals showed second tier 26:0 Lyso-PC values greater than the borderline cut-off of ≥0.24 μM that in routine screening would have resulted in a request for repeat testing. None of the samples from AGS affected individuals showed second tier results ≥0.4 μM which would result in a referral to a speciality care center as a positive result for X-ALD. The value of the second tier C26:0 Lyso-PC in AGS individuals (mean 0.17 μM, IC95% 0.13–0.20) also showed intermediate levels between controls (mean 0.08 μM, IC95% 0.08–0.08) and ALD individuals (mean 0.54 μM, IC95% 0.42–0.66) (Figure 2).
Multivariate linear regression analysis analyzing the factors that influence the value of C26:0 Lyso-PC (1st and 2nd tiers) identified higher median values of IFN signature in blood spot and time from sample (blood spot) to analysis of C26:0 Lyso-PC as positive influencing factors, and age of disease onset as a negative influencing factor (p<0.01) (Supplemental Tables 2 and 3).
VLCFA were performed on all 19 newborn screening samples (Supplemental Table 4) and post-natal VLCFA were measured in a total of 56 plasma post-natal samples from 36 AGS individuals, including 9 individuals from the newborn screening cohort for whom post-natal samples were available. None of the samples exhibited a pattern of VLCFA that is consistent with the diagnosis of a peroxisomal disorder (i.e. a triad of elevated C26:0 and C26/C22 and C24/C22 ratios) (Supplemental Table 4). Further peroxisomal studies on banked fibroblasts from AGS affected individuals didn’t exhibit evidence of a generalized peroxisomal disorder.
Discussion
Based on the experience of a single AGS newborn with TREX1 mutations detected during newborn screening for X-ALD (unpublished data, individual LD_1179.0), we designed the current study to explore the relevance of C26:0 Lyso-PC, very long chain fatty acids, and interferons to AGS in the newborn and post-natal periods. We find that 13 of 19 newborn blood spots from infants with AGS had first tier C26:0 Lyso-PC levels above the 0.4uM threshold that is commonly used in X-ALD newborn screening. AGS individuals showed higher mean values of first tier C26:0 Lyso-PC than controls but lower than X-ALD individuals (Figure 2). We find no further evidence of very long chain fatty acid disturbances to suggest an underlying peroxisomal dysfunction in plasma or fibroblast testing in these or additional AGS individuals. Thus it is presumed that the finding of elevated C26:0 Lyso-PC in AGS subjects represents a false positive for X-ALD screening. We speculate that elevated C26:0 Lyso-PC, and perhaps elevations in other phosphatidylcholines, may be a marker of an undefined downstream inflammatory pathway in AGS pathophysiology 31; 32.
Elevated interferon-α levels have long been recognized as a hallmark of AGS13. The IFN signature, a measure of expression of interferon signaling genes, is considered a biomarker for the disease 15. Of the newborn blood spot samples available for testing, 14 of 19 samples had elevated IFN signatures on neonatal blood spots (Figure 1). In the five individuals without an IFN signature at birth, all individuals had RNASEH2B mutations; however, not all individuals with RNASEH2B mutations causing AGS have negative IFN signatures 15. Limitations to this approach are that none of the blood spots were from a patient with an ADAR1 mutation and that there are small numbers of patients with the individual genes, in particular RNASEH2A, RNASEH2C and IFIH1 mutations for which a single affected individual was affected.
It is important to note that the IFN signatures in all but one neonatal blood spot sample in this study were tested retrospectively. Although length of time the blood spots were stored did not correlate with IFN signatures in this small sample, prospective testing of neonatal blood spots cards could have been more sensitive.
Demonstration of elevated IFN response gene expression as well as C26:0 Lyso-PC elevations in a subset of AGS individuals at birth indicates that interferon activation in these individuals precedes the onset of symptoms by several months, and that the elevation of phosphatidylcholines is likely consistent with this early inflammatory response. This finding suggests that it might be possible to detect AGS in infants prior to clinical disease manifestations. Whether this will help prevent neurologic injury if an effective therapy were to be identified remains to be established, but emerging therapies in AGS add increasing urgency to an early diagnosis. Thus, as X-ALD newborn screening is adopted, practitioners should be aware of the potential for incidental identification of AGS affected infants; if peroxisomal testing is inconclusive, the possibility of AGS should be considered.
Highlights.
Aicardi Goutières Syndrome (AGS) is a heritable interferonopathy associated with systemic autoinflammation causing interferon (IFN) elevation, central nervous system calcifications, leukodystrophy and severe neurologic sequelae.
An infant with TREX1 mutations was recently found to have abnormal C26:0 lysophosphatidylcholine (C26:0 Lyso-PC) in a newborn screening platform for X-linked Adrenoleukodystrophy, prompting analysis of this analyte in retrospectively collected samples from individuals affected by AGS.
Thirteen out of 19 (68%) children with AGS had elevations of first tier C26:0 Lyso-PC (>0.4μM), that would have resulted in a second screen being performed in a two tier screening system for X-linked adrenoleukodystrophy (X-ALD).
This suggests that inflammatory markers in AGS can be identified in the newborn period, before symptom onset.
