Abstract
Primary coenzyme Q10 (CoQ10; MIM# 607426) deficiencies are an emerging group of inherited mitochondrial disorders with heterogonous clinical phenotypes. Over a dozen genes are involved in the biosynthesis of CoQ10, and mutations in several of these are associated with human disease. However, mutations in COQ5 (MIM# 616359), catalyzing the only C-methylation in the CoQ10 synthetic pathway, have not been implicated in human disease.
Here, we report three female siblings of Iraqi-Jewish descent, who had varying degrees of cerebellar ataxia, encephalopathy, generalized tonic-clonic seizures and cognitive disability. Whole exome and subsequent whole genome sequencing identified biallelic duplications in the COQ5 gene, leading to reduced levels of CoQ10 in peripheral white blood cells of all affected individuals and reduced CoQ10 levels in the only muscle tissue available from one affected proband. CoQ10 supplementation led to clinical improvement and increased the concentrations of CoQ10 in blood.
This is the first report of primary CoQ10 deficiency caused by loss of function of COQ5, with delineation of the clinical, laboratory, histological and molecular features, and insights regarding targeted treatment with CoQ10 supplementation.
Keywords: COQ5, CoQ10, Cerebellar ataxia, encephalopathy, personalized medicine, next generation sequencing
Introduction
Coenzyme Q (ubiquinone) is a lipophilic redox molecule, essential for diverse cellular roles, including as an electron carrier, regulator of transition pores, and a substrate for pyrimidine nucleotide synthesis. It also acts as an antioxidant and a modulator of fatty acid β-oxidation. Indeed, it links FAO to mitochondrial respiratory chain through electron transfer flavoprotein (ETF) and ETF:ubiquinone oxidoreductase (ETFQO). Following the characterization of their yeast counterparts, the human Coenzyme Q gene family was shown to include over 13 genes involved in the biosynthesis of CoQ10; at least eight of these are associated with human disorders (Doimo, et al., 2014).
Diseases of CoQ10 deficiency have been reported in over 100 individuals worldwide. Their heterogeneous clinical features are consistent with one of five major phenotypes (Quinzii, et al., 2014): 1) Encephalopathy, associated with mitochondrial myopathy and myoglobinuria (Boitier, et al., 1998; Ogasahara, et al., 1989; Salviati, et al., 2012; Sobreira, et al., 1997); 2) Severe infantile disease with multi-systemic involvement (Desbats, et al., 2015; Dinwiddie, et al., 2013; Quinzii, et al., 2006; Rotig, et al., 2000); 3) Ataxia with cerebellar atrophy, the most common form, variably associated with seizures, cognitive impairment, muscle weakness, neuropathy and hypogonadism (Gironi, et al., 2004; Lagier-Tourenne, et al., 2008; Musumeci, et al., 2001); 4) Nephropathy, with or without sensory hearing loss (Ashraf, et al., 2013; Diomedi-Camassei, et al., 2007; Heeringa, et al., 2011; McCarthy, et al., 2013); and 5) Isolated myopathy (Gempel, et al., 2007; Horvath, et al., 2006; Lalani, et al., 2005). In the past decade, next-generation sequencing techniques have facilitated molecular diagnosis of these patients, enabling further elucidation of genotype-phenotype correlations (Yubero, et al., 2014). Of the numerous human mitochondrial disorders, CoQ10 deficiency is considered the only currently treatable disorder, since patients often respond to oral CoQ10 supplementation (DiMauro, et al., 2007; Garrido-Maraver, et al., 2014).
While several genes in the CoQ10 biosynthesis pathway have been implicated in primary CoQ10 deficiency (MIM 607426), no human phenotype attributable to COQ5 mutations has been described. Coq5 is an important enzyme that catalyzes the only C-methylation step involved in the synthesis of Coq10 in yeast and mammalian cells (Baba, et al., 2004; Barkovich, et al., 1997; Nguyen, et al., 2014), and has been shown to assemble with a multi-subunit Coq polypeptide complex (He, et al., 2014). It is expressed in multiple tissues but highest in liver, lung, placenta, and skeletal muscle.
We report for the first time patients with encephalomyopathy, cerebellar ataxia and additional findings in a multiplex non-consanguineous family with CoQ10 deficiency due to a biallelic duplication in COQ5.
Methods
Patients
Patients and family members were enrolled in NIH protocol 76-HG-0238, “Diagnosis and Treatment of Patients with Inborn Errors of Metabolism or Other Genetic Disorders”, approved by the Institutional Review Board of the National Human Genome Research Institute (NHGRI). The patients gave written, informed consent. Clinical evaluations were performed at the Pediatric Neurology Unit at the Edmond and Lily Safra Children’s Hospital, Sheba Medical Center, Tel-Hashomer, Israel.
Single-Nucleotide Polymorphism Arrays
Single-nucleotide polymorphism (SNP) genotyping was performed on genomic DNA with the use of the HumanOmniExpress DNA Analysis BeadChip (Illumina). SNPs were analyzed using GenomeStudio software (Illumina) (Peiffer, et al., 2006; Steemers and Gunderson, 2007).
Next Generation Sequencing
For whole exome sequencing, the library preparation method was performed using the SureSelect Human All-Exon System version 1.0 (Agilent Technologies). Sequencing was performed with the use of the Genome Analyzer IIx sequencer (Illumina). For whole genome sequencing, the library preparation method was performed using the Illumina TruSeq Nano. Sequencing was performed with the Illumina HiSeq X Ten sequencer (Illumina). Variants were filtered and visualized using the graphical software tool VarSifter v1.5 (Teer, et al., 2012).
Biochemistry
Blood samples were drawn at baseline and at different time points during CoQ supplementation into acid citrate dextrose tubes and were submitted for CoQ10 analyte measurement in leukocytes by a clinical laboratory (https://mnglabs.com). A piece of a muscle biopsy from one of the affected patients was flash-frozen and subjected to CoQ10 measurement by a clinical laboratory using high performance liquid chromatography (http://www.rgbmgl.org). Other pieces were fixed in formalin and embedded in paraffin for cytochrome c oxidase staining.
PCR and Sanger sequencing
For PCR and dideoxy sequencing, specific primers were designed (sequences available upon request) to amplify the regions of interest. Direct sequencing of the PCR products was performed using BigDye 3.1 Terminator chemistry (Applied Biosystems) and analyzed on an ABI3130xl genetic analyzer (Applied Biosystems). Data were evaluated using Sequencher v5.0 software (Gene Codes Corporation, Ann Arbor, MI). Variants identified were submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/).
Copy Number Analysis
Copy number analysis by quantitative PCR (qPCR) was performed using 3 TaqMan probes in the duplicating area (Hs00355860_cn, Hs06353017_cn and Hs00837086_cn), 2 flanking the duplicated area (Hs01474184_cn and Hs07018271_cn) and an endogenous control probe in the GAPDH gene (Hs00894322_cn). All the probes were purchased from Applied Biosystems (Foster City, CA). All the qPCR experiments were performed in triplicate as described (Vilboux, et al., 2011) on an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems). The comparative Ct method was used to determine the copy number (Livak and Schmittgen, 2001).
Analysis of COQ5 expression in fibroblasts
Fibroblasts from III.6 were grown as explants from skin biopsies in 10% FBS in DMEM (Malicdan, et al., 2015). Control fibroblasts were purchased from ATCC. Total RNA was isolated from fibroblasts with the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) and treated with a DNase kit (DNA-free) according to the manufacturer’s protocol (Life Technologies, Carlsbad, CA, USA). RNA concentration and purity were assessed and first strand cDNA was synthesized using a high capacity RNA-to-cDNA kit (Applied Biosystems). Reverse Transcriptase reaction products were amplified using specific primers (sequences available upon request) and loaded into 2% agarose gel. Sanger sequencing of the excised bands was performed as mentioned above. Quantitative real-time PCR (qRTPCR) was performed using TaqMan Gene Expression Master Mix reagent (Applied Biosystems) and carried out on an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems) for total COQ5 mRNA expression (Taqman probes ID). Power SYBR Green PCR master mix (Applied Biosystems, Woolston, Warrington, UK) and qPCR machine with standard qPCR parameters were used to analyze the expression of the different COQ5 isoforms. Results were analyzed with the comparative CT method as described (Livak and Schmittgen, 2001).