Additionally, since C26:0 Lyso-PC screening is currently used in X-ALD newborn screening panels, clinicians should be alert to the fact that AGS infants may present as false positives during X-ALD screening.
Acknowledgments
We thank the individuals and their families. NN, GH, and AV are supported by the Myelin Disorders Bioregistry Project. GH was supported by the Delman Fund for Pediatric Neurology and Education. TA was supported by the Institute of Health III, Spain (CM14/00081 and CV15/00021). CT was supported by the NIH Undiagnosed Diseases Program. CS is supported by a NHMRC Postgraduate Research Scholarship and the Flora Suttie Neurogenetics Fellowship. This publication was supported by Award Number UL1TR000075 from the NIH National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Center for Advancing Translational Sciences or the National Institutes of Health. KV was supported by K23 NS087151.
Footnotes
Authorship Contributions:
JO, AV, MM, RJ and AM developed experimental procedures and carried out the experiments carried out in this manuscript. NN, GH, AC, TA and AV analyzed the data and wrote the manuscript. AI, MSV, SO, DT, FG, CT, NU and CS phenotyped and diagnosed participants. HGD, TA performed statistical analysis and designed the figures. KV and RGM were involved in early conception of the project and provided critical review of the manuscript.
Conflicts of Interest: AV has research efforts supported by Eli-Lilly, Shire Pharmaceuticals, Illumina, and Gilead Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Crow YJ, Chase DS, Lowenstein Schmidt J, Szynkiewicz M, Forte G, Gornall HL, Oojageer A, Anderson B, Pizzino A, Helman G. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. American Journal of Medical Genetics Part A. 2015;167:296–312. doi: 10.1002/ajmg.a.36887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vanderver A, Prust M, Tonduti D, Mochel F, Hussey HM, Helman G, Garbern J, Eichler F, Labauge P, Aubourg P. Case definition and classification of leukodystrophies and leukoencephalopathies. Molecular genetics and metabolism. 2015;114:494–500. doi: 10.1016/j.ymgme.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC. Mutations in the gene encoding the 3′–5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nature genetics. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 4.Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nature genetics. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 5.Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nature genetics. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nature genetics. 2012;44:1243–1248. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rice GI, del Toro Duany Y, Jenkinson EM, Forte GM, Anderson BH, Ariaudo G, Bader-Meunier B, Baildam EM, Battini R, Beresford MW. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nature genetics. 2014;46:503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crow YJ, Rehwinkel J. Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Human molecular genetics. 2009;18:R130–R136. doi: 10.1093/hmg/ddp293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stetson DB. Endogenous retroelements and autoimmune disease. Current opinion in immunology. 2012;24:692–697. doi: 10.1016/j.coi.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aicardi J, Goutieres F. A progressive familial encephalopathy in infancy with calcifications of the basal ganglia and chronic cerebrospinal fluid lymphocytosis. Annals of neurology. 1984;15:49–54. doi: 10.1002/ana.410150109. [DOI] [PubMed] [Google Scholar]
- 11.Crow YJ. Aicardi-goutieres syndrome. 2014. [DOI] [PubMed] [Google Scholar]
- 12.Rice G, Patrick T, Parmar R, Taylor CF, Aeby A, Aicardi J, Artuch R, Montalto SA, Bacino CA, Barroso B. Clinical and molecular phenotype of Aicardi-Goutieres syndrome. The American Journal of Human Genetics. 2007;81:713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.GoutièRes F, Aicardi J, Barth PG, Lebon P. Aicardi-Goutières syndrome: An update and results of interferon-α studies. Annals of neurology. 1998;44:900–907. doi: 10.1002/ana.410440608. [DOI] [PubMed] [Google Scholar]
- 14.Blau N, Bonafe L, Krageloh-Mann I, Thony B, Kierat L, Hausler M, Ramaekers V. Cerebrospinal fluid pterins and folates in Aicardi-Goutieres syndrome: a new phenotype. Neurology. 2003;61:642–647. doi: 10.1212/01.wnl.0000082726.08631.e7. [DOI] [PubMed] [Google Scholar]
- 15.Rice GI, Forte GM, Szynkiewicz M, Chase DS, Aeby A, Abdel-Hamid MS, Ackroyd S, Allcock R, Bailey KM, Balottin U. Assessment of interferon-related biomarkers in Aicardi-Goutieres syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: a case-control study. The Lancet Neurology. 2013;12:1159–1169. doi: 10.1016/S1474-4422(13)70258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desanges C, Lebon P, Bauman C, Vuillard E, Garel C, Cordesse A, Oury JF, Crow Y, Luton D. Elevated interferon-alpha in fetal blood in the prenatal diagnosis of Aicardi-Goutieres syndrome. Fetal Diagn Ther. 2006;21:153–155. doi: 10.1159/000089067. [DOI] [PubMed] [Google Scholar]