For western blots, cell lysates were prepared using RIPA buffer supplemented with protease inhibitors, and lysed using 30 strokes with a Dounce homogenizer. Mitochondrial fractions were prepared using the Mitochondria Isolation Kit for Culture Cells (Abcam) per the manufacturer’s instructions. Samples were electrophoresed on a 4%–12% Bis-Tris gel and blotted onto a 0.2 μm nitrocellulose membrane. Membranes were blocked with Odyssey® Blocking Buffer (PBS), washed and incubated with primary goat anti-human COQ5 antibody (Santa Cruz Biotechnology), mouse anti-beta actin antibody (Abcam), followed by appropriate IRDye 680RD or IRDye 800CW-conjugated secondary antibodies (Li-Cor Biosciences). Antibodies were diluted in Odyssey® Blocking Buffer (PBS).
Nuclear and cytosolic fractions were prepared using the rapid, efficient, and practical (REAP) method (Suzuki, et al., 2010). Cells were scraped into 1x PBS, pelleted, and lysed in 0.1% NP40-PBS buffer. Samples were then pelleted briefly and the supernatant was taken as the cytosolic fraction. The pellet was resuspended in 0.1% NP40-PBS buffer, spun down once more, and the resulting nuclear pellet was dissolved in 1x Laemmli Sample Buffer (Bio-Rad) and used as the nuclear fraction. Samples were electrophoresed as above.
Oxygen consumption measurement in human fibroblasts
Human fibroblasts were measured for the mitochondrial oxygen consumption rate (OCR) by using the Seahorse XF24 Instrument (Seahorse Bioscience, Billerica, MD). Wild-type control and patient fibroblast cells were plated on XF-24 plates 48 hours before the analysis at 20,000 cells per well and grown in DMEM which contained 25 mM glucose, 10% FBS and 1% MEM Non-Essential amino acid at 37°C with 5% CO2. Twenty minutes prior to the analysis, the culture medium was replaced by unbuffered DMEM XF Base medium (Seahorse Bioscience) at pH 7.4 contained with 5 mM glucose, 2 mM GlutaMAX and 1mM sodium pyruvate, and the plates were incubated at 37°C in a CO2-free incubator. The mitochondrial OCR was measured under the following conditions: (1) basal, and after the addition of (2) 1.25 μM oligomycin, (3) 0.2 μM of the uncoupler, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) and (4) 1 μM of rotenone together with 1.8 μM antimycin A. For each condition three cycles of mixing (150 s), waiting (120 s), and measuring (210 s) were performed. This series was repeated following each injection. Mitochondrial high resolution respirometry in fibroblasts, reflecting the in vivo state of the respiratory chain and oxidative phosphorlyation function, was performed by a clinical laboratory (https://mnglabs.com) (Kenyon and Moraes, 1997; Rossignol, et al., 1999).
Muscle biopsy and biochemical measurements
Needle muscle biopsy was processed for muscle histochemistry as previously described (Mozaffar and Pestronk, 2000; Pestronk, et al., 1992). Cryostat sections histochemical stains of rapidly frozen muscle included hematoxylin and eosin, Gomori trichrome, nicotinamide adenine dinucleotide (NADH), cytochrome oxidase (COX), succinate dehydrogenase (SDH), Sudan black (lipid) and adenosine tri-phosphatase (ATPase, pH 9.4, 4.6, and 4.3). Type 2C muscle fiber staining for myosin ATPase activity at pH 4.3 was intermediate between types 1 (dark) and 2 (light) (Brooke and Kaiser, 1970). A total of 197 fibers were counted and the type 2C fiber frequency was defined as the percentage of 2C fibers in a random cross-section photomicrograph field. Analysis of CoQ10 levels in the muscle was performed by a clinical diagnostic laboratory. For measuring the activity of individual mitochondrial complexes, we followed our published methods (Kirby, et al., 2007; Taylor, et al., 1993).
Clinical scoring
Treatment with CoQ supplementation (Ubiquinol; 15 mg/kg/d divided into 3 dosages) was given to the 3 affected patients for a period of six months. We used the ICARS (The International Cooperative Ataxia Rating Scale) (Trouillas, et al., 1997) to assess the effect of CoQ supplementation on the various aspects of their cerebellar dysfunction by performing the test at baseline, and 3 and 6 months after initiation of treatment. Due to technical issues, the treatment was stopped by the parents two weeks before the assessment at 6 months.
The ICARS is an outcome measure that was created in 1997 by the Committee of the World Federation of Neurology with the goal of standardizing the quantification of impairment due to cerebellar ataxia. The scale is scored out of 100 with 19 items and 4 subscales of postural and gait disturbances, limb ataxia, dysarthria, and oculomotor disorders. Higher scores indicate higher levels of impairment.
Results
Patients
Three female siblings (Fig. 1A), born to non-consanguineous parents of Iraqi-Jewish descent, presented to our center with varying degrees of cerebellar ataxia, encephalopathy, generalized tonic clonic seizures and developmental delay. The proband (III.4), the fourth of seven children, presented in early childhood with short stature and moderate delay in motor and cognitive milestones, followed by non-progressive cerebellar ataxia, dysarthria, mild-moderate cognitive disability, and behavioral problems including impulsivity, short attention span and oppositional characteristics. At approximately 12 years of age, she had myoclonic jerks which were more noticeable during voluntary motor movements. At age 17, she experienced several generalized tonic-clonic seizures. Epilepsy and myoclonus were well controlled by valproic acid combined with low dose clonazepam. On physical examination, the patient showed no dysmorphic features. Her head circumference was 54.5cm (35th percentile), and her height was 150cm (−2 Standard Deviations). She had mild horizontal nystagmus, slow saccades with saccadic pursuit and apraxic gaze, normal fundoscopy, dysarthric cerebellar speech, dysmetria in finger-to-nose test, a mild postural and intention tremor, ataxic gait and inability to perform tandem walking. She also showed mild lower limb spasticity with brisk tendon joint reflexes and negative Babinski sign.
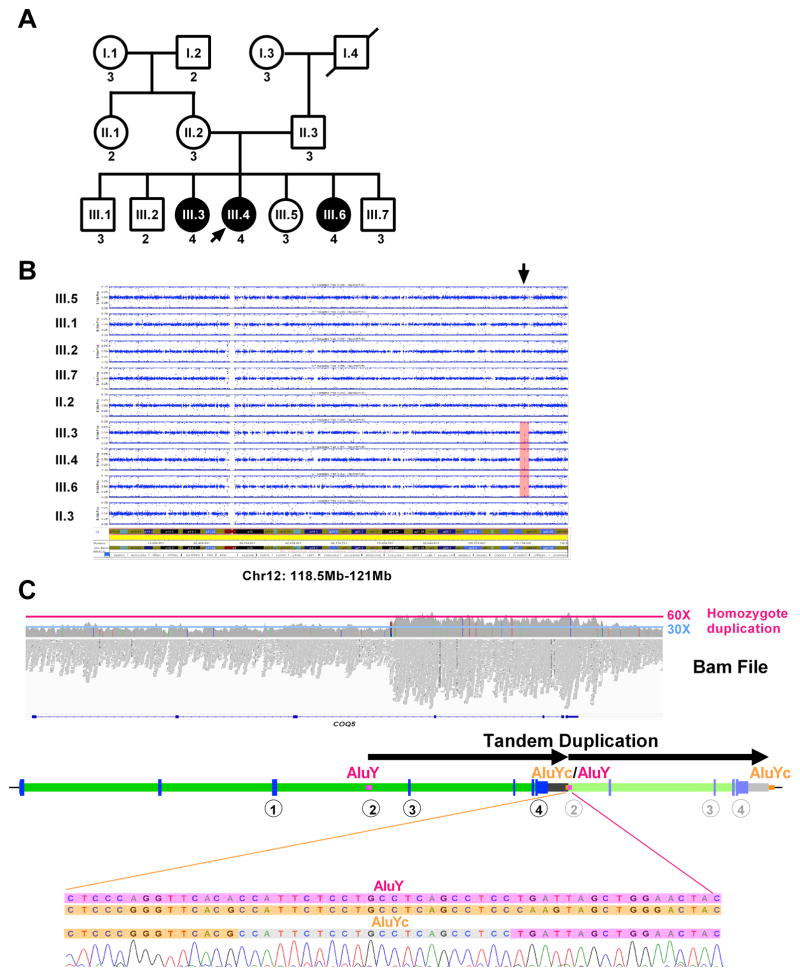
Fig. 1. Patients included in the study.