- 17.Le Garrec M, Doret M, Pasquier JC, Till M, Lebon P, Buenerd A, Escalon J, Gaucherand P. Prenatal diagnosis of Aicardi-Goutieres syndrome. Prenat Diagn. 2005;25:28–30. doi: 10.1002/pd.881. [DOI] [PubMed] [Google Scholar]
- 18.Wood E, Rosenbaum P. The gross motor function classification system for cerebral palsy: a study of reliability and stability over time. Dev Med Child Neurol. 2000;42:292–296. doi: 10.1017/s0012162200000529. [DOI] [PubMed] [Google Scholar]
- 19.Crow YJ, Chase DS, Lowenstein Schmidt J, Szynkiewicz M, Forte GM, Gornall HL, Oojageer A, Anderson B, Pizzino A, Helman G, et al. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am J Med Genet A. 2015;167A:296–312. doi: 10.1002/ajmg.a.36887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tortorelli S, Turgeon CT, Gavrilov DK, Oglesbee D, Raymond KM, Rinaldo P, Matern D. Simultaneous Testing for 6 Lysosomal Storage Disorders and X-Adrenoleukodystrophy in Dried Blood Spots by Tandem Mass Spectrometry. Clin Chem. 2016;62:1248–1254. doi: 10.1373/clinchem.2016.256255. [DOI] [PubMed] [Google Scholar]
- 21.Theda C, Gibbons K, DeFor TE, Donohue PK, Golden WC, Kline AD, Gulamali-Majid F, Panny SR, Hubbard WC, Jones RO. Newborn screening for X-linked adrenoleukodystrophy: further evidence high throughput screening is feasible. Molecular genetics and metabolism. 2014;111:55–57. doi: 10.1016/j.ymgme.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turgeon CT, Moser AB, Morkrid L, Magera MJ, Gavrilov DK, Oglesbee D, Raymond K, Rinaldo P, Matern D, Tortorelli S. Streamlined determination of lysophosphatidylcholines in dried blood spots for newborn screening of X-linked adrenoleukodystrophy. Mol Genet Metab. 2015;114:46–50. doi: 10.1016/j.ymgme.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 23.Lagerstedt SA, Hinrichs DR, Batt SM, Magera MJ, Rinaldo P, McConnell JP. Quantitative determination of plasma c8–c26 total fatty acids for the biochemical diagnosis of nutritional and metabolic disorders. Mol Genet Metab. 2001;73:38–45. doi: 10.1006/mgme.2001.3170. [DOI] [PubMed] [Google Scholar]
- 24.Hubbard WC, Moser AB, Liu AC, Jones RO, Steinberg SJ, Lorey F, Panny SR, Vogt RF, Jr, Macaya D, Turgeon CT, et al. Newborn screening for X-linked adrenoleukodystrophy (X-ALD): validation of a combined liquid chromatography-tandem mass spectrometric (LC-MS/MS) method. Mol Genet Metab. 2009;97:212–220. doi: 10.1016/j.ymgme.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 25.Moser AB, Kreiter N, Bezman L, Lu S, Raymond GV, Naidu S, Moser HW. Plasma very long chain fatty acids in 3,000 peroxisome disease patients and 29,000 controls. Ann Neurol. 1999;45:100–110. doi: 10.1002/1531-8249(199901)45:1<100::aid-art16>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 26.Roscher A, Molzer B, Bernheimer H, Stockler S, Mutz I, Paltauf F. The cerebrohepatorenal (Zellweger) syndrome: an improved method for the biochemical diagnosis and its potential value for prenatal detection. Pediatr Res. 1985;19:930–933. doi: 10.1203/00006450-198509000-00013. [DOI] [PubMed] [Google Scholar]
- 27.Wanders RJ, Denis S, Ruiter JP, Schutgens RB, van Roermund CW, Jacobs BS. Measurement of peroxisomal fatty acid beta-oxidation in cultured human skin fibroblasts. J Inherit Metab Dis. 1995;18(Suppl 1):113–124. doi: 10.1007/BF00711434. [DOI] [PubMed] [Google Scholar]
- 28.Zenger-Hain J, CDAaRWB . In: New Developments in Fatty Acid Oxidation. CPMTK, editor. New York: Wiley-Liss; 1992. pp. 399–407. [Google Scholar]
- 29.Peters TJ, Muller M, De Duve C. Lysosomes of the arterial wall. I. Isolation and subcellular fractionation of cells from normal rabbit aorta. J Exp Med. 1972;136:1117–1139. doi: 10.1084/jem.136.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleiss JLLB, Paik MC. Statistical Methods for Rates and Proportions. New York: John Wiley and Sons; 2003. [Google Scholar]
- 31.Luczaj W, Domingues P, Domingues MR, Pancewicz S, Skrzydlewska E. Phospholipidomic Analysis Reveals Changes in Sphingomyelin and Lysophosphatidylcholine Profiles in Plasma from Patients with Neuroborreliosis. Lipids. 2016 doi: 10.1007/s11745-016-4212-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song J, Guan M, Zhao Z, Zhang J. Type I Interferons Function as Autocrine and Paracrine Factors to Induce Autotaxin in Response to TLR Activation. PLoS One. 2015;10:e0136629. doi: 10.1371/journal.pone.0136629. [DOI] [PMC free article] [PubMed] [Google Scholar]