(A) Family pedigree. Patients who are affected are shown in filled circles. Male individuals are represented by squares, while female individuals are represented by circles. (B) SNP data analysis of all members of the family shows a single homozygous region, from 118.5Mb-121Mb on chromosome 12 (hg19) (arrow).
(C) Whole genome sequencing on III.4 was inconclusive, but analysis of Bam file shows an increase of the number of reads in the candidate locus on chromosome 12 between positions ~120,940,150 and ~120,949,950, with an average number of reads of ~60X compared to the rest of the region (~30X). A cartoon below the Bam file shows the duplication in tandem, breakpoints confirmed by Sanger sequencing), between an AluYc (chr12:120939934–120940228) and an AluY (chr12:120949733–120950042). The encircled numbers below the gene show the positions of the different Taqman probes used for testing the copy number of the duplicated region, which is shown in (A) as the number below each box or circle.
Brain MRI at 8 and 16 years of age showed mild non-progressive cerebellar atrophy; EEG revealed normal background activity and infrequent generalized polyspike-wave (PSW) discharges; nerve conduction study and cardiac echocardiogram were both normal. Complete blood count, electrolyte levels, CPK, liver and renal function tests, carnitine and acyl-carnitine, copper, ceruloplasmin, thyroid function tests, lactate, pyruvate, ammonia, blood amino acid profile, VLCFA, phytanic acid, homocysteine, isoelectric focusing of transferrin, alpha –FP, quantitative immunoglobulin levels, vitamin E levels, as well as urine for protein and organic acids, were within normal limits.
The proband’s older sister (III.3) presented in early childhood (following identification of cerebellar ataxia in her younger sister) with mild static gait ataxia, mild dysarthria, and borderline intelligence (requiring special school only at high school). In her 20’s, the patient had myoclonic jerks followed by a single generalized tonic-clonic seizure at the age 22; there was no seizure recurrence on low dose valproic acid, which was later replaced by lamotrigine. The patient gave birth to a healthy baby with no signs of deterioration during her pregnancy. Upon physical examination, she showed mild gait ataxia with inability to perform tandem walking, mild dysmetria and oculomotor apraxia and horizontal nystagmus. A brain MRI showed mild cerebellar atrophy and EEG showed infrequent generalized PSW discharges.
The youngest of the affected siblings, III.6, exhibited mild motor delay followed by mild learning difficulties in school, characterized by slow response rate, relatively poor verbal skills and poor writing. She was first examined at the age of 14 years, when mild cerebellar ataxia with abnormal tandem walking were identified, as well as brisk deep tendon reflexes, negative Babinski, no tremor or dysmetria but mild cerebellar dysarthria and horizontal nystagmus with hypometric saccades.
Next generation sequencing identifies duplication in COQ5
Because of the Iraqi-Jewish origin of both parents, a homozygote recessive mode of inheritance was suspected. A SNP array study, performed on the parents and all the siblings revealed several areas of homozygous regions (Supp. Table S1), conforming consanguinity in the family; however, there was one homozygous region, from 118.5Mb-121Mb on chromosome 12 (Chr37) (Fig. 1B), which was observed only in the affected children. Whole exome sequencing (WES), performed on patient III.4, did not identify a potential pathogenic variant (data not shown). Whole genome sequencing (WGS) was then pursued on the same individual, but variant analysis again failed to reveal potential pathogenic variant. Analysis of the Bam files showed a significant increase in the number of reads in the candidate locus on chromosome 12 between positions ~120,940,150 and ~120,949,950. The average number of reads in this region (~60X) compared to the rest of the region (~30X) suggested the presence of 4 copies, i.e., a homozygote duplication) (Fig. 1C).
Sanger sequencing confirmed the breakpoints of this duplication and showed that it was in tandem (Fig. 1C). This duplication, exactly 9,590bp [Chr12(GRCh37):120940098–120949687], is between an AluYc (chr12:120939934–120940228) and an AluY (chr12:120949733–120950042); these two Alus have 84% identity, potentially making this duplication the result of an unbalanced crossing-over between these two Alus. The duplication spanned the last 4 exons of the COQ5 gene. COQ5 encodes for 2-hexaprenyl-6-methoxy-1,4-benzoquinone methyltransferase, a C-methyltransferase required in the biosynthesis of CoQ10. This duplication does not alter the original COQ5 ORF and is located about 1kb after the DNA sequence that encodes the COQ5 3′UTR (Fig. 1C).
Copy number (CN) analysis, performed on all the family members for whom DNA was available, revealed that the duplication segregated with the disease based on a homozygote recessive model. Indeed, all the affected individuals were homozygous for the duplication (CN = 4), the parents were both carriers (CN = 3), and all the unaffected members were either carriers or homozygous not duplicated (CN = 2) (Fig. 1A). The duplication was analyzed in a cohort of 92 healthy individual from the same ethnic background, and 3 carriers were identified. These results reveal that, in this population, the duplication has a minor allele frequency of approximately 1.5%.
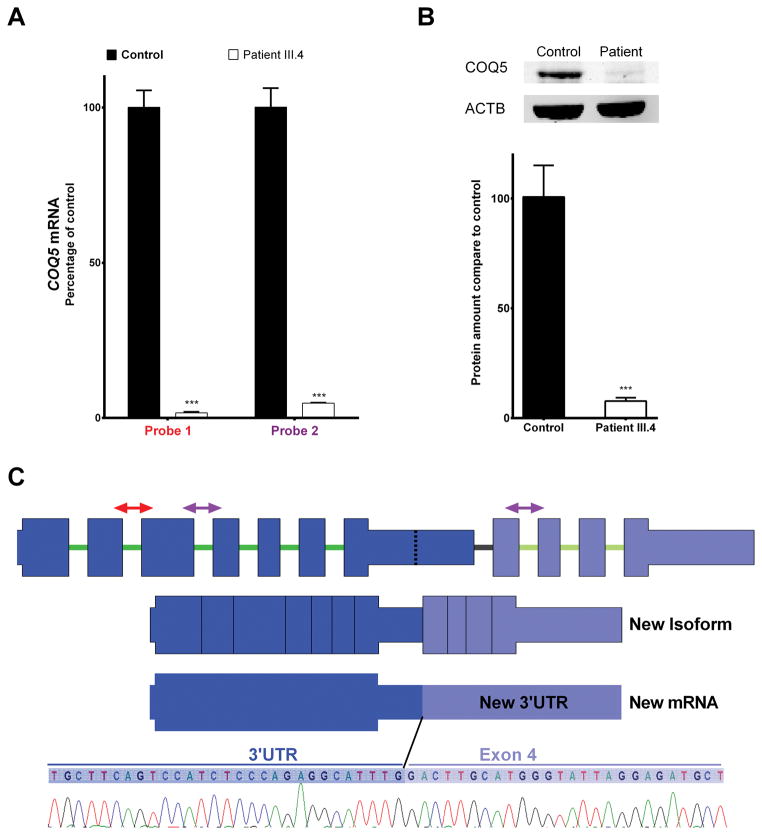
To investigate the potential effect of this duplication on COQ5, we first measured the mRNA expression level. In patient fibroblasts, qRT-PCR using two different probe sets revealed COQ5 mRNA at a level of only 2–4% of that in control fibroblasts (Fig. 2A). We then demonstrated reduced COQ5 protein expression in patient fibroblasts by western blotting (Fig. 2B). Sequencing of the cDNA demonstrated that part of the 3′UTR has been spliced off and replaced by the duplicated part of the gene (exon 4 to 7), generating a longer alternative 3′UTR (Fig. 2C).
Fig. 2. Effects of COQ5 mutation in RNA and protein from fibroblasts.
(A) Quantitative real-time PCR results for the expression of three COQ5 mRNA isoforms in fibroblasts from patient compared with control. Values are percentage expression of COQ5 in patient fibroblasts compared with control fibroblasts, normalized to POLR2A (error bars represent S.D., n=3). (B) Quantification of the expression of COQ5 protein in patient fibroblasts and control fibroblasts. Values are relative expression of COQ5 normalized to the loading control ACTB (error bars represent S.D., n=3 from two independent experiments). Next to the histogram is the immunoblot of fibroblast whole cell lysates of patient and control, showing the decreased expression of COQ5 in the patient. (C) A diagram of the known COQ5 isoform (NM_032314.3) is shown, and regions amplified by qPCR analysis in (A) are marked with arrows (red represents Probe 1, and violet represents Probe 2). Below the normal COQ5 isoform is a diagram depicting a new isoform generated. Sequencing of the cDNA from fibroblasts of patient III.4 shows an abnormal 3′UTR because of an abnormal splicing event: part of the original 3′UTR has been spliced off and replaced by the duplicated part of the gene (Exon 4 to 7), depicted in the figure as “New mRNA”. At the bottom of the figure is a chromatogram that shows the nucleotide sequence between the 3′UTR and the duplicated part of the gene, beginning from exon 4.
COQ10 levels are reduced in patient’s leukocytes and muscle tissue
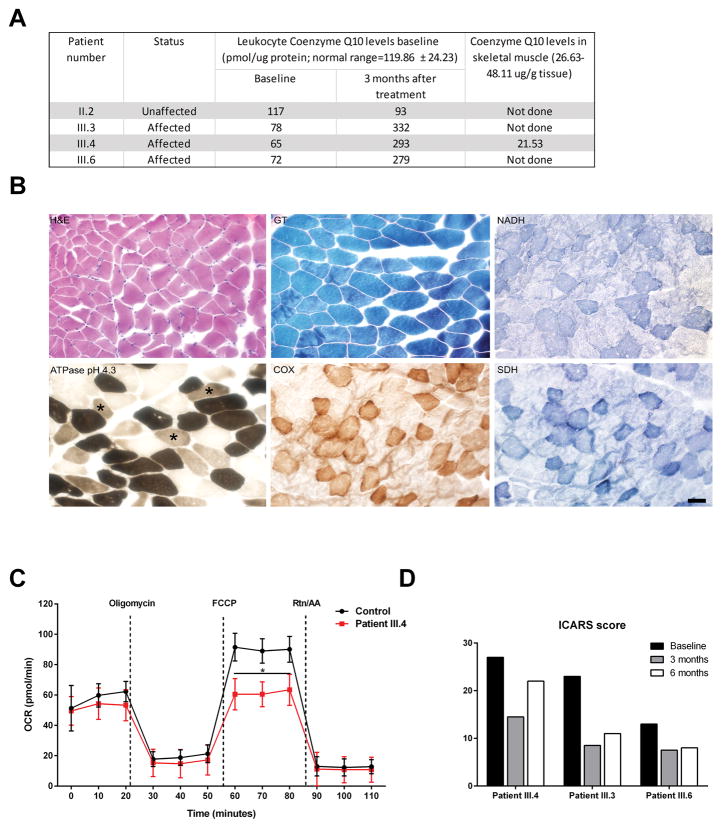
To determine whether the COQ5 deficiency could affect CoQ10 synthesis, we examined CoQ10 levels of affected and unaffected family members. CoQ10 levels were mildly reduced in all three affected patients (III.3, III.4, and III.6), as compared to their unaffected mother (II.2) (Fig. 3A). We then collected a muscle specimen from III.4, that showed increased type 2C muscle fibers (Fig. 3B), which comprised ~34% of all the myofibers; in comparison, type 2C fibers usually comprise 0–6.5% of the total number of fibers in unaffected young women (Staron, et al., 2000). There were no signs of myopathy, ragged red fibers, mitochondrial dysfunction or structural abnormalities on histochemical analysis (Fig. 3B), but Type 2C muscle fibers were 34% the total (normal in young women is 0–6.5% [4]). CoQ10 levels obtained from the skeletal muscle biopsy was reduced, confirming primary CoQ10 deficiency (Fig. 3A; full blot shown as Supp. Figure S1).
Fig. 3. COQ5 mutation reduces CoQ10 levels and lead to abnormal mitochondrial respiration.
(A) Table showing CoQ10 levels in leukocytes from patients III.3, III.4, and III.6. Baseline levels and levels at 3 months after initiation of oral CoQ10 treatment are shown. Range of normal values was the average from 14 individuals with no known mitochondrial defect or mutations in the COQ genes. Level of Coenzyme Q10 levels in skeletal muscle in II.4 is also shown. (B) Representative muscle histopathology slides showing hematoxylin and eosin (H&E), Gomori trichrome (GT), nicotinamide adenine dinucleotide (NADH), cytochrome oxidase (COX) and succinate dehydrogenase (SDH) stains. The morphology of muscle fibers appears normal. Myosin adenosine tri-phosphatase (ATPase) activity staining at pH 4.3 identifies type 2C (immature) muscle fibers with an intermediate staining intensity between dark, type 1 (slow, oxidative) and light, type 2 (fast, glycolytic) fibers. Many type 2C muscle fibers are seen (asterisks). Scale bar represents 50 μm.
(C) Cellular oxygen consumption rates (OCR) for fibroblast cell lines from wild-type (WT) and COQ5 patient (III.4) were measured by XF24 extracellular flux analysis. Oligomycin (1.25μM), carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP, 0.2μM), and antimycin A (AA,1.8μM) together with Rotenone (Rtn,1μM) were injected after 20,50 and 80 minutes, respectively (n 4 plates; * p. value<0.05).
(D) Clinical scoring (ICARS) before and during CoQ10 treatment.
Measurement of oxygen consumption and activity of the respiratory complex chain in human fibroblasts
To understand the metabolic effects in the mitochondria due to the COQ5 mutation, we used the Seahorse analyzer to measure the cellular oxygen consumption rate (OCR) in cultured fibroblasts. The mutation in COQ5 significantly reduced uncoupled respiration compared to normal (Fig. 3C). Similarly, high resolution respirometry in fibroblasts showed a reduced uncoupling ratio and increased net routine flux control ratio, consistent with decreased reserve capacity (Supp. Table S2). In addition, the phosphorylation respiratory control ratio was increased, indicating that a higher proportion of oxidative phosphorylation is activated to drive ATP synthesis. We also measured the activity of the individual complexes of the respiratory complex chain including the combined activity of II and III, on mitochondrial fractions isolated from dermal fibroblasts, following our published methods (Kirby, et al., 2007; Taylor, et al., 1993). Our analysis showed that there is a measurable defect in Complex II+III activity in the cells, using an assay that relies on endogenous CoQ10 to shuttle electrons from succinate to cytochrome c (Supp. Table S3).
Clinical outcome of CoQ10 supplementation in patients with defects in COQ5
The pre-treatment ICARS scoring was consistent with the relative clinical severity of the disorder for each patient; the more affected patient (III.4) scored 27/100 and the least affected patient (III.6) scored 13/100. After 3 months of CoQ10 treatment, the scores of all three patients decreased significantly (patient 1: 27 to 14.5, patient 2: 13 to 7.5, and patient 3: 23 to 8.5). In addition, the patients appeared to have a quicker response rate during conversation and better alertness under the CoQ10 supplementation. A follow-up visit was obtained six months after initiation of treatment, and two weeks after halting CoQ10 treatment. Clinical assessment showed slight deterioration in the scoring results compared to the 3-months’ assessment rates (patient 1: −22, patient 2: −8, patient 3: −11) (Fig. 3D).
Discussion
We describe for the first time primary CoQ10 deficiency caused by loss of function of COQ5, manifesting with varying degrees of cerebellar ataxia, myoclonic generalized tonic clonic seizures and cognitive disability in three female siblings.
Through careful manual analysis of next generation sequencing results, we identified a tandem duplication affecting the last four exons of COQ5, possibly the result of homologous recombination between two Alus (AluY and AluYc). Although the duplication is located about 1kb after the gene and affects neither the coding sequence nor the 3′UTR of the gene, qPCR showed a nearly total absence of expression of COQ5. Sequencing of the residual COQ5 mRNA revealed that the duplication creates an abnormal isoform due to an abnormal splicing event, where the original 3′UTR is partially deleted and fused with duplicated exons (exons 4 to 7), thus creating a new 3′UTR that appears abnormally long.
Our CNV analysis on 92 unaffected Iraqi-Jewish controls may be much higher than anticipated; this could be due to the fact that the samples came from a single area. We approximate the MAF to be 1.5%, indicating that CoQ5 deficiency remain underdiagnosed. The combination of manual or automated CNV analysis and next generation sequencing can help detecting this new variant and therefore can potentially provide diagnosis to some patients with idiopathic cerebellar ataxia.
Our study defines a new subtype of primary CoQ10 deficiency due to a loss-of-function mutation in COQ5. We have demonstrated that modest reduction in CoQ10levels were present in peripheral blood leukocytes of affected patients while in skeletal muscles CoQ10 levels were much more reduced. The overall effect on CoQ10 levels, especially in skeletal muscle tissue, are consistent with the importance of COQ5 in ubiquinone synthesis (Chen, et al., 2013; Yen, et al., 2016), and confirm the finding that destabilization of the COQ5-containing protein complex in human cells reduces CoQ10 levels (Yen, et al., 2016). It is important to note, however, that although the CoQ10 levels low, there is still some amount of CoQ10 produced in leukocytes and muscle. We hypothesize that in the affected individuals in this study, the respiration rate is limited by the number of CoQ10 molecules within the mitochondrial inner membrane. However, because the defect in the synthesis of CoQ10 is partial, there is only a 33% reduction the number of CoQ10 molecules, which can be sufficient to sustain the basal respiration rate in fibroblasts but not the maximum respiration rate.
In addition to the biochemical abnormality of CoQ10 deficiency, the finding of increased frequency of type 2C fibers in morphologically normal muscle in our proband is also consistent with CoQ10 deficiency as previously reported (Sommerville, et al., 2013), where the increase in the number of type 2C fibers was thought to be due to inhibition of fiber maturation. Type 2C muscle fibers represent immature fibers, as they are abundant in fetal muscle and normally decline to less than 5% of fibers by age 1 year (Brooke and Kaiser, 1970). Increased type 2C fiber frequency after the age of 1 year occurs in active myopathic disorders with prominent muscle fiber regeneration.
In terms of clinical features, our patients had variable degrees of cerebellar ataxia as their main neurological feature; other neurological symptoms were more variable. The most affected patient had mild to moderate cognitive impairment and a seizure disorder, while the least affected sib functioned in the low-to-normal intelligence range and never had seizures or an abnormal EEG. Cerebellar ataxia with cerebellar atrophy is the most common of the five phenotypes described with primary CoQ10 deficiency and is found mainly in COQ8 (ADCK3) deficiency, defined also as autosomal recessive cerebellar ataxia 2 (ARCA2), but can also be seen in several secondary CoQ deficiency syndromes (Balreira, et al., 2014; Chamova, et al., 2012; Maruyama, et al., 2014; Quinzii, et al., 2005). The significant cerebellar involvement in both primary and secondary CoQ10 deficiency conditions may be related to lower content of endogenous CoQ10 in the cerebellum compared to other brain regions, as demonstrated in the rat (Naini, et al., 2003).
In addition to cerebellar ataxia and atrophy, other features described in CoQ10 deficiency syndromes include seizure disorder, dystonia, migraines, cognitive disability and exercise intolerance. Patients in previous reports and in the current study have not shown extra CNS symptoms or signs, except for occasional exercise intolerance, despite reduced levels of CoQ10 in other tissues (Lagier-Tourenne, et al., 2008; Mignot, et al., 2013; Mollet, et al., 2008).
High dose oral supplementation of CoQ10 has been effective in primary (Ashraf, et al., 2013; Doimo, et al., 2014; Heeringa, et al., 2011; Montini, et al., 2008; Salviati, et al., 2005) and secondary CoQ10 deficiency syndromes (Musumeci, et al., 2001; Quinzii, et al., 2005), and in cerebellar ataxia-related syndromes (whether with or without a molecular diagnosis) (Lo, et al., 2015; Pineda, et al., 2010). We started a therapeutic trial in our three affected patients soon after the molecular diagnosis was made, while all were in their 20’s. The dosage used was 15 mg/kg/d divided into three doses. Even though our patients’ ataxia severity is currently mild to moderate with baseline scores ranging from 13–27/100, after 3 months of treatment their scoring was significantly reduced and ranged from 14.5/100 in the most severely affected patient and to 7.5/100 in the least severely affected. Unfortunately, the three sisters stopped their CoQ10 intake 2 weeks before the second assessment at 6 months. It is possible that the relatively higher score, showing worsening from the previous exam after 6 months of treatment (8–22/100) instead of the expected further improvement, may be attributable to the fact that they did not take the CoQ10 supplementation for 2 weeks before the second assessment. This assumption is consistent with similar experience with treatment tapering previously reported in the literature (Salviati, et al., 2012). The ICARS score, which is the more detailed scoring system used for ataxia assessment, was used in patients with ADCK3 mutations, and significant improvement in the ataxia score was detected after 3 months of treatment (Montero, et al., 2007), as in our patients. Based on the significant change noticed at three months, we recommended our patients to resume the CoQ10 treatment in the current dosage, and will even consider increasing the dosage if no side effects are noticed and an attenuation in the clinical response is seen.
Variability of the clinical response to CoQ10 supplementation in the different syndromes, and within the same syndrome in different cohorts, may be related to therapeutic dosages chosen, the pharmaceutical formulation employed, the severity of the underlying illness and organs involved, the time elapsed from clinical presentation to treatment onset (with significant tissue damage already present in late treated patients), or poor tissue delivery of CoQ10 into the CNS. In addition, many other environmental and even epigenetic factors likely modulate the response to treatment (Acosta, et al., 2016; Desbats, et al., 2015; Fernandez-Ayala, et al., 2013; Trevisson, et al., 2011). It is recommended to institute treatment as early as possible, since only minimal recovery is possible once damage in critical organs (such as the kidney or the CNS) is established (Acosta, et al., 2016). The non-cerebellar symptoms in our patients were much more difficult to assess because their seizures were already well-controlled by anti-epileptic drugs (AED’s) and the timeframe of the treatment trial did not allow us to perform a repeated neuropsychological assessment. Nonetheless, we obtained a clinical impression of improvement in both cognitive and behavioral features.
In conclusion, primary CoQ10 deficiency due to COQ5 mutations is a treatable condition. Response to CoQ10 supplementation might be influenced by the timing of treatment, as we saw in the patients in this study. Early diagnosis by next generation sequencing in populations showing mild forms of cerebellar ataxia and static encephalomyopathy will be helpful in defining this disorder further, and evaluating the importance of early CoQ10 supplementation.
Supplementary Material
Acknowledgments
The authors wish to thank the patients and their family for their cooperation and support. We also thank NISC for assisting in the initial whole exome sequencing analysis for one of the probands included in the study. This study is supported in part by the National Institutes of Health Intramural Research Program of the National Human Genome Research Institute and the Common Fund, Office of the Director; the NIH grants RO1-N5021328-030 and RO1OD010944-05; and the DOD grant, PR150585P1. RWT is supported by the Wellcome Centre for Mitochondrial Research (203105/Z/16/Z), the Medical Research Council (MRC) Centre for Translational Research in Neuromuscular Disease, Mitochondrial Disease Patient Cohort (UK) (G0800674), the Lily Foundation and the UK NHS Highly Specialised Service for Rare Mitochondrial Disorders of Adults and Children.
Footnotes
Competing Interests Statement
The authors declare that they have no competing financial interests.
References
- Acosta MJ, Vazquez Fonseca L, Desbats MA, Cerqua C, Zordan R, Trevisson E, Salviati L. Coenzyme Q biosynthesis in health and disease. Biochim Biophys Acta. 2016;1857(8):1079–85. doi: 10.1016/j.bbabio.2016.03.036. [DOI] [PubMed] [Google Scholar]
- Ashraf S, Gee HY, Woerner S, Xie LX, Vega-Warner V, Lovric S, Fang H, Song X, Cattran DC, Avila-Casado C, Paterson AD, Nitschke P, Bole-Feysot C, Cochat P, Esteve-Rudd J, Haberberger B, Allen SJ, Zhou W, Airik R, Otto EA, Barua M, Al-Hamed MH, Kari JA, Evans J, Bierzynska A, Saleem MA, Bockenhauer D, Kleta R, El Desoky S, Hacihamdioglu DO, Gok F, Washburn J, Wiggins RC, Choi M, Lifton RP, Levy S, Han Z, Salviati L, Prokisch H, Williams DS, Pollak M, Clarke CF, Pei Y, Antignac C, Hildebrandt F. ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J Clin Invest. 2013;123(12):5179–89. doi: 10.1172/JCI69000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba SW, Belogrudov GI, Lee JC, Lee PT, Strahan J, Shepherd JN, Clarke CF. Yeast Coq5 C-methyltransferase is required for stability of other polypeptides involved in coenzyme Q biosynthesis. J Biol Chem. 2004;279(11):10052–9. doi: 10.1074/jbc.M313712200. [DOI] [PubMed] [Google Scholar]
- Balreira A, Boczonadi V, Barca E, Pyle A, Bansagi B, Appleton M, Graham C, Hargreaves IP, Rasic VM, Lochmuller H, Griffin H, Taylor RW, Naini A, Chinnery PF, Hirano M, Quinzii CM, Horvath R. ANO10 mutations cause ataxia and coenzyme Q10 deficiency. Journal of neurology. 2014;261(11):2192–8. doi: 10.1007/s00415-014-7476-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkovich RJ, Shtanko A, Shepherd JA, Lee PT, Myles DC, Tzagoloff A, Clarke CF. Characterization of the COQ5 gene from Saccharomyces cerevisiae. Evidence for a C-methyltransferase in ubiquinone biosynthesis. J Biol Chem. 1997;272(14):9182–8. doi: 10.1074/jbc.272.14.9182. [DOI] [PubMed] [Google Scholar]
- Boitier E, Degoul F, Desguerre I, Charpentier C, Francois D, Ponsot G, Diry M, Rustin P, Marsac C. A case of mitochondrial encephalomyopathy associated with a muscle coenzyme Q10 deficiency. J Neurol Sci. 1998;156(1):41–6. doi: 10.1016/s0022-510x(98)00006-9. [DOI] [PubMed] [Google Scholar]
- Brooke MH, Kaiser KK. Muscle fiber types: how many and what kind? Arch Neurol. 1970;23(4):369–79. doi: 10.1001/archneur.1970.00480280083010. [DOI] [PubMed] [Google Scholar]
- Chamova T, Florez L, Guergueltcheva V, Raycheva M, Kaneva R, Lochmuller H, Kalaydjieva L, Tournev I. ANO10 c.1150_1151del is a founder mutation causing autosomal recessive cerebellar ataxia in Roma/Gypsies. Journal of neurology. 2012;259(5):906–11. doi: 10.1007/s00415-011-6276-6. [DOI] [PubMed] [Google Scholar]
- Chen SW, Liu CC, Yen HC. Detection of suppressed maturation of the human COQ5 protein in the mitochondria following mitochondrial uncoupling by an antibody recognizing both precursor and mature forms of COQ5. Mitochondrion. 2013;13(2):143–52. doi: 10.1016/j.mito.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Desbats MA, Vetro A, Limongelli I, Lunardi G, Casarin A, Doimo M, Spinazzi M, Angelini C, Cenacchi G, Burlina A, Rodriguez Hernandez MA, Chiandetti L, Clementi M, Trevisson E, Navas P, Zuffardi O, Salviati L. Primary coenzyme Q10 deficiency presenting as fatal neonatal multiorgan failure. Eur J Hum Genet. 2015;23(9):1254–8. doi: 10.1038/ejhg.2014.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S, Quinzii CM, Hirano M. Mutations in coenzyme Q10 biosynthetic genes. J Clin Invest. 2007;117(3):587–9. doi: 10.1172/JCI31423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinwiddie DL, Smith LD, Miller NA, Atherton AM, Farrow EG, Strenk ME, Soden SE, Saunders CJ, Kingsmore SF. Diagnosis of mitochondrial disorders by concomitant next-generation sequencing of the exome and mitochondrial genome. Genomics. 2013;102(3):148–56. doi: 10.1016/j.ygeno.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diomedi-Camassei F, Di Giandomenico S, Santorelli FM, Caridi G, Piemonte F, Montini G, Ghiggeri GM, Murer L, Barisoni L, Pastore A, Muda AO, Valente ML, Bertini E, Emma F. COQ2 nephropathy: a newly described inherited mitochondriopathy with primary renal involvement. J Am Soc Nephrol. 2007;18(10):2773–80. doi: 10.1681/ASN.2006080833. [DOI] [PubMed] [Google Scholar]
- Doimo M, Desbats MA, Cerqua C, Cassina M, Trevisson E, Salviati L. Genetics of coenzyme q10 deficiency. Mol Syndromol. 2014;5(3–4):156–62. doi: 10.1159/000362826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Ayala DJ, Guerra I, Jimenez-Gancedo S, Cascajo MV, Gavilan A, Dimauro S, Hirano M, Briones P, Artuch R, De Cabo R, Salviati L, Navas P. Survival transcriptome in the coenzyme Q10 deficiency syndrome is acquired by epigenetic modifications: a modelling study for human coenzyme Q10 deficiencies. BMJ Open. 2013;3(3) doi: 10.1136/bmjopen-2012-002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Maraver J, Cordero MD, Oropesa-Avila M, Fernandez Vega A, de la Mata M, Delgado Pavon A, de Miguel M, Perez Calero C, Villanueva Paz M, Cotan D, Sanchez-Alcazar JA. Coenzyme q10 therapy. Mol Syndromol. 2014;5(3–4):187–97. doi: 10.1159/000360101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gempel K, Topaloglu H, Talim B, Schneiderat P, Schoser BG, Hans VH, Palmafy B, Kale G, Tokatli A, Quinzii C, Hirano M, Naini A, DiMauro S, Prokisch H, Lochmuller H, Horvath R. The myopathic form of coenzyme Q10 deficiency is caused by mutations in the electron-transferring-flavoprotein dehydrogenase (ETFDH) gene. Brain. 2007;130(Pt 8):2037–44. doi: 10.1093/brain/awm054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gironi M, Lamperti C, Nemni R, Moggio M, Comi G, Guerini FR, Ferrante P, Canal N, Naini A, Bresolin N, DiMauro S. Late-onset cerebellar ataxia with hypogonadism and muscle coenzyme Q10 deficiency. Neurology. 2004;62(5):818–20. doi: 10.1212/01.wnl.0000113719.67643.b7. [DOI] [PubMed] [Google Scholar]
- He CH, Xie LX, Allan CM, Tran UC, Clarke CF. Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants. Biochim Biophys Acta. 2014;1841(4):630–44. doi: 10.1016/j.bbalip.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeringa SF, Chernin G, Chaki M, Zhou W, Sloan AJ, Ji Z, Xie LX, Salviati L, Hurd TW, Vega-Warner V, Killen PD, Raphael Y, Ashraf S, Ovunc B, Schoeb DS, McLaughlin HM, Airik R, Vlangos CN, Gbadegesin R, Hinkes B, Saisawat P, Trevisson E, Doimo M, Casarin A, Pertegato V, Giorgi G, Prokisch H, Rotig A, Nurnberg G, Becker C, Wang S, Ozaltin F, Topaloglu R, Bakkaloglu A, Bakkaloglu SA, Muller D, Beissert A, Mir S, Berdeli A, Varpizen S, Zenker M, Matejas V, Santos-Ocana C, Navas P, Kusakabe T, Kispert A, Akman S, Soliman NA, Krick S, Mundel P, Reiser J, Nurnberg P, Clarke CF, Wiggins RC, Faul C, Hildebrandt F. COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J Clin Invest. 2011;121(5):2013–24. doi: 10.1172/JCI45693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath R, Schneiderat P, Schoser BG, Gempel K, Neuen-Jacob E, Ploger H, Muller-Hocker J, Pongratz DE, Naini A, DiMauro S, Lochmuller H. Coenzyme Q10 deficiency and isolated myopathy. Neurology. 2006;66(2):253–5. doi: 10.1212/01.wnl.0000194241.35115.7c. [DOI] [PubMed] [Google Scholar]
- Kenyon L, Moraes CT. Expanding the functional human mitochondrial DNA database by the establishment of primate xenomitochondrial cybrids. Proc Natl Acad Sci U S A. 1997;94(17):9131–5. doi: 10.1073/pnas.94.17.9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby DM, Thorburn DR, Turnbull DM, Taylor RW. Biochemical assays of respiratory chain complex activity. Methods Cell Biol. 2007;80:93–119. doi: 10.1016/S0091-679X(06)80004-X. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Tazir M, Lopez LC, Quinzii CM, Assoum M, Drouot N, Busso C, Makri S, Ali-Pacha L, Benhassine T, Anheim M, Lynch DR, Thibault C, Plewniak F, Bianchetti L, Tranchant C, Poch O, DiMauro S, Mandel JL, Barros MH, Hirano M, Koenig M. ADCK3, an ancestral kinase, is mutated in a form of recessive ataxia associated with coenzyme Q10 deficiency. Am J Hum Genet. 2008;82(3):661–72. doi: 10.1016/j.ajhg.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani SR, Vladutiu GD, Plunkett K, Lotze TE, Adesina AM, Scaglia F. Isolated mitochondrial myopathy associated with muscle coenzyme Q10 deficiency. Arch Neurol. 2005;62(2):317–20. doi: 10.1001/archneur.62.2.317. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lo RY, Figueroa KP, Pulst SM, Lin CY, Perlman S, Wilmot G, Gomez C, Schmahmann J, Paulson H, Shakkottai VG, Ying S, Zesiewicz T, Bushara K, Geschwind M, Xia G, Subramony SH, Ashizawa T, Kuo SH. Coenzyme Q10 and spinocerebellar ataxias. Mov Disord. 2015;30(2):214–20. doi: 10.1002/mds.26088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malicdan MCV, Vilboux T, Stephen J, Maglic D, Mian L, Konzman D, Guo J, Yildirimli D, Bryant J, Fischer R, Zein WM, Snow J, Vemulapalli M, Mullikin JC, Toro C, Solomon BD, Niederhuber JE, Gahl WA, Gunay-Aygun M, Progra NCS. Mutations in human homologue of chicken talpid3 gene (KIAA0586) cause a hybrid ciliopathy with overlapping features of Jeune and Joubert syndromes. Journal of Medical Genetics. 2015;52(12):830–839. doi: 10.1136/jmedgenet-2015-103316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama H, Morino H, Miyamoto R, Murakami N, Hamano T, Kawakami H. Exome sequencing reveals a novel ANO10 mutation in a Japanese patient with autosomal recessive spinocerebellar ataxia. Clinical genetics. 2014;85(3):296–7. doi: 10.1111/cge.12140. [DOI] [PubMed] [Google Scholar]
- McCarthy HJ, Bierzynska A, Wherlock M, Ognjanovic M, Kerecuk L, Hegde S, Feather S, Gilbert RD, Krischock L, Jones C, Sinha MD, Webb NJ, Christian M, Williams MM, Marks S, Koziell A, Welsh GI, Saleem MA Group RtUSS. Simultaneous sequencing of 24 genes associated with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol. 2013;8(4):637–48. doi: 10.2215/CJN.07200712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignot C, Apartis E, Durr A, Marques Lourenco C, Charles P, Devos D, Moreau C, de Lonlay P, Drouot N, Burglen L, Kempf N, Nourisson E, Chantot-Bastaraud S, Lebre AS, Rio M, Chaix Y, Bieth E, Roze E, Bonnet I, Canaple S, Rastel C, Brice A, Rotig A, Desguerre I, Tranchant C, Koenig M, Anheim M. Phenotypic variability in ARCA2 and identification of a core ataxic phenotype with slow progression. Orphanet J Rare Dis. 2013;8:173. doi: 10.1186/1750-1172-8-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet J, Delahodde A, Serre V, Chretien D, Schlemmer D, Lombes A, Boddaert N, Desguerre I, de Lonlay P, de Baulny HO, Munnich A, Rotig A. CABC1 gene mutations cause ubiquinone deficiency with cerebellar ataxia and seizures. Am J Hum Genet. 2008;82(3):623–30. doi: 10.1016/j.ajhg.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero R, Pineda M, Aracil A, Vilaseca MA, Briones P, Sanchez-Alcazar JA, Navas P, Artuch R. Clinical, biochemical and molecular aspects of cerebellar ataxia and Coenzyme Q10 deficiency. Cerebellum. 2007;6(2):118–22. doi: 10.1080/14734220601021700. [DOI] [PubMed] [Google Scholar]
- Montini G, Malaventura C, Salviati L. Early coenzyme Q10 supplementation in primary coenzyme Q10 deficiency. N Engl J Med. 2008;358(26):2849–50. doi: 10.1056/NEJMc0800582. [DOI] [PubMed] [Google Scholar]
- Mozaffar T, Pestronk A. Myopathy with anti-Jo-1 antibodies: pathology in perimysium and neighbouring muscle fibres. J Neurol Neurosurg Psychiatry. 2000;68(4):472–8. doi: 10.1136/jnnp.68.4.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musumeci O, Naini A, Slonim AE, Skavin N, Hadjigeorgiou GL, Krawiecki N, Weissman BM, Tsao CY, Mendell JR, Shanske S, De Vivo DC, Hirano M, DiMauro S. Familial cerebellar ataxia with muscle coenzyme Q10 deficiency. Neurology. 2001;56(7):849–55. doi: 10.1212/wnl.56.7.849. [DOI] [PubMed] [Google Scholar]
- Naini A, Lewis VJ, Hirano M, DiMauro S. Primary coenzyme Q10 deficiency and the brain. Biofactors. 2003;18(1–4):145–52. doi: 10.1002/biof.5520180217. [DOI] [PubMed] [Google Scholar]
- Nguyen TP, Casarin A, Desbats MA, Doimo M, Trevisson E, Santos-Ocana C, Navas P, Clarke CF, Salviati L. Molecular characterization of the human COQ5 C-methyltransferase in coenzyme Q10 biosynthesis. Biochim Biophys Acta. 2014;1841(11):1628–38. doi: 10.1016/j.bbalip.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasahara S, Engel AG, Frens D, Mack D. Muscle coenzyme Q deficiency in familial mitochondrial encephalomyopathy. Proc Natl Acad Sci U S A. 1989;86(7):2379–82. doi: 10.1073/pnas.86.7.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer DA, Le JM, Steemers FJ, Chang W, Jenniges T, Garcia F, Haden K, Li J, Shaw CA, Belmont J, Cheung SW, Shen RM, Barker DL, Gunderson KL. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16(9):1136–48. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestronk GJ, Kaiser KK, Brooke MH. ATPase stain in muscle histochemistry. Muscle Nerve. 1992;15(2):258. [PubMed] [Google Scholar]
- Pineda M, Montero R, Aracil A, O’Callaghan MM, Mas A, Espinos C, Martinez-Rubio D, Palau F, Navas P, Briones P, Artuch R. Coenzyme Q(10)-responsive ataxia: 2-year-treatment follow-up. Mov Disord. 2010;25(9):1262–8. doi: 10.1002/mds.23129. [DOI] [PubMed] [Google Scholar]
- Quinzii C, Naini A, Salviati L, Trevisson E, Navas P, Dimauro S, Hirano M. A mutation in para-hydroxybenzoate-polyprenyl transferase (COQ2) causes primary coenzyme Q10 deficiency. Am J Hum Genet. 2006;78(2):345–9. doi: 10.1086/500092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinzii CM, Emmanuele V, Hirano M. Clinical presentations of coenzyme q10 deficiency syndrome. Mol Syndromol. 2014;5(3–4):141–6. doi: 10.1159/000360490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinzii CM, Kattah AG, Naini A, Akman HO, Mootha VK, DiMauro S, Hirano M. Coenzyme Q deficiency and cerebellar ataxia associated with an aprataxin mutation. Neurology. 2005;64(3):539–41. doi: 10.1212/01.WNL.0000150588.75281.58. [DOI] [PubMed] [Google Scholar]
- Rossignol R, Malgat M, Mazat JP, Letellier T. Threshold effect and tissue specificity - Implication for mitochondrial cytopathies. Journal of Biological Chemistry. 1999;274(47):33426–33432. doi: 10.1074/jbc.274.47.33426. [DOI] [PubMed] [Google Scholar]
- Rotig A, Appelkvist EL, Geromel V, Chretien D, Kadhom N, Edery P, Lebideau M, Dallner G, Munnich A, Ernster L, Rustin P. Quinone-responsive multiple respiratory-chain dysfunction due to widespread coenzyme Q10 deficiency. Lancet. 2000;356(9227):391–5. doi: 10.1016/S0140-6736(00)02531-9. [DOI] [PubMed] [Google Scholar]
- Salviati L, Sacconi S, Murer L, Zacchello G, Franceschini L, Laverda AM, Basso G, Quinzii C, Angelini C, Hirano M, Naini AB, Navas P, DiMauro S, Montini G. Infantile encephalomyopathy and nephropathy with CoQ10 deficiency: a CoQ10-responsive condition. Neurology. 2005;65(4):606–8. doi: 10.1212/01.wnl.0000172859.55579.a7. [DOI] [PubMed] [Google Scholar]
- Salviati L, Trevisson E, Rodriguez Hernandez MA, Casarin A, Pertegato V, Doimo M, Cassina M, Agosto C, Desbats MA, Sartori G, Sacconi S, Memo L, Zuffardi O, Artuch R, Quinzii C, Dimauro S, Hirano M, Santos-Ocana C, Navas P. Haploinsufficiency of COQ4 causes coenzyme Q10 deficiency. J Med Genet. 2012;49(3):187–91. doi: 10.1136/jmedgenet-2011-100394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira C, Hirano M, Shanske S, Keller RK, Haller RG, Davidson E, Santorelli FM, Miranda AF, Bonilla E, Mojon DS, Barreira AA, King MP, DiMauro S. Mitochondrial encephalomyopathy with coenzyme Q10 deficiency. Neurology. 1997;48(5):1238–43. doi: 10.1212/wnl.48.5.1238. [DOI] [PubMed] [Google Scholar]
- Sommerville RB, Zaidman CM, Pestronk A. Coenzyme Q10 deficiency in children: frequent type 2C muscle fibers with normal morphology. Muscle Nerve. 2013;48(5):722–6. doi: 10.1002/mus.23837. [DOI] [PubMed] [Google Scholar]
- Staron RS, Hagerman FC, Hikida RS, Murray TF, Hostler DP, Crill MT, Ragg KE, Toma K. Fiber type composition of the vastus lateralis muscle of young men and women. J Histochem Cytochem. 2000;48(5):623–9. doi: 10.1177/002215540004800506. [DOI] [PubMed] [Google Scholar]
- Steemers FJ, Gunderson KL. Whole genome genotyping technologies on the BeadArray platform. Biotechnol J. 2007;2(1):41–9. doi: 10.1002/biot.200600213. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Bose P, Leong-Quong RY, Fujita DJ, Riabowol K. REAP: A two minute cell fractionation method. BMC research notes. 2010;3:294. doi: 10.1186/1756-0500-3-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RW, Birch-Machin MA, Bartlett K, Turnbull DM. Succinate-cytochrome c reductase: assessment of its value in the investigation of defects of the respiratory chain. Biochim Biophys Acta. 1993;1181(3):261–5. doi: 10.1016/0925-4439(93)90030-5. [DOI] [PubMed] [Google Scholar]
- Teer JK, Green ED, Mullikin JC, Biesecker LG. VarSifter: visualizing and analyzing exome-scale sequence variation data on a desktop computer. Bioinformatics. 2012;28(4):599–600. doi: 10.1093/bioinformatics/btr711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevisson E, DiMauro S, Navas P, Salviati L. Coenzyme Q deficiency in muscle. Curr Opin Neurol. 2011;24(5):449–56. doi: 10.1097/WCO.0b013e32834ab528. [DOI] [PubMed] [Google Scholar]
- Trouillas P, Takayanagi T, Hallett M, Currier RD, Subramony SH, Wessel K, Bryer A, Diener HC, Massaquoi S, Gomez CM, Coutinho P, Ben Hamida M, Campanella G, Filla A, Schut L, Timann D, Honnorat J, Nighoghossian N, Manyam B. International Cooperative Ataxia Rating Scale for pharmacological assessment of the cerebellar syndrome. The Ataxia Neuropharmacology Committee of the World Federation of Neurology. J Neurol Sci. 1997;145(2):205–11. doi: 10.1016/s0022-510x(96)00231-6. [DOI] [PubMed] [Google Scholar]
- Vilboux T, Ciccone C, Blancato JK, Cox GF, Deshpande C, Introne WJ, Gahl WA, Smith AC, Huizing M. Molecular analysis of the Retinoic Acid Induced 1 gene (RAI1) in patients with suspected Smith-Magenis syndrome without the 17p11.2 deletion. PLoS One. 2011;6(8):e22861. doi: 10.1371/journal.pone.0022861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HC, Liu YC, Kan CC, Wei HJ, Lee SH, Wei YH, Feng YH, Chen CW, Huang CC. Disruption of the human COQ5-containing protein complex is associated with diminished coenzyme Q10 levels under two different conditions of mitochondrial energy deficiency. Biochim Biophys Acta. 2016;1860(9):1864–76. doi: 10.1016/j.bbagen.2016.05.005. [DOI] [PubMed] [Google Scholar]
- Yubero D, Montero R, Artuch R, Land JM, Heales SJ, Hargreaves IP. Biochemical diagnosis of coenzyme q10 deficiency. Mol Syndromol. 2014;5(3–4):147–55. doi: 10.1159/000362390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.